Abstract
DEXD/H-box RNA helicases couple ATP hydrolysis to RNA remodeling by an unknown mechanism. We used x-ray crystallography and biochemical analysis of the human DEXD/H-box protein DDX19 to investigate its regulatory mechanism. The crystal structures of DDX19, in its RNA-bound prehydrolysis and free posthydrolysis state, reveal an α-helix that inserts between the conserved domains of the free protein to negatively regulate ATPase activity. This finding was corroborated by biochemical data that confirm an autoregulatory function of the N-terminal region of the protein. This is the first study describing crystal structures of a DEXD/H-box protein in its open and closed cleft conformations.
RNA helicase activity is involved in all aspects of RNA metabolism, including transcription, pre-mRNA splicing, ribosome biogenesis, nuclear export, translation initiation and termination, RNA degradation, viral replication, and viral RNA detection. The DEXD/H-box RNA helicases couple hydrolysis of ATP to cycles of RNA binding and release that typically result in non-processive RNA duplex unwinding (1) or disruption of RNP3 complexes (2, 3). These proteins interact in a non-sequence-specific manner with the phosphoribose backbone of single-stranded RNA. DEXD/H-box RNA helicases contain two α/β-RecA-like domains that both feature conserved sequence motifs involved in RNA binding and ATP hydrolysis (4, 5). Accessory proteins are involved in the regulation of RNA binding and ATPase activities, although no general mechanism has been demonstrated.
The DDX19 member of the DEXD/H-box RNA helicase family performs an essential function in mRNA nuclear export by remodeling RNP particles during passage of mRNA through the nuclear pore complex (3, 6). Dbp5, the yeast orthologue of DDX19 (7, 8), causes displacement of the RNP constituent, Mex67, thereby preventing re-entry of mRNA into the nucleus (9). Dbp5 is also involved in translation termination (10). A specific function has been assigned to the ADP-bound form of Dbp5, which displaces the RNA-binding protein Nab2, an event that is required for mRNA export (3). In vivo, Dbp5 is activated by the nuclear pore complex-associated protein, Gle1 (11, 12).
Crystal structures of DEXD/H-box proteins show two-lobed proteins with the nucleotide binding site located in the lower part of the cleft separating the conserved domains and the RNA binding site across the upper cleft opening (13–17). DEXD/H-box helicases in general share little homology in their coding sequences upstream of the conserved domain-1. The N-terminal extension of DDX19, however, shares significant homology with that of DDX25/GRTH, a testis-specific protein that is essential for spermatogenesis (18), supporting a functional significance for this sequence. Herein, we present a crystal structure of human DDX19 that shows the ADP-bound protein with an α-helical segment of the N-terminal extension wedged between the core domains, preventing cleft closure. In the structure of the ADPNP-bound protein in complex with RNA, this α-helix has moved out of the way to allow formation of a functional ATPase site. Our biochemical evidence supports a model for DDX19 autoregulation where the N-terminal extension helix prevents ATP hydrolysis, unless the N terminus of the protein is displaced by RNA binding, allowing cleft closure to bring key side chains into position for catalysis.
EXPERIMENTAL PROCEDURES
Human DDX19 cDNA was obtained from the Mammalian Gene Collection (MGC; accession number BC003626) provided by the National Institute of Health. The sequences encoding DDX191–479 (full-length protein), DDX1954–475, and DDX1992–475 were amplified by PCR and inserted into pNIC28-Bsa4 by ligation-independent cloning. The expression constructs included a tobacco etch virus protease-cleavable N-terminal hexahistidine tag. Protein expression in Escherichia coli strain BL21(DE3) (DDX191–479), BL21(DE3) gold pRARE2 (DDX1954–475), or BL21(DE3) R3 pRARE (DDX1992–475) was done in Terrific Broth medium supplemented with 8 g/liter glycerol and 50 μg/ml kanamycin, induction with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside, and culture at 18 °C for 20 h. Cell pellets were resuspended in 50 mm HEPES, pH 7.8, 500 mm NaCl, 10 mm imidazole, 10% glycerol, 0.5 mm TCEP, and Complete EDTA-free protease inhibitor (Roche Applied Science). Cells were lysed by a freeze/thaw cycle followed by the addition of benzonase (Novagen) and sonication (Sonics VibraCell).
