Abstract
Topoisomerase IV (Topo IV), an essential ATP-dependent bacterial type II topoisomerase, transports one segment of DNA through a transient double-strand break in a second segment of DNA. In vivo, Topo IV unlinks catenated chromosomes before cell division and relaxes positive supercoils generated during DNA replication. In vitro, Topo IV relaxes positive supercoils at least 20-fold faster than negative supercoils. The mechanisms underlying this chiral discrimination by Topo IV and other type II topoisomerases remain speculative. We used magnetic tweezers to measure the relaxation rates of single and multiple DNA crossings by Topo IV. These measurements allowed us to determine unambiguously the relative importance of DNA crossing geometry and enzymatic processivity in chiral discrimination by Topo IV. Our results indicate that Topo IV binds and passes DNA strands juxtaposed in a nearly perpendicular orientation and that relaxation of negative supercoiled DNA is perfectly distributive. Together, these results suggest that chiral discrimination arises primarily from dramatic differences in the processivity of relaxing positive and negative supercoiled DNA: Topo IV is highly processive on positively supercoiled DNA, whereas it is perfectly distributive on negatively supercoiled DNA. These results provide fresh insight into topoisomerase mechanisms and lead to a model that reconciles contradictory aspects of previous findings while providing a framework to interpret future results.
Keywords: DNA, magnetic tweezers, single molecule, topology, segregation
Topological stress associated with excess supercoiling and knotted or interlinked DNA is resolved by an essential and conserved class of enzymes, the topoisomerases (1–4). Topoisomerases fall into two broad categories. Type I topoisomerases transiently cut a single strand of duplex DNA permitting them to relax overwound, positively supercoiled DNA [(+)sc DNA], and underwound, negatively supercoiled DNA [(−)sc DNA]. Type II topoisomerases catalyze the ATP-dependent passage of one segment of double-stranded DNA (the transfer or T segment) through a second transiently cleaved segment of duplex DNA (the gate or G segment) (3). Type II topoisomerases can both relax and generate supercoils and, importantly, can unknot and unlink DNA. Cellular DNA topology is maintained through the concerted activity of these two classes of topoisomerases (5).
Topo IV, a bacterial type II topoisomerase, is primarily responsible for unlinking replicated chromosomes in vivo (5–7). A summary of the in vivo roles and enzymatic cycle of Topo IV is presented in supporting information (SI) Fig. S1. In vitro, Topo IV relaxes (+)sc DNA faster than (−)sc DNA. Paradoxically, the relaxation rate asymmetry is variable, ranging from a factor of ≈20 in ensemble measurements to a much larger value in single-molecule measurements (8–10). In contrast with Topo IV, the homologous yeast topoisomerase II (Topo II) relaxes positive and negative supercoils at the same rate (8). However, human topoisomerase IIα preferentially relaxes (+)sc DNA (11), suggesting that asymmetric supercoil relaxation by type II topoisomerases is a general phenomenon.
A chiral-sensing model, in which the binding of juxtaposed G and T segments by Topo IV depends on the angle between them, has been proposed to account for asymmetric supercoil relaxation (8–10) (Fig. 1A). Plectonemes in (+)sc DNA form a left-handed superhelix with DNA segments juxtaposed at an acute angle (αL < 90°), whereas plectonemes in (−)sc DNA form a right-handed superhelix with DNA segments juxtaposed at an obtuse angle (αR > 90°) (Fig. 1A). The chiral-sensing model proposes that asymmetric supercoil relaxation results from Topo IV-binding G and T segments juxtaposed at an acute preferred crossing angle (α0 < 90°), which is more likely in (+)sc DNA than in (−)sc DNA. Although the chiral-sensing model is consistent with the majority of available data (8–10), it has not been tested directly.
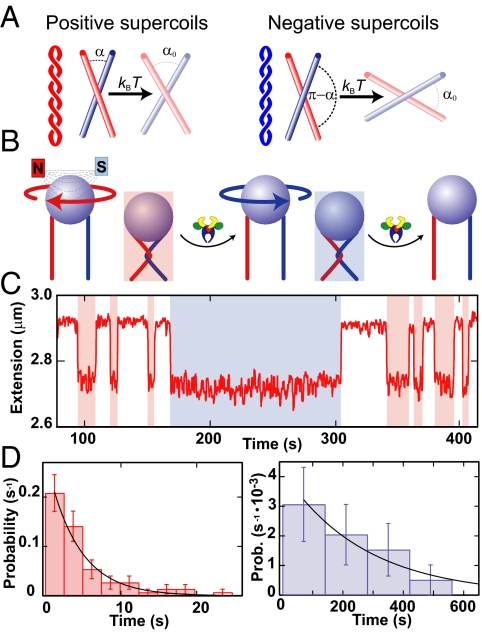
Fig. 1.
