Abstract
The reversible phosphorylation of tyrosyl residues in proteins is a cornerstone of the signaling pathways that regulate numerous cellular responses. Protein tyrosine phosphorylation is controlled through the concerted actions of protein-tyrosine kinases and phosphatases. The goal of the present study was to unveil the mechanisms by which protein tyrosine dephosphorylation modulates secretion. The acrosome reaction, a specialized type of regulated exocytosis undergone by sperm, is initiated by calcium and carried out by a number of players, including tyrosine kinases and phosphatases, and fusion-related proteins such as Rab3A, α-SNAP, N-ethylmaleimide-sensitive factor (NSF), SNAREs, complexin, and synaptotagmin VI. We report here that inducers were unable to elicit the acrosome reaction when permeabilized human sperm were loaded with anti-PTP1B antibodies or with the dominant-negative mutant PTP1B D181A; subsequent introduction of wild type PTP1B or NSF rescued exocytosis. Wild type PTP1B, but not PTP1B D181A, caused cis SNARE complex dissociation during the acrosome reaction through a mechanism involving NSF. Unlike its non-phosphorylated counterpart, recombinant phospho-NSF failed to dissociate SNARE complexes from rat brain membranes. These results strengthen our previous observation that NSF activity is regulated rather than constitutive during sperm exocytosis and indicate that NSF must be dephosphorylated by PTP1B to disassemble SNARE complexes. Interestingly, phospho-NSF served as a substrate for PTP1B in an in vitro assay. Our findings demonstrate that phosphorylation of NSF on tyrosine residues prevents its SNARE complex dissociation activity and establish for the first time a role for PTP1B in the modulation of the membrane fusion machinery.
Exocytosis is a highly regulated membrane fusion process consisting of multiple stages that culminate in the attachment of secretory vesicles to the plasma membrane followed by the opening of fusion pores (1). Many molecules belonging to, or associated with, the fusion machinery have been identified and characterized. These molecules include integral components of the vesicle (R-SNAREs) and of the plasma membrane (Q-SNAREs), in addition to soluble factors such as N-ethylmaleimide-sensitive factor (NSF),2 α-SNAP, Munc18, and complexin (2–4). SNAREs form a superfamily of proteins that assemble into tightly packed helical bundles (5). Assembly of Q- and R-SNAREs in trans (in opposite membranes) is thought to pull the fusing membranes closely together, which results in bilayer fusion. Disassembly of fusion-incompetent, cis (in the same membrane) SNARE complexes requires the concerted action of α-SNAP and NSF (6, 7). It was believed that NSF is constitutively active to guarantee the constant regeneration of free SNAREs. In the last few years, however, we have learned that NSF activity is regulated by post-translational modifications (8–12).
The acrosome is a large secretory vesicle that overlies the nucleus in the apical region of the mature sperm head (13). In response to physiological or pharmacological stimuli, the acrosome undergoes a special type of calcium-dependent exocytosis (the acrosome reaction, AR), which is a prerequisite for fertilization. The AR proceeds through a sequential set of events initiated when Rab3A is activated by calcium to trigger NSF/α-SNAP-dependent disassembly of cis SNARE complexes. Interestingly, NSF appears to be dormant in resting cells, but its activity is de-repressed when sperm are challenged with AR inducers (14). Later on, monomeric SNAREs re-associate in loose trans complexes until an efflux of calcium from the intra-acrosomal store promotes synaptotagmin- and SNARE-dependent membrane fusion (reviewed in Refs. 15 and 16).
Tyrosine phosphorylation is controlled by the coordinated actions of protein-tyrosine kinases and phosphatases (PTPs). The widely expressed PTP1B is the prototype of the superfamily of PTPs and belongs to the non-transmembrane subfamily 1 of intracellular PTPs (17, 18). PTP1B consists of a single N-terminal phosphatase domain (321 residues) and a regulatory C-terminal domain (114 residues). Using structural insights derived from biochemical and crystallographic data, scientists have identified several residues crucial for substrate recognition and catalysis. Two such residues are Cys215 and Asp181. The catalytic activity of PTP1B is lost or severely diminished, without detrimental effects on its affinity for substrates, when Cys215 is mutated to Ser/Ala or Asp181 to Ala (19). These mutant forms are known as substrate traps.
The precise role of protein tyrosine dephosphorylation on membrane fusion remains unexplored. We and others have inferred the modulation of the AR by PTPs from results achieved with pharmacological inhibitors (see Ref. 20 and references therein). However, data on the identity of the PTPs and substrates involved are lacking. We have determined the role and site of action of PTPs, particularly PTP1B, in secretion by employing antibodies, photosensitive inhibitors, and a substrate-trapping mutant in a functional assay using permeabilized human sperm as the model for exocytosis. We identified a new role for PTP1B, that of indirectly catalyzing cis SNARE complex disassembly through the dephosphorylation of NSF. In defining this role, we also explained the regulation of NSF activity in sperm through its interaction with PTP1B.
EXPERIMENTAL PROCEDURES
Reagents—Recombinant streptolysin O (SLO) was obtained from Dr. Bhakdi (University of Mainz, Mainz, Germany). Spermatozoa were cultured in Human Tubal Fluid media (Irvine Scientific, Santa Ana, CA) supplemented with 0.5% bovine serum albumin (HTF media). The mouse monoclonal anti-PTP1B (clone FG6–1G, purified IgG2a) (21) was initially a gift from Dr. N. Tonks (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY); we subsequently purchased the same antibody from Oncogene Science, Inc. (Cambridge, MA). The mouse monoclonal anti-phosphotyrosine antibody (clone 4G10, subclass IgG2b) was from Upstate Biotechnology Inc. (Lake Placid, NY). The rabbit polyclonal anti-NSF (whole serum) and the mouse monoclonal anti-synaptobrevin 2 (clone 69.1, purified IgG) were from Synaptic Systems (Göttingen, Germany). An anti-Rab3A antibody (rabbit polyclonal, purified IgG) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase-conjugated goat antimouse IgG was from Kierkegaard & Perry Laboratories, Inc. (Gaithersburg, MD). Horseradish peroxidase-conjugated goat anti-rabbit IgG was from Jackson ImmunoResearch (West Grove, PA). The Escherichia coli strain TKB1, which harbors an inducible Elk tyrosine kinase domain under the control of the tryptophan operon, was from Stratagene (La Jolla, CA). O-Nitrophenyl-EGTA-acetoxymethyl ester (NP-EGTA-AM) and BAPTA-AM were from Molecular Probes (Eugene, OR). α-Bromo-4-hydroxyacetophenone (PTP Inhibitor I) was from Calbiochem (La Jolla, CA). Prestained molecular weight markers were from Boston BioProducts Inc. (Worcester, MA). Glutathione-Sepharose was from GE Healthcare and nickel-nitrilotriacetic acid-agarose from Qiagen GmbH (Hilden, Germany). All other chemicals were reagent or analytical grade and were purchased from Sigma or ICN Biochemicals, Inc. (Aurora, OH).
Recombinant Proteins—Two expression plasmids encoding amino acids 1–321 of PTP1B were kindly provided by Dr. N. Tonks: the substrate-trapping mutant PTP1B D181A fused to glutathione S-transferase was in pGEX-3X (GE Healthcare) and the wild type PTP1B fused to His6 was in pET21b (Stratagene). Two plasmids encoding NSF were used for protein expression: pQE9 (Qiagen) was generously provided by Dr. S. Whiteheart (University of Kentucky, Lexington, KY) and pET28a (Stratagene) was a kind gift from Dr. D. Fasshauer (Max-Planck Institute for Biophysical Chemistry, Göttingen, Germany). To obtain tyrosine-phosphorylated NSF in E. coli, NSF in pET28a was transformed into TKB1 cells; protein expression and tyrosine phosphorylation were carried out following Stratagene's instructions. A pQE9 construct encoding α-SNAP was also from Dr. S. Whiteheart. Plasmid pGEX-2T containing the cDNA-encoding human Rab3A was provided by Dr. P. Stahl (Washington University, St. Louis, MO). The light chain of wild type TeTx and BoNT/B fused to His6 (pQE3, Qiagen) were generously provided by Dr. T. Binz (Medizinische Hochschule Hannover, Hannover, Germany). The light chain of catalytically dead TeTx fused to His6 (TeTx E234Q in pET28a) was a gift from Dr. F. Zilly (Max-Planck Institute for Biophysical Chemistry, Göttingen, Germany).
