Abstract
Aminopeptidase A (APA; EC 3.4.11.7) is a membrane-bound zinc metalloprotease cleaving in the brain the N-terminal aspartyl residue of angiotensin II to generate angiotensin III, which exerts a tonic stimulatory effect on the central control of blood pressure in hypertensive animals. We docked the specific APA inhibitor, glutamate phosphonate, in the three-dimensional model of the mouse APA ectodomain in the presence of Ca2+. In the S1 subsite of this model, the Ca2+ atom was coordinated with Asp-213, Asp-218,y and Glu-215 and three water molecules, one of which formed a hydrogen bond with the carboxylate side chain of the inhibitor. We report here that the carboxylate side chain of glutamate phosphonate also formed a hydrogen bond with the alcohol side chain of Thr-348. Mutagenic replacement of Thr-348 with an aspartate, tyrosine, or serine residue led to a modification of the hydrolysis velocity, with no change in the affinity of the recombinant enzymes for the substrate GluNA, either in the absence or presence of Ca2+. In the absence of Ca2+, the mutations modified the substrate specificity of APA, which was nevertheless restored by the addition of Ca2+. An analysis of three-dimensional models of the corresponding Thr-348 mutants revealed that the interaction between this residue and the inhibitor was abolished or disturbed, leading to a change in the position of the inhibitor in the active site. These findings demonstrate a key role of Thr-348 in substrate specificity of APA for N-terminal acidic amino acids by insuring the optimal positioning of the substrate during catalysis.
Aminopeptidase A (APA; EC 3.4.11.7)3 is a 160-kDa homodimeric type II membrane-bound monozinc aminopeptidase also activated by Ca2+ (1, 2). It specifically cleaves the N-terminal glutamyl or aspartyl residue from peptide substrates, such as angiotensin II and cholecystokinin-8, in vitro (3, 4). APA is strongly expressed in many tissues, including the brush border of intestinal and renal epithelial cells, and in the vascular endothelium (5). APA has also been detected in several brain nuclei involved in controlling body fluid homeostasis and cardiovascular function (6, 7). Studies with specific and selective APA inhibitors (8) have shown that APA cleaved the N-terminal aspartyl residue of brain angiotensin II to generate angiotensin III in vivo (9) and that brain angiotensin III exerts a tonic stimulatory action on the control of blood pressure in hypertensive animals (10, 11). Thus, the inhibition of brain APA with specific and selective inhibitors normalizes blood pressure in conscious spontaneously hypertensive rats or hypertensive deoxycorticosterone acetate salt rats (10–12), suggesting that brain APA constitutes an interesting candidate target for the treatment of certain forms of hypertension (13–15). This justifies the development of potent and selective APA inhibitors crossing the blood-brain barrier after oral administration for use as centrally acting antihypertensive agents. To achieve this goal, the study of the organization of the APA active site has been pursued.
Using the crystal structure of leukotriene-A4 hydrolase (EC 3.3.2.6) (16) as a template and the functional data collected from site-directed mutagenesis studies on APA (17–22), we previously built a three-dimensional model of the mouse APA ectodomain from residues 79 to 559 (23), including the active site of the enzyme. In this model, the zinc atom is coordinated by the two histidine residues (His-385 and His-389) of the consensus sequence HEXXH, Glu-408 and a water molecule, as previously shown (21, 22). We then docked a potent and selective APA inhibitor, 4-amino-4-phosphobutyric acid (glutamate phosphonate) into the three-dimensional model of the APA active site. This inhibitor behaves as a transition state pseudoanalog, in which the replacement of the substrate scissile amide bond with a phosphonic acid group mimics the tetrahedral transition state (24). Analysis of the APA three-dimensional model complexed with glutamate phosphonate showed that Tyr-471 interacted with the inhibitor, providing support for previous demonstrations that this residue is essential for stabilizing the transition state of catalysis in APA (19). This model also provided evidence of an interaction between the N-terminal amine of glutamate phosphonate and the Glu-352 and Glu-215 residues of APA, which are responsible for the exopeptidase specificity of this enzyme (20, 25). We then introduced Ca2+ into this three-dimensional model. We identified the Ca2+ ligands in the S1 subsite: Asp-213 (corresponding to Asp-221 in human APA (26)) and Asp-218 (27).
We also visualized another residue in the S1 subsite, Thr-348, which interacts with the side chain of the inhibitor. We characterized here the role of Thr-348 in substrate/inhibitor binding and substrate specificity of APA for N-terminal acidic amino acid residues further, through a combination of molecular modeling and site-directed mutagenesis studies. We replaced Thr-348 by a serine, tyrosine, or aspartate residue. We checked that the mutated enzymes displayed similar processing and had a similar subcellular distribution to wild type APA. We then biochemically and kinetically characterized the purified recombinant wild type and mutated enzymes and determined their sensitivity to Ca2+ and various inhibitors. We also evaluated the influence of the mutations on APA substrate specificity. The effects of the mutations on the three-dimensional model of APA were followed in parallel.
EXPERIMENTAL PROCEDURES
Materials
Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs Inc. and were used according to the manufacturer's instructions. The Expand high fidelity Taq polymerase PCR system was purchased from Roche Applied Science. The liposomal transfection reagent, Lipofectamine 2000, the pcDNA 3.1-His vector, and the monoclonal anti-Xpress antibody were purchased from Invitrogen. The monoclonal anti-α-tubulin antibody and the horseradish peroxidase-conjugated goat anti-mouse antibody was purchased from Sigma-Aldrich. Immobilized cobalt affinity columns (Talon) were obtained from Clontech (Heidelberg, Germany). The synthetic substrate, α-l-glutamyl-β-naphthylamide (GluNA), was purchased from Bachem (Bunderdorf, Switzerland).
Methods
Modeling of the Mutated APA + Ca2+ System—The protein + ligand + Zn2+ + Ca2+ system was investigated as previously described (27); we checked the stability of the system by carrying out several rounds of energy minimization followed by short molecular dynamics runs, using a molecular mechanics program and simulation protocol similar to those described in a previous study (23). The refined whole model of APA + glutamate phosphonate + Zn2+ + Ca2+ + water obtained in this way was used for all of the subsequent calculations. The mutants were obtained by changing only the side chains of the amino acid residue 348 and subjecting the corresponding structures to a new cycle of refinement.