Single DNA crossing assay. (A) Crossing geometry for left- and right-handed supercoils. The crossing angle is the clockwise angle between the top and bottom DNA segments. Plectonemes in (+)sc DNA (red) impose an acute angle on juxtaposed DNA segments, whereas plectonemes in (−)sc DNA (blue) impose an obtuse crossing angle on juxtaposed DNA segments. The chirality-sensing model is illustrated with reference to the single crossings. For a preferred crossing angle, α0 < 90°, a small thermal angular fluctuation (δL = α0 − α) is required to relax a left-handed crossing, whereas a larger, hence less probable, fluctuation (δR = 180°−α0 − α), is required to relax a right-handed crossing. (B) Diagram of the single-crossing experimental geometry (not to scale). A paramagnetic bead (blue) is tethered to the surface of a capillary by two DNA molecules (red and blue). Magnets above the capillary apply a constant vertical force on the bead. Rotating the magnets by one clockwise (negative) turn imposes a left-handed crossing and reduces the height of the bead (red shading). Topo IV relaxes the crossing, returning the bead to its original height. The process is repeated by rotating the magnets in the counterclockwise (positive) direction, imposing a right-handed crossing (blue shading), which is subsequently relaxed. (C) Single DNA crossing relaxation measurements. The extension of a doubly tethered bead (red line) is displayed as a function of time for 3.5-μm DNA tethers separated by 0.73 μm at a force of 0.87 pN. Left-handed DNA crossings were relaxed rapidly (red shading), whereas the right-handed crossing took longer to unlink (blue shading). (D) Normalized distribution of relaxation times for left-handed crossings (Left, red bars) and right-handed crossings (Right, blue bars) were fit with single exponentials, P(t) = τ−1exp(−t/τ). The characteristic relaxation times were τL = 2.7 ± 0.6 s (χ2ν = 0.6), and τR = 270 ± 130 s (χ2ν = 0.2), and their ratio was τL/τR = 0.010 ± 0.005. Fig. S3 displays an example of relaxation times obtained for a different tether geometry and force.
To test the chiral-sensing model directly, we measured the relaxation rate of single left- and right-handed DNA crossings by Topo IV. Combining these measurements with angular probability distributions of DNA crossings from Monte Carlo (MC) simulations, we determined the preferred crossing angle for Topo IV (α0 = 85.5 ± 0.4°). This value of the preferred crossing angle could not account for observed supercoil relaxation rate asymmetries within the framework of the chiral-sensing model. High-resolution measurements of (±)sc DNA relaxation by Topo IV revealed that asymmetric supercoil relaxation results primarily from differences in processivity.
Results and Discussion
Single DNA Crossing Assay.
The single DNA crossing assay (Fig. 1B) is similar to described “braiding” assays in which two dsDNA strands attached between a bead and a surface are wound around each other multiple times (8, 9, 12). Imposing a single crossing between the DNA molecules offers important advantages over the braided geometry. There is a large extension change with the first crossing, whereas subsequent crossings result in smaller extension changes (8, 12). Furthermore, the crossing angle between juxtaposed DNA segments in the single-crossing geometry depends on the applied force, the spacing between, and length of the DNA tethers. By controlling these factors, the crossing-angle distribution can be varied over a wide range. A left-handed crossing is imposed by rotating the bead one full turn (360°) in the clockwise direction. Rotating the bead in the counterclockwise direction imposes a right-handed crossing (Fig. 1 A and B). The left- and right-handed crossing angles are symmetrically spaced with respect to 90°, i.e., the two crossing angles are supplementary (Fig. 1A). The complete crossing-angle distribution can be obtained from MC simulations (12).
Single DNA crossings permit a sensitive test of the preferred crossing angle for Topo IV. The only difference between left- and right-handed crossings is the crossing angle; in every other respect the crossings are identical. Consequently G segment binding rates will be the same for both left- and right-handed crossings. T segment binding rates, and hence relaxation rates, will depend on the crossing angle, provided that T segment binding is the rate-limiting step. Because single crossings are unlinked in one catalytic cycle, measurements can be made at relatively high enzyme concentrations, ensuring that T segment, rather than G segment, binding is rate limiting. If the preferred crossing angle is <90°, left-handed crossings would be relaxed faster than the equivalent right-handed crossings. Furthermore, the ratio of left- to right-handed relaxation times would be inversely proportional to the deviation of the imposed crossing angles from 90°. The ratio would be unity at an imposed angle of 90° and would decrease for increasing deviations of the imposed crossing angles from 90°.
