Abstract
Spinal muscular atrophy (SMA) is a motor neuron disease caused by the loss of survival motor neuron-1 (SMN1). A nearly identical copy gene, SMN2, is present in all SMA patients, which produces low levels of functional protein. Although the SMN2 coding sequence has the potential to produce normal, full-length SMN, ∼90% of SMN2-derived transcripts are alternatively spliced and encode a truncated protein lacking the final coding exon (exon 7). SMN2, however, is an excellent therapeutic target. Previously, we developed bifunctional RNAs that bound SMN exon 7 and modulated SMN2 splicing. To optimize the efficiency of the bifunctional RNAs, a different antisense target was required. To this end, we genetically verified the identity of a putative intronic repressor and developed bifunctional RNAs that target this sequence. Consequently, there is a 2-fold mechanism of SMN induction: inhibition of the intronic repressor and recruitment of SR proteins via the SR recruitment sequence of the bifunctional RNA. The bifunctional RNAs effectively increased SMN in human primary SMA fibroblasts. Lead candidates were synthesized as 2′-O-methyl RNAs and were directly injected in the central nervous system of SMA mice. Single-RNA injections were able to illicit a robust induction of SMN protein in the brain and throughout the spinal column of neonatal SMA mice. In a severe model of SMA, mean life span was extended following the delivery of bifunctional RNAs. This technology has direct implications for the development of an SMA therapy, but also lends itself to a multitude of diseases caused by aberrant pre-mRNA splicing.
INTRODUCTION
Spinal muscular atrophy (SMA) is the second most common autosomal recessive disorder with an incidence of 1 in 6000 (1) and a carrier frequency of one in 35 (2). SMA is caused by the loss of motor neurons and the subsequent atrophy of voluntary muscle groups. Mutations in the gene SMN1 have been linked to SMA development. SMN is ubiquitously expressed in all tissues and is a critical factor in a variety of RNA pathways. The best characterized role for SMN involves its role in the UsnRNP maturation pathway (3).
In humans, there is a copy gene called SMN2 that primarily produces an alternatively spliced isoform and low levels of full-length protein. The alternative splicing event is due to a silent C to T nucleotide transition 6 nucleotides within exon 7 (4), altering an important SF2/ASF enhancer sequence within this region (5). Though SMN1 and SMN2 encode identical proteins, the silent nucleotide transition initiates an alternative splicing event, which is the default splice variant for SMN2, resulting in a dysfunctional and unstable protein (6). Therefore in the case of SMA, patients have a loss of the SMN1 gene with an increased reliance on the lower levels of full length from SMN2.
SF2/ASF, a serine–arginine (SR)-rich protein, has been shown to interact with an enhancer that overlaps the critical C/T transition in SMN exon 7 and the protein is unable to bind the region in the SMN2 context (5). The C/T transition not only disrupts the SF2/ASF enhancer, but also it creates a novel hnRNPA1-dependent splicing silencer (7). Other SR or SR-like proteins such as hTra2β1, SRp30c, RBMY and hnRNP-G have all been shown to associate either directly or indirectly with SMN exon 7 (8–10). There are also several cis-acting negative regulatory regions that have been identified that surround SMN exon 7, including a putative negative element upstream of exon 7 called Element 1 (E1), and several additional negative elements including the Extended Inhibitory Context (11–15).
Since all SMA patients retain SMN2, and this gene has the capacity to encode a full-length protein compared with SMN1, the SMN2 gene is an attractive target for potential therapies. A variety of compounds have been used to increase SMN protein levels by modulating SMN2 gene transcription, splicing or protein stability (16–21). In addition, there have been many antisense technologies used to specifically increase SMN protein expression from the SMN2 transcript (11,22–28). These strategies maintain endogenous SMN regulation since the various molecular agents target the native SMN2 transcripts. Recent antisense targets have been identified flanking SMN2 exon 7 (11,26), which add to the number of regulating signals that work in unison, resulting in the overall splice decision. One negative regulator was identified 10 nucleotides downstream of exon 7, called ISSN1 (26), whereas another was found to be upstream of exon 7 between nucleotides −67- and −112 designated E1 (11).
Several nucleic acid-based SMA therapeutics have been developed in recent years, which are designed to promote exon 7 inclusion, and thus increase SMN protein (22,23). Oligonucleotide strategies have been described that functionally inhibit the exon 8 splice acceptor site, thereby increasing utilization of the upstream exon 7 splice acceptor site (25,29). Novel peptide-nucleic acids that comprise an exon 7 antisense domain and a synthetic splicing-activation domain peptide were shown to increase SMN2 exon 7 inclusion (24). Similar methods utilizing bifunctional RNAs have also been shown to increase full-length SMN transcripts as well as SMN protein levels and to derive their name from the presence of two domains: an antisense RNA sequence specific to the target RNA and an untethered RNA segment that serves as a binding platform for splicing factors (30–32). Negatively acting bifunctional RNAs that recruit negatively acting proteins to an intron/exon boundary successfully redirected pre-mRNA splice site decisions (33,34). In this report, we identify a class of bifunctional RNAs that were designed to elevate SMN levels through two modes of action: recruitment of splicing factors; and inhibition of negatively regulating sequences through the antisense component of the bifunctional RNAs. To this end, we genetically verified the identity of a putative intronic repressor and developed bifunctional RNAs that target an inhibitory sequence upstream of exon 7, called E1. Two RNA-binding factors were identified that formed a complex on the E1 region, PTB and FUSEBP. These factors have been shown to be negative regulators in other genetic systems (35–39), further linking the E1 region as a negative regulator of SMN2 exon 7. Bifunctional RNAs were initially characterized in cell culture, and lead candidates were identified that were administered into animal models of SMA. Single-RNA injections were able to illicit a robust induction of SMN protein in the brain and throughout the spinal column of neonatal SMA mice. In a severe model of SMA, mean lifespan was extended following the delivery of bifunctional RNAs. This technology has direct implications for the development of an SMA therapy, but also lends itself to a multitude of diseases caused by aberrant pre-mRNA splicing.
RESULTS
Deletion of E1 increases full-length expression from SMN2
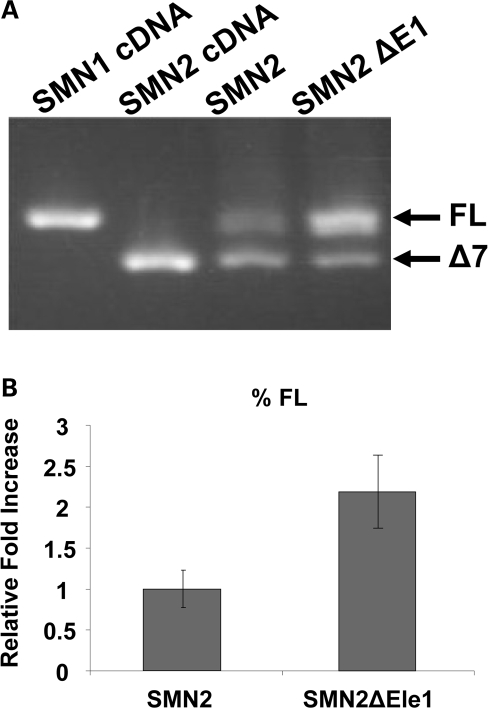
The previously described bifunctional RNAs contained an antisense sequence that would anneal to a critical regulatory region within exon 7 (31). As a means to enhance the effectiveness of a new class of bifunctional RNAs, we hypothesized that targeting an intronic regulatory element that inhibited the inclusion of SMN2 exon 7 with the antisense component of a bifunctional RNA would result in enhanced full-length expression. The two mechanisms of action that could lead to enhanced full-length expression are the inhibition of the repressor via the antisense component, and increasing the local concentration of positively acting SR protein via the SR-binding motifs. A previous report identified a potential negative regulatory element 67 nucleotides 5′ of SMN exon 7, referred to as E1 (11). E1 was characterized only in a heterologous exon-trapping vector system; therefore, we first sought to confirm the activity of this region in a more native genetic context. A small deletion comprised of the E1 region was engineered into an otherwise wildtype genomic SMN2 minigene, and RNA expression patterns were analyzed by RT–PCR. Deletion of the E1 regulatory region dramatically altered SMN2 splicing such that the majority of SMN2-derived transcripts were full length compared with the normally low levels of full-length transcript (Fig. 1A). Quantitation of RT–PCR confirmed an ∼2-fold increase of exon 7 inclusion when E1 was deleted (Fig. 1B, t-test P > 0.009).
