Abstract
Saccharomyces cerevisiae MPH1 was first identified as a gene encoding a 3′ to 5′ DNA helicase, which when deleted leads to a mutator phenotype. In this study, we isolated MPH1 as a multicopy suppressor of the dna2K1080E helicase-negative lethal mutant. Purified Mph1 stimulated the endonuclease activities of both Fen1 and Dna2, which act faithfully in the processing of Okazaki fragments. This stimulation required neither ATP hydrolysis nor the helicase activity of Mph1. Multicopy expression of MPH1 also suppressed the temperature-sensitive growth defects in cells expressing dna2Δ405N, which lacks the N-terminal 405 amino acids of Dna2. However, Mph1 did not stimulate the endonuclease activity of the Dna2Δ405N mutant protein. The stimulation of Fen1 by Mph1 was limited to flap-structured substrates; Mph1 hardly stimulated the 5′ to 3′ exonuclease activity of Fen1. Mph1 binds to flap-structured substrate more efficiently than to nicked duplex structures, suggesting that the stimulatory effect of Mph1 is exerted through its binding to DNA substrates. In addition, we found that Mph1 reversed the inhibitory effects of replication protein A on Fen1 activity. Our biochemical and genetic data indicate that the in vivo suppression of Dna2 defects observed with both dna2K1080E and dna2Δ405N mutants occur via stimulation of Fen1 activity. These findings suggest that Mph1 plays an important, although not essential, role in processing of Okazaki fragments by facilitating the formation of ligatable nicks.
Lagging strand DNA synthesis requires the orchestrated actions of many proteins and can be divided into several distinct enzymatic steps (1–4). First, the polymerase (pol)2 α-primase complex synthesizes RNA-DNA primers on the template DNA that are recognized by replication factor C. This complex loads proliferating cell nuclear antigen onto DNA, which acts as a processivity factor tethering pol δ to primer ends (5–7). This series of reactions leads to a polymerase switch in which the pol α-primase complex at primer ends is displaced and replaced by pol δ. Okazaki fragments are then elongated by pol δ until they encounter downstream Okazaki fragments (2, 8–10). Pol δ continues to synthesize DNA by displacing the 5′ termini of downstream Okazaki fragments, which generate 5′ RNA-DNA flap structures (11). These flaps are then cleaved by structure-specific nucleases that lead to the generation of ligatable nicks, which are sealed by DNA ligase converting the noncontiguous lagging strands to a contiguous DNA chain (12, 13).
A number of biochemical studies have shown that flap structures are removed by the concerted action of Dna2 endonuclease/helicase and Fen1 (flap endonuclease 1) (14–16). DNA2, first identified as a gene that complemented a yeast temperature-sensitive mutant defective in the elongation stage of DNA replication, was shown to encode a protein with structure-specific endonuclease and 5′ to 3′ helicase activities (17–21). RAD27, encoding yeast Fen1, is a gene showing strong mutator phenotype and genome instability when inactivated. Fen1 participates in a variety of DNA transactions, including Okazaki fragment processing, due to its endonuclease, gap endonuclease, and exonuclease activities (12, 22–28). Fen1 can cleave flap structures efficiently to generate ligatable nicks, especially on short 5′ flap structures (<20 nt). However, Fen1 is not effective in cleaving replication protein A (RPA) bound or secondary structured flaps in vitro (14, 29). These findings suggest that in vivo Fen1 acts on flap DNA by first loading onto the 5′-ends of flap structures and then migrating to the junction of single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA), a process referred to as a “tracking mechanism” (30). In contrast, Dna2 can remove secondary structured flaps through the combined action of its helicase/endonuclease activities; its helicase activity can unwind intramolecular base-paired hairpin-like flap structures, thus facilitating the cleavage of unwound ssDNA through its endonuclease activity. RPA can also assist in the removal of relatively long DNA flaps by Dna2 (14). RPA, which can bind to long flaps (>27 nt), inhibits the action of Fen1 in vitro while stimulating the Dna2-catalyzed cleavage of long flap by recruiting Dna2 (14). Thus, RPA is dynamically removed by Dna2 and then Fen1 cleaves the remaining flaps (15, 31). The Pif1 5′ to 3′ helicase activity is implicated in accelerating the growth of long flap structures by rapidly displacing downstream 5′ flap, thereby increasing the chances that RPA can bind before Fen1 acts (33). These considerations indicate that Dna2 and Fen1 remove flap structures by their sequential action, and their activities are influenced by other proteins like RPA and Pif1.
Saccharomyces cerevisiae MPH1 was first identified as a mutator phenotype 1 gene (34), and the mph1Δ null mutant displayed increased mutation rates and sensitivity to a variety of DNA-damaging agents (35). Based on genetic studies, it is thought that MPH1 functions in an error-free DNA damage bypass pathway that requires homologous recombination genes (36). It was also shown that Mph1 has DNA-dependent ATPase activity and translocates on ssDNA in the 3′ to 5′ direction (37). Recently, it was reported that overexpression of Mph1 increased gross chromosomal rearrangements by partially inhibiting homologous recombination through its interaction with RPA (38). These data suggest that Mph1 is important in maintaining the integrity of DNA.
In addition to the proteins described above, the processing of Okazaki fragment is affected by other proteins that stimulate the endonucleases that cleave flap structures. Mgs1, which has DNA-dependent ATPase and DNA-annealing activities, markedly stimulates the endonuclease activity of Fen1 on flap DNA substrates in an ATP-dependent manner (39). Bloom and Werner helicases interact directly with hFen1 and stimulate its endonuclease activity (40, 41).
The detection of auxiliary proteins that stimulate the activities of Dna2 or Fen1 is likely to reveal important redundant pathways involved in the processing of Okazaki fragments. Moreover, their discovery may reveal novel pathways used to maintain genome integrity, since DNA replication is closely linked to genome stability. In order to identify novel factors that influence Okazaki fragment processing, we have carried out multicopy suppressor screens with dna2 helicase-negative mutant strains. We introduced multicopy plasmids containing randomly inserted genomic DNA fragments into the dna2K1080E mutant yeast strain and analyzed the inserts that suppressed the lethal phenotype of this mutant. This screen resulted in the identification of MPH1 as a multicopy suppressor. To understand the biochemical mechanism of their suppression and its role in Okazaki fragment processing, we purified the Mph1 protein and investigated its interactions with Fen1 and Dna2 both in vivo and in vitro. We found that Mph1 enhanced the endonuclease activities of Fen1 and Dna2 in vitro, and this stimulation did not require ATPase/helicase activities of Mph1. Our results indicate that Mph1 participates in Okazaki fragment processing by increasing the rate of cleavage of flap substrate, thereby facilitating the production of ligatable nicks.
EXPERIMENTAL PROCEDURES
Enzymes and Nucleotides—The restriction endonucleases, polynucleotide kinase, and T4 DNA ligase were purchased from Enzynomics (Daejeon, Korea). The oligonucleotides used in this study were commercially synthesized from Genotech (Daejeon, Korea). ATP was obtained from Sigma. ATPγS was from Roche Applied Science. [γ-32P]ATP (3,000 Ci/mmol) was purchased from Izotop (Budapest, Hungary). Yeast Dna2 and Fen1 were purified as described (14, 42).
Preparation of Helicase and Nuclease Substrates—All substrates were prepared as described previously (14), and the sequences of oligonucleotides used are listed in Table 1. Briefly, one of the two oligonucleotides in a partial duplex DNA substrate was 5′-labeled with [γ-32P]ATP and polynucleotide kinase according to the manufacturer's protocol and then annealed to the other oligonucleotide. For a flap or a nicked duplex substrate, a downstream oligonucleotide was labeled as described above and then annealed with the template and upstream oligonucleotides at a molar ratio of 1:3:5. All annealed substrates were purified by polyacrylamide gel electrophoresis.