The plasmid constructs pQE9 encoding His6-NSF and pQE3 encoding His6-BoNT/B were transformed into E. coli M15pRep4 (Qiagen) and protein expression was induced 4 h at 30 °C with 1 mm isopropyl β-d-thiogalactoside (IPTG). DNAs encoding wild type His6-TeTx and α-SNAP were transformed into E. coli XL-1 Blue (Stratagene) and induced overnight at 20 °C with 0.2 mm IPTG. All proteins encoded by cDNAs contained in pET vectors were expressed in E. coli BLR(DE3) (Stratagene) by inducing with 0.25 mm IPTG (3 h at 25 °C for TeTx E234Q, 3 h at 30 °C for NSF), except PTP1B wild type that was induced with 0.1 mm IPTG overnight at 22 °C. Purification of His6-tagged recombinant proteins was carried out according to Qiagen's instructions, except that 0.5 mm ATP, 5 mm MgCl2, and 2 mm β-mercaptoethanol were added to all buffers involved in the purification of His6-NSF. The His6 tag was cleaved from NSF by incubation with thrombin during dialysis to remove imidazole. Thrombin activity was stopped with 2 mm phenylmethylsulfonyl fluoride.
DNAs encoding Rab3A and PTP1B D181A were transformed into E. coli BL21 (Stratagene). We induced the expression of Rab3A with 0.5 mm IPTG overnight at 22 °C, and that of PTP1B D181A with 0.1 mm IPTG overnight at 22 °C. Recombinant proteins were purified on glutathione-Sepharose following standard procedures. Purified Rab3A was prenylated and loaded with guanosine 5′-O-3-thiotriphosphate as described (22).
Recombinant protein concentrations were determined by the Bio-Rad Protein assay in 96-well microplates. Bovine serum albumin was used as a standard and the results were quantified on a Bio-Rad 3550 Microplate Reader.
Human Sperm Sample Preparation for AR Assays and Western Blotting—Human semen samples were obtained from normal healthy donors. Semen was allowed to liquify for 30–60 min at 37 °C. We used a swim-up protocol to isolate highly motile sperm. Sperm concentrations were adjusted to 5–10 × 106/ml before incubating for at least 2 h under capacitating conditions (HTF, 37 °C, 5% CO2, 95% air). Permeabilization was accomplished as described (22). Briefly, washed spermatozoa were resuspended in cold phosphate-buffered saline containing 1.5 units/ml SLO for 15 min at 4 °C. Cells were washed once with phosphate-buffered saline and resuspended in ice-cold sucrose buffer (250 mm sucrose, 0.5 mm EGTA, 20 mm Hepes-K, pH 7) containing 2 mm DTT. For AR assays, we added inhibitors and stimulants sequentially as indicated in the figures legends, and incubated for 10–15 min at 37 °C after each addition. When indicated, we preloaded SLO-permeabilized sperm with photo-inhibitable reagents before incubating in the presence of inhibitors and/or calcium, carrying out all procedures in the dark. Photolysis was induced after the last incubation by exposing twice (1 min each time) to an UV transilluminator, mixing, and incubating for 5 min at 37 °C. Sperm were spotted on Teflon-printed slides, air dried, and fixed/permeabilized in ice-cold methanol for 1 min. Acrosomal status was evaluated by staining with FITC-coupled Pisum sativum (FITC-PSA) according to Ref. 23. At least 200 cells were scored using an upright Nikon microscope equipped with epifluorescence optics. Basal (no stimulation, “control”) and positive (0.5 mm CaCl2, corresponding to 10 μm free Ca2+, “Ca2+”) controls were included in all experiments. Acrosomal exocytosis indexes were calculated by subtracting the number of spontaneously reacted spermatozoa from all values and expressing the results as a percentage of the AR observed in the positive control. Data were evaluated using one-way analysis of variance. The Tukey-Kramer post hoc test was used for pairwise comparisons. Differences were considered significant at the p < 0.05 level.
For Western blotting, SLO-permeabilized sperm (5–10 × 106 cells) were treated or not with 300 nm PTP1B D181A, 27 nm wild type PTP1B or both, and incubated for 30 min at 37 °C. Cells were lysed in cold 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5 mm Na3VO4, 2 mm EDTA, a protease inhibitor mixture (P2714, Sigma), 5 μg/ml trypsin inhibitor, 0.5 mm phenylmethylsulfonyl fluoride, 1% sodium deoxycholate, 1% Triton X-100, and 0.1% SDS. We sonicated sperm (2 × 15 s) and let proteins diffuse into the lysis buffer for 30 min at 4 °C. These whole cell detergent extracts were clarified by centrifugation at 12,000 × g for 5 min and electrophoresed immediately or stored at –20 °C.
Disassembly of the SNARE Complex—Brain cortices from adult male Sprague-Dawley rats were homogenized in 5 mm Hepes buffer, pH 7.4, containing 0.32 m sucrose and 1 mm EDTA using a glass Teflon homogenizer (8 strokes at 800 rpm). The homogenates were centrifuged at 1,000 × g for 5 min and the pellets re-extracted in 1 volume of buffer. Following a second centrifugation at 1,000 × g for 5 min, the supernatants were pooled and spun at 17,500 × g for 15 min. The crude synaptosomal pellets were washed once by centrifugation and stored at –20 °C until use (24). For a typical disassembly reaction, synaptosomal protein (2–4 μg) was incubated in a 20-μl volume containing 20 mm Hepes-NaOH, pH 7.8, 100 mm KCl, 1% (v/v) glycerol, 1 mm DTT, 2 μm TeTx or BoNT/B, 8 μm α-SNAP, 600 nm NSF or phospho-NSF, 2.5 mm ATP, and 2 mm MgCl2. In control reactions, 10 mm EDTA was added to chelate the MgCl2 and thus prevent ATP hydrolysis by NSF. Reactions were carried out for 15 min at 37 °C and terminated by addition of SDS sample buffer containing 50 mm Tris-HCl, pH 6.8, 4% (w/v) SDS, 12% (w/v) glycerol, 100 mm DTT, and 0.01% (w/v) Serva Blue G and incubated for an additional 5 min at 95 °C before separation by SDS-PAGE (25) and immunoblotting with the anti-synaptobrevin antibody.
SDS-PAGE and Western Blots—Prior to electrophoresis, sperm proteins were dissolved in SDS sample buffer containing 0.0625 m Tris-HCl, pH 6.8, 2% (w/v) SDS, 10% (w/v) glycerol, 5% (v/v) β-mercaptoethanol, and 0.05% bromphenol blue by incubating for 5 min at 95 °C, separated on polyacrylamide slab gels (26), and transferred to 0.22-μm nitrocellulose membranes (Hybond, GE Healthcare). Nonspecific reactivity was blocked by incubation for 1 h at room temperature with 3% gelatin dissolved in washing buffer (phosphate-buffered saline, pH 7.6, 0.1% Tween 20). Blots were incubated with the anti-phosphotyrosine antibody (0.1 μg/ml in blocking solution) for 2 h at room temperature or overnight at 4 °C. Anti-NSF blots were processed similarly to the anti-phosphotyrosine, except that the blocking solution contained 3% skim milk and 1% polyvinylpyrrolidone (40,000 average Mr, ICN) instead of gelatin, and that the primary antibody was diluted 1:6,000 in 3% skim milk. For brain samples, blots were blocked in 5% skim milk and the anti-synaptobrevin 2 antibody was used at 0.05 μg/ml in blocking solution. Horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit IgGs were used as secondary antibodies (0.25 μg/ml) with 1-h incubations. Excess first and second antibodies were removed by washing 5 × 7 min in washing buffer. Detection was accomplished with a chemiluminescence system (Western Lightning, PerkinElmer Life Sciences, Boston, MA) and subsequent exposure to HyperFilm™ ECL High performance (GE Healthcare) or CL-XPosure (Pierce, Tecnolab, Buenos Aires, Argentina) chemiluminescence film for 1–10 min.
Phospho-NSF Dephosphorylation by PTP1B—NSF was tyrosine phosphorylated in E. coli TKB1 following a two-step protocol. First, IPTG induced the expression of NSF; second, indoleacrylic acid elicited the synthesis of the Elk tyrosine kinase, which phosphorylated NSF in vivo. His6-phospho-NSF was purified with nickel-nitrilotriacetic acid-agarose; tyrosine phosphorylation was confirmed by Western blot.
Phospho-NSF (310 nm) was incubated with 1 μg/ml wild type or D181A PTP1B in a buffer containing 50 mm Hepes-Na, pH 7.4, 150 mm NaCl, 1 mm EDTA, 3 mm DTT, and protease inhibitors in a final reaction volume of 30 μl. Incubations were carried out at 30 °C in a water bath. At the times indicated reactions were stopped with SDS-sample buffer and the products analyzed by anti-phosphotyrosine and anti-NSF Western blotting.