Figure 1.
E1 is a negative regulator of SMN 2 exon 7 inclusion. (A) RT–PCR of SMN minigenes (1 µg) transfected into HeLa cells, and total RNA was isolated after 24 h of transfection. Bands of exon 7 and Δ7 are shown by the use of previously published SMN minigene-specific primers. (B) Quantification of RT–PCR performed in triplicate on transfected cells utilizing a Cy3 fluorescent primer pair specific to SMN minigene; independent experiments repeated three times. This quantification shows an ∼2-fold increase of splicing to exon 7 when E1 is deleted. P-value ≤ 0.009.
PTB and FUSE-BP interact with E1
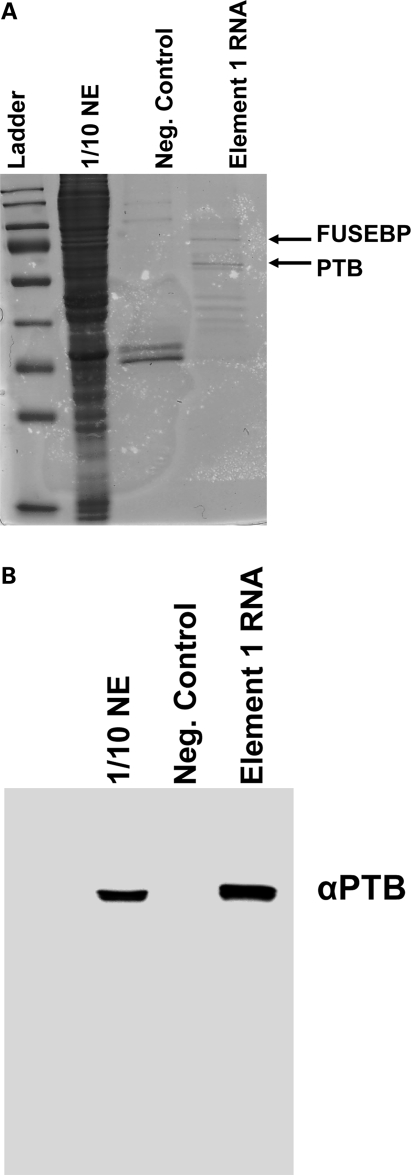
To extend the genetic analysis and identify potential regulatory proteins that mediate the repressive activity of E1, RNA affinity chromatography was performed with a biotinylated synthetic RNA consisting of the entire E1 region. E1 RNA or a similarly sized control RNA was incubated in vitro with splicing-competent HeLa nuclear extract, bound fractions were washed extensively and RNA-associated factors were resolved on a denaturing gel (Fig. 2A). Two bands were specifically enriched in the E1 reactions compared with control reactions that used a similarly sized control RNA (Fig. 2A). The two specific bands were excised and analyzed by MALDI-TOF. Two previously described RNA-binding proteins were identified: PTB and FUSEBP (Fig. 2A). As a confirmatory measure for this complex, an additional RNA affinity chromatography was performed and the gel was probed with an anti-PTB antibody, confirming the MALDI-TOF results (Fig. 2B). These results are consistent with E1 functioning as a negative regulator, as PTB has been shown to function as a potent repressor of splicing in a variety of cellular contexts (35,40). Additionally, FUSE-BP has been implicated in the regulation of mRNA expression including GAP-43 (38), c-myc and TF-IIH (41–43). Collectively, these results provide genetic and biochemical evidence that E1 is a bona fide negative regulator of SMN2 exon 7 inclusion. This evidence validates the strategy of targeting E1 with bifunctional RNAs as a means to further enhance SMN2 exon 7 inclusion.
Figure 2.
Identification of PTB and FUSE-BP proteins bound to E1. (A) RNA affinity chromatography with subsequent Coommassie blue stain identified at least two specific proteins interacting with E1 but not with control RNA. The indicated bands were excised from the gel, and MALDI-TOF identified PTB and FUSE-BP as the two unique bands. (B) Confirmation of PTB interaction with E1. RNA-protein affinity chromatography was performed, and the western blot was developed using an anti-PTB-specific antibody. PTB specifically interacts with E1 RNA and not with the negative control RNA.
Development of E1-targeted bifunctional RNAs
To identify an optimal antisense region that would effectively block E1 activity, a series of overlapping antisense RNAs ranging from 19–24 nucleotides in length were generated that sequentially progressed through the entire E1 sequence. However, none of the sequential antisense RNAs effectively inhibited E1 activity extensively (data not shown). An additional antisense RNA was designed that consisted of two non-sequential target sequences that effectively flank the entirety of the E1 region (Fig. 3). This strategy is similar to RNAs that were developed to functionally inhibit splice site recognition and to promote exon skipping in the dystrophin gene (44). Unlike the initial E1 antisense RNAs, the non-sequential antisense RNA effectively stimulated SMN protein levels (data not shown). On the basis of this antisense sequence, two bifunctional RNAs were generated that contained binding platforms for SF2/ASF or hTra2β. These are well-described splicing factors that promote SMN2 exon 7 inclusion (5,8) and functioned efficiently in the first generation of bifunctional RNAs. Additionally, a U7 stem-loop was included within the bifunctional RNA sequences since it has been shown that antisense effects can be increased by including a U7 snRNA-derived stem loop (25,44–46). The bifunctional RNA sequences were cloned into the pMU2 vector under the expression control of a U6 or CMV promoter (31). A separate CMV-driven green fluorescent protein (GFP) is present on the vector and allows the detection of transfected cells.
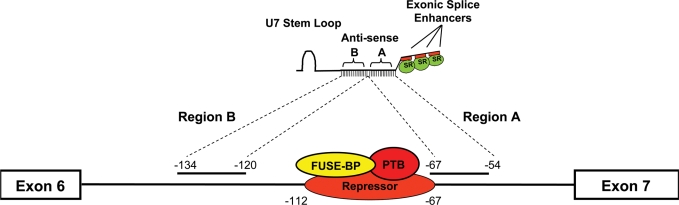
Figure 3.
Schematic of E1-specific bifunctional RNAs. The organization of the bifunctional RNA is illustrated with the split antisense domain targeting the sequences flanking E1, with the non-specific linker sequence and three tandem repeats of high-affinity exonic splice enhancer sequences for either hTra2β1 or SF2/ASF. In addition, the schematic indicates the orientation of the U7-sm-opt sequence found on the plasmid-derived bifunctional RNAs.