Cloning and Site-directed Mutagenesis—The mouse cDNA encoding APA was used as a template for the generation of mutants by PCR-based site-directed mutagenesis, as previously described (28). Two overlapping regions of the cDNA were amplified separately, using two flanking oligonucleotides: A (5′-TTAATACGACTCACTATAGGGA-3′; corresponding to nucleotides 862–883) as a forward primer and B (5′-GAATCCTAAGATAGAGGCCCGGAG-3′; corresponding to nucleotides 3215–3238) as a reverse primer, together with two overlapping oligonucleotides corresponding to nucleotides 2045–2065 and containing the mutated residue at the appropriate position (C1D1 for Ala-348, C2D2 for Asp-348, C3D3 for Ser-348, and C4D4 for Tyr-348). The forward primers were: C1, 5′-GATTTTGGCGCCGGCGCCATG-3′; C2, 5′-GATTTTGGCGACGGCGCCATG-3′; C3, 5′-GATTTTGGCTCAGGCGCCATG-3′; and C4, 5′-GATTTTGGCTACGGCGCCATG-3′. The reverse primers were: D1, 5′-CATGGCGCCGGCGCCAAAATC-3′; D2, 5′-CATGGCGCCGTCGCCAAAATC-3′; D3, 5′-CATGGCGCCTGAGCCAAAATC-3′; and D4, 5′-CATGGCGCCGTAGCCAAAATC-3′.
The underlined bases encode the new amino acid residue and replace the naturally occurring codon at position 348. Nucleotide numbering is as for the mouse APA sequence (29) deposited in the GenBank™/EBI data base (accession number M29961). The products of the first two rounds of amplification (A-D1–4 and B-C1–4) were used as the template for a second PCR with the two flanking oligonucleotides, A and B. For all PCRs, high fidelity Taq polymerase (Roche Applied Science) (1 unit) was used (25 cycles: 94 °C for 30 s, 54 °C for 45 s, and 72 °C for 2 min). The final 2376-bp PCR product was digested with HindIII and EcoRV, and the resulting 1505-bp HindIII-EcoRV fragment containing the mutation was used to replace the corresponding nonmutated region (HindIII-EcoRV) of the full-length APA cDNA. The presence of the mutation and the absence of nonspecific mutations were confirmed by automated sequencing on an Applied Biosystems 377 DNA sequencer with dye deoxyterminator chemistry.
Cell Culture, Establishment of Stable CHO-K1 Cell Lines Producing Wild type and Mutated His-APAs, and Purification of Recombinant His-APAs—CHO-K1 (American Type Culture Collection, Manassas, VA) cells were maintained in Ham's F-12 medium supplemented with 7% fetal calf serum, 0.5 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen). The cells were transfected with 1 μg of plasmid containing the polyhistidine-tagged wild type or a polyhistidine-tagged mutated APA cDNA, using Lipofectamine 2000 (Invitrogen), and stable cell lines producing the polyhistidine-tagged wild type and mutated His-APAs were established as previously described (18). Stably transfected CHO cells were harvested, and a crude membrane preparation was obtained, as previously described (18). Wild type and mutated His-APAs were purified from the solubilized crude membrane preparation by metal affinity chromatography with a metal chelate resin column (Talon Co2+), as previously described (18). The purity of the final preparation was assessed by SDS-PAGE in 8% polyacrylamide gels, as described by Laemmli (30). The proteins were stained with Coomassie Brilliant Blue R-250. We routinely obtain a purified protein >80% pure with an overall yield of about 14% and a purification factor of 10. Protein concentrations were determined with the BCA protein assay kit (Pierce), using bovine serum albumin (BSA) as the standard.
Western Blotting—Purified wild type and mutated His-APAs were resolved by 8% SDS-PAGE, and the proteins were transferred to a polyvinylidene difluoride membrane in 25 mm Tris/glycine buffer, pH 8.3, supplemented with 20% (V/V) methanol. His-tagged recombinant proteins were detected with the anti-Xpress antibody (dilution 1:5000), and loading control was done with the anti-α-tubulin antibody (dilution 1: 4000). The resulting immune complex was then detected with the ECL Plus Western blotting detection system (GE Healthcare).
Immunofluorescence Analysis of Stably Transfected CHO Cells—CHO cells producing wild type and mutated His-APAs were dispensed (25,000 cells) onto 14-mm-diameter coverslips. The cells were cultured for 24 h in Ham's F-12 medium in a humidified atmosphere of 5% CO2, 95% air. They were then fixed and permeabilized by incubation for 5 min in 100% ice-cold methanol. The cells were rinsed three times in 0.1 m PBS, pH 7.4, and then saturated by incubation with 5% BSA for 30 min at room temperature. They were then incubated with a 1:1000 dilution of rabbit polyclonal anti-(rat APA) serum (31) in 0.5% BSA in PBS for 2 h at room temperature. The coverslips were washed three times with cold PBS and then incubated with a 1:1000 dilution of cyanin 3-conjugated polyclonal anti-rabbit antibody in 0.5% BSA in PBS for 2 h at room temperature. The coverslips were washed three times with PBS and once with water and mounted in Mowiol. The cells were examined with a Leica TCS SP II (Leica Microsystems, Heidelberg, Germany) confocal laser scanning microscope equipped with an argon/krypton laser and configured with a Leica DM IRBE inverted microscope. Cyanin 3 fluorescence was detected after 100% excitation at 568 nm. The images (1024 × 1024 pixels) were obtained with a 63× magnification oil-immersion objective. Each image corresponded to a cross-section of the cell.