Two-dimensional Gel Electrophoresis—Recombinant phospho-NSF (10–20 μg) and frozen sperm pellets (150 × 106 cells) were suspended in a rehydration solution containing 8 m urea, 2% CHAPS, 0.002% bromphenol blue, 0.5 mm Na3VO4, and a protease inhibitor mixture. Each sample was sonicated at 4 °C using 2 × 15-s bursts; insoluble material was removed by centrifugation. Aliquots (150 μl) containing ∼150 μg of solubilized sperm protein were mixed with 1 μl of carrier ampholytes (Pharmalyte pH 3–10) and loaded onto immobilized pH-gradient (IPG, Immobiline DryStrip gels, Amersham Biosciences) strips (7 cm, linear pH 3–10). The IPG strips were then covered in mineral oil and left to rehydrate for 15 h at 20 °C; 100 mm DTT was added just prior to the run. Isoelectric focusing was performed at 20 °C using an Ettan IPGphor isoelectric focusing system (Amersham Biosciences), with a program of stepped voltages (30 min at 500 V, 30 min at 1000 V, and 100 min at 5000 V). Following focusing, the IPG strips were immediately incubated in 10 ml of equilibration buffer (30% glycerol, 2% SDS, 6 m urea, 50 mm Tris-HCI, pH 8.8, and 0.002% bromphenol blue) supplemented with 100 mg of DTT for 15 min at room temperature, followed by fresh equilibration buffer (10 ml) containing 250 mg of iodoacetamide for a further 15 min at room temperature. The strips were then placed on top of 8% polyacrylamide slab gels (1 mm thick, 10 × 10 cm) and held in place using 0.5% agarose prepared in SDS-running buffer. Proteins were then resolved at constant current (10–15 mA), electrotransferred onto nitrocellulose membranes, and immunoblotted sequentially using anti-NSF and anti-phosphotyrosine antibodies. Blots were stripped prior to reprobing in a solution containing 0.1 n HCl (30 min at room temperature with vigorous shaking), followed by two 10-min washes in washing buffer, and a final 10-min wash in double-distilled water.
RESULTS
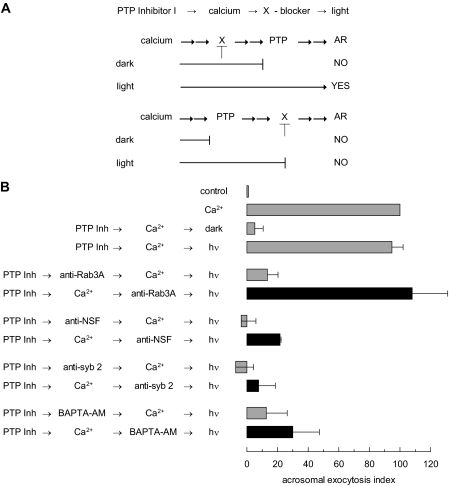
PTP Activity Is Required after Rab3 and before NSF during the AR—We resorted to the photosensitive PTP Inhibitor I to determine the steps governed by protein tyrosine dephosphorylation during the AR. This compound binds covalently to the catalytic domain of SHP-1 and PTP1B, blocking their enzymatic activity. The inhibition is reversed by irradiation with UV light (27). We reasoned that the AR can be modeled to fit in a linear signaling pathway operating through the sequence below (14), where single arrows mean there is one step between the proteins connected, and double arrows mean that the number of steps taking place between the connected proteins is unknown: calcium →→ Rab3 →→ α-SNAP/NSF → SNAREs (cis complex disassembly → monomeric proteins →→ reassembly in trans) →→ intra-acrosomal calcium efflux →→ SNAREs (full zippering in trans) →→ exocytosis.
If PTPs catalyzed any of those unknown steps, PTP Inhibitor I would halt the AR at the point when the activity of the first PTP was required, because the sequence is linear, for as long as the system was kept in the dark. Photoreversion of the inhibition caused by PTP Inhibitor I should resume exocytosis. Indeed, when permeabilized sperm were pre-loaded with PTP Inhibitor I, calcium failed to elicit exocytosis when the system was protected from light. Exocytosis was restored upon illumination (Fig. 1B). These data confirm that PTPs are required for sperm exocytosis and also demonstrate the applicability of PTP Inhibitor I to our study.
FIGURE 1.
PTP activity is required after Rab3A and before NSF during the AR. A, a schematic representation of the use of PTP Inhibitor I in permeabilized sperm. Shown is the sequence of addition of reagents PTP Inhibitor I → calcium → X-blocker → light. The long lines under the scheme represent the farthest step reached by calcium under each condition. The two possible scenarios: X required before or after PTP, and their different outcomes when tubes are illuminated, AR YES or NO, are depicted. B, SLO-permeabilized spermatozoa were loaded with 0.25 mm PTP Inhibitor I (PTP Inh) for 10 min at 37 °C to prevent all PTP activity. AR was subsequently initiated by adding 0.5 mm CaCl2 (10 μm free calcium, estimated by MAXCHELATOR, a series of program(s) for determining the free metal concentration in the presence of chelators). After a 10-min incubation at 37 °C to allow exocytosis to proceed to the first PTP Inhibitor I-sensitive step, sperm were treated with antibodies that recognize Rab3A (20 μg/ml), NSF (1:300), synaptobrevin 2 (20 μg/ml, anti-syb2), or with BAPTA-AM (10 μm). All procedures were carried out in the dark. The tubes were illuminated with UV light to reverse the block on PTPs at the end of the incubation period (hv), and the samples were incubated for 5 min to promote exocytosis (PTP Inh → Ca2+ → antibody/BAPTA-AM → hv, black bars). Sperm were fixed and AR was measured by FITC-PSA binding as described under “Experimental Procedures.” Several controls were run (gray bars): inhibitory effect of PTP Inhibitor I in the dark (PTP Inh → Ca2+ → dark); and the recovery upon illumination (PTP Inh → Ca2+ → hv); and the inhibitory effect of the antibodies and BAPTA-AM when present throughout the experiment (PTP Inh → antibody/BAPTA-AM → Ca2+ → hv). The data were normalized as described under “Experimental Procedures.” Actual percentages of reacted sperm for control and Ca2+ ranged between 10–41 and 17–53%, respectively. The data represent the mean ± S.E. of at least three independent experiments.
PTP Inhibitor I serves to place the requirement for fusion-related factors to steps occurring before or after the first PTP Inhibitor I-sensitive step. Briefly, PTP Inhibitor I allows calcium to prepare the fusion machinery up to the point when the first PTP is required. An inhibitor of step X (see schematic in Fig. 1A) is then added and the tubes illuminated. Resistance to the X-blocker, reflected in unaffected exocytosis, implies that its target is required upstream of PTP activity (Fig. 1A, top). Sensitivity to the X-blocker, revealed by no exocytosis, means its target is located after the PTP Inhibitor I-sensitive step (Fig. 1A, bottom). In the control condition, the AR should be prevented when the X-blocker is added prior to the inducer (calcium in this case) and maintained throughout the experiment (not shown in schematic). We asked whether the PTP-dependent step takes place before or after Rab3A by blocking the function of this small G protein with a specific antibody. The anti-Rab3A antibody inhibited exocytosis when added before, but not after, challenging with calcium. In contrast, antibodies against NSF and synaptobrevin 2 were able to abrogate exocytosis even when added after calcium. The acrosome is a calcium store and intra-acrosomal calcium release is mandatory for exocytosis. The calcium chelator BAPTA conjugated to an AM group accumulates inside the acrosome in permeabilized sperm, preventing the AR. When combined with PTP Inhibitor I, BAPTA-AM displayed the same behavior than the anti-NSF and anti-synaptobrevin antibodies (Fig. 1B, black bars). These results suggest that PTP activity is required after Rab3A, but before NSF, SNAREs, and intra-acrosomal calcium, during the signaling cascade leading to the AR, allowing us to update the original sequence as follows: calcium →→ Rab3 →→ PTP → substrate-P → substrate →→ α-SNAP/NSF → SNAREs (cis complex disassembly → monomeric proteins →→ reassembly in trans) →→ intra-acrosomal calcium efflux →→ SNAREs (full zippering in trans) →→ exocytosis.
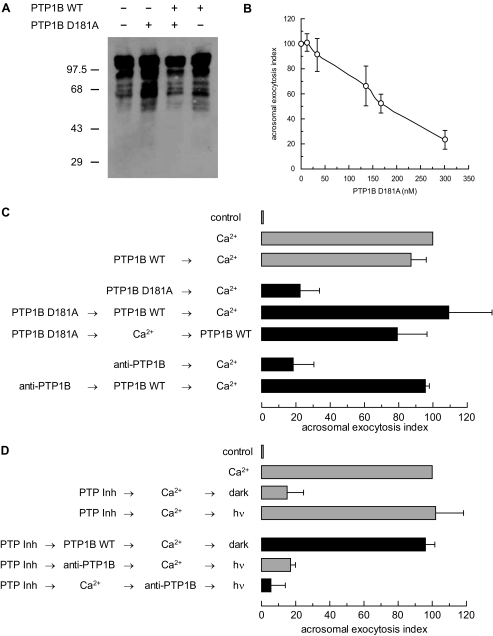
PTP1B Is Required for Exocytosis in a Human Sperm AR Model—We have shown that PTP activity is necessary for the AR (Fig. 1 and Ref. 20). Because one of the PTPs targeted by the photosensitive PTP Inhibitor I is PTP1B, and the enzyme has been detected in ejaculated human and epididymal mouse (20) and rat (28) sperm, we asked whether this phosphatase might modulate protein tyrosine phosphorylation in human sperm. Full-length PTP1B contains 435 amino acids, although a shorter variant containing the entire catalytic domain is typically used for biochemical studies. To test the effect of this domain, and that of the substrate-trapping mutant D181A, on the extent of tyrosine phosphorylation of proteins, we expressed these constructs in E. coli, introduced them into SLO-permeabilized sperm, and immunoblotted cell lysates with an anti-phosphotyrosine antibody (Fig. 2A). The level of tyrosine phosphorylation in cells treated with wild type PTP1B was comparable with that in untreated controls, as was the case when PTP1B was overexpressed in COS7 cells (19). Overall protein tyrosine phosphorylation increased in response to PTP1B D181A; the increase dissapeared with the wild type enzyme. These data suggest that PTP1B D181A binds to and protects sperm proteins from dephosphorylation by endogenous PTP1B. They also suggest that recombinant, wild type PTP1B competes with the D181A mutant for sperm substrates.