Topo IV Asymmetrically Unlinks Single Left- and Right-Handed DNA Crossings.
To test the hypothesis that Topo IV binds T and G DNA segments juxtaposed at an acute angle, we measured the unlinking times for single left- and right-handed DNA crossings by Escherichia coli Topo IV. A micrometer-sized magnetic particle was tethered to the surface of a glass capillary by two DNA molecules (Fig. 1B) (8, 12). Magnets above the capillary applied a constant upward force and provided rotational control on the bead. One rotation (360°) of the magnets imposed a single DNA crossing, which reduced the extension of the bead (Fig. 1C). The binding geometry of the double tether was determined in situ by fitting the extension versus rotation curve with a geometric function to determine the spacing between DNA tethers (2e) and to ensure that they were parallel (Fig. S2) (8).
Single-crossing unlinking measurements were made at Topo IV concentrations ranging from 0.2 to 2 nM, at 1 mM ATP. Single crossings were unlinked after a variable relaxation time, returning the bead to its original height (Fig. 1C and Fig. S3A). Crossings were not unlinked unless both Topo IV and ATP were present. In a typical experiment repeated cycles of 1–5 relaxation events of each DNA crossing handedness were measured. The relaxation time distributions for both left- and right-handed crossings were well fit by single exponentials (Fig. 1D and Fig. S3B). On average, left-handed crossings were relaxed faster than the equivalent right-handed crossings. The ratio of left- to right-handed characteristic relaxation times, τL/τR, provides a measure of the relaxation rate asymmetry arising from different T segment capture rates, provided that the T segment binding is rate limiting. Several lines of evidence suggest that this was the case. (i) The concentration of Topo IV (≈0.2–2 nM) exceeded the Kd for DNA measured under similar conditions (≈0.15 nM) (13). (ii) The ratio of relaxation times for a given tether geometry and force was independent of enzyme concentration. (iii) Relaxation times were exponentially distributed (Fig. 1D and Fig. S3B), indicating a single rate-limiting step. (iv) Left- and right-handed relaxation times were proportional to the applied force, suggesting that both unlinking rates were sensitive to the crossing-angle distribution. Together, these observations indicate that T segment binding was the single rate-limiting step in the relaxation of single DNA crossings. Therefore, the ratio of single-crossing relaxation times reflects the relative rates of T segment capture and hence the relative probability of obtaining the preferred crossing angle in left- and right-handed crossings.
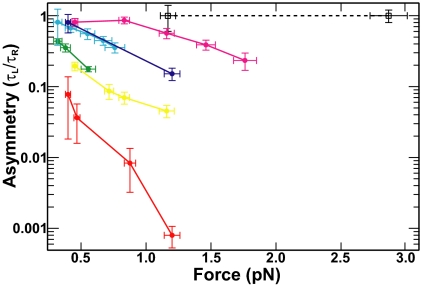
Whereas the relaxation rate asymmetry (τL/τR) was always less than unity, it was not constant (0.0008 ± 0.0003 < τL/τR < 0.9 ± 0.1) (Fig. 2). At low forces and large tether separations, for which both left- and right-handed crossing angles were close to 90°, the ratio approached unity (Fig. 2). As the force was increased, or the tether separation decreased, both of which increased the deviation of the crossing angles from 90°, the ratio decreased. These results provide qualitative support for a preferred crossing angle <90°. In experiments with Saccharomyces cerevisiae Topo II, the ratio of relaxation times was 1.0 ± 0.3 independent of the force (Fig. 2), consistent with symmetric relaxation of positive and negative supercoils (8, 14) and a preferred crossing angle of 90° (15, 16).
Fig. 2.
Single DNA crossing relaxation rate asymmetry. Single DNA crossing relaxation rate asymmetry (τL/τR) as a function of force for Topo IV (solid colored lines and filled circles) and S. cerevisiae Topo II (dashed black line and open squares) was plotted on a semilogarithmic axis. Error bars represent the propagation of statistical uncertainties. Lines between points are provided to guide the eye. Each tether is characterized by the ratio of the tether separation to the length of the tethers (2e/L0). The 2e/L0 values are: 0.21 (red), 0.27 (yellow), 0.34 (green), 0.44 (cyan), 0.50 (blue), 0.56 (magenta), and 0.25 (black dashed line).
Topo IV Binds DNA Segments Juxtaposed at 85°.
