Abstract
Mutations in the coding region of hepatocyte nuclear factor 4α (HNF4α), and its upstream promoter (P2) that drives expression in the pancreas, are known to lead to maturity-onset diabetes of the young 1 (MODY1). HNF4α also controls gluconeogenesis and lipid metabolism in the liver, where the proximal promoter (P1) predominates. However, very little is known about the role of hepatic HNF4α in diabetes. Here, we examine the expression of hepatic HNF4α in two diabetic mouse models, db/db mice (type 2, insulin resistant) and streptozotocin-treated mice (type 1, insulin deficient). We found that the level of HNF4α protein and mRNA was decreased in the liver of db/db mice but increased in streptozotocin-treated mice. Because insulin increases the activity of sterol regulatory element-binding proteins (SREBP)-1c and -2, we also examined the effect of SREBPs on hepatic HNF4α gene expression and found that, like insulin, ectopic expression of SREBPs decreases the level of hepatic HNF4α protein and mRNA both in vitro in primary hepatocytes and in vivo in the liver of C57BL/6 mice. Finally, we use gel shift, chromatin immunoprecipitation, small interfering RNA, and reporter gene analysis to show that SREBP2 binds the human HNF4α P1 promoter and negatively regulates its expression. These data indicate that hyperinsulinemia down-regulates HNF4α in the liver through the up-regulation of SREBPs, thereby establishing a link between these two critical transcription factor pathways that regulate lipid and glucose metabolism in the liver. These findings also provide new insights into diabetes-associated complications such as fatty liver disease.
Hyperinsulinemia results in down regulation of HNF4α gene expression in the liver via SREBPs, which could contribute to hepatic steatosis (fatty liver) during diabetes.
Hepatocyte nuclear factor 4α (HNF4α) (NR2A1) is a highly conserved member of the nuclear receptor superfamily of ligand-dependent transcription factors (1,2). Expression of the gene is driven by two distinct promoters, the P1 promoter that drives expression of splice variants HNF4α1-6 in the liver, kidney, and intestine/colon and the P2 promoter that drives expression of splice variants HNF4α7-9 in the intestine/colon, stomach, and β-cells of the pancreas but notably not the adult liver or kidney. Hundreds of HNF4α target genes have been identified in liver, pancreas, and colon (3,4,5; also see http://burgundy.cmmt.ubc.ca/cgi-bin/tfe/home.pl). In the liver, some of these targets are genes involved in glucose [PEPCK (PCK1), l-pyruvate kinase (l-PK, PKLR), glucose-6-phosphatase (G6Pase, G6PC)] and xenobiotic and drug metabolism (e.g. CYP7A1, CYP3A4, and CYP2D6). HNF4α is also known for its regulation of genes involved in lipid transport, especially those encoding apolipoproteins (APOA1, APOA2, APOA4, APOB, APOC2, APOC3, and APOC4) that are linked to circulating levels of cholesterol and triglycerides and hence to atherosclerosis.
Inherited mutations in the coding region of the HNF4A gene lead to maturity-onset diabetes of the young 1 (MODY1), a disease characterized by a loss of insulin secretion from the pancreatic β-cells in response to glucose (7). The effect of the MODY1 mutations are presumably on the pancreatic form of HNF4α because single-nucleotide polymorphisms in the P2 promoter that drives β-cell expression have also been linked to MODY1 (see Ref. 8 and references therein); no such mutations have been identified in the P1 promoter that drives expression in the liver. Consequently, several studies have investigated the role of pancreatic HNF4α in diabetes (5,9,10), but much less is known about the role of hepatic HNF4α in diabetes.
Sterol regulatory element-binding proteins (SREBPs) are also well characterized transcription factors that regulate genes involved in cholesterol and fatty acid synthesis and metabolism (11,12,13). There are two SREBP genes, SREBP1 (SREBF1) and SREBP2 (SREBF2), and two SREBP1 splice variants, SREBP1a and SREBP1c. SREBP1c activates genes involved in fatty acid synthesis, whereas SREBP2 activates genes that regulate cholesterol biosynthesis, and SREBP1a is involved in both pathways (14). In cells with high sterol levels, SREBPs are bound by insulin-inducing gene (Insig)-1 and -2a together with SREBP cleavage-activating protein (SCAP) in the rough endoplasmic reticulum membrane. Upon sterol deprivation, SREBPs are cleaved from the membrane to liberate the amino termini [SREBP1(N) and SREBP2(N)] that translocate to the nucleus where they bind sterol response elements (SREs) in the promoters of target genes such as fatty acid synthetase (FAS, Fasn), LDL receptor (LDLR, Ldlr), stearoyl-coenzyme A desaturase 1 (SCD-1, Scd1), and HMG coenzyme A reductase (HMGCR, Hmgcr) (14,15).
SREBPs are involved not only in the sterol-mediated regulation of lipogenic enzymes but also in the insulin-mediated regulation of genes involved in carbohydrate metabolism (16,17,18,19). Insulin not only increases SREBP1 transcription in the liver (20,21,22) but also promotes SREBP1c cleavage and increases lipogenesis in the liver by inhibiting Insig2a expression (23,24). Furthermore, ectopic expression of a dominant-negative mutant of SREBP1c blocks the insulin-dependent induction of genes enhancing lipogenesis in the liver (25), indicating that SREBPs are key downstream regulators of insulin action.
Despite the importance of both HNF4α and SREBPs in carbohydrate and lipid metabolism, there are only a few reports of cross talk between these two transcription factor pathways on the protein level (26,27,28) and none that examine the regulation of HNF4α gene expression by SREBPs. In the current study, we identify an inverse correlation between HNF4α and SREBP expression in the livers of db/db type 2 and streptozotocin (STZ)-induced type 1 diabetic mice and show that insulin and ectopic expression of SREBPs lead to lower levels of hepatic HNF4α mRNA and protein both in vitro and in vivo. Finally, we show that SREBP2 acts directly on the human HNF4Α P1 promoter, suggesting that our results may be clinically relevant. These results not only identify a conserved link between the insulin, SREBP, and HNF4α pathways in the liver but also have potentially important implications for complications associated with diabetes, such as hepatic steatosis (fatty liver).
Results
Inverse correlation between insulin levels and the expression of HNF4α in the liver of diabetic mice
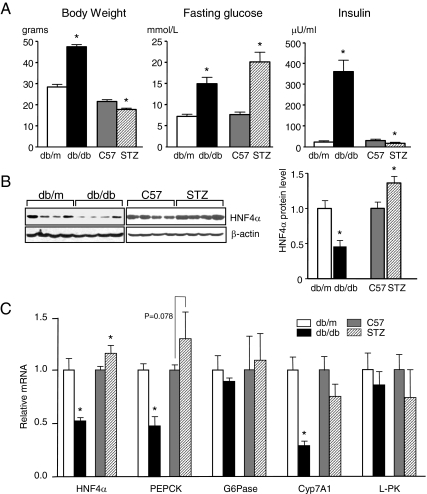
The level of HNF4α protein in the livers of db/db mice, a model for type 2 diabetes characterized by obesity, hyperglycemia, and hyperinsulinemia, was compared with that of STZ-treated mice, a model for type 1 diabetes characterized by hyperglycemia and insulin deficiency (Fig. 1A). Whereas there was a slight increase in HNF4α protein in the liver of STZ mice compared with C57BL/6 control mice, there was a marked decrease in the liver of db/db mice compared with db/m controls (Fig. 1B). The change in HNF4α protein levels was paralleled by a change in HNF4α mRNA in the livers of the two diabetic models as well as by a change in certain HNF4α target genes such as PEPCK and Cyp7A1 (Fig. 1C). Interestingly, not all HNF4α target genes decreased in db/db mice, G6Pase and l-PK, for example, were unchanged. In contrast, as observed previously, there was an increase in PEPCK expression in STZ-treated animals, which could be due to elevated levels of the coactivator PGC1α, which has been shown to coactivate HNF4α-mediated expression of PEPCK (29). However, it must be kept in mind in this and subsequent experiments that all of these target genes are regulated by many transcriptional activators in addition to HNF4α (e.g. SREBPs, C/EBP, and FoxO1 plus at least 12 others for PEPCK) that may also be responsive to insulin (16). Complex mechanisms of regulation could explain why not all HNF4α target gene levels correspond precisely with HNF4α protein levels.