TABLE 1.
Oligonucleotide sequences used in this study
| Number | Sequence (5′-3′) |
|---|---|
| 1 | GGGCTCACGTGGTCGACGCTGGAGGTGATCACCAGATGATTGCTAGGCATGCTTTCCGCAAGAGAACGGGCGTCTGCGTACCCGTGCAG (89 nt)a |
| 2 | CAGCGTCGACCACGTGAGCCC (21 nt) |
| 3 | CTGCACGGGTACGCAGACGCC (21 nt) |
| 4 | CGAACAATTCAGCGGCTTTAACCGGACGCTCGACGCCATTAATAATGTTTTC (52 nt) |
| 5 | GAAAACATTATTAATGGCGTCGAGCTAGGCACAAGGCGAACTGCTAACGG (50 nt) |
| 6 | CCGTTAGCAGTTCGCCTTGTGCCTA (25 nt) |
| 7 | CCGTTAGCAGTTCGCCTTGTGCCTAG (26 nt) |
| 8b | GCGCATGTGCGTTCCATTTAGTTCAAGCCGCAGCGGCTTGAACCGGACGCTCGACGCCATTAATAATGTTTTC (73 nt) |
| 9 | GCTCGACGCCATTAATAATGTTTTC (25 nt) |
The numbers in parentheses indicate the length of each oligonucleotide.
Underlined sequences form hairpin structures.
Construction of Mph1 Expression Vectors—For overexpression of Mph1 in yeast, the ADH1 promoter was used. This was PCR-amplified from pRS323(ADH)-FLAG using two oligonucleotides (5′-TCC CCG CGG GAT ATC CTT TTG TTT CCG GG-3′ and 5′-GGC CGC GGC CGC GAG TTG ATT GTA TGC TTG GTA-3′), and the SacII-NotI fragment of the PCR product was cloned into pRS325 and pRS424 plasmids to prepare pRS325(ADH) and pRS424(ADH), respectively. The open reading frame of MPH1 was amplified from a plasmid containing MPH1 using two oligonucleotides (5′-GCG GCC GCA TGG CTA GTG CAG ATG ATT A-3′ and 5′-CTG CAG TCA AAA ATC AGA ATC TGA GC-3′). The NotI-PstI fragment of the PCR fragment was cloned into pRS325(ADH) to obtain pRS325(ADH)-MPH1. The NotI-PstI fragment from pRS325(ADH)-MPH1 was subcloned to pRS424(ADH) to make pRS424(ADH)-MPH1. To express hexahistidine and the FLAG-tagged Mph1, pRS424(ADH) was digested first with ClaI, and the resulting recessed ends were filled in with Klenow and then digested with KpnI. Similarly, pFastBac-Hta-FLAG-MPH1 (see below) was digested with RsrII, and the recessed end was filled in with Klenow, followed by digestion with KpnI. The fragment containing His6-FLAG-MPH1 was cloned into the digested pRS424(ADH)vector, resulting in pRS424(ADH)-HF-MPH1. For construction of a vector expressing the His6-FLAG-tagged protein in insect cells, two oligonucleotides containing the FLAG peptide sequence (5′-TCG ACT TGA CTA CAA GGA CGA TGA CGA TAA GAG C-3′ and 5′-GGC CGC TCT TAT CGT CAT CGT CCT TGT AGT CAA G-3′) were annealed, and the products were cloned into pFastBac-HTa vector, resulting in pFastBac-Hta-FLAG. pFastBac-Hta-FLAG-MPH1 was made by cloning the NotI-Kpn1 fragment of the PCR product amplified from pET28-MPH1 using 5′-GCG CGG CCG CAT GGC TAG TGC AGA TGA TTA C-3′ and 5′-GCG GTA CCT CAA AAA TCA GAA TCT GAG CC-3′. To make the mph1K113E mutant DNA, in vitro mutagenesis was carried out using the EZchange™ site-directed mutagenesis kit (Enzynomics) using the oligonucleotides (5′-GCC ATC CCA ACG GGT ATG GGT GAA ACG TTC ATT GCC AG-3′ and 5′-CTG GCA ATG AAC GTT TCA CCC ATA CCC GTT GGG ATG GC-3′), according to the manufacturer's protocol.
Screening Multicopy Suppressors of dna2K1080E Mutant—A yeast genomic DNA library inserted into a pYEp13 multicopy plasmid (ATCC27323) was transformed into YJA1B (MATα ade2-101 ura3-52 lys2-801trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 (pRS316-DNA2)) containing the pRS314-dna2K1080E plasmid. Transformants were grown in SD without histidine, leucine, and tryptophan for 24 h at 30 °C, followed by replica plating onto the same SD medium supplemented with 0.1% 5-fluoroorotic acid. The plates were incubated for an additional 3–4 days. The colonies grown were transferred to liquid medium, and total genomic DNA was prepared and used to transform Escherichia coli by electroporation, from which plasmids were isolated. To confirm multicopy suppression, recovered plasmids were retransformed into the YJA1B strain and examined for their ability to support growth of mutant cells. Double-checked plasmids were analyzed by sequencing to identify genomic DNA fragments inserted in the plasmid. One of the analyzed plasmids contained the MPH1 gene. To confirm that MPH1 is a multicopy suppressor, the open reading frame of MPH1 alone was cloned into a 2-μm origin-based pRS plasmid series and expressed under the ADH1 promoter in mutant cells.
Drop Dilution Assay—Transformants were inoculated in appropriate medium (2 ml) and grown for 16–36 h. Each saturated culture was diluted with distilled water to a density of 1 × 107 cells/ml, and serially diluted samples (5–10 μl) were then spotted on appropriate medium and incubated for 3–5 days at the indicated temperatures. Where 5-fluoroorotic acid and control plate were used, inoculation was performed in the medium containing uracil to allow cells to spontaneously lose pRS316-Dna2 (containing the URA3 gene as a marker) during growth before spotting (35, 39).
Purification of Recombinant Mph1 Proteins—Baculoviruses expressing the N-terminally histidine-FLAG-tagged Mph1 protein (wild type or K113E mutant) were used to infect Sf9 insect cells (2 liters; 1 × 106 cells/ml) for 60 h at 27 °C in Sf-900 II serum-free medium (Invitrogen); cells were harvested by centrifugation, resuspended in 50 ml of lysis buffer (25 mm Hepes-NaOH, pH 7.6, 500 mm NaCl, 1 mm DTT, 1 mm EDTA, 10% glycerol, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 0.1 mm benzamidine, 2 μg/ml leupeptin, 0.1 μg/ml anti-pain, and 1 μg/ml pepstatin A), and sonicated. Crude extracts were cleared by centrifugation at 18,000 rpm for 1 h using the Hanil A50S-8 rotor, and the supernatant was mixed with 0.5 ml of anti-FLAG M2-agarose beads (Sigma) pre-equilibrated with the lysis buffer for 2 h. The beads were collected and washed three times with 40 ml of buffer A (25 mm Hepes-NaOH, pH 7.6, 1 mm DTT, 1 mm EDTA, 10% glycerol, 0.01% Nonidet P-40) containing 800 mm NaCl and once with buffer A containing 300 mm NaCl. Bound proteins were eluted three times with 0.5 ml of buffer A containing 300 mm NaCl and 0.1 mg/ml 1× FLAG peptide (Sigma) with rocking at 4 °C for 30 min. The initial two eluates were combined and loaded onto a Heparin-Sepharose Fast Flow column (3 ml; GE Healthcare) by gravity, followed by elution with a 6-ml linear gradient of NaCl from 0.1 to 1 m in buffer A (flow rate, 0.1 ml/min). Mph1 was eluted in 550–750 mm NaCl. BSA was added to the active fractions (final concentration, 1 mg/ml) to stabilize the enzyme, which was then frozen at –80 °C. The protein concentrations of Mph1 fractions were quantified by SDS-PAGE analysis, followed by Coomassie Brilliant Blue staining, and the protein band intensity was determined using BSA as the standard (Bio-Rad).