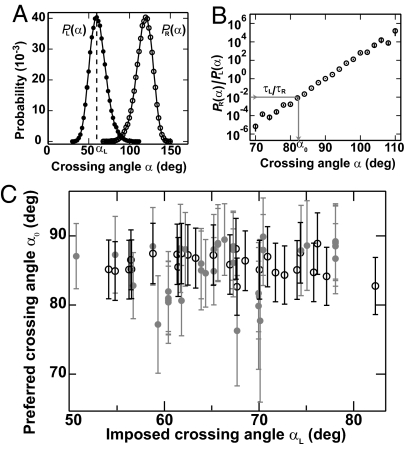
By combining relaxation measurements with crossing-angle probability distributions [P(α)] obtained from MC simulations of single DNA crossings (12), we determined the preferred crossing angle for Topo IV. The analysis was similar to that developed by Stone et al. (9). The preferred crossing angle (α0) was the angle at which the measured relaxation rate asymmetry ratio (τL/τR) was equal to the ratio of the angular probability distributions: τL/τR = PR(α0)/PL(α0) (Fig. 3 A and B). In other words, at the preferred crossing angle, the ratio of right- and left-handed crossing-angle probabilities is equal to the ratio of relaxation times. We obtained an independent determination of the preferred crossing angle from each measurement of the relaxation rate asymmetry (τL/τR), combined with the computed crossing-angle probability distributions (9, 12) (SI Text). The average preferred crossing angle was α0 = 85.5 ± 0.4°. Calculated preferred crossing angles were independent of applied force, protein concentration, and tether geometry (Fig. 3C). Moreover, a narrow distribution of preferred crossing angles (Fig. 3C) was obtained from rate asymmetry values that varied over 3 orders of magnitude (Fig. 2). The remarkable consistency of the preferred crossing angle measured over wide ranges of protein concentration, applied force, imposed DNA crossing angle, and relaxation rate asymmetries, provides additional evidence that T segment capture was the rate-limiting step. Statistically identical results were obtained with Topo IV from two independent sources (Fig. 3C).
Fig. 3.
Preferred crossing angle (α0) for Topo IV. (A) Angular probability distribution functions, P(α), for a single left- (filled circles) and right-handed (open circles) DNA crossing from MC simulations (12) for the tether geometry and force given in Fig. 1. The angular distribution for the right-handed crossing PR(α) was derived from the distribution for the left-handed crossing PL(α) through the relation PR(α) = PL(180° − α). The imposed DNA crossing angle, αL [corresponding to the peak of PL(α)] ≈ 59°. (B) Ratio of right-handed to left-handed probability from A plotted on a semilogarithmic axis. The preferred crossing angle, α0, is obtained from the relation τL/τR = PR(α0)/PL(α0), as illustrated by the gray arrow on the graph. The measured relaxation rate ratio for this tether geometry and force (Fig. 1) was τL/τR = 0.010 ± 0.005, which corresponds to a preferred crossing angle α0 = 82° ± 1.2°. Fig. S3 C and D provides an example of the determination of the preferred crossing angle for a larger crossing angle. (C) Preferred crossing angles (α0) for two different Topo IV preparations (open and filled circles) plotted versus the imposed left-handed crossing angle, αL. The combined average preferred crossing angle was α0 = 85.5° ± 0.4° (mean ± SEM). Error bars are a combination of measurement and statistical uncertainties added in quadrature.
The value of α0 ≈ 85° is close to the symmetric value of 90°, for which there would be no difference in relaxation rates. Indeed, MC simulations (9) of crossing-angle distributions for plasmid DNA suggest that for α0 ≈ 85° (+)sc DNA would be relaxed only ≈2-fold faster than (−)sc DNA, significantly less than the measured ratio of at least 20 (8–10) and therefore inconsistent with the chiral-sensing model.
Supercoil Relaxation Asymmetry Is Caused by Differences in Processivity.