Figure 1.
Down-regulation of HNF4α gene expression in the liver of db/db mice. A, Body weight, fasting glucose, and plasma insulin levels in male diabetic db/db mice (n = 8), their lean nondiabetic heterozygous littermates (db/m) (n = 8), STZ-induced (40 mg/kg) diabetic mice (n = 12), and C57BL/6J (C57) untreated controls (n =12). Plotted are means ± sd (*, P < 0.05). B, IB analysis of whole-cell liver extracts (50 μg/lane) probed with anti-HNF4α and β-actin antibodies; left, IB representative of eight to 12 mice in each group; right, IBs from all mice were quantified, normalized to β-actin, and plotted as mean ± sd (*, P < 0.05). C, Quantitative RT-PCR detecting HNF4α and target gene mRNA levels in the livers of db/db, STZ-induced diabetic mice and their respective controls (db/m and C57). Plotted are means ± sd normalized to β-actin with control mice (db/m or C57) arbitrarily set to 1.0 (*, P < 0.05).
Insulin decreases hepatic HNF4α gene expression in vitro
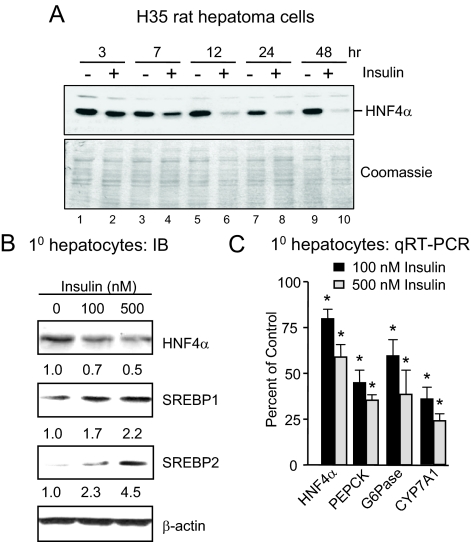
Because the db/db and STZ mice (and type 1 and 2 diabetes in humans, respectively) both have elevated blood glucose levels but significantly different levels of plasma insulin, we examined the effect of insulin on the expression of the endogenous HNF4α gene using cell-based systems. A liver cell line known to respond well to insulin, rat hepatoma H35 cells, showed a time-dependent decrease in HNF4α protein levels upon insulin treatment (Fig. 2A). Primary rat hepatocytes also showed a dose-dependent decrease in HNF4α protein upon insulin treatment (Fig. 2B). Because the decrease in HNF4α protein levels in H35 cells occurred only after several hours of delay (lane 4, HNF4α signal at 7 h is only slightly lower than the untreated control), we hypothesized that the primary effect of the insulin treatment was on the expression of the HNF4α gene rather than the stability of the HNF4α protein. And indeed, there was a decrease in HNF4α mRNA levels in the primary hepatocytes upon insulin treatment, as well as a decrease in HNF4α target genes PEPCK, G6Pase, and Cyp7A1 (Fig. 2C). Because others have observed an increase in SREBP1c levels in insulin-treated STZ mice (23,30), we also examined the levels of the mature form of SREBP1c and SREBP2 in the primary hepatocytes and saw a dose-dependent increase of both factors with insulin treatment (Fig. 2B).
Figure 2.
Down-regulation of HNF4α gene expression in hepatocytes by insulin. A, IB analysis with anti-HNF4α antibodies of whole cell extracts (15 μg/lane) from H35 rat hepatoma cells, cultured in serum-free medium for 18 h, and then treated with or without insulin (5 μg/ml) for the times indicated. Equal loading was verified by Coomassie staining. B, IB analysis of whole-cell extracts (50 μg/lane) from primary rat hepatocytes, treated with insulin as indicated for 24 h, and probed with anti-HNF4α, SREBP1, SREBP2, and β-actin antibodies. The mature forms of both SREBPs (SREBP1a and -1c and SREBP2) are detected. Numbers below each blot correspond to signals normalized to β-actin. All panels are representative of three independent experiments. C, Quantitative RT-PCR detecting HNF4α and target gene mRNA levels in primary rat hepatocytes treated with insulin for 24 h. Plotted is percentage of the untreated control from three independent experiments with duplicate samples normalized to β-actin (*, P < 0.05).
Fasting/refeeding regulates hepatic HNF4α expression in vivo
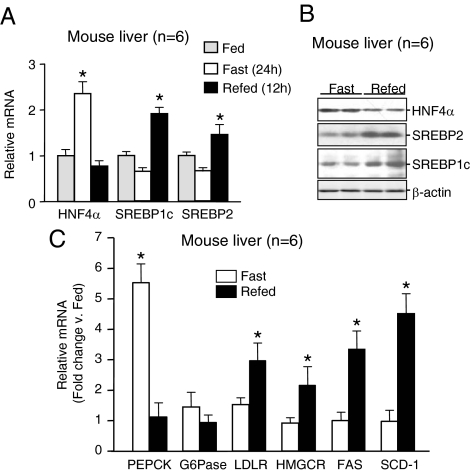
To recapitulate the in vitro effects of insulin in vivo under physiological conditions, we fasted C57BL/6 mice for 24 h and then refed them for an additional 12 h. We observed an appreciable increase in HNF4α mRNA levels in the livers of fasted animals when insulin levels are presumably low and a significant decrease back to basal levels upon refeeding when insulin levels are expected to be elevated (Fig. 3A). We also observed a concomitant decrease in HNF4α protein levels upon refeeding (Fig. 3B) and a corresponding decrease in the HNF4α target gene PEPCK (Fig. 3C). Finally, upon refeeding, we noted elevated levels of the mature forms of SREBP1c and SREBP2 proteins (Fig. 3B) as well as mRNA levels of SREBPs (Fig. 3A) and their target genes LDLR, HMGCR, FAS, and SCD-1 (Fig. 3C). Although similar results regarding insulin effects on HNF4α mRNA and SREBP protein levels have been observed previously (23,29,30,31,32), our results show for the first time, in the same liver samples, the inverse relationship between the levels of HNF4α and SREBP upon fasting and refeeding. [Similar results were also observed when the experiments were repeated with 6 h of refeeding after a 24-h fast (supplemental Fig. S1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org.)] Together with the decrease in HNF4α levels upon treatment of the hepatocytes with insulin (Fig. 2) and the disparate levels of HNF4α in the db/db and STZ mice (Fig. 1), these results suggest that insulin, HNF4α mRNA, and SREBP protein levels might be linked in a hierarchical fashion.
Figure 3.