Cleared and filtered lysates were loaded onto HiTrap chelating HP columns (GE Healthcare) pre-equilibrated with buffer 1 (30 mm HEPES, pH 7.5, 500 mm NaCl, 10 mm imidazole, 10% glycerol, and 0.5 mm TCEP). The columns were washed with buffer 1 containing 25 mm imidazole. Bound protein was eluted with buffer 1 containing 500 mm imidazole and loaded onto a HiLoad 16/60 Superdex-200 column (GE Healthcare) pre-equilibrated with buffer 2 (30 mm HEPES, pH 7.5, 500 mm NaCl, 10% glycerol, and 0.5 mm TCEP). Fractions containing DDX19 protein were pooled, and the concentration of TCEP was adjusted to 2 mm. The N-terminal hexahistidine tag was removed by incubation of DDX19 proteins with His-tagged tobacco etch virus protease at a molar ratio of 30:1 overnight at room temperature and subsequent passage over a 1-ml HisTrap HP column in buffer 1. The ADP-bound DDX19 apoenzyme structure (PDB entry 3EWS) was obtained with uncleaved protein. The purified proteins were concentrated into buffer 2 using Vivaspin (Sartorius) centrifugal concentrators. Aliquots were flash-frozen and stored at –80 °C. All proteins were verified by time-of-flight mass spectrometry analysis.
For the ADP-bound DDX1954–475 enzyme (PDB entry 3EWS), needle-like crystals of DDX19 were grown using vapor diffusion at 4 °C by mixing 0.1 μl of protein solution (18.3 mg/ml) including 20 mm ADP, 10 mm MgCl2, and 0.1 μl of reservoir solution containing 17% polyethylene glycol 10000, 0.1 m ammonium acetate, 0.1 m Bis-Tris, pH 5.5. Crystals appeared after 8 days and continued to grow for one more week. Reservoir solution supplemented with 20% glycerol was used as cryo-solution and added directly to the drop. Crystals were mounted and flash-frozen in liquid nitrogen. Diffraction data to 2.7 Å resolution were collected at Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY), Berlin, Germany (beamline BL14-2). Data were processed with XDS (19). The structure was solved by Phaser (20) using PDB entry 1FUU as a search model. Domains were searched for separately and were edited according to sequence alignment with CHAINSAW (21) before molecular replacement. The structure was refined with Phenix (22). TLS parameters were refined using individual domains as rigid groups. Model building was done using Coot (23). For further details, see supplemental Table S1.
Crystals of DDX1954–475 in complex with RNA and Mg-ADPNP were obtained by vapor diffusion in sitting drops incubated at 4 °C by mixing 0.1 μl of protein solution (20 mg/ml) including 10-molar excess of decauracil ssRNA, ADPNP, and MgCl2 and 0.2 μl of reservoir solution containing 14% polyethylene glycol monomethyl ether 2000, 0.25 m trimethylamine n-oxide, 0.1 m Tris, pH 8. Crystals appeared after 7 days. Reservoir solution supplemented with 25% glycerol was used as cryosolution and added directly to the drop. Crystals were mounted and flash-frozen in liquid nitrogen. Diffraction data to 2.7 Å resolution were collected at the European Synchrotron Radiation Facility (ESRF) Grenoble, France (beamline ID-29). Data were integrated and scaled using XDS. One monomer was located in the asymmetric unit. The structure was solved by Phaser using the previous DDX19 structure (PDB entry 3EWS) as a search model. Domains were searched for separately. The structure was refined with RefMac5 (24). TLS parameters were refined using individual domains as rigid groups. Model building was done using Coot. For further details, see supplemental Table S1.
Geometry of the models was analyzed with Molprobity (25) Sequence alignments were obtained using ESPript (26). The figures were created using Pymol (27).
ATPase activities of the DDX19 protein constructs (0.5 μm; in 30 mm HEPES, pH 7.5, 2 mm ATP, 5 mm MgCl2, 75 mm NaCl, 2 mm TCEP) were measured using the EnzChek phosphate assay kit (Invitrogen) at 22 °C. The assay was performed in either the presence or the absence of RNA (poly(U) RNA or poly(I)-poly(C) (Sigma-Aldrich)) and in the presence and absence of 250 units of benzonase (Merck). All presented data represent means ± S.D. of five independent determinations.
RESULTS AND DISCUSSION
In an effort to determine the mechanism of DDX19 action and of its regulation, we determined the crystal structures of human DDX19 (residues Glu54–Glu475, encompassing the two conserved domains) as a complex with ADP and as a ternary complex with ADPNP and RNA (Fig. 1; see supplemental Table S1 for details of data collection and refinement statistics). Both structures revealed the canonical DEXD/H-box RNA helicase N-terminal ATPase and C-terminal helicase domains in the typical arrangement, with the two domains coming together to form the nucleotide binding cleft.
FIGURE 1.