Nuclease Assay and Helicase Assay—Standard nuclease assays of Fen1 and Dna2 were performed in reaction mixtures (20 μl) containing 25 mm Tris-HCl, pH 7.8, 5 mm MgCl2, 2 mm DTT, 0.25 mg/ml BSA, and 15 fmol of DNA substrate. Unless stated otherwise, 50 mm NaCl was used for each reaction. Proteins were diluted in buffer (25 mm Hepes-NaOH, pH 7.6, 500 mm NaCl, 1 mm DTT, 0.25 mg/ml BSA, 0.01% Nonidet P-40, 10% glycerol) before their addition to reaction mixtures. The reactions were incubated at 30 °C for 15 min and halted with 6× stop solution (60 mm EDTA, pH 8.0, 40% (w/v) sucrose, 0.6% SDS, 0.25% xylene cyanol, 0.25% bromphenol blue). Reaction products were subjected to electrophoresis for 40 min at 150 V through 10% polyacrylamide gel in 1× TBE (89 mm Tris-base, 89 mm boric acid, and 2 mm EDTA); gels were dried on DEAE-cellulose paper and autoradiographed. The resolved DNA products were quantified using a PhosphorImager (Amersham Biosciences). Helicase assays were performed using the same conditions as described in the above nuclease assay but in the presence of 5 mm ATP.
Gel Mobility Shift Assay—Reaction mixtures (20 μl) containing 25 mm Tris-HCl, pH 7.8, 50 mm NaCl, 2 mm DTT, 0.25 mg/ml BSA, 15 fmol of DNA substrate with or without 5 mm MgCl2 and with or without 5 mm ATP (or ATPγS) were incubated at 30 °C for 15 min, followed by the addition of glycerol and bromphenol blue to 10% (v/v) and 0.05% (w/v), respectively. Reaction products were subjected to electrophoresis through prerun 6% polyacrylamide gels for 1.5 h in 0.5× TBE at 4 °C (39). Gels were dried on DEAE-cellulose paper and then autoradiographed. Levels of nucleoprotein complexes formed were quantified using a PhosphorImager (Amersham Biosciences).
RESULTS
Overexpression of MPH1 Suppresses the Lethal Phenotype of dna2K1080E—To identify novel factors involved in Okazaki fragment processing, extensive multicopy suppressor screens were carried out with several dna2 mutant strains. The mutant strains used were defective in the processing of DNA flap structures generated from 5′-end regions of Okazaki fragments in the pol δ-catalyzed displacement reaction. The rationale for this screen was that it might reveal additional proteins that participate in Okazaki fragment maturation by (i) replacing one of the defective activities of Dna2, (ii) stimulating the flap-cleaving enzymes, including Dna2 and Fen1, or (iii) repairing DNA structures resulting from defects in flap processing. Multicopy suppressor screens were performed with the helicase-negative dna2K1080E mutant in the hope of isolating a helicase gene that could substitute for the helicase function of Dna2. This effort resulted in the isolation of one clone that harbored a library plasmid containing the full-length open reading frame of the MPH1 gene.
To confirm that MPH1 was responsible for suppression, the open reading frame of MPH1 was amplified from the library plasmid and positioned under the constitutive promoter of the gene encoding alcohol dehydrogenase 1 (ADH1) in the pRS325 multicopy vector (pRS325pADH-MPH1) and expressed in the dna2K1080E mutant strain. Overexpression of MPH1 driven by either the ADH1 promoter or its natural promoter suppressed the lethal phenotype of dna2K1080E (Fig. 1A). Since the Mph1 protein has intrinsic ATPase and 3′ to 5′ DNA helicase activities, we examined whether they were required for the observed suppression (37). For this purpose, we mutated lysine 113 to glutamic acid in the Walker A motif of MPH1 that is responsible for binding to the terminal phosphate of ATP (35). Interestingly, overexpression of the mutated version of MPH1 also suppressed the lethality of dna2K1080E, as observed with the wild type MPH1. This result suggests that the helicase activity of Mph1 is dispensable for suppression of the dna2K1080E mutation.
FIGURE 1.
Overexpression of MPH1 suppresses the lethal phenotype of dna2K1080E. A, drop dilution assay. The YJA1B (dna2::HIS3, pRS316-DNA2; described under “Experimental Procedures”) strain containing pRS314-dna2K1080E was transformed with plasmids, as indicated. Wild type MPH1 or helicase-negative mutant mph1K113E was expressed in the multicopy plasmid (pRS325) under the ADH1 promoter (pADH-MPH1 and pADH-mph1K113E, respectively). The library clone identified to contain MPH1 (library MPH1) and pRS325-DNA2 (DNA2) is a positive control, and empty pRS325 vector (none) was also transformed as the negative control. Transformants were inoculated and grown in liquid SD lacking histidine, leucine, and tryptophan (SD(–HLW)) until saturation and spotted in duplicate onto SD(–HLW) in the absence or presence of (+FOA)5′-fluoroorotic acid with a 5-fold serial dilution, and cells were then incubated for 4 days at 30 °C. B, Mph1 protein was purified (described under “Experimental Procedures”) from Sf9 insect cells. The peak fraction (700 fmol) was subjected to 8% SDS-PAGE and stained with Coomassie Brilliant Blue R. Mw, molecular weight marker. C, helicase assays were performed with two different substrates, partial double-stranded DNA with 3′ single-stranded or 5′ single-stranded overhang as illustrated at the top. The circled numbers denote the oligonucleotide described in Table 1. The asterisk in the substrate indicates the 32P-labeled end. Reaction conditions used are as follows: 30 mm Hepes-NaOH, pH 7.6, 25 mm NaCl, 2 mm ATP, 2.5 mm MgCl2, 1 mm DTT, 0.25 mg/ml BSA, and 15 fmol of DNA substrate. Reaction mixtures were incubated at 30 °C. The amount of unwound DNA is indicated at the bottom of the gel. B, boiled substrate.
To gain more information about the mechanism by which MPH1 suppressed DNA2 mutated strain, we isolated the recombinant Mph1 protein. Its expression in E. coli resulted in mostly insoluble and severely degraded protein preparations. For this reason, recombinant baculoviruses were prepared that expressed Mph1 with N-terminally fused His6 and FLAG tags and were used to infect Sf9 cells. Extracts prepared from infected insect cells yielded soluble recombinant HF-Mph1 protein after two column purification steps (Fig. 1B), as described under “Experimental Procedures.” The recombinant Mph1 protein possessed helicase activity on partial double strand substrates with a 3′ single-stranded DNA but not with a 5′ single-stranded DNA (Fig. 1C), consistent with the previous report that Mph1 purified from yeast translocates in the 3′ to 5′ direction (37).
Mph1 Stimulates the Endonuclease Activity of Fen1—RPA and Mgs1, both identified as multicopy suppressors of dna2 mutants, markedly stimulated the endonuclease activity of Dna2 and Fen1, respectively (14, 39). To investigate whether Mph1 utilized a similar mechanism, we examined the structure-specific nuclease activity of Fen1 in the absence (Fig. 2A, lanes 3–7) or presence (Fig. 2A, lanes 8–12) of Mph1 in a time course experiment (Fig. 2, A and B). Mph1 markedly enhanced the activity of Fen1 when a 27-nt 5′ flap substrate was used (Fig. 2A). The purified Mph1 protein alone was devoid of any nuclease activity (Fig. 2A, lane 2). Maximal stimulation (15.3-fold) was observed at the earliest time point examined (Fig. 2B).