Inverse relationship between HNF4α gene expression and SREBPs in the livers of fasted and refed mice. A, Quantitative RT-PCR detecting HNF4α SREBP1c and SREBP2 mRNA levels in the livers of 8-wk-old male C57BL/6J mice fed normal chow, fasted for 24 h, or refed for 12 h after a 24-h fast (n = 6). B, IB analysis of whole-cell extracts (80 μg/lane) from the livers of C57BL/6J mice treated as in A probed with anti-HNF4α, SREBP2, SREBP1, and β-actin antibodies. Six mice per group were analyzed; representative results from two mice are shown. C, Quantitative RT-PCR detecting HNF4α and SREBP target gene mRNA levels in the livers of C57BL/6J mice treated as in A. A and C, Plotted are means ± sd normalized to β-actin. Statistically significant differences (*, P < 0.05) relative to the fed controls, arbitrarily set to 1.0, are indicated. See supplemental Fig. S1 for similar results from another experiment performed with a 6-h refeeding period.
Ectopic expression of SREBPs decrease HNF4α expression in vitro and in vivo
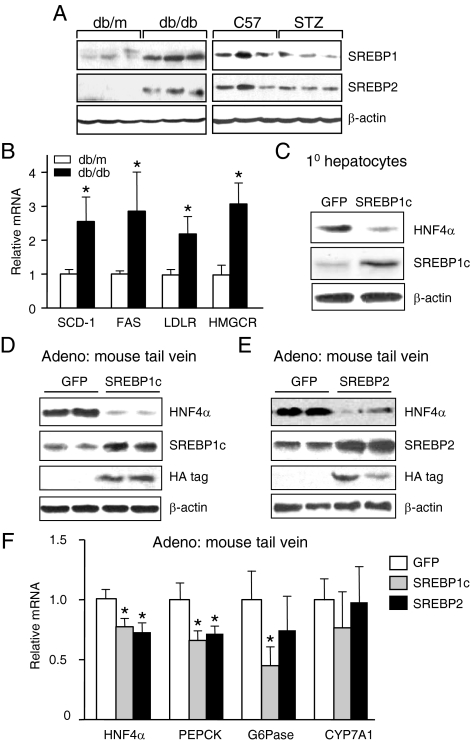
To determine whether elevated levels of insulin in db/db mice, and after refeeding in normal mice, decrease HNF4α mRNA levels via an increase in SREBP protein, we first verified that the livers of our db/db mice had elevated levels of the mature SREBP proteins and that the STZ mice had reduced levels (Fig. 4A) (supplemental Fig. S2 shows a concomitant change in SREBP1c and SREBP2 mRNA levels). These effects have been noted previously for SREBP1, although not SREBP2 (22,30,33). We also showed that the db/db mice had elevated levels of SREBP target genes SCD-1, FAS, LDLR, and HMGCR (Fig. 4B). To determine a direct causal relationship between SREBPs and HNF4α expression, we ectopically expressed the active form of SREBP1c [SREBP1c(N)] in primary rat hepatocytes using a recombinant adenovirus and observed a corresponding decrease in the expression of endogenous HNF4α protein (Fig. 4C). We next performed in vivo experiments, injecting adenoviral vectors expressing the active forms of the SREBPs [SREBP1c(N) or SREBP2(N)] into mouse tail veins and also observed a decrease in endogenous HNF4α protein in the liver (Fig. 4, D and E). Elevated SREBP protein levels were confirmed with antibodies to both the native proteins and the hemagglutinin (HA) tag used in the recombinant virus (Fig. 4, D and E). Finally, we measured the level of HNF4α mRNA in the same mice and likewise observed a small but significant decrease in the level of HNF4α mRNA as well as a decrease in the HNF4α targets PEPCK and G6Pase, although not CYP7A1 (Fig. 4F). These results indicate that both in vitro and in vivo SREBP1c and SREBP2 down-regulate the expression of the HNF4α gene in the rodent liver.
Figure 4.
Ectopic expression of SREBPs in mouse liver down-regulates HNF4α expression. A, IB analysis of whole-cell extracts (80 μg/lane) from livers of diabetic (db/db, STZ) and nondiabetic (db/m, C57) male mice probed with antibodies to SREBP1, SREBP2, and β-actin. Shown are representative results from three mice (n = 8 per group). See supplemental Fig. S2 for SREBP1c and SREBP2 mRNA levels, which parallel the protein levels. B, Quantitative RT-PCR detecting SREBP target gene mRNA levels in the livers of db/db and db/m mice. Plotted are the means ± sd of eight mice per group normalized to β-actin with the control mice (db/m) arbitrarily set to 1.0 (*, P < 0.05). C, IB analysis of whole-cell extracts (50 μg/lane) from primary rat hepatocytes 24 h after infection with Ad-HA-SREBP1c(N) (SREBP1c) or Ad-GFP (GFP) (multiplicity of infection = 20). Data are representative of three independent experiments. D and E, IB analysis of whole-cell extracts (80 μg/lane) from livers of 8-wk-old male C57BL/6J mice 7 d after iv infection with recombinant adenovirus Ad-GFP (GFP) (D and E), Ad-SREBP1c(N) (SREBP1c) (D), or Ad-SREBP2(N) (SREBP2) (E) (1 × 109 plaque-forming units). Blots were probed with anti-HNF4α, SREBP1, SREBP2, HA (SREBP1c and SREBP2 adenoviruses contain an HA tag), and β-actin antibodies. Shown are results from two representative animals of six per group. F, Quantitative RT-PCR detecting HNF4α and target gene mRNA levels in the livers of mice injected with the recombinant adenovirus [Ad-SREBP1c(N) and Ad-SREBP2(N)] as described in D and E. Plotted are means ± sd of six mice per group normalized to β-actin except the GFP control, which contained 12 mice. Statistically significant differences relative to Ad-GFP, arbitrarily set to 1.0, are indicated (*, P < 0.05).
SREBP2 down-regulates the expression of the human HNF4α P1 promoter in response to insulin
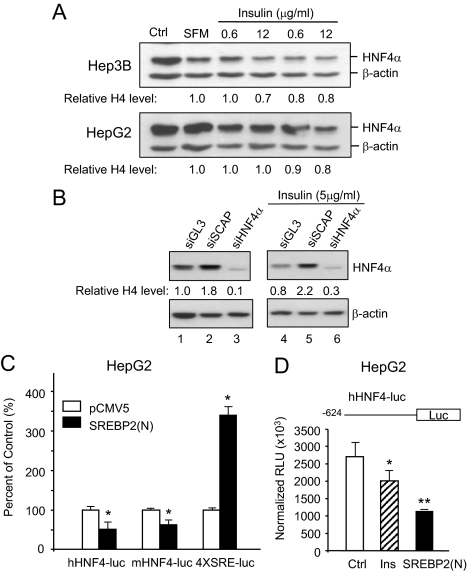
To determine whether the results we observed in the rodent systems are applicable to humans, we ectopically expressed SREBP2(N) in the human liver cell line HepG2 using the recombinant adenovirus and observed a decrease in the levels of the endogenous HNF4α protein, just as we did in the rodent systems (data not shown). We next examined the effect of insulin on the expression of HNF4α in HepG2 cells as well as another human liver cell line, Hep3B. The results show a small but reproducible decrease in HNF4α protein levels in these cell lines (Fig. 5A). To our knowledge, this is the first report of a decrease in HNF4α levels in human liver cells in response to insulin. To determine whether the SREBP pathway is involved in this decrease, we inactivated it by using small interfering RNA (siRNA) to knock down SCAP, the protease that generates the active form of the SREBPs. In HepG2 cells, we consistently observed an increase in endogenous levels of HNF4α protein in response to siSCAP both in the absence and the presence of insulin treatment (Fig. 5B, lanes 2 and 5), suggesting that SREBPs are required for the decrease in HNF4α expression upon insulin treatment.
Figure 5.