Enzyme Assay—Enzyme assays were carried out for wild type and mutated His-APA in 50 mm Tris-HCl buffer, pH 7.4, with or without 4 mm CaCl2, by monitoring the rate of hydrolysis of GluNA, as previously described (8). The sensitivity to Ca2+ of purified wild type and mutated His-APAs was determined by incubating 0.5 mm GluNA with 0 or 4 mm CaCl2 in a final volume of 100 ml of 50 mm Tris-HCl buffer, pH 7.4. All of the assays were performed in black 96-well plates (solid black 96-well flat-bottomed nontreated plates with no lid; Corning Costar). All of the enzymatic studies were performed such that substrate hydrolysis rates were maintained below 10% (initial rate conditions). The progress curves were monitored by following the increase in fluorescence at 460 nm (lex = 330 nm), induced by the release of the β-naphthylamine fluorogenic part of the substrate by APA. Fluorescence signals were monitored by counting photons with a spectrophotometer (Fusion™ universal microplate analyzer; Packard) equipped with a temperature control device and a plate shaker. The kinetic parameters (Kmand kcat) were determined from Michaelis-Menten plots, using Enzfitter software (Biosoft™), with a final concentration of substrate (GluNA) from 0.005 to 2 mm. For evaluation of the substrate specificity of APA in the absence or presence of Ca2+, the rate of hydrolysis of two additional substrates, LeuNA and LysNA, was determined under the same experimental conditions as described for GluNA. The sensitivity of wild type and mutated His-APAs to inhibition by glutamate thiol (GluSH), lysine thiol (LysSH), and methionine thiol (MetSH) was determined by establishing dose-dependent inhibition curves for a final GluNA concentration of 0.5 mm, in the presence of 0 or 4 mm CaCl2. Because these compounds are linear competitive inhibitors, their Ki were calculated from the formula Ki = IC50/(1+ [GluNA]/Km).
Statistical Analysis—Statistical comparisons were carried out with Student's unpaired t test. Differences were considered significant if p was less than 0.05.
RESULTS
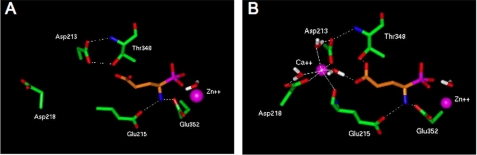
Modeling of APA Mutants in the Absence of Ca2+ and in the Presence of the APA Inhibitor Glutamate Phosphonate—The three-dimensional model of wild type APA complexed with glutamate phosphonate obtained in the absence of Ca2+ showed the presence of a hydrophilic pocket containing two aspartate residues (Asp-213 and Asp-218) and a threonine residue (Thr-348), corresponding to the S1 subsite (Fig. 1A) (27). The acidic side chain of the inhibitor is pointing toward this pocket and the Cα amine of Thr-348 is engaged in one hydrogen bond with the acidic side chain of Asp-213. Moreover, the Thr-348 alcohol side chain is engaged in two balanced hydrogen bonds, one with the carboxylate side chain of Asp-213 and the other with the carboxylate side chain of glutamate phosphonate. In the absence of Ca2+, this interaction is skewed toward the carboxylate side chain of Asp-213.
FIGURE 1.
Organization of the S1 subsite of APA. The three-dimensional model of the APA active site complexed with the inhibitor glutamate phosphonate (in orange) without (A) or with calcium (B) revealed a hydrophilic pocket corresponding to the S1 subsite (amino acids in green).
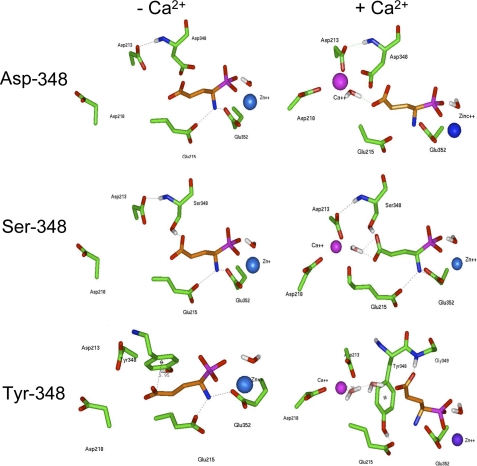
We investigated the role of Thr-348, by replacing this residue with a serine, aspartate, or tyrosine residue and constructing the corresponding three-dimensional active site models. The three-dimensional active site model of the Ser-348 mutated APA (Fig. 2) is similar to that of wild type APA, as described above, except that the Ser-348 hydroxyl group is hydrogen-bonded only with the carboxylate side chain of glutamate phosphonate. In the three-dimensional active site model of the Asp-348 mutated APA (Fig. 2), only the hydrogen bond between the Asp-348 backbone and the Asp-213 side chain is maintained, because the carboxylate side chain of Asp-348 now interacts with a water molecule. Greater disturbance of the binding pocket is observed in the three-dimensional model of Tyr-348 mutated APA (Fig. 2), because of the presence of an aromatic group involved in a pi-anion interaction with the carboxyl end of the inhibitor (32). In this case, the position of the inhibitor differs from that in the wild type.
FIGURE 2.
Molecular modeling of the S1 subsite of mutated APAs. Three-dimensional model of Asp-348, Ser-348, and Tyr-348 mutated APAs complexed with the inhibitor glutamate phosphonate (in orange) without or with calcium.
Modeling of APA in the Presence of Ca2+ and in the Presence of the APA Inhibitor Glutamate Phosphonate—In the three-dimensional model of wild type or Ser-348 mutated APAs in the presence of Ca2+ and glutamate phosphonate (Figs. 1B and 2), the alcohol side chain of Thr- or Ser-348 in the S1 subsite establishes a hydrogen bond with the carboxylate side chain of glutamate phosphonate. Moreover, the nitrogen of the Cα amine moiety of residue 348 interacts with Asp-213 (27). The Ca2+ atom is thus connected to glutamate phosphonate through a water molecule. In the three-dimensional model of Asp-348 mutated APA (Fig. 2), the glutamate phosphonate-water-Ca2+ link is maintained, but the carboxylate side chain of Asp-348 displaces the water molecule away from the Ca2+ coordination shell. As in the three-dimensional model of the Tyr-348 mutated APA obtained in the absence of Ca2+, in the presence of Ca2+ (Fig. 2), the phenol ring of Tyr-348 strongly modifies the binding pocket and, consequently, the position of the inhibitor; Tyr-348 establishes a hydrogen bond with the carboxylate side chain of Glu-215, whereas a new hydrogen bond is created between the Gly-349 backbone and glutamate phosphonate, which is now held away from the Ca2+ atom through a network of two water molecules.