FIGURE 2.
Protein tyrosine dephosphorylation by PTP1B is required for calcium-triggered exocytosis. A, SLO-permeabilized human sperm were treated with 300 nm PTP1B D181A, 27 nm wild type PTP1B, or both for 30 min at 37 °C. Sperm were lysed and proteins extracted and analyzed by Western blot using the anti-phosphotyrosine 4G10 antibody as probe. Protein from 1 × 106 cells was loaded per lane. Mr standards (×103) are indicated on the left. B, SLO-permeabilized sperm were treated at 37 °C for 15 min with increasing concentrations of purified PTP1B D181A. AR was subsequently initiated by adding 0.5 mm CaCl2 and incubating for 15 min at 37 °C. C, SLO-permeabilized human sperm were treated for 10 min at 37 °C in the presence of 300 nm PTP1B D181A or 3.3 nm anti-PTP1B antibodies. Wild type PTP1B (27 nm) was added when indicated and incubated for 10 min at 37 °C. AR was initiated with calcium as in B. Control (gray bar) demonstrated that the AR was unperturbed by 27 nm wild type (WT) PTP1B (PTP1B WT → Ca2+). D, SLO-permeabilized spermatozoa loaded with 0.25 mm PTP Inhibitor I (PTP Inh) as indicated in the legend to Fig. 1, were treated with 27 nm wild type PTP1B. AR was subsequently initiated with calcium as in B. All procedures were carried out in the dark. Additionally, PTP Inhibitor I-loaded sperm were treated as described in the legend to Fig. 1, except that 3.3 nm anti-PTP1B antibodies were used instead of anti-Rab3A, anti-synaptobrevin 2, or anti-NSF. Several controls were run (gray bars): inhibitory effect of PTP Inhibitor I in the dark (PTP Inh → Ca2+ → dark); and the recovery upon illumination (PTP Inh → Ca2+ → hv); and the inhibitory effect of the anti-PTP1B antibodies when present throughout the experiment (PTP Inh → anti-PTP1B → Ca2+ → hv). Sperm were fixed and AR was measured by FITC-PSA binding. The data were normalized as described under “Experimental Procedures.” Actual percentages of reacted sperm for control and Ca2+ ranged between 10–30 and 18–48%, respectively. The data represent the mean ± S.E. of at least three independent experiments.
When added to permeabilized human sperm, PTP1B D181A prevented exocytosis in a dose-dependent fashion (Fig. 2B). The mutant PTP inhibits the AR because, although it binds to the substrates of PTP1B, it is unable to dephosphorylate them efficiently. Thus, mutant and substrate become locked in a stable, “dead-end” complex. Wild type PTP1B overcame the D181A mutant (Fig. 2C) and PTP Inhibitor I (see Fig. 1) protected from light (Fig. 2D) blocks on the AR, possibly by competition and/or displacement.
When introduced into permeabilized sperm, an anti-PTP1B antibody prevented calcium-triggered exocytosis, reinforcing the notion that endogenous PTP1B was necessary for secretion (Fig. 2C). Addition of wild type PTP1B reversed the block. When tested in combination with PTP Inhibitor I in an experimental setting similar to that depicted in Fig. 1, the anti-PTP1B antibody inhibited the AR whether it was added before or after calcium (Fig. 2D). This indicates that the AR cannot be resumed upon photoreversion of the effect of PTP Inhibitor I when the PTP1B of the sperm is unavailable. Taken together, these results demonstrate the requirement for PTP1B in sperm exocytosis and are consistent with our proposition that the PTP targeted by the photoreversible inhibitor is PTP1B.
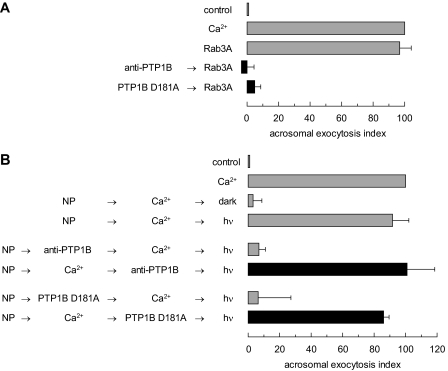
PTP1B Activity Is Required after Rab3A and before Intra-acrosomal Calcium Release during the AR—Having established the requirement for PTP1B in the AR, we set out to investigate where in the signaling cascade does dephosphorylation by this phosphatase take place. First, we made use of the fact that prenylated, GTPγS-loaded, recombinant Rab3A induces an exocytotic response of a magnitude comparable with that of calcium (29). Pretreatment with the anti-PTP1B antibody or PTP1B D181A inhibited Rab3A-triggered AR by >90% (Fig. 3A). These data indicate that Rab3A relies on PTP1B to achieve acrosomal exocytosis in human sperm.
FIGURE 3.
PTP1B is required for human sperm AR after Rab3A but before intra-acrosomal calcium efflux. A, SLO-permeabilized human sperm were treated for 15 min at 37 °C in the presence of 3.3 nm anti-PTP1B antibodies or 300 nm PTP1B D181A. Acrosomal exocytosis was evaluated by FITC-PSA binding after an additional 15-min incubation at 37 °C in the absence or presence of 0.5 mm CaCl2 or 300 nm GTPγS-bound Rab3A (Rab3A). Controls (gray bars) included AR triggered by 300 nm GTPγS-bound Rab3A. B, SLO-permeabilized human sperm were loaded with 10 μm NP-EGTA-AM for 10 min at 37 °C to chelate intra-acrosomal calcium. AR was subsequently initiated by adding 0.5 mm CaCl2. After a further 10 min at 37 °C to allow exocytosis to proceed up to the intra-acrosomal calcium-sensitive step, sperm were treated for 10 min at 37 °C with anti-PTP1B antibodies or PTP1B D181A as in A. These procedures were carried out in the dark. UV photolysis of the chelator was induced at the end of the incubation period (NP → Ca2+ → anti-PTP1B/PTP1B D181A → hv, black bars). Sperm were fixed and AR was measured by FITC-PSA binding as described under “Experimental Procedures.” Several controls were run (gray bars): inhibitory effect of NP-EGTA-AM in the dark (NP → Ca2+ → dark); and the recovery upon illumination (NP → Ca2+ → hv); and the inhibitory effect of the antibodies and PTP1B D181A when present throughout the experiment (NP → anti-PTP1B/PTP1B D181A → Ca2+ → hv). The data were normalized as described under “Experimental Procedures.” Actual percentages of reacted sperm for control and Ca2+ ranged between 8–23 and 15–34%, respectively. The data represent the mean ± S.E. of at least three independent experiments.
Second, we resorted to NP-EGTA-AM, a photolabile calcium chelator that reversibly blocks the AR by sequestering intra-acrosomal calcium (30). The experimental protocol is similar to that described for PTP Inhibitor I, and can be summarized with the same scheme depicted in Fig. 1A, except that “intra-acrosomal calcium release” should be read instead of “PTP.” Preloading permeabilized sperm with NP-EGTA-AM in the dark allows for the AR inducer to prepare the fusion machinery up to the point when intra-acrosomal calcium release is required. Subsequently, the inhibitors to test are added and the chelated calcium is released with UV light. Neither the anti-PTP1B antibody nor PTP1B D181A inhibited the AR (Fig. 3B, black bars). Taken together, these data indicate that the step catalyzed by PTP1B takes place after Rab3A and prior to intra-acrosomal calcium efflux, and are in agreement with the results achieved with the PTP Inhibitor I in combination with the anti-Rab3A antibody and BAPTA-AM (Fig. 1B).