SREBP2 down-regulates the expression of the human HNF4α P1 promoter. A, IB analysis of whole-cell extracts (15 μg/lane) from human Hep3B and HepG2 cells, treated with increasing amounts of insulin for 24 h, and probed with anti-HNF4α and β-actin antibodies. Relative amounts of HNF4α protein normalized to β-actin compared with the serum-free media (SFM) control are indicated. Ctrl, Control in serum-containing media. Data are representative of three independent experiments. B, IB analysis of whole-cell extracts (25 μg/lane) from HepG2 transfected with three different siRNAs (siSCAP, siGL3 against luciferase as a negative control, and siHNF4α as a positive control) in serum-free media for 18 h followed by insulin treatment (5 μg/ml) for 24 h. Blots were probed with anti-HNF4α and β-actin antibodies. Relative HNF4α protein amounts normalized to β-actin are indicated. Data are representative of at least two independent experiments. (Supplemental Fig. S3 shows the expected decrease in SCAP mRNA levels upon siRNA.) C and D, HepG2 cells transiently cotransfected with HNF4α P1 promoter luciferase constructs (human, hHNF4-luc; mouse, mHNF4-luc) or control reporter (4xSRE-luc) and SREBP2(N) expression plasmid or pCMV5 empty vector control for 48 h or treated with insulin (5 μg/ml) (Ins) for 18 h after a 6-h preteatment without serum. A β-gal plasmid served as a transfection control. Plotted are means ± sd of normalized relative light units (RLU) (D) or percentage of empty vector control (C) from duplicate samples from one representative experiment of at least three that were performed (*, P < 0.05; **, P < 0.01).
To determine whether SREBPs directly regulate the HNF4α promoter (P1) that regulates expression in the liver, we used a reporter gene assay with the human and mouse HNF4α P1 promoters linked to luciferase; cotransfection into HepG2 cells with an expression vector for SREBP2(N) decreased transcription of both HNF4α P1 promoter constructs (Fig. 5C, hHNF4-luc and mHNF4-luc, respectively). Insulin also decreased the luciferase activity from the human P1 promoter, although not as well as the transfected SREBP2 (Fig. 5D). In contrast, SREBP2(N) activated a control reporter construct containing four SREs (4XSRE-luc), demonstrating a specific inhibition of the HNF4α P1 promoter (Fig. 5C).
SREBP2 is recruited to the human HNF4α P1 promoter
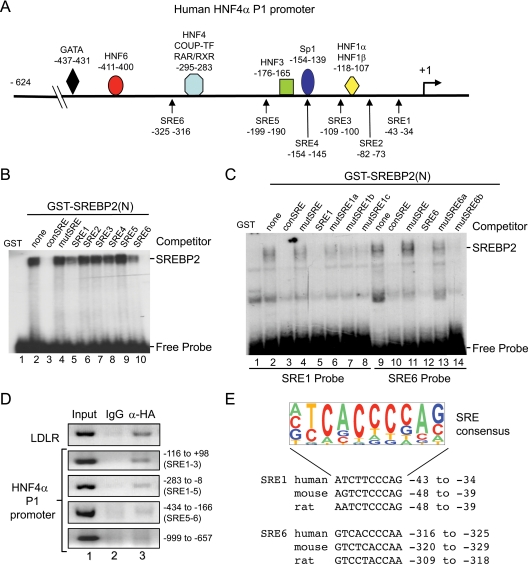
To investigate further the effect of SREBP2 on HNF4α expression, we searched the human HNF4α P1 promoter for potential SREs. We identified six such sites (SRE1-6) distributed throughout the proximal promoter region, particularly between −200 and +1 (Fig. 6A). We assayed the ability of SREBP2(N) to bind these elements in vitro by performing a gel shift assay with a purified glutathione S-transferase (GST). SREBP2(N) fusion protein and found that SREBP2(N) binding to a consensus SRE was specifically competed by two of the six elements (SRE1 and SRE6) (Fig. 6B, lanes 5 and 10). We also observed direct, specific binding to both of those elements (Fig. 6C). To determine whether SREBP2 also binds the endogenous HNF4α P1 promoter, we performed a chromatin immunoprecipitation (ChIP) assay in HepG2 cells and found that ectopically expressed HA-tagged SREBP2(N) is recruited to regions corresponding to SRE1 but not SRE6 (Fig. 6D). Finally, we verified that the SRE1 and SRE6 sites are conserved in both mouse and rat (Fig. 6E), suggesting that our results on the human HNF4α P1 promoter are applicable to the rodent promoters as well.
Figure 6.
SREBP2 is recruited to the human HNF4α P1 promoter. A, Schematic diagram of the human HNF4α P1 promoter from −624 to +1 adapted from Ref. 6 with positions indicated of known transcription factor binding sites and putative SREs (SRE1-6) identified using EMBOSS applications Profit and Prophecy and published SREs. Numbers are nucleotides relative to the transcription start site (+1). EMSA of 32P-labeled consensus SRE (B) and potential SREs from the human HNF4α P1 promoter (SRE1 and SRE6) (C) incubated for 20 min at room temperature with GST or GST-SREBP2(N) fusion protein (2–6 μg/reaction). Competitor, 100-fold molar excess of unlabeled wild-type (conSRE) or mutated SRE oligos. See supplemental Table S2 for sequence of oligonucleotides. SREBP2-DNA complex and free probe are indicated. Shown are representative autoradiograms from 5% native polyacrylamide gels from one of three independent experiments. D, ChIP assay of HepG2 cells infected with Ad-SREBP2(N) for 36 h. Shown are ethidium-bromide-stained 1% agarose gels of PCR products amplified from DNA immunoprecipitated with normal rabbit IgG (control) or anti-HA antibody with primers for the human LDLR or HNF4A P1 promoter as indicated. Results are representative of two to three independent experiments. Sequences of primers are given in supplemental Table S1. E, Sequence logo of SRE consensus generated from published SREs using WebLogo (www.weblogo.berkeley.edu) and alignment of SRE1 and SRE6 in human, mouse, and rat HNF4α P1 promoters. The size of the letters corresponds to the frequency with which the nucleotide is found at that position. Nucleotides relative to +1 are given. [SRE 2–5 also showed significant conservation, although they did not bind GST-SREBP2(N), not shown].
Discussion
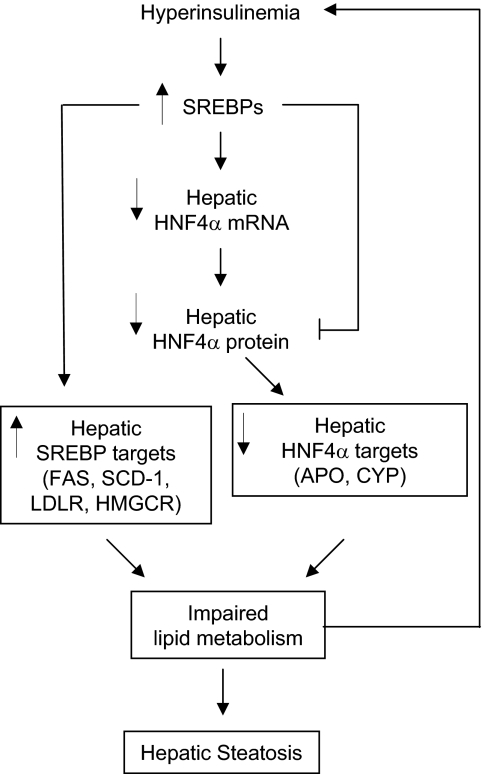
Despite the fact that both HNF4α and SREBPs are known to be major players in hepatic lipid and carbohydrate metabolism and are linked to diabetes and insulin signaling, potential interactions between these pathways have not been thoroughly explored. Similarly, the role of hepatic HNF4α in diabetes is ill defined. Here, we link the insulin, SREBP, and HNF4α pathways by showing an inverse relationship between HNF4α gene expression and activation of SREBP proteins in the livers of db/db, STZ, and fasted/refed mice and insulin-treated hepatocytes (Figs. 1–3). We also show that SREBPs down-regulate the expression of the hepatic HNF4α P1 promoter both in vitro and in vivo (Figs. 4 and 5) and investigate the mechanism of that repression, which appears to be conserved from rodents to humans (Fig. 6). These results lead us to propose a regulatory cascade that may have important implications for diabetes mellitus (Fig. 7).