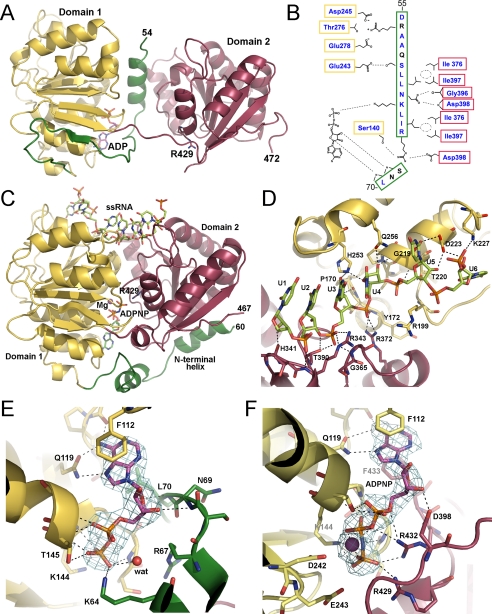
Structure of human DDX19. A, overview of DDX19 with ADP bound and the N-terminal flanking helix in the central cleft. The Arg429 side chain that acts as an arginine finger is presented as sticks. B, schematic representation of the cleft-inserted helix with the two conserved domains of the protein, shown in the same view as in panel A. Residues that are conserved in DDX25 are shown in blue. C, overview of the DDX19-RNA complex, with Mg-ADPNP bound in the central cleft. The Arg429 side chain is presented as sticks. D, detail of the RNA binding site of the DDX19-RNA complex. E, detail of the nucleotide binding site in the open conformation, with the electron density (2Fobs – Fcalc) for ADP rendered at 1.5 σ. F, detail of the nucleotide binding site in the RNA complex, with the electron density (2Fobs – Fcalc) for Mg-ADPNP rendered at 1.5 σ. In all panels, the conserved domain-1 (yellow), the conserved domain-2 (red), and the N-terminal flanking sequence (green) are indicated.
The structure of RNA-free DDX19 bound to ADP revealed a surprising feature (Fig. 1A). The section Asp55–Ser68 of the N-terminal extension, which was partly disordered in the RNA complex, folded into an α-helix that occupied the central cleft between the two lobes. The helix extended from the RNA binding site at its N terminus to the nucleotide binding site, where it contacted the phosphate groups of ADP (Fig. 1, B and E). The helix is predicted to disrupt the ATP binding site as it displaces the arginine finger (Arg429) that is essential for the ATPase reaction to occur (Fig. 1, A, C, E, and F) (6, 28). A similar placement of a secondary structural element has never been observed in a DEXD/H-box protein.
The Asp55–Ser68 α-helix makes a significant number of side chain interactions with both conserved domains (Fig. 1B). The loop leading out of the C terminus of the α-helix hydrogen bonds with two hydroxyls of the nucleotide ribose (the backbone amide of Asn69 with the 3′ hydroxyl and the backbone amide of Leu70 with the ring oxygen), suggesting selective binding to nucleotides at that site (Fig. 1, B and E). The significance of these interactions is supported by the conservation of the sequence of this N-terminal flanking segment, as well as the side chains it makes contacts with, in the DEXD/H-box RNA helicase, DDX25/GRTH (Fig. 1B and supplemental Fig. S1).
The structure of the ternary complex with a decauracil (U10) mRNA mimic and the non-hydrolyzable nucleotide, ADPNP, shows that both lobes contribute to nucleotide and RNA binding (Fig. 1C, D, and F and supplemental Fig. S2). Six bases of the RNA molecule are visible in the electron density, positioned across the top of the cleft; this mode of binding is consistent with the mechanism of RNA binding predicted for other DEXD/H-box RNA helicases, as determined by the structures of the complexes of RNA with DDX48/elF4AIII (12, 13) and Vasa (14). Also common to the mechanisms of the DEXD/H-box RNA helicase interactions with RNA is a kink that the proteins introduce in the RNA backbone between U4 and U5.
DDX19 differs from DDX48 and Vasa in the binding to RNA downstream of the kink. In the DDX48 and Vasa structures, U5 and U6 remain stacked with each other on the 3′ side of the kink, whereas in DDX19, the U6 base is rotated through ∼150 degrees around the sugar-phosphate backbone (Fig. 1D). This conformation is stabilized by hydrogen-bonding interactions between the 2′-ribose hydroxyl of U6 and the ε-amide of Lys227 and between the uracil base and the Lys202 and Asp223 side chains. Structure overlays suggest that a similar flipped conformation of U6 could not be accommodated by either DDX48 or Vasa.