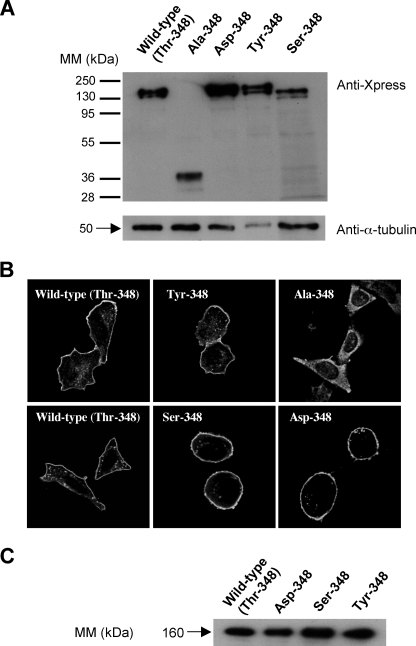
Site-directed Mutagenesis, Expression, and Purification of Recombinant Wild type and Mutant His-APAs—We investigated the role of Thr-348 in substrate/inhibitor binding and substrate specificity by replacing this residue with an alanine, serine, aspartate, or tyrosine residue by site-directed mutagenesis. Wild type and mutated His-APAs were then stably expressed, and the recombinant enzymes were purified by metal affinity chromatography, as previously described (18). We first checked that the mutations did not affect the production and processing of the recombinant proteins. For this purpose, we analyzed by Western blot wild type and mutated His-APAs transitory transfected in CHO cells. We showed that all recombinant proteins, except the Ala-348 mutant, displayed a major mature 160-kDa form and a minor immature 140-kDa form. The Ala-348 mutant displayed a truncated form of 40-kDa (Fig. 3A), probably reflecting the cleavage of this misfolded mutated protein. In an additional set of experiments (data not shown) performed on CHO cells stably expressing the Ala-348 mutant, we also detected the presence of a truncated form of 40-kDa, but only when cells were treated with proteasome inhibitor, MG132 (16 μm, cell treatment for 16 h), suggesting that the mutation induces the cleavage of a nonmature unglycosylated form of APA rapidly degraded by the proteasome machinery.
FIGURE 3.
Expression of histidine-tagged wild type and mutated mouse recombinant APAs. A, CHO cells transitory transfected with wild type or mutated His-APA constructs were analyzed by SDS-PAGE 8% and Western blotting with a mouse anti-X-press antibody. The membrane was then stripped and reprobed with a mouse anti-α-tubulin. B, transfected CHO cells stably expressing wild type and mutated His-APA constructs were fixed and immunolabeled with a rabbit polyclonal anti-(rat-APA) serum, which was detected with a cyanin 3-conjugated anti-rabbit secondary antibody. Immunofluorescence was observed by confocal microscopy. The bar indicates 20 μm. C, crude membrane preparations were solubilized and subjected to metal affinity chromatography, as previously described (18). Material eluted (0.25 μg of protein) from the column was analyzed by SDS-PAGE 7.5% and Western blotting with a mouse anti-X-press antibody. Immune complexes were detected by enhanced chemiluminescence. MM, molecular mass.
Moreover, we investigated the subcellular distribution of wild type and mutant His-APAs in stably transfected CHO cells by immunofluorescence staining with a rabbit polyclonal anti-rat APA antibody and a cyanin-3-conjugated anti-rabbit secondary antibody. Confocal microscopy analysis of CHO cells stably expressing wild type and mutated His-APA constructs showed that APA was located at the plasma membrane, except for the Ala-348 mutant APA, which remained trapped in an intracellular network possibly corresponding to the endoplasmic reticulum (Fig. 3B). We therefore did not carry out enzymatic characterization of this mutant. Finally, Western blot analysis of purified wild type and mutated His-APAs stably expressed in CHO cells showed that all proteins displayed a mature 160-kDa form (Fig. 3C).
Enzymatic Activity of Purified Wild type and Mutated His-APAs—Because APA activity is enhanced by Ca2+, we evaluated the enzymatic activity of purified wild type and mutated APAs in the presence or absence of 4 mm Ca2+, using GluNA as a substrate (Table 1). In the absence of Ca2+, the replacement of Thr-348 by a serine residue led to a significant increase in substrate hydrolysis, by a factor of 1.5 compared with wild type His-APA. By contrast, the level of enzymatic activity detected for the Asp-348 and Tyr-348 mutants was significantly lower, corresponding to 40 and 13% of wild type His-APA activity levels, respectively. In the presence of 4 mm Ca2+, the enzymatic activity of Ser-348 remained significantly (2 times) higher than that of the wild type His-APA, whereas those of Asp-348 and Tyr-348 APA were significantly lower, corresponding to 11 and 50% of wild type His-APA activity, respectively. The presence of 4 mm Ca2+ enhanced the enzymatic activity of wild type, Ser-348, Asp-348, or Tyr-348 His-APA by factors of 8, 10, 2, and 30, respectively (Table 1).
TABLE 1.