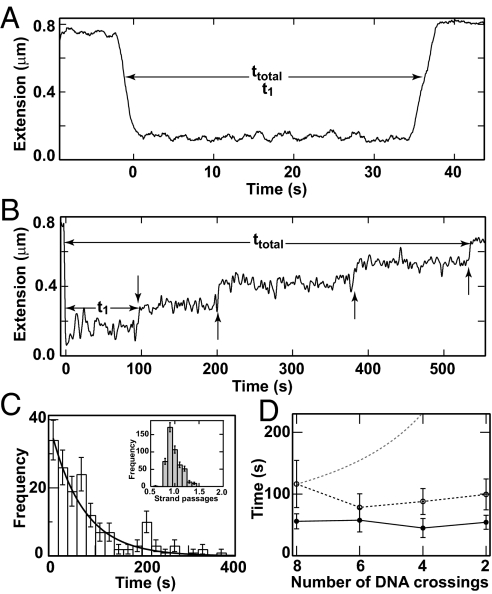
To address the apparent contradiction between the slight asymmetry predicted by the nearly symmetric preferred crossing angle and the large asymmetry observed in supercoil relaxation rates, we made high-resolution measurements of supercoil relaxation by Topo IV (10, 17). Supercoil relaxation measurements were analogous to the single-crossing measurements: 8–12 positive or negative supercoils were alternately introduced in a single 3.6-kb torsionally constrained DNA molecule held at constant force (≈0.4 pN) (Fig. 4 A and B) (see Materials and Methods). As observed (9, 10), positive supercoils were relaxed in one, or rarely two, rapid bursts of multiple strand passages (Fig. 4A). In contrast, negative supercoils were relaxed in individual steps of precisely one strand passage (Fig. 4 B and C), indicating distributive relaxation of negative supercoils.
Fig. 4.
Supercoil relaxation by Topo IV. (A) Relaxation of positive supercoils. Extension data were recorded at 25 Hz and median-filtered over 45 points. The extension of the DNA molecule (solid line) decreased rapidly as eight positive supercoils were introduced. All eight supercoils were relaxed in a single burst of activity ≈30 s after they were introduced. The initial time (t1) and the total relaxation time (ttotal) are indicated. (B) Relaxation of negative supercoils. The extension of the DNA molecule (solid line) rapidly decreased as eight negative supercoils were introduced. Subsequently, individual strand passage events relaxed pairs of supercoils (vertical arrows). (C) Normalized distribution of waiting times for the relaxation of negative supercoils was binned and fit to a single exponential, P(t) = τ−1exp(−t/τ), (line) (χ2ν = 1.25). (Inset) Distribution of strand passages per binding event on (−)sc DNA. (D) Average time between relaxation events as a function of the number of DNA crossings. The same data from C (open circles and solid line) are compared with data at a 2-fold higher protein concentration (≈150 pM, filled circles and dashed line). For comparison, a model in which the time between relaxation events is inversely proportional to the number of DNA crossings is shown (thin dashed line).
Supercoil relaxation events were characterized by two parameters: (i) t1, the time interval between the introduction of supercoils and the onset of activity, which was dominated by the protein-binding time under our experimental conditions; and (ii) tT, the total time to relax the supercoils. At a concentration of 75 ± 15 pM Topo IV t1 for negative supercoils was 5.5 ± 0.8 times longer, and tT for 8 supercoils was 21 ± 3 times longer than for positive supercoils. For both (−)sc and (+)sc DNA, the distribution of t1 times was exponentially distributed, indicating a single rate-limiting step. For (−)sc DNA, the distribution of all individual relaxation times was well described by a single exponential (Fig. 4C), suggesting that rebinding occurs after each relaxation cycle. This is further supported by the observation that the average time between relaxation events depended on the enzyme concentration, but not on the number of crossings (Fig. 4D). In experiments in which 4–12 positive supercoils were relaxed by Topo IV, the initial waiting time (t1) was similarly independent of the initial number of DNA crossings. These observations suggest that Topo IV binds the end of the plectoneme. If Topo IV bound DNA crossings at random, the binding time would vary as the inverse of the number of crossings, as illustrated in Fig. 4D. Because the binding time was insensitive to the number of DNA crossings for both positive and negative supercoils, Topo IV likely binds the distal end of the plectoneme. This is consistent with electron micrographs showing Topo IV bound to the ends of plectonemes (18) and single-molecule force-induced unbinding experiments showing that Topo IV stabilizes the distal end of plectonemes (13).
Processivity Model of Supercoil Relaxation by Topo IV.
Our data suggest that the preferential relaxation of positive supercoils stems from differences in the processivity of (+)sc versus (−)sc DNA relaxation. Previous reports have established that Topo IV relaxes positive supercoils with a processivity of at least 81 strand passages (9). Here, we show that Topo IV relaxes negative supercoils in a perfectly distributive fashion (Fig. 4C Inset), in agreement with previous estimates (10). We propose a processivity model for supercoil relaxation that provides a coherent explanation for the discrepancies in relaxation asymmetry measured in ensemble and single-molecule experiments (10).