FIGURE 2.
Mph1 stimulates the endonuclease activity of Fen1 and Dna2. A, the Fen1 endonuclease activity (described under “Experimental Procedures”) was determined after incubation for various periods in the presence or absence of Mph1. The schematic structure of the double-flap substrate is shown at the top. B, the amount of product formed in reactions described in A was plotted against the time of incubation. C, the YJA1B strain carrying the pRS315-dna2K1080E plasmid was transformed with the indicated plasmids. Wild type MPH1 or helicase-negative mutant mph1K113E was expressed in the multicopy plasmid (pRS424) under the ADH1 promoter (pADH-HFMPH1 and pADH-HFmph1K113E, respectively). HF, the tandem array of His6 and FLAG tags. FEN1 was also expressed in the pRS424 plasmid under its natural promoter. The empty pRS424 vector containing only the ADH1 promoter (pADH) was transformed as a negative control. Transformants were inoculated and grown in liquid SD(–HLW) medium until saturation and spotted in duplicate onto SD(–HLW) plates in the absence or presence (+FOA) of 5′-fluoroorotic acid in 5-fold serial dilutions. Cells were then incubated for 4 days at 30 °C. HF, tandem array of His6 and FLAG tags on the N terminus of MPH1. D, Dna2 endonuclease activity (described under “Experimental Procedures”) was measured in the presence of increasing (0, 10, 25, and 50 fmol) amounts of Mph1. The schematic structure of the single 5′ flap substrate used is shown at the top.
If Mph1 suppressed the lethality of dna2K1080E solely by stimulating the action of Fen1, we would expect that overexpression of FEN1 should suppress the dna2K1080E mutant. As predicted, multicopy expression of FEN1 suppressed the dna2K1080E mutant (Fig. 2C, second row). Since it is possible that the presence of affinity tags on proteins can abolish their enzymatic activities or other associated functions, we examined whether the N-terminally tagged Mph1 maintained its in vivo ability to suppress the dna2K1080E mutation. As shown in Fig. 2C (third and fourth rows), expression of both wild type and K113E mutant-tagged proteins still suppressed the growth defect of the dna2K1080E mutant. Since overexpression of FEN1 also rescued the temperature-sensitive growth defect of dna2Δ405N and dna2-1 in addition to dna2K1080E, the stimulation of Fen1 activity appears to be a general mechanism for the suppression of any defective dna2 (39, 43). These findings are consistent with the notion that the most critical function of Dna2 is dependent on its endonuclease activity.
Mph1 Stimulates the Endonuclease Activity of Dna2—Since overexpression of the dna2K1080E mutant allele itself alone resulted in the growth of this mutant,3 we examined the possibility that the stimulation of the endonuclease activity of Dna2K1080E protein (the only enzymatic activity of this Dna2 mutant protein) was responsible for the observed suppression (Fig. 2D). Nuclease assays were performed with wild type Dna2 in the presence of increasing levels of Mph1 but in the absence of ATP. As shown in Fig. 2D, Mph1 markedly stimulated the endonuclease activity of Dna2 (∼17-fold). The stimulation increased in proportion to the level of Mph1 added up to 25 fmol (12.5-fold molar excess over Dna2 used). The increase in nuclease activity was not due to the Mph1 preparation alone (Fig. 2D, lane 6).
Stimulation of Fen1 and Dna2 by Mph1 Is Not ATP-dependent—Overexpression of both wild type and the ATPase-negative mph1 mutant suppressed the growth defect of the dna2K1080E mutant to a similar extent as shown in Fig. 1A. If suppression occurred through the stimulation of Fen1 or Dna2 endonuclease activity, Mph1 should stimulate those proteins regardless of the presence/absence of ATP. In order to examine this, nuclease assays were performed in the presence or absence of ATP. As shown in Fig. 3, the Mph1 stimulation of Fen1 action was the same in the presence and absence of ATP. These results demonstrate that ATP binding, ATP hydrolysis, and the DNA helicase activity of Mph1 play no role in the stimulating the flap endonuclease activity. An additional band noted between the substrate and the cleavage product was formed only in the presence of ATP (Fig. 3A, lanes 2 and 6–8) and not in its absence (Fig. 3A, lanes 10 and 14–16) or in reactions with ATPγS (data not shown). These results suggest that formation of this band required ATP hydrolysis by Mph1. This product migrated exactly to the position detected with the boiled substrate (Fig. 3A, lane 17), suggesting that it originated from the displacement of the downstream oligonucleotide by the helicase activity of Mph1. Mph1 is likely to translocate through the duplex DNA, displacing any 5′ flap DNA encountered, although it translocates on a 3′ to 5′ helicase in assays with the 3′ single-stranded DNA tail, as shown in Fig. 1C.
FIGURE 3.
Stimulation of the endonuclease activity of Fen1 and Dna2 by Mph1 do not require ATP. A, endonuclease assays were carried out for various time periods in the absence or presence of 5 mm ATP, Fen1, and Mph1, where indicated. B, boiled substrate. B, the amount of cleaved product formed in reactions described in A was plotted against the incubation period. C, the rate of Dna2 endonucleolytic activity was measured in the absence or presence of Mph1 and ATP. The amounts of ATP, Dna2, and Mph1 added were as indicated. B, boiled substrate. D, the amount of product formed in reactions described in C was plotted against the incubation period.
Like Fen1, ATP did not significantly alter the stimulation of Dna2 activity by Mph1 (Fig. 3, C and D), although its presence slightly decreased the cleavage efficiency, and longer products were formed. Their cleavage is probably due to the fact that (i) ATP (5 mm) inhibits the endonuclease activity of Dna2 and (ii) Dna2 itself translocated along the flap in the presence of ATP and yielded longer cleavage products. As described above with Fen1 (Fig. 3A), the Mph1-catalyzed displacement of the labeled oligonucleotide was detected (Fig. 3C, lanes 8 and 14–16). Thus, in vitro and in vivo results indicate that ATP plays no role in the Mph1 stimulation of the activities of Fen1 and Dna2.
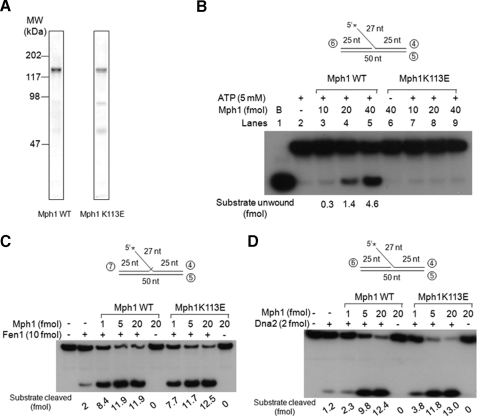
Walker A Motif-mutated Mph1 Stimulates Endonuclease Activity of Fen1 and Dna2—To further verify that the stimulation of Fen1 and Dna2 by Mph1 does not require ATP, we purified mutant Mph1 (Mph1K113E) devoid of helicase/ATPase activities by changing the lysine 113 in the Walker A motif to glutamic acid (35). This mutation appears to increase the susceptibility of the enzyme to proteolytic degradation, since preparations of Mph1K113E always contained more degradation products than the wild type protein (Fig. 4A).
FIGURE 4.