Figure 7.
Regulation of hepatic HNF4α expression by insulin: involvement of SREBPs and proposed liver impairments. Hyperinsulinemia commonly found in type 2 diabetes leads to increased levels of activated SREBPs in the liver (reviewed in Refs. 16, 48, and 49), which in turn down-regulate the expression of the HNF4α P1 promoter (this study). Lower HNF4α protein results in decreased activation of HNF4α target genes, which in concert with increased expression of SREBP target genes, could lead to dysregulation of lipid metabolism and eventually to hepatic steatosis (fatty liver), a common complication of type 2 diabetes. SREBP proteins also negatively affect the function of the hepatic HNF4α protein (26,27,28). Accumulation of lipids in the liver can lead to insulin resistance, triggering a feed-forward effect and aggravating the disease. See text for additional details and a discussion of the role of other factors in the insulin pathway and the effects on carbohydrate metabolism.
Link between the insulin, SREBP, and HNF4α pathways and complications of diabetes
Elevated plasma insulin levels, such as those in type 2 diabetic patients and db/db mice, lead to elevated levels of the mature form of the SREBP proteins and its target genes (this study) (19,22,25,30,34). They also lead to lower HNF4α levels in the liver and suppression of its target genes (this study). Although there is one previous report of elevated levels of HNF4α protein and mRNA in STZ-treated rats being normalized by insulin treatment, the level of HNF4α in db/db mice was not examined (35). Furthermore, ours is the first report to investigate the mechanism underlying the down-regulation of HNF4α expression by insulin, which we show is due, at least in part, to a direct action of SREBPs on the hepatic HNF4α P1 promoter. This insulin-SREBP-HNF4α regulatory cascade is reinforced by protein-protein interactions between SREBPs and HNF4α that interfere with the HNF4α-PGC1α complex that plays a critical role in up-regulating PEPCK gene expression during fasting (26,27,28). Furthermore, because the effect of SREBPs on HNF4α protein levels appeared to be even more marked than on the mRNA levels (Figs. 4), SREBPs may also directly regulate HNF4α protein as well as mRNA levels, although additional studies are required to confirm this result and elucidate the mechanism behind it.
The insulin-SREBP-HNF4α pathway may offer insight into some of the most common complications during the onset of diabetes, namely hepatic steatosis and coronary heart disease. Both insulin resistance and SREBPs have been linked to nonalcoholic fatty liver disease (36). Although this is undoubtedly due in a large part to many of the direct targets of SREBPs that lead to fatty acid and cholesterol synthesis, the data presented here indicate that HNF4α and its target genes might also be involved. For example, HNF4α regulates most of the apolipoprotein genes involved in lipid transport and metabolism (1,4). Therefore, an increase in lipid synthesis in the liver caused by elevated SREBPs, along with a decrease in lipid export caused by decreased expression of HNF4α and its target genes, could lead to lipid accumulation and a fatty liver. Such an involvement of HNF4α in hepatic steatosis is supported by the fact that the liver-specific knockout of HNF4α also showed an accumulation of lipids in the liver (37).
It is intriguing to speculate that the dysregulation of the HNF4α P1 promoter by the insulin-SREBP pathway could also be a factor in the higher incidence of atherosclerosis and cardiovascular disease among diabetic patients who tend to have low high-density lipoprotein (HDL) levels (38). Because HNF4α positively regulates the expression of the APOA1, APOA2, APOA4, and APOC3 genes, all of which encode protein components of HDL, and negatively regulates the SR-BI (Scarb1) gene, which encodes the receptor for HDL uptake in the liver (37), decreased HNF4α could lead to lower serum HDL levels in type 2 diabetic patients, just as in the liver-specific HNF4α knockout mice (37). Lower serum ApoA2, ApoC3, and HDL levels have indeed been observed in certain MODY1 patients (39,40). Finally, our results could also help explain the paradox of normal or high HDL levels in certain type 1 diabetic patients with cardiovascular disease (41). If those patients have elevated hepatic HNF4α levels, such as those observed in the STZ mice, this could lead to elevated serum HDL by increased production of the apolipoproteins and a decreased expression of the HDL receptor SR-BI. Although additional studies are required to confirm the role of hepatic HNF4α in diabetes, hepatic steatosis and atherosclerosis in humans, the results presented here indicate that these avenues of investigation may be worth pursuing.
The overall effect of the insulin-SREBP-HNF4α pathway on carbohydrate metabolism in the liver, and its subsequent effect on diabetic complications, is less clear. For example, both PEPCK and G6Pase, two key enzymes in hepatic gluconeogenesis, are down-regulated in response to insulin by a variety of other factors in addition to HNF4α, including SREBPs, C/EBP, glucocorticoid receptor, and FoxO1 (16,42,43,44,45). In contrast to an increased accumulation of lipids in the liver that would exacerbate the diabetic condition, a decrease in PEPCK and G6Pase would help lower blood glucose levels and hence ameliorate the effects of diabetes.
Effects of additional factors on the insulin-SREBP- HNF4α pathway
The model presented in Fig. 7 is admittedly narrowly focused on just the two factors examined in this study. The insulin pathway is in fact much more complex and known to affect several different signaling pathways and transcription factor networks. Additional factors will impact the final affect of insulin in vivo and may also affect the insulin-SREBP-HNF4α pathway described here. For example, FoxO1 (also known as FKHR) is another well characterized player in carbohydrate and lipid metabolism in the liver that, like HNF4α, stimulates the expression of genes involved in hepatic gluconeogenesis, such as PEPCK and G6Pase (42,43,44). FoxO1 has also been shown to interact with the DNA-binding domain of HNF4α, thereby inhibiting its ability to bind DNA and activate transcription (46). In response to insulin, FoxO1 is phosphorylated and exported from the nucleus, thereby eliminating its repressive effect on HNF4α (46) and other targets (47). Another potential target of FoxO1 is the SREBP1c gene (SREBF1), although it is not clear if activated FoxO1 increases (42) or decreases (44) its expression. Hence, one mechanism by which insulin may increase the expression of SREBP1c is via phosphorylation of FoxO1 (others include SCAP, Insig, LXR, and Sp1) (48,49). The results presented here suggest that the increase in SREBP1c would then lead to a decrease in HNF4α mRNA and subsequently protein. Although this scenario raises the question of the relevance of the relief of HNF4α from repression by phosphorylation of FoxO1 (46) (if HNF4α protein levels are reduced by insulin, a lack of inhibition of HNF4α DNA binding by FoxO1 becomes irrelevant), it could explain in part why overexpression of nuclear FoxO1 leads to hepatic steatosis (42); it represses HNF4α function, which is known to lead to a fatty liver (37).
Another player in the insulin pathway is protein kinase C lambda (PKCλ) which has been shown to increase SREBP1c expression in response to insulin (50). We have shown previously that classical PKC phosphorylates HNF4α and causes translocation to the cytoplasm and subsequent degradation of HNF4α protein (51). It will be of interest, therefore, to determine whether in response to insulin PKCλ has a similar effect on HNF4α. Indeed, activation of PKCλ, or a related kinase, could explain why the effect of insulin on the HNF4α protein level appears to be somewhat greater than on the RNA level (Fig. 2).