The supercoil relaxation asymmetry is the ratio of times to relax n negative and positive plectonemes. For positive supercoils t+ = tb + 0.5ntc where tb is the binding time and tc ≈ 0.4 s is the time to relax a single crossing (8–10). The factor of 0.5 arises because each strand passage event removes two plectonemes. The relaxation time corresponds to a single binding event followed by processive relaxation of the DNA crossings. In contrast, for negative supercoils, t− = 0.5n(βtb + tc), where β = 5.5 ± 0.8 is the measured ratio of initial binding times for positive and negative supercoils. This expression reflects the necessity of Topo IV to rebind after each strand passage of (−)sc DNA. The resulting relaxation rate asymmetry is
Plots of R as a function of the number of DNA crossings (n) and the binding time (tb) are displayed in Fig. S4A. As discussed in the SI Text, this model is consistent with previous asymmetry measurements and suggests that the discrepancy between single-molecule and ensemble measurements is caused by differences in experimental conditions. In ensemble assays, the relaxation of supercoiled plasmid DNA, containing on the order of 10–25 DNA crossings, is measured at a relatively high concentration of Topo IV. Conversely, in single-molecule assays the relaxation of tens to hundreds of DNA crossings is measured at low enzyme concentrations. These disparate experimental regimes understandably yield different results for an enzyme that displays a large difference in processivity as a function of its substrate.
Enzyme processivity reflects kinetic competition between a forward rate and an off-rate. An upper limit for the processivity on (−)sc DNA of 0.08 (P = 0.05) can be estimated (SI Text) from the observation that all ≈500 relaxation events consisted of a single-strand passage (Fig. 4C Inset). The ≈6-fold difference in initial strand passage rates (t1−1) on (±)sc DNA suggests that the kinetic competition step must occur after the initial strand passage. If the kinetic competition step occurred before the initial strand passage, the difference in initial relaxation times (t1) on (±)sc DNA would be comparable with the difference in processivity, a ≈100-fold larger difference than observed (Fig. 4).
The large conformational change in the DNA that occurs after the initial strand passage is the most likely candidate for the kinetic competition step. At this step the enzyme is bound to a highly unstable loop of DNA that rapidly refolds into a plectonemic loop. We propose that on (+)sc DNA Topo IV is stabilized, and plectoneme refolding is favored, by the binding of the C-terminal domains (CTDs), which have been shown to be required for chirality discrimination (19). On (−)sc DNA, the interaction of the CTDs with the DNA may be destabilized during the large conformational rearrangement, resulting in the release of Topo IV. The chiral bending and twisting of DNA by Topo IV and its CTDs (13, 20) may also contribute to the differences in binding stability and rates of plectoneme refolding on (+)sc and (−)sc DNA (Fig. S4B). Removing the CTDs from Topo IV increases the Kd for linear DNA by a factor of ≈2, demonstrating their importance in DNA binding (19). Moreover, cross-linking experiments suggest that Topo IV adopts a more open configuration on (−)sc DNA compared with (+)sc DNA (21), lending further support to a model in which the enzyme is displaced from (−)sc DNA during the large DNA rearrangements after the initial strand passage (Fig. S4B). The ensemble of evidence favors a model in which the CTDs modulate the processivity of Topo IV in a chiral-dependent manner (Fig. S4). More generally, the CTDs likely modulate the interaction of type II topoisomerases with DNA, permitting the spectrum of topological activities including negative supercoiling by DNA gyrase, asymmetric supercoil relaxation by Topo IV and human type IIα (11), and symmetric supercoil relaxation by yeast Topo II (Fig. 2). The CTDs are poorly conserved among type II topoisomerases (3), and a growing body of evidence implicates them as the determinants of enzyme-specific activities (19, 20, 22). Our results suggest a role for the CTD in modulating the processivity of type II topoisomerases.
In Vivo Implications.
Our results suggest a resolution of the paradox concerning the roles of Topo IV in vivo: how can Topo IV unlink the right-handed crossings of positive catenanes and precatenanes (6, 7, 23–25) yet preferentially relax the left-handed crossings of positive supercoils? The efficient unlinking of positive catenanes is consistent with a preferred crossing angle of ≈85° because the crossing angles of catenanes at low positive catenation densities are broadly distributed with a peak slightly >90° (9, 26). Indeed, Topo IV has been shown to unlink weakly catenated DNA more rapidly than it relaxes (+)sc DNA (6), consistent with a nearly symmetric preferred crossing angle. Discrimination between positive and negative supercoils based solely on the crossing angle, however, would result in a modest asymmetry in relaxation rates. The apparent paradox is resolved by the large processivity difference between positive and negative supercoil relaxation, resulting in preferential relaxation of positive supercoils. An alternative model, in which Topo II interacts with two potential T segments, was developed by Trigueros et al. (16) based on their observation that the catenation/decatenation activity of Topo II depends on the supercoiling sign and density. This 3-segment model provides a qualitative explanation for the observed dependence of decatenation/catenation probability on the supercoiling sign and suggests an elegant solution to the Topo IV paradox. The current experiments cannot address the predictions of this intriguing model, although we are hopeful that future experiments will be able to test this model.