Plasmid-derived E1 bifunctional RNAs increase SMN levels in patient fibroblasts
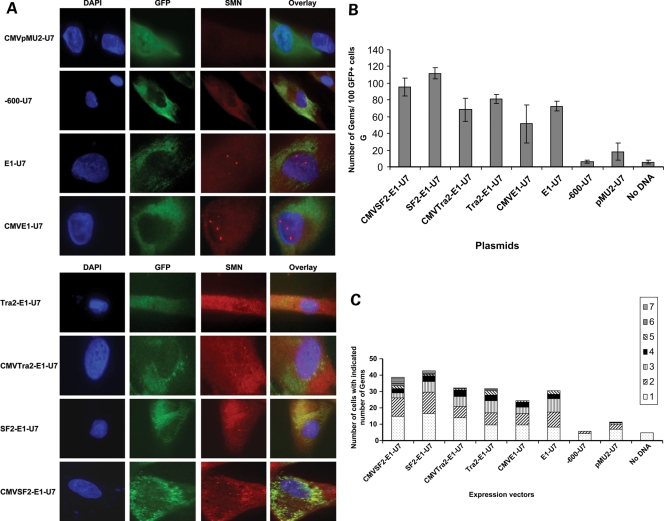
In a first step towards the development of E1-targeted bifunctional RNAs, we transfected primary SMA type 1 patient fibroblasts, 3813 cells, with our pMU2-based plasmids expressing the various E1 RNAs. These cells are a valuable model for assessing bifunctional RNA activity since they lack SMN1 and consequently express very low levels of SMN and contain very few nuclear gems (47). Plasmids were transfected into the 3813 patient fibroblasts and cells visualized 48 h later by indirect immunofluorescence. Plasmid entry was identified by CMV-driven GFP expression and representative pictures are shown (Fig. 4A). Bifunctional RNAs were expressed under the control of the U6 promoter or the CMV promoter (as indicated). In addition to the bifunctional RNAs, control RNAs were also generated, including a similarly sized irrelevant RNA expressed from CMV-pMU2-U7 and an antisense RNA that targeted a region approximately 600 nucleotides upstream relative to exon 7. As expected, neither of these control RNAs increased SMN levels.
Figure 4.
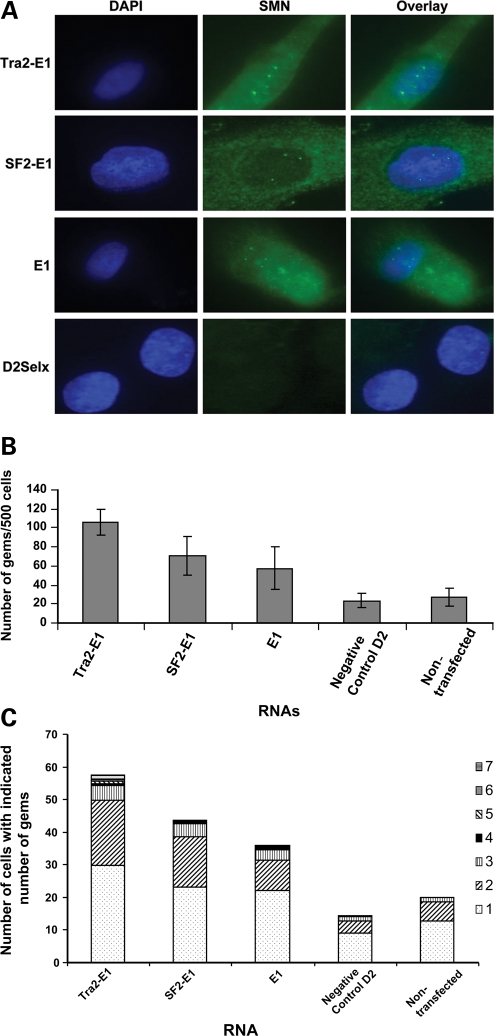
Increase in SMN protein in the presence of plasmid expressed bifunctional RNAs. (A) SMA type 1 fibroblasts (3813 cells) were transiently transfected with 1 µg of plasmid DNA producing the indicated RNAs. Cells were incubated for 48 h and an immunoflourescence was performed. Unless indicated as the CMV promoter, the U6 promoter is driving RNA expression. Transfected cells are identified by GFP expression. Pictures are of representative cells found for each sample. (B) 3813 cells transfected with plasmids were randomly selected and gem numbers compiled. A total of 100 GFP-positive cells were observed and SMN-positive foci in the nucleus were counted (n = 3 and error bars are ±STD). (C) Gem data compiled and now expressed as the number of SMN-positive gems per nucleus (n = 3).
Expression of the E1-bifunctional RNAs or the E1 antisense RNA consistently increased SMN staining in the cytoplasm and in nuclear gems (Fig. 4A). Consistent with these observations, analysis of 100 transfected cells, as determined by GFP expression, demonstrated that gem numbers were significantly increased in cells treated with the bifunctional RNA vectors (Fig. 4B). Control RNAs did not increase the SMN-positive gem numbers above the background of non-transfected cells. However, the antisense-alone RNA did increase the gem number to about 70 nuclear gems in 100 cells (Fig. 4B [all E1-specific RNAs were statistically higher than controls; one-way ANOVA P < 0.0001)]. The E1 bifunctional RNAs increased gem numbers higher than the gem numbers produced from the E1 antisense alone (Fig. 4B); the average of all bifunctional RNAs are ∼30% higher than antisense alone (one-way ANOVA of U6-driven bifunctional RNAs and antisense RNA P = 0.0005, Fig. 4B). This suggests a potential dual mode of action from the E1 bifunctional RNAs: blocking a negative regulatory element in cis (E1) and recruiting positive regulatory factors, SR proteins. Interestingly, the RNAs expressed by a U6 promoter were ∼15–30% higher than the CMV-driven counterparts (Fig. 4B). These results were also tabulated as ‘gems per nucleus’ to demonstrate that a small population of cells did not have an aberrantly high number of gems, thus skewing the results. From these results, the majority of transfected cells expressing the E1 RNAs contained one or two gems per nucleus, consistent with the numbers observed with carrier fibroblasts, 3814 cells (Fig. 4C).
2′-O-methyl-modified bifunctional RNAs increase SMN protein levels in primary patient fibroblasts
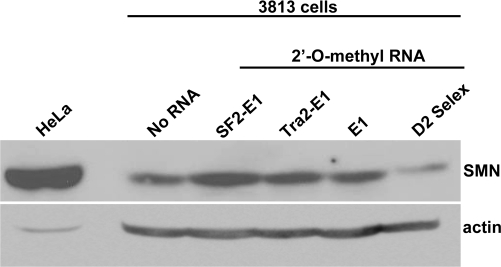
As it is not currently practical to deliver plasmid-derived bifunctional RNAs as an SMA therapeutic, the previous qualitative analysis of bifunctional RNAs served as a proof-of-concept screen to determine whether E1 bifunctional RNAs could increase SMN levels. 2′-O-methyl RNAs are a more tractable therapeutic candidate and were therefore selected as the chemistry to analyze in the subsequent generation of bifunctional RNAs. The addition of the 2′-O-methyl chemistry stabilizes the RNA and allows for sustained activity in vivo. Manufactured RNA/DNA homologues have been used extensively in other disease models with encouraging results (22,23,48–50). To initially verify that the 2′-O-methyl RNAs exhibited similar activities compared with the plasmid-derived RNAs, 2′-O-methyl bifunctional RNAs were transfected into 3813 cells, and SMN protein was visualized by indirect immunofluorescence after 48 h. Following transfection of the bifunctional RNAs or E1 antisense RNA, SMN protein levels increased in the cytoplasm, and gems were more readily detectable (Fig. 5A). Gem counts were performed on cell populations transfected with the various RNAs; however, in these experiments, it was not possible to identify the transfected cells since the RNA molecules were not tagged with a fluorescent moiety. Therefore, a larger number of cells were examined in each transfected population. Five hundred cells were counted in at least three separate experiments and the tabulated results demonstrate that E1 and the E1 bifunctional RNAs significantly increased SMN-containing gems (Fig. 5B, one-way ANOVA P < 0.0001) and that the majority of gem-positive cells contained one to two gems (Fig. 5C). In addition, the bifunctional RNAs increased gems numbers above E1 antisense alone (Fig. 5B, one-way ANOVA P = 0.015). The negative control RNA, D2-Selx, did not induce SMN protein (Fig. 5A). Consistent with these results, the analysis of steady-state levels of SMN protein by western blot in extracts from transfected 3813 cells demonstrates that the 2′-O-methyl bifunctional RNAs elevate total SMN protein levels above untransfected and control-transfected levels (Fig. 6). These data identify RNAs that are capable of elevating SMN levels in a variety of cell-based assays and suggest that these molecules may be capable of increasing SMN levels in the more complex environment of the SMA mouse model.