Walker A motif-mutated Mph1 stimulates endonuclease activity of Fen1 and Dna2. A, the wild type Mph1 and Mph1K113E mutant protein were purified at the same time. Baculovirus expressing each type of protein was used to infect Sf9 insect cells (1L; 1 × 106 cell/ml) (see “Experimental Procedures”), and proteins were purified by a FLAG M2-agarose affinity column followed by heparin-Sepharose Fast Flow. Heparin-Sepharose beads were collected and washed with heparin buffer containing 0.5 m NaCl, and proteins were eluted with the same buffer containing 0.75 m NaCl in place of a linear gradient. The purified wild type Mph1 and Mph1K113E proteins were subjected to 8% SDS-PAGE and stained with Coomassie Brilliant Blue. B, helicase activity was measured with wild type and mutant Mph1 proteins using the standard endonuclease assay condition described under “Experimental Procedures” in the presence of 15 fmol of the flap-structured DNA substrate. B, boiled substrate. C, endonuclease assays were performed with 10 fmol of Fen1 and the indicated amounts of wild type or mutant Mph1 proteins D, endonuclease assays were performed with 2 fmol of Dna2 and the indicated amounts of wild type or mutant Mph1 proteins.
As expected, the mutated protein was devoid of both helicase activity with flap-structured substrate (Fig. 4B, lanes 6–9) and DNA-dependent ATPase activity (data not shown). We examined whether Mph1K113E stimulated the endonuclease activities of Fen1 and Dna2 (Fig. 4, C and D), and no significant differences were observed between wild type and mutant Mph1. These results again demonstrate that the ATPase motif is not required for the stimulation of the two flap-processing endonucleases. Moreover, they support the notion that the in vivo suppression of dna2K1080E by overexpression of MPH1 was due to the stimulation of the nuclease activities of either Fen1 and/or Dna2 and independent of the catalytic activities of Mph1.
Mph1 Reverses the Inhibitory Effects of RPA on the Endonuclease Activity of Fen1—RPA is a relatively abundant nuclear protein that binds to ssDNA intermediates arising during DNA metabolism, such as replication and recombination (44). RPA inhibits the endonuclease activity of Fen1 by binding to a flap DNA and preventing its translocation. We investigated whether Mph1 reversed the inhibition. We first determined the level of RPA required to inhibit the endonuclease activity. Substantial inhibition (>90%) of Fen1 was observed with 6.4 fmol of RPA when we used 10 fmol of Fen1 and 15 fmol of DNA substrate (data not shown).
Fen1 nuclease assays were carried out in the presence of a fixed amount (20 fmol) of RPA and increasing levels of Mph1 (Fig. 5). Under these conditions, 10 fmol of Fen1 alone cleaved 1.1 fmol (Fig. 5, lane 2) of the substrate, and the addition of RPA (20 fmol) inhibited this activity 90% (Fig. 5, compare lanes 2 and 3). However, the inhibition was prevented by the addition of Mph1 (Fig. 5, lanes 4–6). Although the addition of ATP did not alter the amount of cleavage products formed (Fig. 5, lanes 10–15), it activated the helicase activity of Mph1, resulting in the formation of additional bands as observed in Fig. 3. It should be noted that although the Mph1-catalyzed unwinding reaction markedly reduced the level of substrate available for Fen1 action, it did not affect the amount of cleavage products formed (Fig. 5, compare lanes 7 and 16). One possible explanation for this is that the helicase activity of Mph1 displaced the RPA bound to the flap, thereby facilitating its cleavage by Fen1.
FIGURE 5.
Mph1 reverses the inhibitory effects of RPA on the endonuclease activity of Fen1. Endonuclease assays were carried out at various times in the absence or presence of RPA, Fen1, and Mph1, as indicated. The salt (NaCl) concentration used in this assay was 75 mm. Reactions were stopped with 4 μl of 6× stop solution (described under “Experimental Procedures”). Then 1 μl of Proteinase K (1 mg/ml) was added to each reaction, which was further incubated for 15 min at 37 °C. B, boiled substrate.
Mph1 Specifically Stimulates the Endonuclease Activity and Not the Exonuclease Activity of Fen1—As shown in Fig. 2, the activity of Fen1 on physiologically preferred substrates (flap-structured DNA with 5′ flap and 1-nt 3′ flap) was markedly stimulated by Mph1. We also examined their effect using two additional flap substrates: one with no 1-nt 3′ flap and the other containing a secondary structured 5′ flap formed by intra-base pairing positioned in the middle of the flap (Fig. 6). Dna2 cleaved these two flap substrates with equal efficiency (data not shown), whereas Fen1 preferentially cleaved the substrate with a 1-nt 3′ flap (Fig. 6A, compare lanes 3–6 and 14–16). It should be noted that elevated levels (20 fmol) of Fen1 were used with the single 5′ flap and the secondary structured flap substrates. In a time course experiment with these substrates, Mph1 stimulated Fen1 most efficiently with the 1-nt 3′ flap substrate (Fig. 6A, compare lanes 4–6 with lanes 8–10, lanes 14–16 with lanes 18–20, and lanes 24–26 with lanes 28–30). Under our assay conditions, the rate of cleavage of the unstructured single 5′ flap (Fig. 6A, middle) and the secondary structured 5′ flap (right) substrate was similar. Note that two different products were formed with the secondary structured substrate. The upper band (*, 48 nt) was formed by cleavage at the base of the flap, whereas the lower one (**, 20 nt) was derived from a cleavage event in the 5′ ssDNA tail just before the hairpin structure. Although Fen1 cleaved the latter two substrates inefficiently, its activity was enhanced by Mph1 addition (Fig. 6A). Although it is not clear whether this stimulation is physiologically significant, these findings suggest that Mph1 may enhance the ability of Fen1 to process secondary structured flaps by multiple means.
FIGURE 6.
Mph1 specifically stimulates the endonuclease activity but not the exonuclease activity of Fen1. A, the rate of endonucleolytic cleavage measured with three different substrates. The amount of Mph1 used is 32 fmol, and additions (+) or omissions (–) are as indicated in the figure. The single and double asterisks indicated on the right side denote the migration of distinct cleaved products; the single asterisk indicates the product arising after cleavage at the base of the 5′ flap, and the double asterisk indicates the product formed by cleavage at the right before the hairpin. The amount of cleaved products formed, indicated below the panel, is a sum of these two products. B, exonuclease assays were performed with the nicked duplex substrate.
We also tested whether Mph1 stimulated the exonuclease activity of Fen1 using a nicked substrate without flap (Fig. 6B). Unlike its endonuclease activity, the exonuclease activity of Fen1 was not stimulated significantly (maximally 2-fold) by Mph1. Thus, the stimulation of Fen1 by Mph1 is most likely limited to its endonuclease activity.
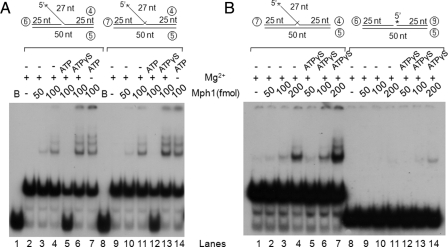
Mph1 Binds to Flap-structured DNAs but Not Nicked Duplex DNAs—We examined whether the stimulation of Fen1 and Dna2 by Mph1 required direct protein-protein interactions. Physical interactions between Mph1 and Fen1 or Dna2 were screened in vivo using the yeast two-hybrid assay, but no interaction was detected (data not shown), suggesting that the interactions may not be stable or may occur indirectly through binding to DNA. To test for the latter possibility, gel mobility shift assays were carried out with several DNA substrates, including those containing a single flap, a double flap, and a nicked duplex. Since Mph1 contains DNA-dependent ATPase and DNA helicase activities, it is likely to bind to these DNA substrates, which could be important for its stimulatory effects. We postulated that differences in its binding affinity to various substrates could contribute to its substrate-specific stimulation. For these reasons, we examined the concentration-dependent binding of Mph1 to various DNA substrates in the absence and presence of ATP. As shown in Fig. 7A, incubation of the single flap substrate with high levels (100 fmol) of Mph1 (Fig. 7A, lane 4) yielded multiple protein-DNA complexes, suggesting that multiple molecules of Mph1 can bind to a single DNA molecule. The presence of the additional 1-nt 3′ flap in the substrate hardly affected the binding efficiency compared with the 5′ single flap substrate (Fig. 7A, compare lanes 2–7 and 9–13). Mph1 bound these DNA substrates in the absence of ATP (lanes 3, 4, 10, and 11) or Mg2+ (lanes 7 and 14). The addition of both ATP and Mg2+ resulted in the disappearance of the Mph1-DNA complex, due to unwinding of the labeled strand (lanes 5 and 12). When ATP was replaced with the nonhydrolyzable ATPγS in the presence of Mg2+, the binding efficiency increased ∼2-fold (Fig. 7A, compare lanes 4 and 11 with lanes 6 and 13, respectively). In contrast to the Mph1-DNA complexes formed with the above flap substrates, the nicked duplex substrate hardly supported the Mph1 binding (Fig. 7B, compare lanes 6 and 7 with lanes 13 and 14). These results suggest that binding of Mph1 to DNA substrates could be critical for its ability to stimulate the nuclease activities of Fen1 and Dna2.