Enzymatic activity of purified recombinant wild type and mutated His-APAs Shown are the enzymatic activity (μmol of GluNA hydrolyzed/min/mg of protein) means ± the standard error from three to six separate experiments with duplicate determinations.
| Recombinant His-APAs | No Ca2+ | 4 mm Ca2+ | Factor of stimulation |
|---|---|---|---|
| Wild type | 1.98 ± 0.12 | 14.67 ± 0.42d | 8 |
| Ser-348 | 3.05 ± 0.15a | 31.54 ± 1.33a,c | 10 |
| Asp-348 | 0.8 ± 0.05a | 1.6 ± 0.14a,d | 2 |
| Tyr-348 | 0.25 ± 0.007b | 7.4 ± 0.13a,d | 30 |
p < 0.05 (corresponding mutated His-APA vs. corresponding wild-type APA)
p < 0.01 (corresponding mutated His-APA vs. corresponding wild-type APA)
Not significant when compared to the corresponding recombinant enzyme activity in the absence of Ca2+
p < 0.05
Kinetic Parameters for Purified Recombinant Wild type and Mutated His-APAs—The enzymatic activities of purified recombinant His-APAs were analyzed by determining the catalytic constants (Km and kcat) in the absence or presence of 4 mm Ca2+, using GluNA as a substrate. The results are summarized in Table 2. In the absence of Ca2+, the replacement of Thr-348 with a serine residue doubled hydrolysis efficiency. By contrast, the replacement of Thr-348 with an aspartate or a tyrosine residue led to decreases in kcat/Km by factors of 2 and 13, respectively. These effects were due to changes in hydrolysis velocity, because the affinities of the mutant His-APAs for GluNA were similar to that of the wild type APA. Indeed, in the absence of Ca2+, the kcat value of Ser-348 APA was twice that of wild type APA, whereas the kcat values of Asp-348 and Tyr-348 APA were one-third and one-tenth, respectively, that of wild type APA. Similarly, in the presence of Ca2+, the replacement of Thr-348 with a serine residue increased the hydrolysis efficiency by a factor of 3. By contrast, the replacement of Thr-348 with an aspartate or a tyrosine residue decreased kcat/Km by factors of 9 and 2, respectively. This was due to changes in hydrolysis velocity, because no significant change in Km value was observed, regardless of the mutant analyzed. Indeed, in the presence of Ca2+, the kcat value of Ser-348 APA was three times higher than that of wild type, whereas those of Asp-348 and Tyr-348 APA were lower than that of wild type APA, by factors of 6 and 2, respectively. In the absence or presence of Ca2+, each mutation induced changes in substrate hydrolysis velocity but not in the affinity of the enzyme for the substrate. Ca2+ increased hydrolysis efficiency by factors of 7 for wild type, 12 for Ser-348, 2 for Asp-348, and 40 for Tyr-348 APA.
TABLE 2.
Kinetic parameters for wild type and mutated APAs Km and kcat values mean ± S.E. from three to nine separate experiments with duplicate determinations.
|
Recombinant APAs
|
No Ca2+
|
4 mm Ca2+
|
||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | s–1 | mm–1·s–1 | μm | s–1 | mm–1·s–1 | |
| Wild type | 1481 ± 60 | 292 ± 10 | 197 | 198 ± 19a | 256 ± 5 | 1319 |
| Ser-348 | 2034 ± 391a | 640 ± 121b | 314a | 159 ± 17a | 575 ± 25b | 3616b |
| Asp-348 | 1136 ± 48a | 92 ± 3c | 81b | 258 ± 25a | 40 ± 2c | 155d |
| Tyr-348 | 1919 ± 102a | 28 ± 4d | 15c | 282 ± 13a | 167 ± 6a | 596b |
Values not significant when compared with the corresponding wild type values
p < 0.05
p < 0.01
p < 0.001
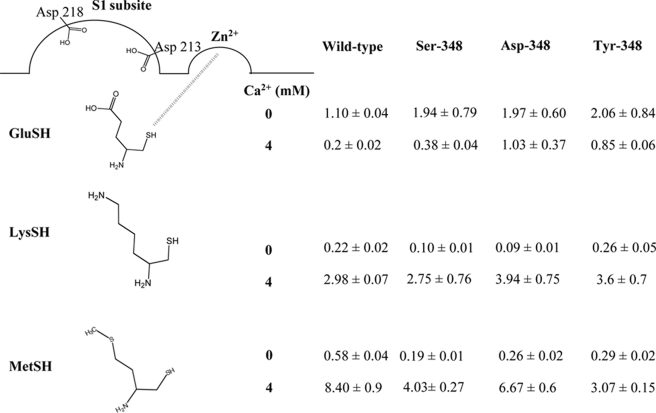
Inhibitory Potencies of Various Classes of Compounds against Purified Recombinant His-APAs—We further characterized the role of Thr-348 by evaluating the inhibitory potencies of various classes of compounds against GluNA (0.5 mm) hydrolysis by wild type and mutated His-APAs in the absence or presence of 4 mm Ca2+. The results are summarized in Table 3.
TABLE 3.
Ki values (μm) for GluSH, LysSH, and MetSH inhibitors with wild type and mutated APAs The Ki values are the means ± S.E. of three to eight experiments with duplicate determinations.
In the absence of Ca2+, the inhibitory potency of GluSH was slightly higher on wild type APA than on Ser-348, Asp-348, and Tyr-348 (by a factor 2). By contrast, LysSH was twice as potent an inhibitor of Ser-348 and Asp-348 APA than of wild type APA. However, the inhibitory potencies of LysSH on Tyr-348 and wild type APA were similar in the absence of Ca2+. Moreover, in the absence of Ca2+, MetSH was 2.2 times, 3 times, and 2 times more potent on Asp-348, Ser-348, and Tyr-348, respectively, than on wild type APA.
In the presence of 4 mm Ca2+, GluSH was 2, 5, and 5 times less potent on Ser-348, Asp-348, and Tyr-348, respectively, than on the wild type enzyme. Moreover, LysSH inhibited wild type APA, Ser-348, Asp-348, and Tyr-348 APA with similar potency. Moreover, in the presence of 4 mm Ca2+, MetSH inhibited wild type APA less strongly than Asp-348 (factor 1.2), Ser-348 (factor 2), and Tyr-348 (factor 2.7) APA.