Figure 5.
Increase in SMN expression after the transfection of 2′-O-methyl-modified bifunctional RNAs. (A) SMA type 1 fibroblasts (3813 cells) were transiently transfected with 100 ng of 2′-O-methyl bifunctional RNAs for 48 h. Immunoflourescence staining was performed and images are of representative cells. (B) Five hundred 3813 cells were randomly counted (n = 4) and the total gem number shown (error bars are ±STD). (C) Gem data compiled and expressed as the number of gems per nucleus (n = 4).
Figure 6.
2′-O-methyl bifunctional RNAs increased total SMN protein levels in SMA fibroblasts. Subconfluent 3813 cells were transfected with 100 ng of the indicated RNAs and subsequently incubated for 48 h. Relative SMN protein levels were determined by western blot. HeLa cellular extracts were included as a size control.
CNS delivery of 2′-O-methyl RNA into SMA mice increases SMN protein levels
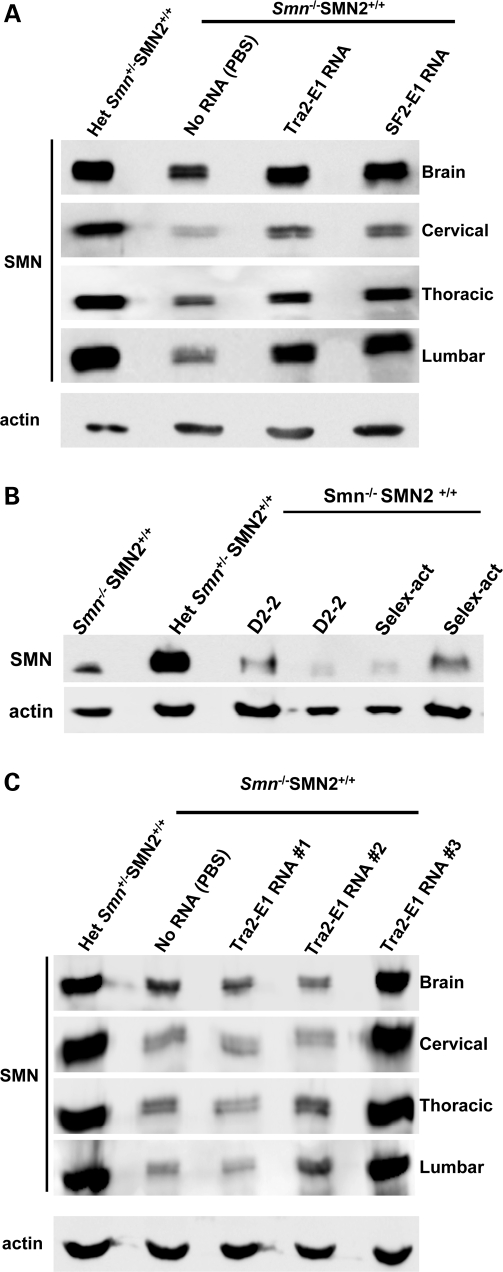
The subsequent experiments utilize a well-described animal model of SMA (51). These animals lack endogenous murine Smn, but express transgenes for human SMN2 and an SMNΔ7 cDNA (Smn−/−; SMN2+/+; SMNΔ7+/+). To deliver the RNAs to the central nervous system (CNS), we performed intra-cerebral ventricular (ICV) injections on 2-day-old SMA mice with the 2′-O-methyl RNAs (30,52). Twenty-four hours after ICV delivery of the RNAs, tissues were collected and SMN protein levels were analyzed (Fig. 7A). The delivery of either modified bifunctional RNA increased SMN protein levels in the brain (Fig. 7A). Additionally, this delivery procedure also likely allowed for the transportation of the RNAs throughout the spinal column as evidenced by the increase in total SMN levels in extracts generated from the cervical, thoracic and lumbar segments of treated mice (Fig. 7A). The analysis of 33 injected animals demonstrated that tissue extracts from 8 out of 13 (Tra2-E1) or 5 out of 10 (SF2-E1) treated animals exhibited a 2-fold or greater increase in SMN protein levels (throughout the CNS, data not shown). Localized delivery and the delivery procedure did not contribute to the SMN induction as evidenced by the low levels of SMN in control RNA-treated animals (Fig. 7B).
Figure 7.
ICV injection of 2′-O-methyl bifunctional RNAs increases SMN protein levels throughout the CNS. (A) Post-natal day (PND) 2 SMA mice were ICV-injected with 4 µg of the modified bifunctional RNAs, SF2-E1 and Tra2-E1 or PBS. Indicated tissues were isolated 24 h after injections. Western blots were done in quadruplicate, and a representative blot is shown. Multiple mice were injected and tested [Tra2-E1 (n = 13); SF2-E1 (n = 10), data not shown]. (B) Control RNAs do not elevate SMN protein levels. Four micrograms of modified control RNAs were delivered via ICV injection into PND 2 SMA mice and brain tissue was isolated 24 h post-injection. SMN protein levels were observed by western blot, which were done in triplicate, and a representative blot is shown. D2-2 and Selex-act are the two previously described RNAs, corresponding to an SMN intron 7 antisense (D2-2) or three tandem repeats of an in vitro affinity-determined binding motif for hnRNP-A1 (Selex-act). (C) ICV injection of Tra2-E1-modified bifunctional RNA increases SMN protein levels 5 days after a single injection. 2 PND SMA mice were ICV-injected with 4 µg of Tra2-E1 RNA. Five days following injection, the indicated tissues were harvested and SMN western blot performed. Western blots were done in quadruplicate and representative blot is shown.
To examine the sustainability of the SMN increase from a single injection, similar single-ICV injections were performed on PND 2 SMA mice; however, tissues were collected 5 days post-injection. The subsequent SMN protein levels were detected by western blot (Fig. 7C). Although not as reproducible, in all animals, we observed an SMN increase in tissue from one out of three animals injected with the Tra2-E1 RNA such that levels were comparable to the unaffected heterozygote animal (Fig. 7C). A lower percentage of animals examined at the 5 day time point exhibited a 2-fold or greater increase in SMN levels; however, two out of nine animals did exhibit a 2-fold increase (data not shown). Similar to the previous injections, bifunctional activity is detected distal from the injection site as evidenced by the increase in SMN levels in various spinal cord sections, including the lumbar section (Fig. 7C). These results suggest that modified RNAs can have a sustained effect on the SMN protein levels. Furthermore, we have shown an increase in the SMN protein in the tissue that is affected by SMA and this increase can be sustained for a period of time.