FIGURE 7.
Mph1 binds to flap-structured DNA and not to nonflap nicked DNA. A, gel mobility shift assays (described under “Experimental Procedures”) were performed with the flap substrate used for endonuclease assays in the presence or absence of 5 mm ATP, ATPγS, and 5 mm MgCl2, as indicated. B, gel mobility shift assays were carried out after incubation of Mph1 with flap and nicked duplex substrates; 5 mm MgCl2 was added in all reactions.
Suppression of dna2 Mutations by MPH1 Occurs via Stimulation of Fen1—In parallel with screening for a multicopy suppressor of dna2K1080E mutant, we also screened for a multicopy suppressor of the temperature-sensitive dna2Δ405N mutant. MPH1 was isolated in three independent screens using this mutant strain (data not shown).
To evaluate whether suppression occurred through the stimulation of Dna2Δ405N or Fen1, nuclease assays were performed with the Dna2Δ405N protein. As shown in Fig. 8A, Mph1 barely stimulated the endonuclease activity of Dna2Δ405N protein. Therefore, the stimulation of Dna2 by Mph1 requires the presence of the N-terminal domain of Dna2. This N-terminal region may be required for the DNA-mediated interaction between Dna2 and Mph1. These results suggest that suppression of the temperature sensitivity of dna2Δ405N by MPH1 probably occurs through the stimulation of Fen1 and not Dna2. To further confirm this, drop dilution assays were carried out with dna2Δ405N cells transformed with plasmids overexpressing either the wild type or mutant mph1K113E. As controls, pRS314-DNA2 (DNA2 cloned in a centromeric plasmid with its native promoter) and pRS424-FEN1 (FEN1 cloned in multicopy plasmid with its native promoter) were used. We found that temperature sensitivity of dna2Δ405N was suppressed by overexpression of both wild type and its Walker A motif-mutated mph1K113E (Fig. 8B). Overexpression of FEN1 also restored the temperature-sensitive growth defect of the dna2Δ405N mutant cells. These genetic results, coupled with the biochemical finding described above, indicate that suppression of dna2Δ405N occurs solely by elevated levels of Fen1 activity. In conclusion, our in vivo and in vitro data suggest that Mph1 participates in Okazaki fragment processing by stimulating the activities of Fen1 and Dna2, which facilitate a more efficient formation of ligatable nicks during lagging strand synthesis.
FIGURE 8.
Overexpression of MPH1 suppresses the temperature sensitivity of dna2Δ405N mutant, but Mph1 protein does not stimulate the endonuclease activity of Dna2Δ405N protein. A, endonuclease assays were carried out with two different levels of Dna2Δ405N and the indicated amounts of Mph1. B, YPH499 (MATa, ade2-101, ura3-52, lys2-801, trp-Δ63, his3-Δ200, leu2-Δ1, and GAL+) and YJA2 (dna2Δ405N in YPH499 genetic background) strains were transformed with plasmids, as indicated. The wild type (WT) and dna2Δ405N strains were transformed with empty pRS424 vector containing only the ADH1 promoter as positive and negative control (pADH in WT and pADH dna2Δ405N, respectively) in the first rows. A tagged version of wild type MPH1 and helicase-negative mph1K113E were expressed in pRS424 plasmid under the ADH1 promoter in the YJA2 strain (pADH-HFMPH1 and pADH-HFmph1K113E), and nontagged MPH1 was also expressed (pADH-MPH1) in the same way. As other positive controls, DNA2 and FEN1 were expressed in pRS314 (centromeric plasmid) and pRS424, respectively, under their natural promoters. Transformants were inoculated and grown in liquid SD(–W) medium until saturation and spotted in duplicates onto SD(–W) plates using 10-fold serial dilutions. Cells were then incubated for 3 days at 25 and 37 °C.
DISCUSSION
In this study, MPH1 was identified as a multicopy suppressor of the dna2K1080E mutant. To understand the biochemical mechanism contributing to this effect, we isolated highly purified Mph1, which contains 3′ to 5′ helicase activity (37). Purified Mph1 markedly stimulated the endonuclease activities of both Fen1 and Dna2 in vitro (19- and 20-fold, respectively). This stimulation, however, was not dependent on the helicase/ATPase activities of Mph1, since the Mph1K113E mutant protein also was as effective as wild-type Mph1. These effects on the two critical Okazaki fragment-processing enzymes are consistent with our in vivo findings that overexpression of MPH1 can suppress growth defects of both dna2K1080E and dna2Δ405N mutants. We noted that the stimulation of Dna2 by Mph1 depends on the presence of the intact N-terminal domain of Dna2 (see below for further discussion). The helicase-negative Dna2K1080E protein does not resolve secondary structure flap substrates efficiently, because this process requires the coordinated action of both helicase and endonuclease activities of Dna2 (21). We found that the lethal phenotype of the dna2K1080E mutant was suppressed by overexpression of dna2K1080E,4 indicating that high levels of Dna2 endonuclease activity alone can resolve secondary structured flaps. We speculate that in the presence of excess levels of its endonuclease activity, Dna2 can rapidly capture and degrade the single-stranded DNA that is transiently formed by melting of the secondary structure (a process also referred to as “breathing”). Overexpression of Fen1 also suppressed the dna2K1080E mutant (Fig. 2C). A plausible explanation for these findings is that any potential secondary structures formed in the long flap (produced by displacement DNA synthesis catalyzed by pol δ) may not be maintained stably in equilibrating flaps in which the 5′ and 3′ flap regions compete dynamically for base pairing with template DNA (30). If this were the case, most flaps, including those with the potential to form secondary structures, would be cleaved by the presence of excess Fen1. In support of this notion, hairpin structures formed in nonequilibrating flap are slowly cleaved by Fen1 in vitro, and importantly, this cleavage rate can be increased by the addition of Mph1. Therefore, secondary structured flaps, unless highly stable, can be processed by the presence of high levels of Fen1 or by lower levels of Fen1 in the presence of Mph1, which stimulates its activity. The secondary structured flap, present in equilibrating flaps, can be removed more rapidly by Fen1 if the endonuclease activity of Dna2 is available, as is the case with the mutant dna2K1080E and dna2Δ405N strains. The double flaps mutually stimulate Dna2-catalyzed cleavage of each strand of flap, which results in rapid shortening of both flaps, which in turn facilitates formation of double flap structure with a 1-nt 3′ flap, a preferred substrate for Fen1 (45). We believe that the cellular levels of Fen1 and Dna2 are not adequate to process all secondary structured flaps generated in vivo in cells devoid of the Dna2 helicase activity. Under such conditions, we suggest that the helicase activity of Dna2 becomes essential. Based on this hypothesis, we suggest that the stimulation of the two nucleases by overexpression of Mph1 would have the same consequences in vivo (i.e. they render the helicase activity of Dna2 dispensable). This hypothesis may explain why the Dna2 homologs from fission yeast5 or humans (45) lack helicase activity. Our biochemical and genetic data, however, indicate that the suppression of the Dna2 defect by Mph1 is due to the stimulation of Fen1 activity and not of Dna2, since overexpression of Mph1 also suppressed the dna2Δ405N mutant. In vitro, the endonuclease activity of this mutated Dna2 was not stimulated by Mph1, indicating that stimulation occurs by a specific protein-protein interaction between the N-terminal domain of Dna2 and Mph1.