Repression of the HNF4α P1 promoter by SREBPs
Although SREBPs are generally thought of as transcriptional activators, they can also inhibit transcription of certain promoters (12). Here we identify two conserved SREs in the HNF4α P1 promoter (SRE1 and SRE6) that are bound by SREBP2(N) in vitro. ChIP analysis confirmed that SREBP2 is recruited to the P1 promoter in the region of SRE1 in live cells, although binding to the SRE6 region was not observed (Fig. 6). We propose that SREBPs bind the HNF4α P1 promoter via SRE1 (and possibly other regions) and either recruit histone deacetylases, as observed previously on other promoters (52), and/or interfere with the binding of other transcription factors that bind in that region, as it does with Sp1 on the PEPCK promoter (53) and FoxO1 on the IRS-2 promoter (54). Indeed, mutation of the SRE1 site in the human HNF4α P1 promoter decreased basal activation of the promoter as well as repression by SREBP2 (data not shown). This suggests that another as yet unidentified positive acting factor may bind in this region and be competed off by SREBP2, although additional studies will be required to confirm this hypothesis. Whatever the mechanism of down-regulation of the HNF4α P1 promoter by insulin and SREBPs, it appears to be conserved between rodents and humans and should have important implications for the progression of diabetes and associated complications.
Materials and Methods
Cell culture and reagents
Human hepatocellular carcinoma/hepatoblastoma cell lines HepG2 and Hep3B and rat hepatoma H35 cells were maintained in DMEM plus 10% fetal bovine serum (FBS) (vol/vol) and HEK293 cells in DMEM plus 5% FBS. Primary hepatocytes were isolated from 8- to 10-wk-old Sprague Dawley male rats by a two-step perfusion with type IV collagenase (55). The isolated hepatocytes were seeded on rat-tail collagen I-coated plates in RPMI 1640 medium supplemented with 10% FBS (vol/vol). All media contained 100 U/ml penicillin and 100 μg/ml streptomycin unless indicated otherwise. Cells were cultured at 37 C in 5% CO2. Recombinant human insulin (I9278; Sigma-Aldrich, St. Louis, MO) was used for HepG2 and H35 cells; insulin purified from bovine pancreas was used for primary hepatocytes (I6634; Sigma-Aldrich).
Animal treatment
In accordance with the guidelines of the Protection of Laboratory Animals, all animal procedures were approved by the Institutional Animal Care and Use Committee of the Peking University Health Science Center where the C57BL/6J mice were bred. The mice were maintained under a 12-h light, 12-h dark cycle and fed a standard laboratory chow and tap water ad libitum. Obese male 4-month-old db/db mice (C57BLKS/JLepr) were used as the type 2 diabetes model with age-matched, heterozygous littermates (db/m mice) as nondiabetic controls. Type 1 diabetes was induced in 8-wk-old male C57BL/6J mice by daily ip injections of STZ (40 μg/g body weight) (Sigma Aldrich) for 5 d; the animals were killed 1 wk later. The control group received an equal volume of vehicle (0.1 mm sodium citrate). Levels of fasting (6 h) blood glucose and plasma insulin from the saphenous vein were measured according to standard protocols (ACCU-CHEK II from Roche Diagnostics, Indianapolis, IN; I125-linked immunosorbent assay kit from Fu Rui, Beijing, China). For adenoviral treatment, 8-wk old male C57BL/6J mice were injected with Ad-GFP, Ad-SREBP-1c(N), or Ad-SREBP-2(N) through the tail vein (1 × 109 plaque-forming units diluted in 0.1 ml saline) and killed by CO2 asphyxiation 7 d later. Liver tissues were dissected, immediately snap-frozen in liquid nitrogen, and stored at −80 C.
Adenovirus purification and infection
Recombinant adenovirus expressing the HA-tagged N-terminal, constitutively active portion of SREBP1c [Ad-SREBP1c(N)] or SREBP2 [Ad-SREBP2(N)] (19,52,56) was amplified in HEK293 cells, purified on cesium chloride gradients, and concentrated with Sephadex-G-25M columns. The viral titers (plaque-forming units) were determined in HEK293 cells using standard procedures. Comparable amounts of Ad-GFP were used as a control (57). Primary rat hepatocytes were infected with adenovirus at 20 multiplicity of infection about18 h after plating and harvested 24 h later.
Whole-cell extracts and immunoblot (IB) analysis
Mouse liver (∼100 mg) was homogenized in 1 ml Triton lysis buffer [150 mm NaCl, 1% Triton X-100, 100 mm Tris-HCl (pH 7.4), 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml pepstatin A), sonicated in 3-sec pulses at an intensity of 200 W/cm2 (Sonicator model JY99-2D; Ningbo Scientz Biotech Co., Ningbo, China), centrifuged at 12,000 rpm for 10 min at 4 C (Micromax RF refrigerated microcentrifuge; Thermo IEC, Waltham, MA), and the supernatants were recovered. Primary rat hepatocytes and HepG2, Hep3B, and H35 cells were rinsed in PBS before lysing with RIPA buffer (Santa Cruz Biotechnology, Santa Cruz, CA). Equivalent amounts of lysates (15–80 μg/lane) were analyzed by 10% SDS-PAGE followed by IB analysis with antibodies against HNF4α (58), SREBP2 (557037; BD Pharmingen, San Diego, CA), SREBP1 (sc-13551; Santa Cruz Biotechnology, Santa Cruz, CA), β-actin (A5316; Sigma), or HA (sc-7392; Santa Cruz) as per the manufacturers’ suggestions. Blots were developed with the appropriate secondary antibody conjugated to horseradish peroxidase and ECL. Nuclear extracts from COS-7 cells overexpressing rat HNF4α2 were used as a marker.
Quantitative real-time PCR analysis
Quantitative real-time PCR (RT-PCR) was performed on reverse-transcribed total RNA isolated with Trizol reagent (Invitrogen, Carlsbad, CA) using SYBR Green I and TaqMan PCR Master Mix with the MX3000P QPCR detection system (Stratagene, La Jolla, CA). The relative amounts of mRNAs were calculated using the comparative threshold cycle method. The sequences of the primers are given in supplemental Table S1.
RNA interference experiment
Duplex siRNAs were introduced into HepG2 cells at about 30–50% confluency using TransIT-TKO transfection reagent as per the manufacturer’s protocol (Mirus Bio Corp., Madison, WI). Each experiment included controls containing the transfection reagent with or without control siRNA against firefly luciferase (see supplemental Table S1 for primer sequences). Cells were lysed using RIPA buffer (Santa Cruz) and analyzed 48 h after transfection.
Plasmid construction and transfection
A luciferase reporter construct containing nucleotides −624 to +89 of the human HNF4α P1 promoter (hHNF4-luc) was generated by inserting into the HindIII and XhoI sites of the pGL3-basic vector (Promega, Madison, WI) a PCR product amplified from human genomic DNA using appropriate primers (see supplemental Table S1). The reporters containing the mouse HNF4α P1 promoter (nucleotides −401 to +17) (mHNF4-luc) (59) and four SREs (SRE-luc) (60) have been described previously. HepG2 cells seeded onto six-well plates were transfected with the reporter constructs (1.0 μg/well) with or without pCMV5-HA-SREBP2N expressing the constitutively active N-terminal fragment of human SREBP2 (52) (0.8 μg) and pRSV-β-gal (0.2 μg) with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Luciferase activities were measured and normalized to that of β-galactosidase 48 h after transfection.