CNS delivery of 2′-O-methyl bifunctional RNA increases weight gain in a more severe model of SMA mice
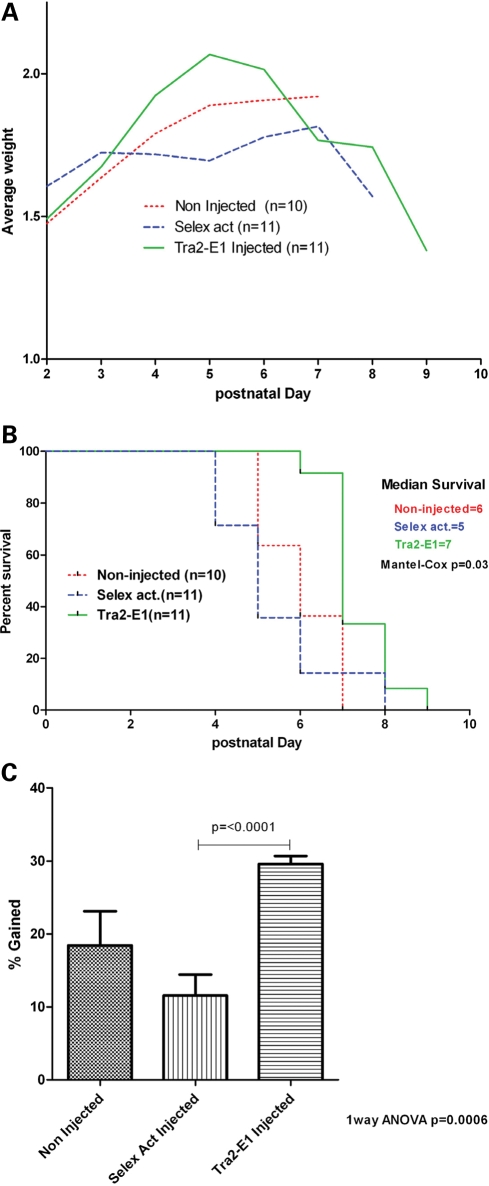
To examine the potential for physiological changes in the SMA mice, we used a more severe mouse model for two reasons: (i) the severe mouse model experimental duration is much shorter than the Δ7 mice and the ability to deliver via repeated ICV injection is limited; (ii) the severity of the model could potentially be a benefit because a small change would be measurable rather than in a less severe model. The severe mouse model SMN2+/+Smn−/− (53) has an average lifespan of <5 days with a very severe form of SMA that phenotypically coincides with the human form. These mice usually do not gain weight significantly after birth and progressively lose weight until death. To examine a potential change in phenotype of these mice, two ICV injections were administered on PND 2 and 4 with 6 µg of RNA of either Tra2-E1 or the negative control ‘Selex’. After the first injection, mice were weighed daily and data compiled (Fig. 8A). Although randomly assigned to each test group, Tra2-E1 mice were lighter at birth than the negative control group, however reached a higher average peak weight on PND 5 than animals in the negative control injection or non-injected groups (Fig. 8A). A Kaplan–Meier curve shows an increased lifespan for the mice ICV-injected with Tra2-E1 (Fig. 8B, Mantel–Cox P = 0.03). Increased weight gain becomes clear when graphing the percent weight gain from PND 2 until peak weight. Mice injected with Tra2-E1 show a significant increase in weight gained from PND 2 to peak weight than either non-injected or ‘Selex’ RNA-injected mice (Fig. 8C, one-way ANOVA P = 0.0006, and t-test Tra2-E1: Selex act P ≤ 0.0001). These data are in agreement with western blot data in the Δ7 mouse model, where the ‘Selex’ RNA treatment did not increase SMN protein levels. This suggests that the action of both Tra2-E1 and the negative control is similar in both mouse models and that bifunctional RNAs can not only elevate SMN levels in the CNS, but can also lessen the severity of the SMA phenotype in SMA mice.
Figure 8.
ICV injection of 2′-O-methyl bifunctional RNA increases weight in a severe mouse model of SMA. (A) 2 PND severe mice were ICV-injected on PND 2, 4 with 6 µg of either Tra2-E1 or Selex activation RNA. Mice were weighed everyday post-first injection and graphed. (B) Kaplan–Meier survival curve depicts a trend towards increased life expectancy for Tra2-E1-injected mice (Mantel–Cox P = 0.03). (C) Percent weight gained post-first injection until maximal weight was graphed. Tra2-E1-injected mice showed a significant weight gain from birth to peak over non-injected and a negative control (Selex act) injected (one-way ANOVA P = 0.0006). There is a significant difference between Tra2-E1- and Selex act-injected mice (t-test P = < 0.0001).
DISCUSSION
In this study, we verified the negative activity of E1 in a more natural context than previously published. In addition, we identified two new proteins as the possible candidates to explain why E1 has a negative effect on SMN2 exon 7 splicing, PTB and FUSE-BP. We used these data as a rational for a new antisense technology directed to disrupt E1. We showed that these RNAs can increase gems, a nuclear hallmark of natural SMN protein distribution, both by plasmid and 2′-O-methyl RNA transfections. Furthermore, upon ICV injection of the 2′-O-methyl bifunctional RNAs into SMA mice, we observed an increase of the SMN protein to levels similar to the heterozygous mice throughout the entire length of the spinal cord, and have the possibility to sustain this effect as long as 5 days after injection with the Tra2-E1 RNA.
The proteins FUSEBP and PTB identified to bind to E1 RNA have been implicated in de-regulating other transcripts. PTB has been identified as a repressor of splicing for α-tropomyosin (54), c-src (37) and FGFr-2 (36), among others. Additionally, FUSEBP has been identified to inhibit TF-IIH (42), and plays a role in c-myc expression (41,55). This supports the data shown here that both FUSEBP and PTB police RNA expression with implication for both being negative regulators.
The use of antisense RNAs in potential therapies is expanding to include many different diseases including Duchenne muscular dystrophy (22,23,25,56–60), and amyotrophic lateral sclerosis and many others (50). These types of translational approaches are moving towards clinical trials and are examples of future directions for other genetic diseases such as SMA (61). In the SMA field, antisense RNAs (22,23,25) and negative bifunctional RNAs (30) can increase SMN protein levels. The later increased SMN protein levels in the brain of SMA mice via ICV injection. In regard to the loop out mechanism we are purposing, we use that statement to describe a general imposition of structural constraints found around the region the antisense is targeting.
ICV delivery has previously been described in a variety of genetic contexts, and this strategy has been used successfully to deliver several viral-based gene vectors (62–64). Additionally, negatively acting bifunctional RNAs have been used in the SMA mouse model and were shown to increase SMN 24 h post-injection in brain extracts (30). This delivery paradigm represents an intriguing possibility for direct delivery to the CNS for therapeutic oligonucleotides, small molecules and viral vectors. As a means to determine the functionality of SMN-inducing compounds in a relevant in vivo context, ICV delivery provides an excellent platform. The severe mouse model data suggest that ICV delivery of bifunctional RNAs can increase the weight gained from birth to peak in a severe model of SMA and suggest a trend towards increasing the lifespan of these mice. The mouse data are in accord with the Δ7 western blot data, where the Selex act RNA did not have an effect on the SMN protein levels in the brain; in the severe mouse model, it did not increase weight gain or lifespan. However, it is clear that additional experimentation will be needed to determine whether the physiology of neonatal SMA animals is a tractable model that can be used to predict the potential benefit of various therapeutic strategies.