Although it is not clear how Mph1 stimulates Fen1 or Dna2 at present, one possible mechanism by which Mph1 stimulates Dna2 and Fen1 is through DNA binding activity of Mph1, which dissociates Dna2 or Fen1 from the cleavage products and thereby helps the nucleases recycle rapidly. This possibility raises a specificity problem. We believe that this is not the case for several reasons. First, the amount of Mph1 should be in excess over substrate DNA in order to stimulate a nuclease. Although most assays in this study were carried out with excess amounts of Mph1, substoichiometric amounts (1 or 5 fmol) of Mph1 and Mph1K113E were sufficient to stimulate activities of Fen1 and Dna2 (see Fig. 4, C and D). Second, we tested unrelated DNA helicases, such as SV40 T antigen and Schizosaccharomyces pombe Pfh1, that also possess DNA binding activity. The two helicases failed to stimulate activity of Fen1 or Dna2 under the same condition (Fig. S1). Third, we performed an experiment in a condition under which recycling was minimized by using the two nucleases in a stoichiometric amount with or in excess of substrate DNA. The result was that Mph1 stimulated the rate of both Fen1 and Dna2 under this condition; it stimulated Fen1 better than Dna2 (Figs. S2 and S3). This is consistent with our finding that Mph1 suppressed Dna2 mutation by stimulating Fen1 activity.
The results presented here, as well as those published by others (35), raise the possibility that Mph1 serves two different physiological functions, one requiring its ATPase/helicase activities and the other not. The helicase-dependent pathway is likely to play a role(s) in maintaining the stability of the genome, since mph1 devoid of ATPase/helicase activity does not restore the mutator phenotype of mph1Δ cells (35). The second function, which does not require the intrinsic biochemical activities of Mph1, is likely to aid in the maturation of Okazaki fragments, as shown in this study. Although the mutator phenotype of mph1Δ was not rescued by expression of mph1K113Q (35), this observation does not exclude the possibility that the stimulation of Fen1 and Dna2 by Mph1 is able to contribute to genome stability in vivo. It is well established that Fen1 is involved in many pathways other than Okazaki fragment processing, including nucleotide excision repair, base excision repair, mismatch repair, and large loop repair (12, 46–48). Mph1 may participate in one of these repair pathways by stimulating the endonuclease activity of Fen1. If this were the case, mph1Δ null mutants should be more sensitive than the helicase-negative mutant to DNA-damaging agents that activate the above repair pathways.
It is noteworthy that Mph1 reversed the inhibition of Fen1 by flap-bound RPA, suggesting that the stimulation of Fen1 by Mph1 may have physiological significance in vivo by virtue of their effect. We found that the stimulation was limited to the structure-specific endonuclease activity of Fen1. Mph1 did not stimulate the 5′ to 3′ exonuclease activity of Fen1. Substrate-specific stimulation was also consistent with the findings that Mph1 binds preferentially to flap substrates. Possible mechanisms by which Mph1 stimulates Fen1 include Mph1 first binding the dsDNA-ssDNA junction (see below for further discussion), which alters the DNA structure to a form more susceptible to Fen1 cleavage or induces a conformational change in Mph1 suitable for the recruitment of Fen1 to DNA. These two scenarios would not require ATP. Mgs1, unlike Mph1 or Werner and Bloom syndrome helicases (40, 41), requires ATP to stimulate Fen1, although ATP hydrolysis is not required for this effect (39). We unexpectedly observed that Mph1 displaced the 5′ flap strand in a flap substrate containing a 5′ single-stranded DNA, although the Mph1 helicase translocates in the 3′ to 5′ direction (37). A plausible way that Mph1 could displace this 5′ single-strand flap is through its ability to translocate along duplex DNA, after loading onto the double-stranded DNA or dsDNA-ssDNA junction. Recently, it was shown that mph1Δ increased the recombination rate in a sgs1Δ background, suggesting that Mph1 acts as an anti-recombinase (36, 37). Furthermore, it is known that the human homologue of Mph1, FANCM, which plays a role in the Fanconi anemia pathway, has branch migration activity (49). In addition, the archaeal homologue, Hef, also has helicase activity in its N-terminal domain and can unwind fork-structured and Holliday junction DNAs (50). Most recently, it was reported that the C-terminal domain of Mph1 is responsible for promoting gross chromosomal rearrangement by partially inhibiting homologous recombination (38). These findings suggest that biochemical activities associated with Mph1 are likely to be important in maintaining genome stability. Currently, the properties of this helicase are under intensive investigation.
Another conceivable role of Mph1 is that it could function with Dna2 in telomere homeostasis. It is well known that maintenance of telomere length requires extensive lagging strand synthesis in order to make up for gradual loss from their ends. In support of this, the involvement of Dna2 with regard to telomere homeostasis was demonstrated in S. cerevisiae (51) and S. pombe (52). In addition, it was reported that Fen1 is also involved in telomere homeostasis (32). Therefore, it is likely that Mph1 is capable of stimulating both nucleases functions in telomere homeostasis. This possibility is under investigation. The further elucidation of the biochemical properties of the Mph1 helicase should help explain how it acts to maintain genome integrity, including telomere. We noted that the Fen1 preparation used in this study appeared to have a feeble flap endonuclease activity during the review process. We found that it possessed approximately one-third the activity of our newly prepared Fen1.
Supplementary Material
This work was supported by Korea Science and Engineering Foundation Grants funded by the Ministry of Education, Science and Technology. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: pol, polymerase; RPA, replication protein A; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; ATPγS, adenosine 5′-O-(thiotriphosphate); DTT, dithiothreitol; BSA, bovine serum albumin; nt, nucleotide(s); SD, synthetic-dropout medium.
C. H. Lee and Y. S. Seo, unpublished data.
C. H. Lee, unpublished observation.
Y.-S. Seo, unpublished observation.