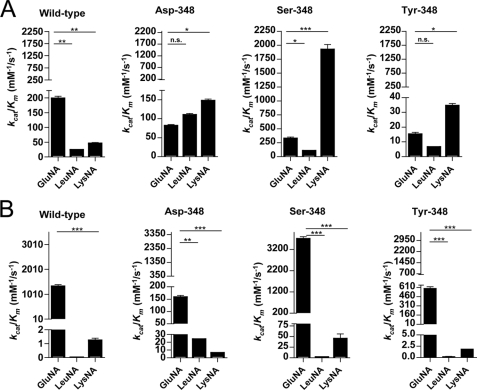
Hydrolysis of Various Substrates by Wild type and Mutated His-APAs—We investigated the effect of the mutations of Thr-348 on APA substrate specificity, by comparing the hydrolysis of different synthetic substrates, GluNA, LysNA, and LeuNA, by purified recombinant wild type and mutated His-APAs (Fig. 4).
FIGURE 4.
Efficiency of hydrolysis of the substrates GluNA, LeuNA and LysNA by the recombinant APAs. Recombinant wild type and mutated APAs hydrolysis efficiencies were evaluated for GluNA, LeuNA and LysNA in the absence (A) or the presence (B) of 4 mm Ca2+. The data shown are the means ± S.E. of three to seven experiments performed in duplicate.
In the absence of Ca2+, wild type APA hydrolyzed GluNA more efficiently than LeuNA and LysNA (by factors of 8 and 4, respectively). Each mutation of residue 348 modified the substrate specificity profile, resulting in the more efficient hydrolysis of LysNA than of GluNA and LeuNA. The most marked difference was that for Ser-348 APA, which hydrolyzed LysNA 20 times more efficiently than LeuNA and 10 times more efficiently than GluNA. An analysis of the kinetic parameters of mutated His-APAs revealed that these effects were due to changes in kcat values (data not shown). The presence of Ca2+ restored the substrate specificity of mutated His-APAs for the acidic substrate.
DISCUSSION
APA belongs to the monozinc aminopeptidase family, in which the presence of Zn2+ is crucial for catalytic activity. A prominent feature of APA is its stimulation by Ca2+, which improves the affinity of APA for N-terminal acidic substrates (2). We previously proposed a putative catalytic mechanism for APA, deduced from site-directed mutagenesis data and from the APA active site three-dimensional model (17, 23, 25), consistent with the catalytic model of thermolysin developed from the results of co-crystallization studies (33). This mechanism is based on nucleophilic attack of the scissile peptide bond by a water molecule (the catalytic water molecule), which must be optimally positioned for polarization. After the formation of the Michaelis complex induced by interactions of the substrate with Glu-215 (25), Glu-352 (20), and the zinc and calcium atoms, the optimal positioning and the polarization of the catalytic water molecule are mediated by its interactions with Glu-386, the zinc atom (21), and Glu-352 (20), resulting in efficient hydrolysis velocity (as shown by kcat).
We recently identified the ligands of Ca2+ in the S1 subsite of APA: Asp-213 and Asp-218 (27). We also demonstrated that these two residues contributed to APA substrate specificity by interacting with the Ca2+, which itself interacts with the carboxylate side chain of the substrate P1 residue via a water molecule. Analysis of the APA active site three-dimensional model, in which was docked glutamate phosphonate in the presence of Ca2+, revealed an interaction between one of the Ca2+ ligands, Asp-213, and Thr-348. Another interaction, between the alcohol side chain of Thr-348 and the carboxylate side chain of glutamate phosphonate, was also observed. We hypothesized that Thr-348 may play a role in substrate binding and therefore may contribute with Ca2+ to the substrate or inhibitor specificity of APA for N-terminal acidic amino acid residues.
We investigated the involvement of Thr-348 in APA substrate binding by replacing this residue, by site-directed mutagenesis, with an alanine, aspartate, tyrosine, or serine residue. We showed, by confocal microscopy analysis of CHO cells stably expressing the wild type or a mutated His-APA construct, that all recombinant APAs, except the mutant Ala-348, were expressed similarly at the plasma membrane, consistent with the correct processing of wild type and mutated enzymes. By contrast, Ala-348 was not expressed at the plasma membrane and remained trapped in an intracellular network possibly corresponding to the endoplasmic reticulum. Western blot analysis of the Ala-348 mutant showed a protein with an apparent molecular mass of 40-kDa, corresponding to a cleaved immature unglycosylated APA, consistent with confocal microscopy analysis and the lack of specific enzymatic activity of the enzyme. Finally, Western blot analysis of purified recombinant His-APAs showed that wild type and mutated His-APAs had an apparent molecular mass of 160-kDa, corresponding to the mature glycosylated APA sorted from the Golgi apparatus, as previously reported (18, 34).
Because Thr-348 interacts with Asp-213, one of the Ca2+ ligands (26, 27), we investigated whether mutations of this residue affected the activation of APA by Ca2+. We observed, as previously described (18), that wild type APA was strongly activated by Ca2+, with enzymatic activity increased by a factor of about 8 when Ca2+ concentration was increased from 0 to 4 mm. The mutagenic replacement of Thr-348 with a serine, aspartate, or tyrosine residue resulted in mutated His-APAs that were also activated by Ca2+. This finding is consistent with the localization of Thr-348 into the APA active site three-dimensional model, showing that it does not directly interact with the Ca2+ ion in the S1 subsite. These results also suggest that Thr-348 mutation has no major effect on the position of Asp-213, as visualized in the three-dimensional models of Thr-348 mutated enzymes. Thus, these mutated enzymes maintained their ability to bind one Ca2+ atom into the S1 subsite and to be activated by Ca2+.
We investigated the functional role of Thr-348 further, by determining the kinetic parameters for wild type and mutated enzymes. The affinity of the mutated enzymes for GluNA was similar to that of the wild type enzyme, in both the absence and presence of Ca2+. However, the replacement of Thr-348 with a serine residue increased the kcat with respect to the value obtained for wild type APA, whereas the replacement of this residue with an aspartate or tyrosine residue decreased kcat. These findings suggested that Thr-348 does not participate in the establishment of the Michaelis complex but is involved in the catalysis. Indeed, analysis of the three-dimensional models of the various mutants showed a slight modification of the position of glutamate phosphonate in the active site, inducing a change in the positioning of Glu-215 and Glu-352 and the distance between Glu-352 and the catalytic water molecule. These changes resulted in modifications of the polarization of this catalytic water molecule, thereby affecting its nucleophilic attack on the substrate scissile peptide bond. This resulted in more efficient hydrolysis of GluNA by Ser-348 and less efficient hydrolysis by Tyr-348 and Asp-348 with respect to wild type.