One outstanding question in the SMA field relates to the temporal requirement of SMN, and when a therapeutic should be administered for maximal effect. Another challenge facing SMA therapy is the necessity for the therapy to cross the blood–brain barrier. Other studies have utilized antisense RNA therapies directly delivered to ventricles in order to overcome this challenge (65). It is also possible to express these small RNAs utilizing recombinant adeno-associated virus (rAAV). This virus is an ideal candidate for gene therapy, as it is replication-deficient and certain AAV serotypes have been shown to have high tropism for both muscles and neurons and can utilize retrograde transport to travel from muscle to neurons in vivo. Previous studies have successfully utilized the retrograde transport ability of a pseudotyped lentivirus to deliver SMN cDNA (66). Although these data have direct implications for SMA, this type of technology has the potential to impact a variety of genetic conditions that are due to aberrant pre-mRNA splicing.
MATERIAL AND METHODS
Cloning
The E1 deletion construct was derived from the SMN2 minigene plasmid, and the following primers (IDT, Coralville, IA, USA) were used to delete E1 by overlapping PCR. The orientation of the E1 region deletion is indicated by a Δ symbol. SMN E1 deletion: 5′-CTT AAT TTC TGA TCA TAT TTT GTT GAA TAA AAT AAG TΔCT ATC TAT ATA TAG CTA TCT ATG TCT ATA TAG C-3′ and 5′-GCT ATA TAG ACA TAG ATA GCT ATA TAT AGA TAG ΔACT TAT TTT ATT CAA CAA TAA ATG ATC AGA AAT TAA G-3′. The U7-Opt-sm sequence was cloned into the pMU2 vector by overlapping PCR with DNA primers (IDT): Frw, 5′-CCG CGG TCC TAG GAG CAT GCT AAA AAA AGG GGT TTT CCG ACC GAA GTC AGA AAA CCT GCT CCA AAA ATT ACT AGT TAA GCT GAT ATC TGA GC-3′; Rev, 5′-GC TCA GAT ATC AGC TTA ACT AGT AAT TTT TGG AGC AGG TTT TCT GAC TTC GGT CGG AAA ACC CCT TTT TTT AGC ATG CTC CTA GGA CCG CGG-3′ paired with plasmid-specific primers previously published (27). pMU2-U7 plasmid was subsequently used in the cloning of the U6 promoter-driven bifunctional clones. To make the CMV-driven bifunctional RNAs, the U6 promoter from the pMU2-U7 plasmid was digested with Pme1 and Sal1; then using CMV-specific primers from the parent EGFP promoter, CMV was subsequently cloned into the sites to make a CMV-pMU2-U7 parent plasmid. The bifunctional clones were generated with annealed complementary pairs of DNA oligonucleotides (IDT) that were cloned into the pMU2-U7 vector between the BamHI and Spe1 sites. Sequences for the E1-targeting clones are as follows: E1, top, 5′-GAT CCC TAT ATA TAG ATA GTT ATT CAA CAA AA-3′, bottom, 5′-CTA GTT TTG TTG AAT AAC TAT CTA TAT ATA GG-3′; Tra2-E1, top, 5′-GAT CCG AAG GAG GGA AGG AGG GAA GGA GGA GAT CTC TAT ATA TAG ATA GTT ATT CAA CAA-3′, bottom, 5′-CTA GTT TTG TTG AAT AAC TAT CTA TAT ATA GAG ATC TCC TCC TTC CCT CCT TCC CTC CTT CG-3′; SF2-E1, top, 5′-GAT CCC ACA CGA CAC ACG ACA CACG AAG ATC TCT ATA TAT AGA TAG TTA TTC AAC AAA A-3′, bottom, 5′-CTA GTT TTG TTG AAT AAC TAT CTA TAT ATA GAG ATC TTC GTG TGT CGT GTG TCG TGT GG-3′.
2′-O-methyl bifunctional RNA
The following modified oligos were modified at every base with 2′-O-methyl groups (IDT): Tra2-E1, 5′-GAA GGA GGG AAG GAG GGA AGG AGG CUA UAU AUA GAU AGU UAU UCA ACA AA-3′; SF2-E1, 5′-CAC ACG ACA CAC GAC ACA CGA CUA UAU AUA GAU AGU UAU UCA ACA AA-3′. Negative control RNAs: D2Selx, 5′-AAG AUU AAA GAG UAA ACG UCC GGU ACC UAG GGA UAG GGA UAG GGA-3′; Selex-act, 5′-UAG GGA UAG GGA UAG GGA-3′. The modified oligos differ from the plasmid-expressed bifunctional RNAs by the lack of a non-specific 6 nucleotide spacer sequence between the antisense domain and the SR recruitment domain, and furthermore do not have the U7-opt-sm sequence.
E1Δ RT–PCR
In vivo splicing assays were performed essentially as described previously. In brief, subconfluent HeLa cells were plated on a 60 mm dish and transfected with 1.0 µg of minigene plasmid using Lipofectamine 2000 per the manufacturer’s directions (Invitrogen). Total RNA was isolated 24 h post-transfection using TRIzol reagent (Invitrogen), and first-strand cDNA synthesis was performed using Super Script II (Invitrogen). To ensure only transfected minigene template was amplified, PCR primers that specifically anneal to the plasmid backbone common to all SMN minigene plasmids were used in the subsequent PCR amplification.
RNA affinity chromatography
The following 5′-biotinylated RNA corresponding to E1 and a negative control RNA, respectively, were ordered (IDT): 5′-GUA AAA UGU CUU GUG AAA CAA AAU ACU UUU UAA CAU CCA UAU AAA-3′, 5′-AGU CCU CAA CUU AGC CUC UAA CUU-3′. Control RNA was obtained (IDT) with 5′ biotinylation. This control was utilized as a negative control for interaction with PTB and was composed of entirely purines: 5′-GAA GAA AGA GAA GAA AGA GAA GAA AGA-3′. RNA affinity chromatography was done as shown previously (23), but briefly, the biotinylated RNAs were incubated with splicing-competent HeLa cell nuclear extract and avidin-agarose beads under splicing conditions, in the presence of heparin as a non-specific competitor. The RNA and any interacting proteins were immunoprecipitated and washed. The proteins were then resolved on a 10% SDS–PAGE gel.
Protein identification
The E1 interacting proteins were visualized utilizing Coommassie blue stain. Two specific bands were excised from the gel and sequenced using MALDI-TOF (University of Missouri Charles W. Gehrke Proteomics Center).
Immunofluorescence imaging
For all immunofluorescence staining, subconfluent type 1 patient fibroblast cells (3813 cells, Coriell Cell Repositories) were transfected in eight-chambered slides (BD Biosciences, Bedford, MA, USA) with first plasmid clones using Lipofectamine 2000 reagent (Invitrogen) or 2′-O-methyl RNA oligos using the same reagent, incubated for 48 h in DMEM supplemented with 10% fetal bovine serum and antibiotics. Transfected cells were fixed with a solution of acetone/methanol (1/1 by volume) and then washed with phosphate-buffered saline (PBS) (Gibco). Fixed and washed cells were then blocked in PBS + 5% BSA and then washed again in PBS. A pooled group of three previously described anti-SMN monoclonal antibodies was added, diluted 1:10 in PBS + 1.5% BSA (17). Cells were washed again in PBS and a secondary monoclonal antibody, an anti-mouse conjugated to Texas red (Jackson), or conjugated to FITC (Sigma) for either the plasmid or 2′-O-methyl-transfected cell, respectively, diluted 1:200 in PBS + 1.5% BSA. After washing in PBS, DAPI was added to each chamber for 5 min, and samples were washed again. Chambers were then fitted with cover slips using mounting media [DABCO, 2.3% (w/v), 10% PBS, 87.7% glycerol] and sealed with nail polish. Microscope images were captured using a Nikon Eclipse E1000 using Meta-Morph software. Cells for gem counting were chosen at random. Each randomly chosen nucleus was examined for nuclear gem accumulation and gems were counted.