The prokaryotic genome is maintained at a constant level of negative supercoiling through the opposing activities of gyrase and Topo I (5). Rapid relaxation of negative supercoils would perturb this topological homeostasis. Consistent with this constraint, Topo IV appears to relax (−)sc DNA in a perfectly distributive manner, similar to bacterial Topo I. Indeed, Topo IV may act in conjunction with Topo I to limit the degree of negative supercoiling because inactivation of Topo IV leads to hypernegative supercoiling similar to the level observed with mutant Topo I (27). Nevertheless, the unconstrained activity of the ≈1,000 Topo IV molecules in the bacterial cell (28) would rapidly relax the genome. This argument supports the conclusion that Topo IV is regulated in both space and time (28). Evidence from localization studies suggests that Topo IV is tightly controlled during the cell cycle, only gaining access to the DNA at later stages of replication (28). However, the slight relaxation of negative supercoils (27) and unknotting by Topo IV (29), which persist in the absence of replication, suggest that some fraction of the Topo IV has access to the DNA continuously (27).
This work explores mechanistic aspects of Topo IV activity and provides measures for comparison with in vivo and in vitro measurements of type II topoisomerase activity. Our results resolve a longstanding ambiguity concerning the relative importance of the binding geometry and enzymatic processivity in chiral discrimination by Topo IV. We anticipate that the mechanistic framework and techniques developed here will be applied to understand the relaxation asymmetry observed in other type II topoisomerases (11) and more generally to probe the geometry and kinetics of DNA synapses.
Materials and Methods
DNA Constructs.
Eleven- and 13.5-kb DNA for single-crossing experiments was constructed as described (8, 30). An additional construct for single-crossing experiments consisted of 10,619 bp of dsDNA generated by PCR between positions 10,700 and 21,319 of pFC94 using PCR primers containing a single biotin or digoxigenin label at their 5′ extremity. DNA (3.6 kb) for supercoil relaxation experiments was constructed as described in ref. 31.
Magnetic Manipulation.
The magnetic tweezers have been described (8, 13). Briefly, a streptavidin-coated magnetic bead was tethered to the surface of a glass capillary mounted on an inverted microscope. A pair of magnets above the capillary applied a vertical force on the magnetic bead. Torque applied by the magnets caused the bead to rotate with the magnet rotation. The 3-dimensional position of the magnetic bead was determined with ≈10-nm accuracy in real time (25 Hz) via video microscopy and custom tracking software.
Decatenation and Supercoil Relaxation Experiments.
Single DNA crossing decatenation experiments were conducted with 0.2–2 nM Topo IV or 10–50 pM yeast Topo II at 22 °C in 25 mM Tris (pH 7.6), 100 mM potassium glutamate, 10 mM magnesium chloride, 0.5 mM DTT, 50 μg/mL BSA, 0.1% Triton X-100, and 1 mM ATP. Topo IV was a gift from N. Crisona and N. Cozzarelli (University of California, Berkeley). Additional Topo IV was purchased from John Innes Enterprises. S. cerevisiae Topo II was a gift from J. M. Berger (University of California, Berkeley). Magnetic beads (Dynal) (1- or 2.8-μm diameter) were tethered to the surface of the glass capillary by two dsDNA molecules (≈11 or 13.5 kb). The extension as a function of magnet rotation was used to ensure that there were only two tethers that were parallel when fully extended, to determine the spacing between them, and to determine the zero of rotation as described in ref. 8 (Fig. S2). The extension of the tether was measured as function of time, and a feedback routine automatically rotated the magnets after each relaxation event. For supercoil relaxation experiments, 1-μm magnetic beads (Dynal) were tethered to the surface via a 3.6-kb torsionally constrained DNA molecule.
Data Processing.
Decatenation of a single DNA crossing produced a large extension change (Fig. 1C), which facilitated data analysis. Each transition was fit to a sigmoid function of the form L0 + ΔL·(1 + expβ(t½ − t)−1, where L0 is the extension when the DNA is crossed, ΔL is the increase in extension associated with relaxation of the crossing, t1/2 is the time at the midpoint of the transition, and β describes the slope of the transition. The time between imposing the crossing and t1/2 was defined as the relaxation time.