EMSA and ChIP assay
EMSA was carried out as previously described (61) with double-stranded oligonucleotide 32P-labeled probes of consensus or putative SREs from the human HNF4α P1 promoter (see supplemental Table S2) and GST fusion proteins [GST-SREBP2(N) and GST control] purified as previously described (52,62). ChIP assays of HepG2 cells infected with Ad-SREBP2(N) for 36 h were performed as previously described (63) with polyclonal anti-HA (Santa Cruz) or normal rabbit IgG. The precipitated DNA was PCR amplified (30–35 cycles) with primers corresponding to various regions of the human HNF4α P1 promoter or the human LDLR (see supplemental Table S1).
Image analysis and statistical analyses
IBs were imaged by the National Institutes of Health Image J program. Quantitative data are expressed as mean ± sd. Statistical significance was evaluated using regression analysis, one-way ANOVA, and unpaired Student’s t test. P < 0.05 was considered statistically significant. For nonquantitative data, the results presented are representative of at least three independent experiments.
Supplementary Material
Acknowledgments
We thank E. Bolotin for help identifying SREs and S. Thorigeirsson for the mHNF4.Luc construct, L. Hawell III and J. R. Evans for help with H35 cells, and T. C. Ta for ChIP efforts.
Footnotes
This work was supported by in part by the grants from the National Natural Science Foundation of China (30570713 and 30630032), the Major National Basic Research Grant of China (2006BC503802) and the “111” Project of China (to Y.Z.) and grants from the National Institutes of Health (R01DK053892 to F.M.S.) and (R01HL77448 to J.S.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online January 29, 2009
Abbreviations: ChIP, Chromatin immunoprecipitation; FBS, fetal bovine serum; GST, glutathione S-transferase; HA, hemagglutinin; HDL, high-density lipoprotein; HNF4α, hepatocyte nuclear factor 4α; IB, immunoblot; MODY1, maturity-onset diabetes of the young 1; SCAP, SREBP cleavage-activating protein; siRNA, small interfering RNA; SRE, sterol response element; SREBP, sterol regulatory element-binding protein; STZ, streptozotocin.
References
- Sladek F, Seidel S 2001 Hepatocyte nuclear factor 4α. In: Burris T, McCabe E, eds. Nuclear receptors and genetic diseases. London: Academic Press; 309–361 [Google Scholar]
- Ribeiro A, Archer A, Le Beyec J, Cattin AL, Saint-Just S, Pinçon-Raymond M, Chambaz J, Lacasa M, Cardot P 2007 Hepatic nuclear factor-4, a key transcription factor at the crossroads between architecture and function of epithelia. Recent Patents Endocr Metab Immune Drug Discov 1:166–175 [Google Scholar]
- Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA 2006 Hepatocyte nuclear factor 4α is essential for embryonic development of the mouse colon. Gastroenterology 130:1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA 2006 Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol 2:2006.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Gao N, Gorski RK, White P, Hardy OT, Rafiq K, Brestelli JE, Chen G, Stoeckert Jr CJ, Kaestner KH 2007 Expansion of adult β-cell mass in response to increased metabolic demand is dependent on HNF-4α. Genes Dev 21:756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis P, Talianidis I 2001 Regulatory mechanisms controlling human hepatocyte nuclear factor 4α gene expression. Mol Cell Biol 21:7320–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI 1996 Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1). Nature 384:458–460 [DOI] [PubMed] [Google Scholar]
- Lehman DM, Richardson DK, Jenkinson CP, Hunt KJ, Dyer TD, Leach RJ, Arya R, Abboud HE, Blangero J, Duggirala R, Stern MP 2007 P2 promoter variants of the hepatocyte nuclear factor 4α gene are associated with type 2 diabetes in Mexican Americans. Diabetes 56:513–517 [DOI] [PubMed] [Google Scholar]
- Miura A, Yamagata K, Kakei M, Hatakeyama H, Takahashi N, Fukui K, Nammo T, Yoneda K, Inoue Y, Sladek FM, Magnuson MA, Kasai H, Miyagawa J, Gonzalez FJ, Shimomura I 2006 Hepatocyte nuclear factor-4α is essential for glucose-stimulated insulin secretion by pancreatic β-cells. J Biol Chem 281:5246–5257 [DOI] [PubMed] [Google Scholar]
- Gupta RK, Vatamaniuk MZ, Lee CS, Flaschen RC, Fulmer JT, Matschinsky FM, Duncan SA, Kaestner KH 2005 The MODY1 gene HNF-4α regulates selected genes involved in insulin secretion. J Clin Invest 115:1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson RB 2003 The SREBP pathway: insights from Insigs and insects. Nat Rev Mol Cell Biol 4:631–640 [DOI] [PubMed] [Google Scholar]
- Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F 2004 SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86:839–848 [DOI] [PubMed] [Google Scholar]
- Shimano H 2001 Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res 40:439–452 [DOI] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL 2003 Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA 100:12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS 2002 SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty K, Hanson RW 2007 Insulin regulation of phosphoenolpyruvate carboxykinase-c gene transcription: the role of sterol regulatory element-binding protein 1c. Nutr Rev 65:S47–S56 [DOI] [PubMed] [Google Scholar]
- Meton I, Egea M, Anemaet IG, Fernandez F, Baanante IV 2006 Sterol regulatory element binding protein-1a transactivates 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene promoter. Endocrinology 147:3446–3456 [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim HI, Kim TH, Im SS, Park SK, Lee IK, Kim KS, Ahn YH 2004 SREBP-1c mediates the insulin-dependent hepatic glucokinase expression. J Biol Chem 279:30823–30829 [DOI] [PubMed] [Google Scholar]
- Becard D, Hainault I, Azzout-Marniche D, Bertry-Coussot L, Ferre P, Foufelle F 2001 Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes 50:2425–2430 [DOI] [PubMed] [Google Scholar]
- Chen G, Liang G, Ou J, Goldstein JL, Brown MS 2004 Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci USA 101:11245–11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansmannel F, Mordier S, Iynedjian PB 2006 Insulin induction of glucokinase and fatty acid synthase in hepatocytes: analysis of the roles of sterol-regulatory-element-binding protein-1c and liver X receptor. Biochem J 399:275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL 1999 Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci USA 96:13656–13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS 2003 Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc Natl Acad Sci USA 100:3155–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty BD, Bobard A, Hainault I, Ferre P, Bossard P, Foufelle F 2005 Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc Natl Acad Sci USA 102:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, Guichard C, Ferre P, Foufelle F 1999 Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci USA 96:12737–12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponugoti B, Fang S, Kemper JK 2007 Functional interaction of hepatic nuclear factor-4 and peroxisome proliferator-activated receptor-γ coactivator 1α in CYP7A1 regulation is inhibited by a key lipogenic activator, sterol regulatory element-binding protein-1c. Mol Endocrinol 21:2698–2712 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Shimano H, Nakagawa Y, Ide T, Yahagi N, Matsuzaka T, Nakakuki M, Takahashi A, Suzuki H, Sone H, Toyoshima H, Sato R, Yamada N 2004 SREBP-1 interacts with hepatocyte nuclear factor-4α and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J Biol Chem 279:12027–12035 [DOI] [PubMed] [Google Scholar]