Western blots
For the PTB western blot, proteins were transferred to PVDF membranes (Millipore), blocked overnight in 3% NFDM in TBST and probed with α-hnRNPI (PTB) antibody (Santa Cruz Biotechnology). For the 2′-O-methyl RNA western blot, subconfluent patient fibroblast cells (3813) plated on 60 mm dishes were transfected with 100 ng of each indicated RNA using Lipofectamine 2000 reagent (Invitrogen). After 48 h of transfection, the cells were collected and proteins were resolved on a 12% SDS–PAGE. The gel was transferred to PVDF membrane (Millipore) and SMN immunoblot was performed using an SMN monoclonal antibody. A goat anti-mouse secondary antibody conjugated to horseradish peroxidase was used to detect the presence of the target protein by chemiluminescence (Pierce). Western blot was performed in triplicate and representative results are shown. For the SMA mouse western blots, injected mice were handled by Animal Care and Use Committee (ACUC) regulations, and tissues indicated were collected at selected time points and immediately placed in liquid nitrogen. Tissue was placed at −80°C until ready for analysis. Roughly 100 mg of tissue was isolated and homogenized in JLB buffer [50 mm Tris–HCl, pH 7.5, 150 mm NaCl, 20 mm NaH2(PO4), 25 mm NaF, 2 mm EDTA, 10% glycerol, 1% Triton x-100 and 1× PIC (Roche, Indianapolis, IN, USA)] and equal amounts of subsequent protein were loaded onto 12% SDS–PAGE. The SMN immunoblot was performed using a mouse SMN-specific monoclonal antibody (BD Biosciences, San Jose, CA, USA) diluted 1:3000 in TBST in 1.5% dry milk. Then blots were visualized by chemiluminescence on a Fujifilm imager, LAS-3000, and the corresponding LAS-3000 Image Reader software. To verify loading, the westerns were then stripped using H2O2 for 30 min at room temperature and re-probed with anti-β-actin rabbit and anti-rabbit HRP. Western blots were done in quadruplicate or more and representative blots are shown.
Animal injections
All animal experiments were carried out in accordance with protocols approved by the Animal Care and Use Committee of the University of Missouri. SMN2+/+, SMNΔ7+/+, Smn−/− mice (51) or SMN2+/+ Smn−/− mice (53) were genotyped at day of birth, designated as day 1, and injected on day 2. Single-intracerebralventricular (ICV) injections were performed on PND 2 neonates as described previously (30,52,67) for the Δ7 mice (Fig. 7). For the severe mouse model, mice were ICV-injected on PND 2 and 4 (Fig. 8). Briefly, mice were immobilized via cryo-anesthesia and injected using microliter-calibrated sterilized glass micropipettes. The injection site was approximately 0.25 mm lateral to the sagittal suture and 0.50–0.75 mm rostral to the neonatal coronary suture. The needles were inserted perpendicular to the skull surface using a fiber-optic light (Boyce Scientific Inc.) to aid in illuminating pertinent anatomical structures. Needles were removed after 15 s of discontinuation of plunger movement to prevent backflow. Mice recovered in 5–10 min in a warmed container until movement was restored. Single injections of 4 µg of each 2′-O-methyl oligonucleotides were delivered via ICV described earlier for Δ7 mice, and severe mice were injected on PND 2 and 4 with 6 µg of RNA (Fig. 8).
FUNDING
This work was supported by grants from FightSMA (C.L.L.) and the National Institutes of Health (C.L.L, R01 NS41584; R01 HD054413).
ACKNOWLEDGEMENTS
We thank John Marston for expert technical assistance and Gordon Lutz for assistance with ICV delivery strategies.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Crawford T.O., Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 2.Feldkotter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;2:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meister G., Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 6.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 7.Kashima T., Rao N., Manley J.L. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 2007;104:3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl Acad. Sci. USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann Y., Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum. Mol. Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- 10.Young P.J., DiDonato C.J., Hu D., Kothary R., Androphy E.J., Lorson C.L. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2beta1. Hum. Mol. Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- 11.Miyajima H., Miyaso H., Okumura M., Kurisu J., Imaizumi K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J. Biol. Chem. 2002;277:23271–23277. doi: 10.1074/jbc.M200851200. [DOI] [PubMed] [Google Scholar]
- 12.Singh N.N., Androphy E.J., Singh R.N. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit. Rev. Eukaryot. Gene Expr. 2004;14:271–285. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.30. [DOI] [PubMed] [Google Scholar]
- 13.Singh N.N., Androphy E.J., Singh R.N. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh N.N., Androphy E.J., Singh R.N. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004;315:381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 15.Singh N.N., Singh R.N., Androphy E.J. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kernochan L.E., Russo M.L., Woodling N.S., Huynh T.N., Avila A.M., Fischbeck K.H., Sumner C.J. The role of histone acetylation in SMN gene expression. Hum Mol. Genet. 2005;14:1171–1182. doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- 17.Wolstencroft E.C., Mattis V., Bajer A.A., Young P.J., Lorson C.L. A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum. Mol. Genet. 2005;14:1199–1210. doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- 18.Mattis V.B., Rai R., Wang J., Chang C.W., Coady T., Lorson C.L. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum. Genet. 2006;120:589–601. doi: 10.1007/s00439-006-0245-7. [DOI] [PubMed] [Google Scholar]
- 19.Brichta L., Hofmann Y., Hahnen E., Siebzehnrubl F.A., Raschke H., Blumcke I., Eyupoglu I.Y., Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 20.Riessland M., Brichta L., Hahnen E., Wirth B. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum. Genet. 2006;120:101–110. doi: 10.1007/s00439-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 21.Sakla M.S., Lorson C.L. Induction of full-length survival motor neuron by polyphenol botanical compounds. Hum. Genet. 2008;122:635–643. doi: 10.1007/s00439-007-0441-0. [DOI] [PubMed] [Google Scholar]
- 22.Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cartegni L., Krainer A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- 25.Madocsai C., Lim S.R., Geib T., Lam B.J., Hertel K.J. Correction of SMN2 pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol. Ther. 2005;12:1013–1022. doi: 10.1016/j.ymthe.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 26.Singh N.K., Singh N.N., Androphy E.J., Singh R.N. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coady T.H., Shababi M., Tullis G.E., Lorson C.L. Restoration of SMN function: delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing. Mol. Ther. 2007;15:1471–1478. doi: 10.1038/sj.mt.6300222. [DOI] [PubMed] [Google Scholar]
- 28.Marquis J., Meyer K., Angehrn L., Kampfer S.S., Rothen-Rutishauser B., Schumperli D. Spinal muscular atrophy: SMN2 pre-mRNA splicing corrected by a U7 snRNA derivative carrying a splicing enhancer sequence. Mol. Ther. 2007;15:1479–1486. doi: 10.1038/sj.mt.6300200. [DOI] [PubMed] [Google Scholar]