References
- 1.Bambara, R. A., Murante, R. S., and Henricksen, L. A. (1997) J. Biol. Chem. 272 4647–4650 [DOI] [PubMed] [Google Scholar]
- 2.Hübscher, U., and Seo, Y. S. (2001) Mol. Cells 12 149–157 [PubMed] [Google Scholar]
- 3.Kao, H. I., and Bambara, R. A. (2003) Crit. Rev. Biochem. Mol. Biol. 38 433–452 [DOI] [PubMed] [Google Scholar]
- 4.Budd, M. E., Tong, A. H., Polaczek, P., Peng, X., Boone, C., and Campbell, J. L. (2005) PLoS Genet. 1 634–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conaway, R. C., and Lehman, I. R. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 2523–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shioda, M., Nelson, E. M., Bayne, M. L., and Benbow, R. M. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 7209–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, J., Uhlmann, F., Gibbs, E., Flores-Rozas, H., Lee, C.-G., Pillips, B., Finkelstein, J., Yao, N., O'Donnell, M., and Hurwitz, J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12896–12901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maga, G., Frouin, I., Spadari, S., and Hübscher, U. (2000) J. Mol. Biol. 295 191–801 [Google Scholar]
- 9.Burgers, P. M. (1988) Nucleic Acids Res. 16 6297–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer, G. A., and Burgers, P. M. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 7506–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgers, P. M. (2009) J. Biol. Chem. 284 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, Y., Kao, H. I., and Bambara, R. A. (2004) Annu. Rev. Biochem. 73 589–615 [DOI] [PubMed] [Google Scholar]
- 13.Turchi, J. J., Huang, L., Murante, R. S., Kim, Y., and Bambara, R. A. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 9803–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae, S. H., Bae, K. H., Kim, J. A., and Seo, Y. S. (2001) Nature 412 456–461 [DOI] [PubMed] [Google Scholar]
- 15.Ayyagari, R., Gomes, X. V., Gordenin, D. A., and Burgers, P. M. (2003) J. Biol. Chem. 278 1618–1625 [DOI] [PubMed] [Google Scholar]
- 16.Kao, H. I., Veeraraghaven, J., Polaczek, P., Campbell, J. L., and Bambara, R. A. (2004) J. Biol. Chem. 279 1501–1524 [DOI] [PubMed] [Google Scholar]
- 17.Kuo, C. L., Huang, C. H., and Campbell, J. L. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 6465–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budd, M. E., and Campbell, J. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 7642–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budd, M. E., Choe, W. C., and Campbell, J. L. (1995) J. Biol. Chem. 270 26766–26769 [DOI] [PubMed] [Google Scholar]
- 20.Bae, S. H., Choe, E., Lee, K. H., Park, K. S., Lee, S. H., and Seo, Y. S. (1998) J. Biol. Chem. 273 26880–26890 [DOI] [PubMed] [Google Scholar]
- 21.Bae, S. H., Kim, D. W., Kim, J., Kim, J. H., Kim, D. H., Kim, H. D., Kang, H. Y., and Seo, Y. S. (2002) J. Biol. Chem. 277 26632–26641 [DOI] [PubMed] [Google Scholar]
- 22.Reagan, M. S., Pittenger, C., Siede, W., and Friedberg, E. C. (1995) J. Bacteriol. 177 364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tishkoff, D. Z., Filosi, N., Gaida, G. M., and Kolodner, R. D. (1997) Cell 88 256–263 [DOI] [PubMed] [Google Scholar]
- 24.Johnson, R. E., Kovvali, G. K., Prakash, L., and Prakash, S. (1995) Science 269 238–240 [DOI] [PubMed] [Google Scholar]
- 25.Johnson, R. E., Kovvali, G. K., Prakash, L., and Prakash, S. (1998) Curr. Genet. 34 21–29 [DOI] [PubMed] [Google Scholar]
- 26.Harrington, J. J., and Lieber, M. R. (1994) EMBO J. 13 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murante, R. S., Huang, L., Turchi, J. J., and Bambara, R. A. (1994) J. Biol. Chem. 269 1191–1196 [PubMed] [Google Scholar]
- 28.Zheng, L., Zhou, M., Chai, Q., Parrish, J., Xue, D., Patrick, S. M., Turchi, J. J., Yannone, S. M., and Shen, B. (2005) EMBO. Rep. 6 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiro, C., Pelletier, R., Rolfsmeier, M. L., Dixon, M. J., Lahue, R. S., Gupta, G., Park, M. S., Chen, X., Mariappan, S. V., and McMurray, C. T. (1999) Mol. Cell. 4 1079–1085 [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., Zhang, H., Veeraraghavan, J., Bambara, R. A., and Freudenreich, C. H. (2004) Mol. Cell. Biol. 24 4049–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae, K. H., Kim, H. S., Bae, S. H., Kang, H. Y., Brill, S., and Seo, Y. S. (2003) Nucleic Acids Res. 31 3006–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parenteau, J., and Wellinger, R. J. (1999) Mol. Cell. Biol. 19 4143–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi, M. L., Pike, J. E., Wang, W., Burgers, P. M., Campbell, J. L., and Bambara, R. A. (2008) J. Biol. Chem. 283 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Entian, K. D., Schuster, T., Hegemann, J. H., Becher, D., Feldmann, H., Güldener, U., Götz, R., Hansen, M., Hollenberg, C. P., Jansen, G., Kramer, W., Klein, S., Kötter, P., Kricke, J., Launhardt, H., Mannhaupt, G., Maierl, A., Meyer, P., Mewes, W., Munder, T., Niedenthal, R. K., Ramezani Rad, M., Röhmer, A., Römer, A., and Hinnen, A. (1999) Mol. Gen. Genet. 262 683–702 [DOI] [PubMed] [Google Scholar]
- 35.Scheller, J., Schürer, A., Rudolph, C., Hettwer, S., and Kramer, W. (2000) Genetics 11 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schürer, K. A., Rudonph, C., Ulrich, H. D., and Kramer, W. (2004) Genetics 166 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prakash, R., Krejci, L., Van Komen, S., Anke Schürer, K., Kramer, W., and Sung, P. (2005) J. Biol. Chem. 280 7854–7860 [DOI] [PubMed] [Google Scholar]
- 38.Banerjee, S., Smith, S., Oum, J. H., Liaw, H. J., Hwang, J. Y., Sikdar, N., Motegi, A., Lee, S. E., and Myung, K. (2008) J. Cell Biol. 181 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, J. H., Kang, Y. H., Kang, H. J., Kim, D. H., Ryu, G. H., Kang, M. J., and Seo, Y. S. (2005) Nucleic Acids Res. 33 6137–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma, S., Sommers, J. A., Wu, L., Bohr, V. A., Hickson, I. D., and Brosh, R. M., Jr. (2004) J. Biol. Chem. 279 9844–9856 [DOI] [PubMed] [Google Scholar]
- 41.Brosh, R. M., Jr., von Kobbe, C., Sommers, J. A., Karmakar, P., Opresko, P. L., Piotorowski, J., Dianova, I., Dianov, G. L., and Bohr, V. A. (2001) EMBO J. 20 5791–5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae, S. H., Kim, J. A., Choe, E., Lee, K. H., Kang, H. Y., Kim, H. D., Kim, J. H., Bae, K. H., Cho, Y., Park, C., and Seo, Y. S. (2001) Nucleic Acids Res. 29 3069–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Budd, M. E., and Campbell, J. L. (1997) Mol. Cell. Biol. 17 2136–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fanning, E., Klimovich, V., and Nager, A. R. (2006) Nucleic Acids Res. 34 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim, J. H., Kim, H. D., Ryu, G. H., Kim, D. H., Hurwitz, J., and Seo, Y. S. (2006) Nucleic Acids Res. 34 1854–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sommer, D., Stith, C. M., Burgers, P. M., and Lahue, R. S. (2008) Nucleic Acids Res. 36 4699–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imoto, S., Bransfield, L. A., Croteau, D. L., Van Houten, B., and Greenberg, M. M. (2008) Biochemistry 47 4306–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolodner, R. D., and Marsischky, G. T. (1999) Curr. Opin. Genet. Dev. 9 89–96 [DOI] [PubMed] [Google Scholar]
- 49.Gari, K., Decaillet, C., Stasiak, A. Z., Stasiak, A., and Constantinous, A. (2008) Mol. Cell. 29 141–148 [DOI] [PubMed] [Google Scholar]
- 50.Komori, K., Hidaka, M., Horiuchi, T., Fujikane, R., Shinagawa, H., and Ishino, Y. (2004) J. Biol. Chem. 279 53175–53185 [DOI] [PubMed] [Google Scholar]
- 51.Choe, W., Budd, M., Imamura, O., Hoopes, L., and Campbell, J. L. (2002) Mol. Cell. Biol. 22 4202–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomita, K., Kibe, T., Kang, H. Y., Seo, Y. S., Uritani, M., Ushimaru, T., and Ueno, M. (2004) Mol. Cell. Biol. 24 9557–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.