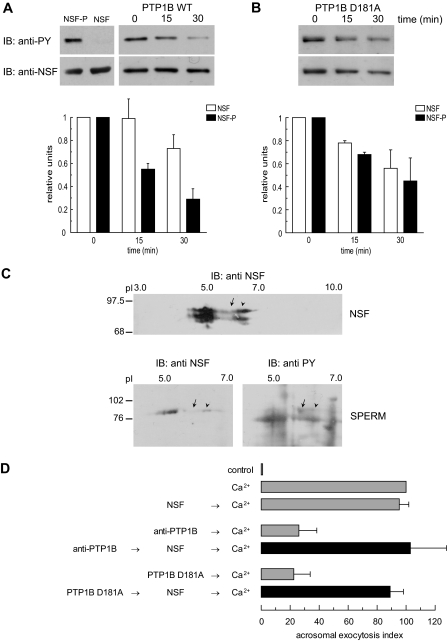
PTP1B Dephosphorylates Tyrosine-phosphorylated NSF—Because both PTP1B and NSF participate in the AR during the same window of time (Fig. 3) (31, 32), we investigated the possibility that PTP1B might directly regulate the phosphorylation status of NSF. We incubated NSF phosphorylated in E. coli TKB1 with PTP1B and analyzed the reaction products by immunoblotting with an anti-phosphotyrosine antibody; an anti-NSF antibody was used as loading control. The phosphotyrosine signal on NSF decreased appreciably after the first 15-min incubation with wild type PTP1B and continued dropping up to 30 min (Fig. 4A). The drop in phosphotyrosine labeling was substantially larger than the fall in the total amount of NSF (which is presumably due to protein degradation/precipitation). NSF remained tyrosine phosphorylated after 30 min incubation with PTP1B D181A (Fig. 4B). These data indicate that NSF is tyrosine phosphorylated by Elk kinase and that phospho-NSF is a novel PTP1B substrate.
FIGURE 4.
PTP1B dephosphorylates tyrosine-phosphorylated NSF. A: top left, recombinant NSF expressed in E. coli BLR(DE3) (0.5 μg) or expressed and tyrosine phosphorylated in E. coli TKB1 (0.5 μg) were resolved on 8% SDS gels, transferred to nitrocellulose, and immunoblotted (IB) with 4G10 (anti-PY, top) and anti-NSF (bottom) antibodies. Top right, recombinant phospho-NSF (310 nm) was incubated with 1 μg/ml recombinant wild type PTP1B. At the indicated time points, aliquots containing 0.7 μg of NSF were mixed with SDS-PAGE sample buffer, proteins were resolved on 8% SDS gels, transferred to nitrocellulose, and immunoblotted with 4G10 (anti-PY, top). The same membranes were probed (without stripping) with anti-NSF (anti-NSF, bottom) antibodies. Shown is an experiment representative of five repetitions; quantification (carried out with Image J, freeware from NIH) of Western blots is depicted as mean ± S.E. from all five replicates below the immunoblots. B, same as in A, except that the substrate trapping mutant was used instead of wild type PTP1B. The experiment was repeated twice. C, two-dimensional electrophoresis of recombinant, tyrosine-phosphorylated NSF expressed in E. coli TKB1 (20μg, NSF) and solubilized sperm proteins (150μg, SPERM). The first dimension was carried out on precast IPG strips and the second dimension on 8% SDS gels. Proteins were transferred to nitrocellulose, and Western blots were performed with anti-NSF and anti-phosphotyrosine (anti-PY) antibodies as described. pH is indicated on top and Mr standards (×103) are indicated on the left. Shown are images representative of two independent experiments. D, SLO-permeabilized human sperm were treated for 15 min at 37 °C in the presence of 3.3 nm anti-PTP1B antibodies or 300 nm PTP1B D181A, followed by an additional 15 min in the presence of 310 nm recombinant NSF (black bars). AR was initiated by adding 0.5 mm CaCl2 and incubating for 15 min at 37 °C. Controls (gray bars) included AR inhibition by 3.3 nm anti-PTP1B antibodies or 300 nm PTP1B D181A (anti-PTP1B/PTP1B D181A → Ca2+); and AR unperturbed by 310 nm NSF (NSF → Ca2+). The data were normalized as described under “Experimental Procedures.” Actual percentages of reacted sperm for control and Ca2+ ranged between 10–38 and 21–52%, respectively. The data represent the mean ± S.E. of at least three independent experiments.
When recombinant NSF expressed in E. coli TKB1 was analyzed by two-dimensional gel electrophoresis followed by Western blot, anti-NSF antibodies detected a protein band with an isoelectric point (pI) similar to that predicted from the human NSF amino acid primary sequence (NP006169, Fig. 4C, top, arrowhead). The band detected by the anti-NSF antibodies extended toward the anode, consistent with the behavior expected for a phosphorylated protein (Fig. 4C, top, arrow). When extracts from capacitated human spermatozoa were run on two-dimensional gels, the anti-NSF antibody detected a protein with pI and electrophoretic mobility similar to those of recombinant NSF (Fig. 4C, bottom left, arrowhead). Stripping and reprobing the membrane with an anti-phosphotyrosine antibody showed that a fraction of NSF molecules was tyrosine phosphorylated (Fig. 4C, bottom right, arrow). The pI of the additional band detected by the anti-NSF antibody toward the anode did not match the predicted pI for this protein and therefore might not correspond to phosphorylated NSF.
Because NSF was required downstream of PTP activity for sperm exocytosis (Fig. 1B) and it served as a PTP1B substrate in vitro (Fig. 4A), we hypothesized that anti-PTP1B antibodies and the mutant PTP1B D181A inhibit the AR because they maintain sperm NSF in a tyrosine-phosphorylated form. If this were the case, recombinant NSF would overcome the inhibition imposed by the PTP1B blockers. Our results confirmed the validity of this hypothesis (Fig. 4D). Based on these results, we modeled the AR to the sequence: calcium →→ Rab3A →→ PTP1B → NSF-P → NSF → SNAREs (cis complex disassembly → monomeric proteins →→ reassembly in trans) →→ intra-acrosomal calcium efflux →→ SNAREs (full zippering in trans) →→ exocytosis.
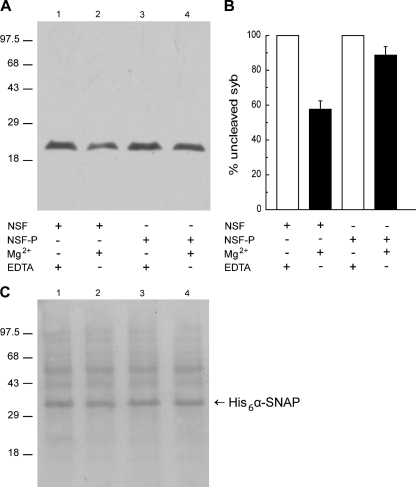
Tyrosine Phosphorylation of NSF Impairs Its Ability to Dissociate SNARE Complexes—The AR sequence depicted earlier predicts that phospho-NSF must be dephosphorylated to accomplish its SNARE complex dissociating activity. We tested the accuracy of this prediction by asking whether phospho-NSF disassembles SNARE complexes. To this end, we adapted an heterologous assay developed to monitor the activity of NSF on SNARE complexes from synaptic vesicles (33). Briefly, we incubated a rat brain synaptosomal fraction (source of native SNARE proteins) with recombinant α-SNAP and NSF or phospho-NSF in the presence of TeTx or BoNT/B. These clostridial neurotoxins cleave synaptobrevin at the same peptide-bond (34). Synaptobrevin is sensitive to proteolytic cleavage as long and is not engaged in ternary SNARE complexes with syntaxin and SNAP-25 (35). As shown in Fig. 5A, an anti-synaptobrevin antibody detected the intact protein under control conditions when Mg2+ was chelated by EDTA and therefore no disassembly occurred (lanes 1 and 3). Over 40% of the synaptobrevin signal was lost when NSF and Mg2+ were added to disassemble ternary SNARE complexes, indicating that the protein was not protected from BoNT/B (Fig. 5A, lane 2). In contrast, when phospho-NSF was used instead of NSF, ∼90% synaptobrevin was protected from proteolytic cleavage, suggesting that the SNARE complexes had not been disassembled (Fig. 5A, lane 4). These data show that phosphorylation of NSF on tyrosine residues render it incapable of dissociating SNARE complexes.
FIGURE 5.
Tyrosine phosphorylation abolishes the ability of NSF to disassemble native SNARE complexes. A, a rat brain preparation enriched in synaptosomes was incubated with 2 μm BoNT/B, 8 μm α-SNAP, 600 nm NSF (lanes 1 and 2), or phospho-NSF (lanes 3 and 4) for 15 min at 37 °C. NSF-driven disassembly rendered synaptobrevin susceptible to cleavage by the light chain of BoNT/B (lane 2), whereas synaptobrevin remained intact when phosho-NSF (NSF-P) was substituted for NSF (lane 4). Control conditions where ATP hydrolysis was prevented by addition of 10 mm EDTA to complex Mg2+ ions are shown on lanes 1 and 3. Reactions were terminated by addition of SDS sample buffer and the amount of intact synaptobrevin 2 was analyzed by Western blot using the 69.1 antibody as probe. Mr standards (×103) are indicated on the left. Shown is a blot representative of three repetitions. BoNT/B and TeTx rendered identical results as did mouse brain membranes instead of rat brain synaptosomes (not shown). B, each band was quantitated by Image J. Shown is the average (±S.E.) relative intensity for each of the experimental conditions from all three replicates. C, Ponceau staining of the membrane shown in A to demonstrate equal protein loads. The electrophoretic mobility of recombinant α-SNAP is indicated with an arrow.