- 29.Lim S.R., Hertel K.J. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem. 2001;276:45476–45483. doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 30.Dickson A., Osman E., Lorson C. A negatively-acting bifunctional RNA increases survival motor neuron in vitro and in vivo. Hum. Gene Ther. 2008 doi: 10.1089/hum.2008.067. August 25. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baughan T., Shababi M., Coady T.H., Dickson A.M., Tullis G.E., Lorson C.L. Stimulating full-length SMN2 expression by delivering bifunctional RNAs via a viral vector. Mol. Ther. 2006;14:54–62. doi: 10.1016/j.ymthe.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Skordis L.A., Dunckley M.G., Yue B., Eperon I.C., Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl Acad. Sci. USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gendron D., Carriero S., Garneau D., Villemaire J., Klinck R., Elela S.A., Damha M.J., Chabot B. Modulation of 5′ splice site selection using tailed oligonucleotides carrying splicing signals. BMC Biotechnol. 2006;6:5. doi: 10.1186/1472-6750-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villemaire J., Dion I., Elela S.A., Chabot B. Reprogramming alternative pre-messenger RNA splicing through the use of protein-binding antisense oligonucleotides. J. Biol. Chem. 2003;278:50031–50039. doi: 10.1074/jbc.M308897200. [DOI] [PubMed] [Google Scholar]
- 35.Ashiya M., Grabowski P.J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 36.Carstens R.P., Wagner E.J., Garcia-Blanco M.A. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7388–7400. doi: 10.1128/mcb.20.19.7388-7400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan R.C., Black D.L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irwin N., Baekelandt V., Goritchenko L., Benowitz L.I. Identification of two proteins that bind to a pyrimidine-rich sequence in the 3′-untranslated region of GAP-43 mRNA. Nucleic Acids Res. 1997;25:1281–1288. doi: 10.1093/nar/25.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grossman J.S., Meyer M.I., Wang Y.C., Mulligan G.J., Kobayashi R., Helfman D.M. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA. 1998;4:613–625. doi: 10.1017/s1355838298971448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Liu W., Grabowski P.J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hao Y.H., Lai L.X., Liu Z.H., Im G.S., Wax D., Samuel M., Murphy C.N., Sutovsky P., Prather R.S. Developmental competence of porcine parthenogenetic embryos relative to embryonic chromosomal abnormalities. Mol. Reprod. Dev. 2006;73:77–82. doi: 10.1002/mrd.20358. [DOI] [PubMed] [Google Scholar]
- 42.Liu J., He L., Collins I., Ge H., Libutti D., Li J., Egly J.M., Levens D. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell. 2000;5:331–341. doi: 10.1016/s1097-2765(00)80428-1. [DOI] [PubMed] [Google Scholar]
- 43.He L., Liu J., Collins I., Sanford S., O’Connell B., Benham C.J., Levens D. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–1044. doi: 10.1093/emboj/19.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goyenvalle A., Vulin A., Fougerousse F., Leturcq F., Kaplan J.C., Garcia L., Danos O. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- 45.Suter D., Tomasini R., Reber U., Gorman L., Kole R., Schumperli D. Double-target antisense U7 snRNAs promote efficient skipping of an aberrant exon in three human beta-thalassemic mutations. Hum. Mol. Genet. 1999;8:2415–2423. doi: 10.1093/hmg/8.13.2415. [DOI] [PubMed] [Google Scholar]
- 46.Gorman L., Suter D., Emerick V., Schumperli D., Kole R. Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc. Natl Acad. Sci. USA. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 48.Alter J., Lou F., Rabinowitz A., Yin H., Rosenfeld J., Wilton S.D., Partridge T.A., Lu Q.L. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat. Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 49.Lu Q.L., Rabinowitz A., Chen Y.C., Yokota T., Yin H., Alter J., Jadoon A., Bou-Gharios G., Partridge T. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc. Natl Acad. Sci. USA. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H., et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 52.Coady T.H., Baughan T.D., Shababi M., Passini M.A., Lorson C.L. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS ONE. 2008;3:e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 54.Ramchatesingh J., Zahler A.M., Neugebauer K.M., Roth M.B., Cooper T.A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol. Cell. Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsushita T., Elliger S., Elliger C., Podsakoff G., Villarreal L., Kurtzman G.J., Iwaki Y., Colosi P. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 1998;5:938–945. doi: 10.1038/sj.gt.3300680. [DOI] [PubMed] [Google Scholar]
- 56.Aartsma-Rus A., Bremmer-Bout M., Janson A.A., den Dunnen J.T., van Ommen G.J., van Deutekom J.C. Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscul. Disord. 2002;12 (Suppl. 1):S71–S77. doi: 10.1016/s0960-8966(02)00086-x. [DOI] [PubMed] [Google Scholar]
- 57.van Deutekom J.C., Bremmer-Bout M., Janson A.A., Ginjaar I.B., Baas F., den Dunnen J.T., van Ommen G.J. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum. Mol. Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- 58.Mann C.J., Honeyman K., Cheng A.J., Ly T., Lloyd F., Fletcher S., Morgan J.E., Partridge T.A., Wilton S.D. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc. Natl Acad. Sci. USA. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aartsma-Rus A., Janson A.A., Kaman W.E., Bremmer-Bout M., van Ommen G.J., den Dunnen J.T., van Deutekom J.C. Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am. J. Hum. Genet. 2004;74:83–92. doi: 10.1086/381039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holzbaur E.L., Howland D.S., Weber N., Wallace K., She Y., Kwak S., Tchistiakova L.A., Murphy E., Hinson J., Karim R., et al. Myostatin inhibition slows muscle atrophy in rodent models of amyotrophic lateral sclerosis. Neurobiol. Dis. 2006;23:697–707. doi: 10.1016/j.nbd.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 61.van Deutekom J.C., Janson A.A., Ginjaar I.B., Frankhuizen W.S., Aartsma-Rus A., Bremmer-Bout M., den Dunnen J.T., Koop K., van der Kooi A.J., Goemans N.M., et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 62.Passini M.A., Macauley S.L., Huff M.R., Taksir T.V., Bu J., Wu I.H., Piepenhagen P.A., Dodge J.C., Shihabuddin L.S., O’Riordan C.R., et al. AAV vector-mediated correction of brain pathology in a mouse model of Niemann-Pick A disease. Mol. Ther. 2005;11:754–762. doi: 10.1016/j.ymthe.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 63.Passini M.A., Bu J., Fidler J.A., Ziegler R.J., Foley J.W., Dodge J.C., Yang W.W., Clarke J., Taksir T.V., Griffiths D.A., et al. Combination brain and systemic injections of AAV provide maximal functional and survival benefits in the Niemann-Pick mouse. Proc. Natl Acad. Sci. USA. 2007;104:9505–9510. doi: 10.1073/pnas.0703509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Passini M.A., Watson D.J., Vite C.H., Landsburg D.J., Feigenbaum A.L., Wolfe J.H. Intraventricular brain injection of adeno-associated virus type 1 (AAV1) in neonatal mice results in complementary patterns of neuronal transduction to AAV2 and total long-term correction of storage lesions in the brains of beta-glucuronidase-deficient mice. J. Virol. 2003;77:7034–7040. doi: 10.1128/JVI.77.12.7034-7040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., Mcalonis-Downes M., Wei H., et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Azzouz M., Le T., Ralph G.S., Walmsley L., Monani U.R., Lee D.C., Wilkes F., Mitrophanous K.A., Kingsman S.M., Burghes A.H., Mazarakis N.D. Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2004;114:1726–1731. doi: 10.1172/JCI22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Passini M.A., Wolfe J.H. Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J. Virol. 2001;75:12382–12392. doi: 10.1128/JVI.75.24.12382-12392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Briese M., Esmaeili B., Sattelle D.B. Is spinal muscular atrophy the result of defects in motor neuron processes? Bioessays. 2005;27:946–957. doi: 10.1002/bies.20283. [DOI] [PubMed] [Google Scholar]
- 69.Rajendra T.K., Gonsalvez G.B., Walker M.P., Shpargel K.B., Salz H.K., Matera A.G. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J. Cell. Biol. 2007;176:831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]