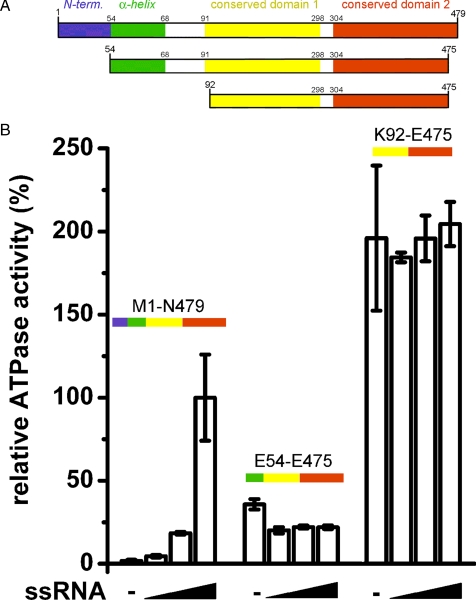
The interactions of the cleft-inserted helix with the conserved domains and with the nucleotide attest to the specificity and selectivity of the positioning of the N-terminal extension and point to a potential regulatory mechanism for the intrinsic ATPase activity of DDX19 (Fig. 1, B and E). To investigate this possibility, we measured the ATPase activities of three different DDX19 protein constructs (Fig. 2A). In the absence of ssRNA, full-length DDX19 (Met1–Asn479) did not hydrolyze appreciable amounts of ATP, but ATPase activity was stimulated to a rate of ∼0.77 ± 0.2 min–1 (S.D.) by 0.5 mg/ml ssRNA (Fig. 2B). The shorter protein construct Glu54–Glu475, lacking the native N terminus but containing the cleft insertion helix, hydrolyzed ATP at an intermediate rate of ∼25% of the ssRNA-stimulated full-length protein. This activity was independent of the presence of ssRNA. The construct Lys92–Glu475, which also lacked the cleft insertion α-helix, displayed the highest turnover of ATP (∼200% of the ssRNA-stimulated full-length protein), again independent of the presence of ssRNA. Further tests showed that the ATPase activities of the DDX19 proteins were not stimulated nor inhibited by the presence of double-stranded RNA (supplemental Fig. S3).
FIGURE 2.
Role of the N-terminal flanking sequence in the regulation of DDX19 ATPase activity. A, schematic diagram of the DDX19 protein constructs used in this study (not drawn to scale). N-term represents the N terminus. B, relative ATPase activities of DDX19 protein constructs in the presence of between 0 and 0.5 mg/ml ssRNA.
Taken together, these results show that the core DEAD and helicase domains (construct Lys92–Glu475) form an active, RNA-independent ATPase. The addition of the cleft-inserted helix (construct Glu54–Glu475) represses the ATPase activity of the core protein but does not confer RNA dependence to it. Further addition of the native N terminus (i.e. resulting in full-length DDX19) completely abolishes the ATPase activity of the protein in the absence of ssRNA. The presence of ssRNA overcomes the complete inhibition of ATPase activity in the full-length protein. The presence of the native N terminus is also required for ssRNA concentration-dependent stimulation of ATPase activity, indicating that ssRNA binding is sufficient to displace the N terminus, release the helix from the cleft, and allow formation of the catalytic triad in the ATP binding site.
RNA- and ATP-induced cleft closure has been shown to occur in solution (29), and the crystal structures of human DDX19 presented here demonstrate how cleft closure can be utilized in the regulation of ATPase activity. Our results suggest a model where an α-helical segment of the N-terminal flanking region is inserted between the core domains in the RNA-free protein. This helix prevents cleft closure and positions the N-terminal flanking sequence in the RNA binding groove or in its proximity. RNA binding then displaces the native N terminus, releasing the helix from the cleft to allow formation of the closed state and subsequent nucleotide hydrolysis. We predict that this mechanism of autoregulation also applies to DDX25, the other DEXD/H-helicase in which the sequence of the cleft insertion helix is highly conserved, and possibly to other family members.
Supplementary Material
Acknowledgments
We gratefully acknowledge the staffs of the BESSY (Berlin, Germany) and ESRF (Grenoble, France) synchrotron radiation facilities. We thank Lovisa Holmberg-Schiavone for initiating this project and Aled Edwards for critical reading of the manuscript.
Author's Choice—Final version full access.