PTP1B Participates in the Priming of the Fusion Machinery, Bringing about the Disassembly of cis SNARE Complexes—If the sequence formulated above is true, then addition of recombinant wild type PTP1B to permeabilized cells should dephosphorylate endogenous NSF and thus disassemble cis SNARE complexes. We designed the next set of experiments to test this hypothesis. During the priming step of the AR, α-SNAP/NSF unpair cis SNARE complexes yielding monomeric SNAREs available for subsequent trans pairing and fusion. Clostridial toxins impair SNAREs assembly into ternary complexes and therefore prevent exocytosis in a variety of cells, including sperm (34, 37). Only monomeric synaptobrevin, but not that engaged in cis, loose or tight trans complexes, is sensitive to TeTx (14, 38). This neurotoxin exhibits zinc-dependent proteolytic activity, and is inactivated by the zinc chelator N,N,N′,N′-tetrakis(2-pyridymethyl)ethylenediamine (TPEN). Therefore we asked if PTP1B disassembled cis SNARE complexes by monitoring the sensitivity of the synaptobrevin to toxin cleavage. When introduced into permeabilized sperm, wild type PTP1B appeared not to perturb calcium-elicited exocytosis (Figs. 2C and 6A, top). Yet, PTP1B was not inert in the assay, as the following experiment shows: first we loaded sperm with wild type PTP1B and TeTx and incubated the mixture under conditions in which the toxin could cleave synaptobrevin; second, we added TPEN to inactivate the toxin and calcium to trigger the AR. Calcium failed to elicit exocytosis because synaptobrevin had been cleaved by TeTx (Fig. 6A, top, black bar). If synaptobrevin was in a monomeric configuration, it was because PTP1B had disassembled cis SNARE complexes. We ran a number of controls to validate this conclusion. Note that when added in the same sequence “toxin → zinc chelator → inducer” but without PTP1B, TeTx could not cleave synaptobrevin, because it was locked in cis complexes, or block exocytosis (Fig. 6A, top, and Ref. 14).
FIGURE 6.
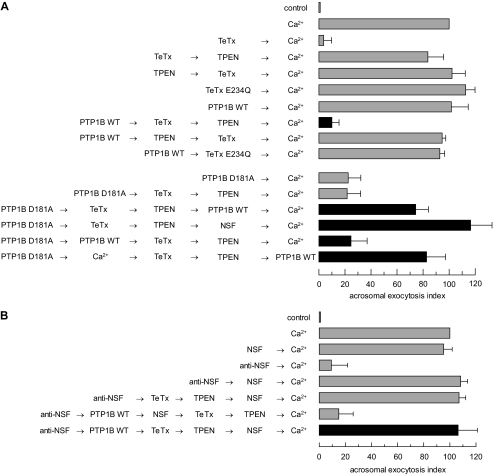
PTP1B elicits cis SNARE complex disassembly through a mechanism that requires NSF. A: top, to investigate whether PTP1B is involved in cis SNARE complex disassembly, which renders synaptobrevin sensitive to TeTx cleavage, SLO-permeabilized sperm were incubated with 27 nm wild type (WT) PTP1B for 10 min at 37 °C followed by 100 nm light chain TeTx and a further 10-min incubation at 37 °C. TeTx activity was subsequently inhibited by adding 2.5 μm TPEN. After 10 min, AR was initiated with 0.5 mm CaCl2 and incubated for 10 min at 37 °C (black bar). Bottom, SLO-permeabilized sperm were incubated with 300 nm PTP1B D181A instead of the wild type enzyme. The D181A block was released with 310 nm NSF (PTP1B D181A → TeTx → TPEN → NSF → Ca2+) or 27 nm wild type PTP1B added before (PTP1B D181A → PTP1B WT → TeTx → TPEN → Ca2+) or after (PTP1B D181A → TeTx → TPEN → PTP1B WT → Ca2+) TeTx (black bars). AR was initiated by adding 0.5 mm CaCl2. Alternatively, calcium was added after PTP1B D181A but before the toxin, TPEN, and wild type PTP1B (PTP1B D181A → Ca2+ → TeTx → TPEN → PTP1B WT, black bar). Sperm were fixed and AR was measured by FITC-PSA binding as described under “Experimental Procedures.” Controls (gray bars) included: AR inhibition by TeTx (TeTx → Ca2+) but not by its inactive mutant TeTx E234Q (43) (TeTx E234Q → Ca2+); impairing of toxin cleavage by TPEN when added before (TPEN → TeTx → Ca2+) or after (TeTx → TPEN → Ca2+) TeTx; unperturbed AR with wild type PTP1B (PTP1B WT → Ca2+); lack of monomeric synatobrevin cleavage by TeTx, despite cis SNARE complex disassembly by PTP1B WT, when added in the presence of TPEN (PTP1B WT → TPEN → TeTx → Ca2+) or when the inactive mutant replaced the wild type toxin (PTP1B WT → TeTx E234Q → Ca2+); and inhibition of the AR by PTP1B D181A in the absence (PTP1B D181A → Ca2+) or presence (PTP1B D181A → TeTx → TPEN → Ca2+) of TeTx. B, to demonstrate that PTP1B relies on NSF to disassemble cis SNARE complexes, SLO-permeabilized sperm were incubated as in A, except that an anti-NSF antibody (1:300) was added before TeTx followed by 310 nm recombinant NSF to rescue the antibody block (anti-NSF → PTP1B WT → TeTx → TPEN → NSF → Ca2+, black bar). AR was triggered as in A. Sperm were fixed and AR was measured by FITC-PSA binding as described under “Experimental Procedures.” Several controls were run (gray bars): unaffected exocytosis in the presence of NSF alone (NSF → Ca2+); inhibition of the AR by the anti-NSF antibody (anti-NSF → Ca2+) and rescue by NSF (anti-NSF → NSF → Ca2+); and lack of effect of TeTx on this rescue (anti-NSF → TeTx → TPEN→NSF→Ca2+). The combination anti-NSF/NSF did not prevent cleavage by TeTx elicited by wild type PTP1B (anti-NSF→PTP1B WT→NSF→TeTx→TPEN→Ca2+). The data were normalized as described under “Experimental Procedures.” Actual percentages of reacted sperm for control and Ca2+ ranged between 11–23 and 20–35%, respectively. The data represent the mean ± S.E. of at least three independent experiments.
We took advantage of the reversibility of the effect of the dead mutant PTP1B D181A on the AR by competition with wild type PTP1B (Fig. 2C) to strengthen the notion that tyrosine dephosphorylation was required for cis SNARE complex disassembly. The sequence of addition PTP1B D181A → TeTx → TPEN → Ca2+ (Fig. 6A, bottom) yields no exocytosis, same as the sequence PTP1B WT → TeTx → TPEN → Ca2+ (Fig. 6A, top). Despite the identical outcome, the mechanisms responsible for the exocytotic block are different in each case. In the first scenario, PTP1B D181A prevented the AR by interfering with the endogenous PTP1B; meanwhile SNAREs were maintained in cis complexes and synaptobrevin was protected from cleavage. In the second, cis complexes were disassembled by recombinant PTP1B WT, synaptobrevin in monomeric configuration was cleaved by TeTx, and therefore the AR was inhibited. This interpretation was substantiated by successful exocytosis achieved when reagents were added in the sequence PTP1B D181A → TeTx → TPEN → PTP1B WT → Ca2+ (Fig. 6A, bottom), indicating that TeTx was unable to cleave its target following incubation with the inactive PTP1B (because synaptobrevin was protected in cis SNARE complexes during the time in which TeTx remained active). Identical results were achieved when NSF replaced wild type PTP1B (PTP1B D181A → TeTx → TPEN → NSF → Ca2+). Wild type PTP1B and NSF rescued exocytosis because they were added after inactivation of TeTx by TPEN. When reagents were supplied in the sequence PTP1B D181A → PTP1B WT → TeTx → TPEN → Ca2+, the inhibition caused by PTP1B D181A was rescued by wild type PTP1B before addition of TeTx and the AR was inhibited (Fig. 6A, bottom). This finding indicates that wild type PTP1B disassembled cis SNARE complexes, because the toxin cut, and was subsequently inactivated, prior to calcium addition, a result that agrees entirely with that depicted in the black bar of the top panel of Fig. 6A. The AR was not prevented by changing the order of addition of reagents so that TeTx was added after calcium and inactivated by TPEN prior to the exocytosis rescue by wild type PTP1B (Fig. 6A, bottom, PTP1B D181A → Ca2+ → TeTx → TPEN → PTP1B WT).