Extension versus time records of supercoil relaxation (Fig. 4 A and B) were smoothed with a 1-s (25-point) second-order Savitzky–Golay filter followed by a 1.8-s (45-point) median filter. Extension changes corresponding to supercoil relaxation events were determined from changes in the running variance calculated over a 1-s window of the smoothed data and the time at which each transition occurred was recorded. Raw extension versus time records were fit with line segments between successive transitions, from which the step time and extension change were determined. Analysis routines were written in IGOR Pro (Wavemetrics).
Supplementary Material
Acknowledgments.
We thank Giuseppe Lia and Elise Praly for DNA constructs, in addition to Omar Saleh, Timothee Lionet, Jean-Pierre Dumas, Jean-Francois Allemand, Alexander Vologodskii, James Berger, Joachim Roca, Anthony Maxwell, and Neil Osheroff for enlightening discussions. We thank Grace Liou, Richard Neuman, Kristina Herbert, Ashley Hardin, and Yeonee Seol for critical reading of the manuscript. This work was supported by the Centre Nationale de la Recherche Science and Association pour la Recherche sur le Cancer. K.C.N. was supported by a long-term fellowship from the Human Frontiers Science Program and by the Intramural Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900574106/DCSupplemental.
References
- 1.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 3.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 4.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: The story of a simple molecular machine. Q Rev Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 5.Espeli O, Marians KJ. Untangling intracellular DNA topology. Mol Microbiol. 2004;52:925–931. doi: 10.1111/j.1365-2958.2004.04047.x. [DOI] [PubMed] [Google Scholar]
- 6.Hiasa H, Marians KJ. Two distinct modes of strand unlinking during θ-type DNA replication. J Biol Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 7.Kato J, et al. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 8.Charvin G, Bensimon D, Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type II topoisomerases. Proc Natl Acad Sci USA. 2003;100:9820–9825. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stone MD, et al. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc Natl Acad Sci USA. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: Implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 12.Charvin G, Vologodskii A, Bensimon D, Croquette V. Braiding DNA: Experiments, simulations, and models. Biophys J. 2005;88:4124–4136. doi: 10.1529/biophysj.104.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charvin G, Strick TR, Bensimon D, Croquette V. Topoisomerase IV bends and overtwists DNA upon binding. Biophys J. 2005;89:384–392. doi: 10.1529/biophysj.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 15.Roca J, Wang JC. The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase II. Genes Cells. 1996;1:17–27. doi: 10.1046/j.1365-2443.1996.01001.x. [DOI] [PubMed] [Google Scholar]
- 16.Trigueros S, Salceda J, Bermudez I, Fernandez X, Roca J. Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J Mol Biol. 2004;335:723–731. doi: 10.1016/j.jmb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Revyakin A, Ebright RH, Strick TR. Single-molecule DNA nanomanipulation: Improved resolution through use of shorter DNA fragments. Nat Methods. 2005;2:127–138. doi: 10.1038/nmeth0205-127. [DOI] [PubMed] [Google Scholar]
- 18.Vologodskii AV, et al. Mechanism of topology simplification by type II DNA topoisomerases. Proc Natl Acad Sci USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 20.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc Natl Acad Sci USA. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crisona NJ, Cozzarelli NR. Alteration of Escherichia coli topoisomerase IV conformation upon enzyme binding to positively supercoiled DNA. J Biol Chem. 2006;281:18927–18932. doi: 10.1074/jbc.M603068200. [DOI] [PubMed] [Google Scholar]
- 22.McClendon AK, et al. Bimodal recognition of DNA geometry by human topoisomerase II α: Preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain. Biochemistry. 2008;47:13169–13178. doi: 10.1021/bi800453h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng H, Marians KJ. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc Natl Acad Sci USA. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zechiedrich EL, Cozzarelli NR. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 25.Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: Biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 26.Vologodskii A, Cozzarelli NR. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zechiedrich EL, et al. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 28.Espeli O, Levine C, Hassing H, Marians KJ. Temporal regulation of topoisomerase IV activity in E. coli. Mol Cell. 2003;11:189–201. doi: 10.1016/s1097-2765(03)00013-3. [DOI] [PubMed] [Google Scholar]
- 29.Deibler RW, Rahmati S, Zechiedrich EL. Topoisomerase IV, alone, unknots DNA in E. coli. Genes Dev. 2001;15:748–761. doi: 10.1101/gad.872301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleh OA, Perals C, Barre FX, Allemand JF. Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004;23:2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lia G, et al. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.