This work was supported by grants from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, the Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 3EWS and 3G0H) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: RNP, ribonucleoprotein; ADPNP, adenosine 5′-(β, γ-imido)triphosphate; ssRNA, single-stranded RNA; PDB, Protein Data Bank; TCEP, tris(2-carboxyethyl)phosphine; Bis-Tris, 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
References
- 1.Lorsch, J. R., and Herschlag, D. (1998) Biochemistry 37 2194–2206 [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky, E., Gross, C. H., Shuman, S., and Pyle, A. M. (2001) Science 291 121–125 [DOI] [PubMed] [Google Scholar]
- 3.Tran, E. J., Zhou, Y., Corbett, A. H., and Wente, S. R. (2007) Mol. Cell 28 850–859 [DOI] [PubMed] [Google Scholar]
- 4.Cordin, O., Banroques, J., Tanner, N. K., and Linder, P. (2006) Gene (Amst.) 367 17–37 [DOI] [PubMed] [Google Scholar]
- 5.Jankowsky, E., and Fairman, M. E. (2007) Curr. Opin. Struct. Biol. 17 316–324 [DOI] [PubMed] [Google Scholar]
- 6.Schmitt, C., von Kobbe, C., Bachi, A., Pante, N., Rodrigues, J. P., Boscheron, C., Rigaut, G., Wilm, M., Seraphin, B., Carmo-Fonseca, M., and Izaurralde, E. (1999) EMBO J. 18 4332–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snay-Hodge, C. A., Colot, H. V., Goldstein, A. L., and Cole, C. N. (1998) EMBO J. 17 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tseng, S. S. L., Weaver, P. L., Liu, Y., Hitomi, M., Tartakoff, A. M., and Chang, T. H. (1998) EMBO J. 17 2651–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund, M. K., and Guthrie, C. (2005) Mol. Cell 20 645–651 [DOI] [PubMed] [Google Scholar]
- 10.Gross, T., Siepmann, A., Sturm, D., Windgassen, M., Scarcelli, J. J., Seedorf, M., Cole, C. N., and Krebber, H. (2007) Science 315 646–649 [DOI] [PubMed] [Google Scholar]
- 11.Murphy, R., and Wente, S. R. (1996) Nature 383 357–360 [DOI] [PubMed] [Google Scholar]
- 12.Weirich, C. S., Erzberger, J. P., Flick, J. S., Berger, J. M., Thorner, J., and Weis, K. (2006) Nature Cell Biol. 8 668–676 [DOI] [PubMed] [Google Scholar]
- 13.Sengoku, T., Nureki, O., Nakamura, A., Satoru, K. I., and Yokoyama, S. (2006) Cell 125 287–300 [DOI] [PubMed] [Google Scholar]
- 14.Bono, F., Ebert, J., Lorentzen, E., and Conti, E. (2006) Cell 126 713–725 [DOI] [PubMed] [Google Scholar]
- 15.Andersen, C. B. F., Ballut, L., Johansen, J. S., Chamieh, H., Nielsen, K. H., Oliveira, C. L. P., Pedersen, J. S., Seraphin, B., Le Hir, H., and Andersen, G. R. (2006) Science 313 1968–1972 [DOI] [PubMed] [Google Scholar]
- 16.Högbom, M., Collins, R., van den Berg, S., Jenvert, R. M., Karlberg, T., Kotenyova, T., Flores, A., Hedestam, G. B. K., and Holmberg Schiavone, L. (2007) J. Mol. Biol. 372 150–159 [DOI] [PubMed] [Google Scholar]
- 17.Schütz, P., Bumann, M., Oberholzer, A. E., Bieniossek, C., Trachsel, H., Altmann, M., and Baumann, U. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufau, M. L., and Tsai-Morris, C. H. (2007) Trends Endocrinol. Metab. 18 314–320 [DOI] [PubMed] [Google Scholar]
- 19.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795–800 [Google Scholar]
- 20.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein, N. (2008) J. Appl. Crystallogr. 40 641–643 [Google Scholar]
- 22.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1948–1954 [DOI] [PubMed] [Google Scholar]
- 23.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov, G. N., Vagin, A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 25.Lovell, S. C., Davis, I. W., Arendall, W. B., III, de Bakker, P. I., Word, J. M., Prisant, M. G., Richardson, J. S., and Richardson, D. C. (2003) Proteins 50 437–450 [DOI] [PubMed] [Google Scholar]
- 26.Gouet, P., Courcelle, E., Stuart, D. I., and Metoz, F. (1999) Bioinformatics (Oxf.) 15 305–308 [DOI] [PubMed] [Google Scholar]
- 27.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA
- 28.Le Hir, H., and Andersen, G. R. (2008) Curr. Opin. Struct. Biol. 18 112–119 [DOI] [PubMed] [Google Scholar]
- 29.Theissen, B., Karow, A. R., Kohler, J., Gubaev, A., and Klostermeier, D. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.