To demonstrate that PTP1B was exerting its effects on the SNARE complex through a mechanism relying on endogenous NSF, we introduced anti-NSF antibodies into permeabilized human sperm before wild type PTP1B and TeTx. Fig. 6B (black bar) shows no inhibition of the exocytic response, which we interpret to mean that synaptobrevin was protected from toxin cleavage. Therefore, our data indicate that PTP1B relies on endogenous NSF to disengage cis SNARE complexes.
DISCUSSION
Protein tyrosine phosphorylation in response to in vitro capacitation is a hallmark of sperm from a number of species (39). Reversible protein tyrosine phosphorylation is also mandatory for the success of the AR. Treatment of human sperm with pharmacological inhibitors of PTPs increases global protein tyrosine phosphorylation and blocks calcium-triggered exocytosis (20). PTPs comprise a large superfamily of related enzymes that play critical roles in the control of a wide array of signaling pathways. The prototypic member of this family is PTP1B. This enzyme has been implicated in cell adhesion, oncogenesis, and metabolic pathways related to diabetes and obesity (17, 40).
PTP1B may play diverse roles in signal transduction because it recognizes distinct substrates in different cellular contexts. This article focuses on the identification of a new function for PTP1B, that of controlling regulated exocytosis. We have extended our initial studies, which were conducted with non-selective PTP inhibitors (20), to investigate how the AR relies on PTP1B activity. We explored the requirement for this phosphatase in sperm exocytosis by using three reagents that target PTP1B. These reagents impaired the function of PTP1B, and therefore prevented the AR, through the following three mechanisms. (i) an anti-PTP1B antibody inhibited the AR because, presumably, it sequesters the endogenous PTP1B, hindering the interaction with its physiological substrates (41). (ii) A photosensitive inhibitor blocked the AR because it binds covalently to the catalytic site of the enzyme, inactivating it (27). (iii) A substrate-trapping mutant prevented the AR through the formation of stable complexes with phosphotyrosine residues that are critical to signaling events, because the trapping impairs the normal interactions between PTP1B substrates and their downstream targets (42). In all cases addition of exogenous PTP1B restored exocytosis (Fig. 2, C and D). We observed an increase in global tyrosine phosphorylation when we introduced PTP1B D181A into permeabilized human sperm, indicating that the mutant PTP was protecting cellular substrates against dephosphorylation by endogenous PTPs. Addition of wild type PTP1B antagonized the effect of the substrate trap and returned protein tyrosine phosphorylation to control levels (Fig. 2A). Taken together, these data point to an essential role of PTP1B in sperm exocytosis.
The cascade of events necessary to bring sperm exocytosis to completion depends on a carefully orchestrated series of steps that include tethering (by Rab3A and effectors) and docking (by neurotoxin-sensitive loose trans SNARE complexes stabilized by complexin) of the acrosome to the plasma membrane, priming of the fusion machinery (by α-SNAP/NSF), mobilization of intravesicular Ca2+ (through inositol 1,4,5-trisphosphate-sensitive channels), activation of synaptotagmin, displacement of complexin, and ultimately, bilayer mixing and fusion (16). Where in this sequence is PTP1B required? We found that PTP1B dephosphorylates proteins that function downstream of Rab3A and prior to intra-acrosomal calcium release (Fig. 3). By applying a complementary strategy, we reduced this window to a step after Rab3A but before or concomitant with NSF (Fig. 1). In white blood cells, the homotypic fusion activity of NSF is repressed until PTP-MEG2 dephosphorylates it (8). Likewise in human sperm the activity of NSF is repressed until an inducer triggers exocytosis (14). We asked if related mechanisms might operate in these two fusion scenarios and if NSF might be the downstream target of the activity of PTP1B during the exocytotic cascade. If this were the case, then PTP1B blockers would prevent the AR because they would maintain sperm NSF in a tyrosine-phosphorylated, and therefore inactive state. In fact, NSF is tyrosine phosphorylated in resting sperm (Fig. 4C). The sole addition of nonphosphorylated NSF rescued exocytosis impaired by PTP1B inhibitors, supporting the notion that the end point of tyrosine dephosphorylation by PTP1B is the activation of NSF (Fig. 4D). Recombinant, tyrosine-phosphorylated NSF serves as a substrate for PTP1B in vitro (Fig. 4A). The hypothesis that tyrosine dephosphorylation impairs the ability of NSF to dissociate SNARE complexes had been put forth based on the observation that phospho-NSF cannot bind α-SNAP (8). In the present study we provide direct evidence that phospho-NSF fails to disassemble native SNARE complexes (Fig. 5). We are currently investigating whether direct protein-protein interactions between NSF and PTP1B also exist in cells. When overexpressed in COS7 and HEK293 cells PTP1B D181A co-precipitated with a 70-kDa tyrosine-phosphorylated protein of unknown identity (19). It is tempting to speculate that this protein might have been NSF. Interestingly, valosin-containing protein, a homolog of NSF, is tyrosine phosphorylated in Rous sarcoma virus-transformed fibroblasts, B lymphocytes (44), and in sperm during capacitation (45).
In resting sperm, SNARE proteins are assembled in fusion-incompetent cis complexes until a stimulant initiates the exocytotic cascade (14). Once exocytosis begins, α-SNAP/NSF unpair cis SNARE complexes to make monomeric SNAREs available for subsequent trans pairing and fusion. Individual SNAREs can be cleaved by botulinum toxin (BoNT) and TeTx, whereas SNAREs engaged in loose trans complexes are only sensitive to BoNTs (46) and those in cis complexes are resistant to all (35). Toxin sensitivity in secretion assays is a reliable read out to investigate the configuration of SNARE proteins (14, 38) and we used it to show that the cis SNAREs present in resting sperm were disassembled by active PTP1B but not by its mutant D181A (Fig. 6A). Calcium depended upon an active PTP1B to sensitize synaptobrevin to TeTx cleavage; in other words, calcium induced cis SNAREs dissociation only in a context of ongoing tyrosine dephosphorylation. As expected, PTP1B did not accomplish cis SNARE complex dissociation directly, but relied on sperm NSF (Fig. 6B). The most likely explanation for these observations is that recombinant PTP1B dephosphorylates sperm NSF. In summary, we were able to show that in permeabilized human sperm, an exocytic experimental model with naturally occurring membranes containing all factors involved in physiological secretion, PTP1B has a novel and positive role modulating the activity of NSF and indirectly driving cis SNARE complexes disassembly, which ultimately leads to membrane fusion. A model depicting our current thinking on the mechanisms underlying these events is shown in Fig. 7.
FIGURE 7.
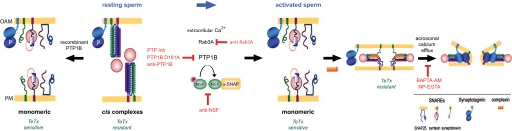
Working model for the biochemical cascade leading to the AR. In resting sperm SNAREs and NSF are inactive, the former engaged in cis complexes and the latter phosphorylated on tyrosine. Upon activation, calcium coming from the extracellular medium enters the cytoplasm and indirectly activates Rab3A, triggering the tethering of the acrosome to the plasma membrane through the assembly of large macromolecular complexes. A reaction taking place during or as a consequence of tethering initiates the activation and/or recruitment of PTP1B to the fusion sites. Next, PTP1B dephosphorylates NSF. This step is inhibited by PTP1B D181A, PTP Inhibitor I, and anti-PTP1B antibodies. Once dephosphorylated, NSF together with α-SNAP, disassembles cis SNARE complexes. Free SNAREs are now able to re-assemble in trans, a process that is facilitated by complexin (36). A local increase in calcium coming from the acrosome through inositol 1,4,5-trisphosphate-sensitive channels activates synaptotagmin and triggers the final steps of membrane fusion, which require SNAREs (presumably in tight trans complexes). The drawings were modified from Ref. 2 and the model from Ref. 36. PM, plasma membrane; OAM, outer acrosomal membrane.
Acknowledgments
We thank M. Furlán and A. Medero for excellent technical assistance, F. Rodriguez for the expression and purification of TeTx E234Q, V. González Polo for the synaptosomal samples and for assistance with the two-dimensional electrophoresis apparatus, Drs. Magadán and Patterson for critical reading of the manuscript, and Drs. Zilly, Fasshauer, Tonks, Stahl, Binz, and Whiteheart for plasmids.
This work was supported by grants from Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina) and Agencia Nacional de Promoción Cientifica y Tecnológica (Argentina).
Footnotes
The abbreviations used are: NSF, N-ethylmaleimide-sensitive factor; AR, acrosome reaction; BAPTA-AM, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester; BoNT/B, botulinum toxin B; FITC-PSA, fluorescein isothiocyanate-coupled Pisum sativum agglutinin; IPTG, isopropyl β-d-thio-galactoside; NP-EGTA-AM, O-nitrophenyl EGTA acetoxymethyl ester; PTP, protein-tyrosine phosphatase; SLO, streptolysin O; TeTx, tetanus toxin; TPEN, N,N,N′,N′-tetrakis(2-pyridymethyl)ethylenediamine; DTT, dithiothreitol; WT, wild type; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; GTPγS, guanosine 5′-O-(thiotriphosphate).