To confirm the functional role of Thr-348, we evaluated the mode of binding of various inhibitors on the wild type and mutated APAs. The three β-amino thiol inhibitors, GluSH, LysSH, and MetSH, interact with the amine-binding site, with the S1 subsite via their acidic, basic, or neutral side chains, and with the zinc via their chelating thiol group. These inhibitors do not behave as analogs of the transition state and thus do not mimic the interaction of the substrate with the enzyme at the transition state. Consequently, the changes in the Ki values induced by the mutations reflect a change in the affinity of the enzyme (Km) for the substrate rather than in the hydrolysis velocity (kcat). In the absence or presence of Ca2+, very modest changes in the Ki values of the various inhibitors were observed for the mutants as compared with wild type. Moreover, in the absence of Ca2+, the inhibition profile for each mutant His-APA was similar to that of the wild type APA (LysSH > MetSH > GluSH). The introduction of Ca2+ restored the specificity of wild type and mutated His-APAs for the acidic inhibitor (GluSH >> LysSH > MetSH), demonstrating that the mutations did not modify the profile of inhibition of APA. These results were consistent with the kinetic parameters showing a role of Thr-348 in the catalysis but not in the substrate recognition at the Michaelis complex level.
We further characterized the role of Thr-348 in substrate specificity, by evaluating the efficiency of hydrolysis (kcat/Km) for the mutant and wild type His-APAs with various substrates differing in terms of the charge on the side chain interacting with the S1 subsite. In the absence of Ca2+, the substrate hydrolysis profile for wild type APA was such that GluNA was hydrolyzed more efficiently than LysNA, which was hydrolyzed more efficiently than LeuNA (GluNA > LysNA > LeuNA).
This profile was modified by Thr-348 substitutions. The substrate hydrolysis profile was LysNA > GluNA > LeuNA for Ser-348 and Tyr-348 and LysNA > LeuNA > GluNA for Asp-348. An analysis of the kinetic parameters of wild type and mutated APAs revealed that the changes in the substrate hydrolysis profile observed for the mutated APAs were due to changes in kcat but not in Km, suggesting that this residue is involved in substrate specificity of APA. In the mutant Ser-348 not only was the profile of substrate hydrolysis modified, but the level of activity was also affected. Indeed, the efficiency of LysNA hydrolysis by the Ser-348 mutant APA was 10 times higher than the efficiency of GluNA hydrolysis by the wild type APA. Thus, in the absence of Ca2+, the Ser-348 mutant behaves as a powerful basic substrate-specific aminopeptidase.
The presence of Ca2+ restored acidic substrate specificity in mutated enzymes, as for the wild type APA (GluNA > LysNA > LeuNA), suggesting that the Ca2+ ion is the most powerful determinant of APA substrate specificity.
Interestingly, the residue corresponding to Thr-348 in the Escherichia coli aminopeptidase N is a methionine (Met-260) located at the entrance of the S1 subsite, as shown in the crystal structure of this enzyme (35). Met-260 is thought to act as a cushion for substrates with N-terminal residues of different sizes, rendering the size of the S1 subsite suitable for the accommodation of substrates with an alanine or tyrosine residue in the P1 position. Thus, this residue located at the entrance of the S1 subsite appears to play an important role in controlling substrate specificity, both in E. coli aminopeptidase N and in APA, although through different mechanisms.
In conclusion, we demonstrated the involvement of Thr-348 in the substrate specificity of APA, especially in absence of Ca2+. This effect is not due to stronger interaction with the carboxylate side chain of the substrate at the Michaelis complex. Instead, it probably involves an interaction of Thr-348 with the acidic side chain of the substrate just after the formation of the Michaelis complex and before the nucleophilic attack on the scissile peptide bond. Indeed, by interacting with the P1 side chain of the substrate, Thr-348 adjusts the position of the substrate in the APA active site, strengthening, together with Glu-215 and Glu-352, the polarization of the catalytic water molecule to optimize the nucleophilic attack on the scissile peptide bond of acidic substrates.
We show here that the substrate specificity of monozinc aminopeptidases, such as APA, depends not only on interactions occurring in the Michaelis complex between the residues of the S1 subsite of the enzyme and the side chain of the substrate but also on the optimal positioning of the substrate during catalysis, thereby optimizing the hydrolysis of the substrate scissile peptide bond. These new insights into S1 subsite organization and the catalytic mechanism of APA conferring substrate specificity on this enzyme provide key information for pharmacophore design for APA inhibitors, paving the way for the development of a new generation of APA inhibitors for use as central-acting antihypertensive agents.
Acknowledgments
We thank Dr. S. Wilk and Dr. D. Healy for providing the APA antiserum.
This work was supported by a grant from the Fondation pour la Recherche Medicale, by the Institut National de la Santé et de la Recherche Medicale, and by the Centre National de la Recherche Scientifique institutes.
Footnotes
The abbreviations used are: APA, aminopeptidase A; CHO, Chinese hamster ovary; GluSH, glutamate thiol; GluNA, α-l-glutamyl-β-naphtylamide; LysSH, lysine thiol; BSA, bovine serum albumin; PBS, phosphate-buffered saline; MetSH, methionine thiol.