- Misawa K, Horiba T, Arimura N, Hirano Y, Inoue J, Emoto N, Shimano H, Shimizu M, Sato R 2003 Sterol regulatory element-binding protein-2 interacts with hepatocyte nuclear factor-4 to enhance sterol isomerase gene expression in hepatocytes. J Biol Chem 278:36176–36182 [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM 2001 Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131–138 [DOI] [PubMed] [Google Scholar]
- Shimomura I, Matsuda M, Hammer RE, Bashmakov Y, Brown MS, Goldstein JL 2000 Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell 6:77–86 [PubMed] [Google Scholar]
- Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA 2004 Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev 18:157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanniman EA, Lambert G, Inoue Y, Gonzalez FJ, Sinal CJ 2006 Apolipoprotein A-IV is regulated by nutritional and metabolic stress: involvement of glucocorticoids, HNF-4α, and PGC-1α. J Lipid Res 47:2503–2514 [DOI] [PubMed] [Google Scholar]
- Ueki K, Kadowaki T, Kahn CR 2005 Role of suppressors of cytokine signaling SOCS-1 and SOCS-3 in hepatic steatosis and the metabolic syndrome. Hepatol Res 33:185–192 [DOI] [PubMed] [Google Scholar]
- Azzout-Marniche D, Becard D, Guichard C, Foretz M, Ferre P, Foufelle F 2000 Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem J 350(Pt 2):389–393 [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Matsuno F, Chowdhury S, Kimura T, Iwase K, Araki E, Shichiri M, Mori M, Takiguchi M 2000 The gene for hepatocyte nuclear factor (HNF)-4α is activated by glucocorticoids and glucagon, and repressed by insulin in rat liver. FEBS Lett 478:141–146 [DOI] [PubMed] [Google Scholar]
- Ahmed MH, Byrne CD 2007 Modulation of sterol regulatory element binding proteins (SREBPs) as potential treatments for non-alcoholic fatty liver disease (NAFLD). Drug Discov Today 12:740–747 [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ 2001 Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21:1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexel H, Aczel S, Marte T, Benzer W, Langer P, Moll W, Saely CH 2005 Is atherosclerosis in diabetes and impaired fasting glucose driven by elevated LDL cholesterol or by decreased HDL cholesterol? Diabetes Care 28:101–107 [DOI] [PubMed] [Google Scholar]
- Pearson ER, Pruhova S, Tack CJ, Johansen A, Castleden HA, Lumb PJ, Wierzbicki AS, Clark PM, Lebl J, Pedersen O, Ellard S, Hansen T, Hattersley AT 2005 Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4α mutations in a large European collection. Diabetologia 48:878–885 [DOI] [PubMed] [Google Scholar]
- Shih DQ, Dansky HM, Fleisher M, Assmann G, Fajans SS, Stoffel M 2000 Genotype/phenotype relationships in HNF-4α/MODY1: haploinsufficiency is associated with reduced apolipoprotein (AII), apolipoprotein (CIII), lipoprotein(a), and triglyceride levels. Diabetes 49:832–837 [DOI] [PubMed] [Google Scholar]
- Valabhji J, Donovan J, McColl AJ, Schachter M, Richmond W, Elkeles RS 2002 Rates of cholesterol esterification and esterified cholesterol net mass transfer between high-density lipoproteins and apolipoprotein B-containing lipoproteins in type 1 diabetes. Diabetes Med 19:424–428 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Han S, Kitamura T, Accili D 2006 Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest 116:2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM 2003 Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423:550–555 [DOI] [PubMed] [Google Scholar]
- Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG 2006 FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem 281:10105–10117 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D 2007 Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab 6:208–216 [DOI] [PubMed] [Google Scholar]
- Hirota K, Daitoku H, Matsuzaki H, Araya N, Yamagata K, Asada S, Sugaya T, Fukamizu A 2003 Hepatocyte nuclear factor-4 is a novel downstream target of insulin via FKHR as a signal-regulated transcriptional inhibitor. J Biol Chem 278:13056–13060 [DOI] [PubMed] [Google Scholar]
- Glauser DA, Schlegel W 2007 The emerging role of FOXO transcription factors in pancreatic β-cells. J Endocrinol 193:195–207 [DOI] [PubMed] [Google Scholar]
- Raghow R, Yellaturu C, Deng X, Park EA, Elam MB 2008 SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol Metab 19:65–73 [DOI] [PubMed] [Google Scholar]
- Shimano H 2007 SREBP-1c and TFE3, energy transcription factors that regulate hepatic insulin signaling. J Mol Med 85:437–444 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Ogawa W, Akimoto K, Inoue H, Miyake K, Furukawa K, Hayashi Y, Iguchi H, Matsuki Y, Hiramatsu R, Shimano H, Yamada N, Ohno S, Kasuga M, Noda T 2003 PKCλ in liver mediates insulin-induced SREBP-1c expression and determines both hepatic lipid content and overall insulin sensitivity. J Clin Invest 112:935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, Parpura V, Paschal BM, Sladek FM 2007 Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol 21:1297–1311 [DOI] [PubMed] [Google Scholar]
- Zeng L, Lu M, Mori K, Luo S, Lee AS, Zhu Y, Shyy JY 2004 ATF6 modulates SREBP2-mediated lipogenesis. EMBO J 23:950–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty K, Wu SY, Chiang CM, Samols D, Hanson RW 2004 SREBP-1c and Sp1 interact to regulate transcription of the gene for phosphoenolpyruvate carboxykinase (GTP) in the liver. J Biol Chem 279:15385–15395 [DOI] [PubMed] [Google Scholar]
- Ide T, Shimano H, Yahagi N, Matsuzaka T, Nakakuki M, Yamamoto T, Nakagawa Y, Takahashi A, Suzuki H, Sone H, Toyoshima H, Fukamizu A, Yamada N 2004 SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat Cell Biol 6:351–357 [DOI] [PubMed] [Google Scholar]
- Seglen PO 1976 Preparation of isolated rat liver cells. Methods Cell Biol 13:29–83 [DOI] [PubMed] [Google Scholar]
- Lu M, Shyy JY 2006 Sterol regulatory element-binding protein 1 is negatively modulated by PKA phosphorylation. Am J Physiol Cell Physiol 290:C1477–C1486 [DOI] [PubMed] [Google Scholar]
- Wang N, Verna L, Liao H, Ballard A, Zhu Y, Stemerman MB 2001 Adenovirus-mediated overexpression of dominant-negative mutant of c-Jun prevents intercellular adhesion molecule-1 induction by LDL: a critical role for activator protein-1 in endothelial activation. Arterioscler Thromb Vasc Biol 21:1414–1420 [DOI] [PubMed] [Google Scholar]
- Sladek FM, Zhong WM, Lai E, Darnell Jr JE 1990 Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev 4:2353–2365 [DOI] [PubMed] [Google Scholar]
- Terai S, Aoki H, Ashida K, Thorgeirsson SS 2000 Human homologue of maid: a dominant inhibitory helix-loop-helix protein associated with liver-specific gene expression. Hepatology 32:357–366 [DOI] [PubMed] [Google Scholar]
- Zeng L, Liao H, Liu Y, Lee TS, Zhu M, Wang X, Stemerman MB, Zhu Y, Shyy JY 2004 Sterol-responsive element-binding protein (SREBP) 2 down-regulates ATP-binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism. J Biol Chem 279:48801–48807 [DOI] [PubMed] [Google Scholar]
- Jiang G, Nepomuceno L, Hopkins K, Sladek FM 1995 Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol Cell Biol 15:5131–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh M, Cole AL, Choi J, Liu Y, Tulchinsky D, Qiao JH, Fishbein MC, Dooley AN, Hovnanian T, Mouilleseaux K, Vora DK, Yang WP, Gargalovic P, Kirchgessner T, Shyy JY, Berliner JA 2004 Role for sterol regulatory element-binding protein in activation of endothelial cells by phospholipid oxidation products. Circ Res 95:780–788 [DOI] [PubMed] [Google Scholar]
- Hwang-Verslues WW, Sladek FM 2007 Nuclear receptor hepatocyte nuclear factor 4α1 competes with oncoprotein c-myc for control of the p21/WAF1 promoter. Mol Endocrinol 22:78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.