References
- 1.Burgoyne, R. D., and Morgan, A. (2003) Physiol. Rev. 83 581–632 [DOI] [PubMed] [Google Scholar]
- 2.Sollner, T. H. (2003) Mol. Membr. Biol. 20 209–220 [DOI] [PubMed] [Google Scholar]
- 3.Jahn, R., Lang, T., and Sudhof, T. C. (2003) Cell 112 519–533 [DOI] [PubMed] [Google Scholar]
- 4.Jahn, R., and Scheller, R. H. (2006) Nat. Rev. Mol. Cell Biol. 7 631–643 [DOI] [PubMed] [Google Scholar]
- 5.Sutton, R. B., Fasshauer, D., Jahn, R., and Brunger, A. T. (1998) Nature 395 347–353 [DOI] [PubMed] [Google Scholar]
- 6.Zhao, C., Slevin, J. T., and Whiteheart, S. W. (2007) FEBS Lett. 581 2140–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, K. M., Thorngren, N. L., Fratti, R. A., and Wickner, W. T. (2005) EMBO J. 24 1775–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynh, H., Bottini, N., Williams, S., Cherepanov, V., Musumeci, L., Saito, K., Bruckner, S., Vachon, E., Wang, X., Kruger, J., Chow, C. W., Pellecchia, M., Monosov, E., Greer, P. A., Trimble, W., Downey, G. P., and Mustelin, T. (2004) Nat. Cell Biol. 6 831–839 [DOI] [PubMed] [Google Scholar]
- 9.Matsushita, K., Morrell, C. N., Cambien, B., Yang, S. X., Yamakuchi, M., Bao, C., Hara, M. R., Quick, R. A., Cao, W., O'Rourke, B., Lowenstein, J. M., Pevsner, J., Wagner, D. D., and Lowenstein, C. J. (2003) Cell 115 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, Y., Man, H. Y., Sekine-Aizawa, Y., Han, Y., Juluri, K., Luo, H., Cheah, J., Lowenstein, C., Huganir, R. L., and Snyder, S. H. (2005) Neuron 46 533–540 [DOI] [PubMed] [Google Scholar]
- 11.Matveeva, E. A., Whiteheart, S. W., Vanaman, T. C., and Slevin, J. T. (2001) J. Biol. Chem. 276 12174–12181 [DOI] [PubMed] [Google Scholar]
- 12.Liu, Y., Cheng, K., Gong, K., Fu, A. K., and Ip, N. Y. (2006) J. Biol. Chem. 281 9852–9858 [DOI] [PubMed] [Google Scholar]
- 13.Yanagimachi, R. (1994) in Mammalian Fertilization, The Physiology of Reproduction (Knobil, E., and Neill, J. D., eds) pp. 189–317, Raven Press, New York
- 14.De Blas, G. A., Roggero, C. M., Tomes, C. N., and Mayorga, L. S. (2005) PLoS Biol 3 e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomes, C. N. (2006) in Molecular Mechanisms of Exocytosis (Regazzi, R., ed) pp. 117–147, Landes Biosciences/Eurekah.com and Springer Science+Business Media, Austin, TX
- 16.Mayorga, L., Tomes C. N., and Belmonte, S. A. (2007) IUBMB Life 59 1–7 [DOI] [PubMed] [Google Scholar]
- 17.Tonks, N. K. (2006) Nat. Rev. Mol. Cell Biol. 7 833–846 [DOI] [PubMed] [Google Scholar]
- 18.Tonks, N. K., and Neel, B. G. (2001) Curr. Opin. Cell Biol. 13 182–195 [DOI] [PubMed] [Google Scholar]
- 19.Flint, A. J., Tiganis, T., Barford, D., and Tonks, N. K. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomes, C. N., Roggero, C. M., De Blas, G., Saling, P. M., and Mayorga, L. S. (2004) Dev. Biol. 265 399–415 [DOI] [PubMed] [Google Scholar]
- 21.Schievella, A. R., Paige, L. A., Johnson, K. A., Hill, D. E., and Erikson, R. L. (1993) Cell Growth & Differ. 4 239–246 [PubMed] [Google Scholar]
- 22.Yunes, R., Michaut, M., Tomes, C., and Mayorga, L. S. (2000) Biol. Reprod. 62 1084–1089 [DOI] [PubMed] [Google Scholar]
- 23.Mendoza, C., Carreras, A., Moos, J., and Tesarik, J. (1992) J. Reprod. Fertil. 95 755–763 [DOI] [PubMed] [Google Scholar]
- 24.Valdez, S. R., Patterson, S. I., Ezquer, M. E., Torrecilla, M., Lama, M. C., and Seltzer, A. M. (2007) Synapse 61 124–137 [DOI] [PubMed] [Google Scholar]
- 25.Schagger, H., and Von Jagow, G. (1987) Anal. Biochem. 166 368–379 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. (1970) Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- 27.Arabaci, G., Guo, X. C., Beebe, K. D., Coggeshall, K. M., and Pei, D. (1999) J. Am. Chem. Soc. 121 5085–5086 [Google Scholar]
- 28.Seligman, J., Zipser, Y., and Kosower, N. S. (2004) Biol. Reprod. 71 1009–1015 [DOI] [PubMed] [Google Scholar]
- 29.Yunes, R., Tomes, C., Michaut, M., De Blas, G., Rodriguez F., Regazzi, R., and Mayorga, L. S. (2002) FEBS Lett. 525 126–130 [DOI] [PubMed] [Google Scholar]
- 30.De Blas, G., Michaut, M., Trevino, C. L., Tomes, C. N., Yunes, R., Darszon, A., and Mayorga, L. S. (2002) J. Biol. Chem. 277 49326–49331 [DOI] [PubMed] [Google Scholar]
- 31.Tomes, C. N., De Blas, G. A., Michaut, M. A., Farre, E. V., Cherhitin, O., Visconti, P. E., and Mayorga, L. S. (2005) Mol. Hum. Reprod. 11 43–51 [DOI] [PubMed] [Google Scholar]
- 32.Michaut, M., Tomes, C. N., De Blas, G., Yunes, R., and Mayorga, L. S. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 9996–10001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otto, H., Hanson, P. I., and Jahn, R. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6197–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiavo, G., Matteoli, M., and Montecucco, C. (2000) Physiol. Rev. 80 717–766 [DOI] [PubMed] [Google Scholar]
- 35.Hayashi, T., McMahon, H., Yamasaki, S., Binz, T., Hata, Y., Sudhof, T. C., and Niemann, H. (1994) EMBO J. 13 5051–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roggero, C. M., De Blas, G. A., Dai, H., Tomes, C. N., Rizo, J., and Mayorga, L. S. (2007) J. Biol. Chem. 282 26335–26343 [DOI] [PubMed] [Google Scholar]
- 37.Tomes, C. N., Michaut, M., De Blas, G., Visconti, P., Matti, U., and Mayorga, L. S. (2002) Dev. Biol. 243 326–338 [DOI] [PubMed] [Google Scholar]
- 38.Giraudo, C. G., Eng, W. S., Melia, T. J., and Rothman, J. E. (2006) Science 313 676–680 [DOI] [PubMed] [Google Scholar]
- 39.Visconti, P. E., Westbrook, V. A., Chertihin, O., Demarco, I., Sleight, S., and Diekman, A. B. (2002) J. Reprod. Immunol. 53 133–150 [DOI] [PubMed] [Google Scholar]
- 40.Simoncic, P. D., McGlade, C. J., and Tremblay, M. L. (2006) Can. J. Physiol. Pharmacol. 84 667–675 [DOI] [PubMed] [Google Scholar]
- 41.Ahmad, F., Li, P. M., Meyerovitch, J., and Goldstein, B. J. (1995) J. Biol. Chem. 270 20503–20508 [DOI] [PubMed] [Google Scholar]
- 42.Tonks, N. K., and Neel, B. G. (1996) Cell 87 365–368 [DOI] [PubMed] [Google Scholar]
- 43.Yamasaki, S., Hu, Y., Binz, T., Kalkuhl, A., Kurazono, H., Tamura, T., Jahn, R., Kandel, E., and Niemann, H. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 4688–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulte, R. J., Campbell, M. A., Fischer, W. H., and Sefton, B. M. (1994) J. Immunol. 153 5465–5472 [PubMed] [Google Scholar]
- 45.Ficarro, S., Chertihin, O., Westbrook, V. A., White, F., Jayes, F., Kalab, P., Marto, J. A., Shabanowitz, J., Herr, J. C., Hunt, D. F., and Visconti, P. E. (2003) J. Biol. Chem. 278 11579–11589 [DOI] [PubMed] [Google Scholar]
- 46.Hua, S. Y., and Charlton, M. P. (1999) Nat. Neurosci. 2 1078–1083 [DOI] [PubMed] [Google Scholar]