References
- 1.Danielsen, E. M., Noren, O., Sjostrom, H., Ingram, J., and Kenny, A. J. (1980) Biochem. J. 189 591-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glenner, G. G., Mc, M. P., and Folk, J. E. (1962) Nature 194 867. [DOI] [PubMed] [Google Scholar]
- 3.Nagatsu, I., Nagatsu, T., Yamamoto, T., Glenner, G. G., and Mehl, J. W. (1970) Biochim. Biophys. Acta 198 255-270 [DOI] [PubMed] [Google Scholar]
- 4.Wilk, S., and Healy, D. (1993) Adv. Neuroimmunol. 3 195-207 [Google Scholar]
- 5.Lodja, Z., and Gossrau, R. (1980) Histochemistry 67 267-290 [DOI] [PubMed] [Google Scholar]
- 6.de Mota, N., Iturrioz, X., Claperon, C., Bodineau, L., Fassot, C., Roques, B. P., Palkovits, M., and Llorens-Cortes, C. (2008) J. Neurochem. 106 416-428 [DOI] [PubMed] [Google Scholar]
- 7.Zini, S., Masdehors, P., Lenkei, Z., Fournie-Zaluski, M. C., Roques, B. P., Corvol, P., and Llorens-Cortes, C. (1997) Neuroscience 78 1187-1193 [DOI] [PubMed] [Google Scholar]
- 8.Chauvel, E. N., Llorens-Cortès, C., Coric, P., Wilk, S., Roques, B., and Fournié-Zaluski, M. C. (1994) J. Med. Chem. 37 2950-2956 [DOI] [PubMed] [Google Scholar]
- 9.Zini, S., Fournie-Zaluski, M. C., Chauvel, E., Roques, B. P., Corvol, P., and Llorens-Cortes, C. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 11968-11973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fournie-Zaluski, M. C., Fassot, C., Valentin, B., Djordjijevic, D., Reaux-Le Goazigo, A., Corvol, P., Roques, B. P., and Llorens-Cortes, C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7775-7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reaux, A., Fournie-Zaluski, M. C., David, C., Zini, S., Roques, B. P., Corvol, P., and Llorens-Cortes, C. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 13415-13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodineau, L., Frugiere, A., Marc, Y., Inguimbert, N., Fassot, C., Balavoine, F., Roques, B., and Llorens-Cortes, C. (2008) Hypertension 51 1318-1325 [DOI] [PubMed] [Google Scholar]
- 13.Ferreira, A. J., and Raizada, M. K. (2008) Hypertension 51 1273-1274 [DOI] [PubMed] [Google Scholar]
- 14.Iturrioz, X., Reaux-Le Goazigo, A., and Llorens-Cortes, C. (2004) Aminopeptidase Inhibitor as Anti-Hypertensive Drugs, Kluwer Academic/Plenum Publishers
- 15.Reaux, A., Iturrioz, X., Vazeux, G., Fournie-Zaluski, M. C., David, C., Roques, B. P., Corvol, P., and Llorens-Cortes, C. (2000) Biochem. Soc. Trans. 28 435-440 [PubMed] [Google Scholar]
- 16.Thunnissen, M. M., Nordlund, P., and Haeggstrom, J. Z. (2001) Nat. Struct. Biol. 8 131-135 [DOI] [PubMed] [Google Scholar]
- 17.Iturrioz, X., Rozenfeld, R., Michaud, A., Corvol, P., and Llorens-Cortes, C. (2001) Biochemistry 40 14440-14448 [DOI] [PubMed] [Google Scholar]
- 18.Iturrioz, X., Vazeux, G., Celerier, J., Corvol, P., and Llorens-Cortes, C. (2000) Biochemistry 39 3061-3068 [DOI] [PubMed] [Google Scholar]
- 19.Vazeux, G., Iturrioz, X., Corvol, P., and Llorens-Cortes, C. (1997) Biochem. J. 327 883-889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazeux, G., Iturrioz, X., Corvol, P., and Llorens-Cortes, C. (1998) Biochem. J. 334 407-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazeux, G., Wang, J., Corvol, P., and Llorens-Cortes, C. (1996) J. Biol. Chem. 271 9069-9074 [DOI] [PubMed] [Google Scholar]
- 22.Wang, J. Y., and Cooper, M. D. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 1222-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozenfeld, R., Iturrioz, X., Maigret, B., and Llorens-Cortes, C. (2002) J. Biol. Chem. 277 29242-29252 [DOI] [PubMed] [Google Scholar]
- 24.Lejczak, B., De Choszczak, M. P., and Kafarski, P. (1993) J. Enzyme Inhib. 7 97-103 [DOI] [PubMed] [Google Scholar]
- 25.Rozenfeld, R., Iturrioz, X., Okada, M., Maigret, B., and Llorens-Cortes, C. (2003) Biochemistry 42 14785-14793 [DOI] [PubMed] [Google Scholar]
- 26.Goto, Y., Hattori, A., Mizutani, S., and Tsujimoto, M. (2007) J. Biol. Chem. 282 37074-37081 [DOI] [PubMed] [Google Scholar]
- 27.Claperon, C., Rozenfeld, R., Iturrioz, X., Inguimbert, N., Okada, M., Roques, B., Maigret, B., and Llorens-Cortes, C. (2008) Biochem. J. 416 37-46 [DOI] [PubMed] [Google Scholar]
- 28.Herlitze, S., and Koenen, M. (1990) Gene (Amst.) 91 143-147 [DOI] [PubMed] [Google Scholar]
- 29.Wu, Q., Lahti, J. M., Air, G. M., Burrows, P. D., and Cooper, M. D. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 993-997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 31.Song, L., Ye, M., Troyanovskaya, M., Wilk, E., Wilk, S., and Healy, D. P. (1994) Am. J. Physiol. 267 F546-F557 [DOI] [PubMed] [Google Scholar]
- 32.Schottel, B. L., Chifotides, H. T., and Dunbar, K. R. (2008) Chem. Soc. Rev. 37 68-83 [DOI] [PubMed] [Google Scholar]
- 33.Matthews, B. W. (1988) Acc. Chem. Res. 21 333-340 [Google Scholar]
- 34.Rozenfeld, R., Muller, L., El Messari, S., and Llorens-Cortes, C. (2004) J. Biol. Chem. 279 43285-43295 [DOI] [PubMed] [Google Scholar]
- 35.Ito, K., Nakajima, Y., Onohara, Y., Takeo, M., Nakashima, K., Matsubara, F., Ito, T., and Yoshimoto, T. (2006) J. Biol. Chem. 281 33664-33676 [DOI] [PubMed] [Google Scholar]