Abstract
Alternative pre-mRNA splicing is a major gene expression regulatory mechanism in metazoan organisms. Proteins that bind pre-mRNA elements and control assembly of splicing complexes regulate utilization of pre-mRNA alternative splice sites. To understand how signaling pathways impact this mechanism, an RNA interference screen in Drosophila S2 cells was used to identify proteins that regulate TAF1 (TBP-associated factor 1) alternative splicing in response to activation of the ATR (ATM-RAD3-related) signaling pathway by the chemotherapeutic drug camptothecin (CPT). The screen identified 15 proteins that, when knocked down, caused the same change in TAF1 alternative splicing as CPT treatment. However, combined RNA interference and CPT treatment experiments indicated that only a subset of the identified proteins are targets of the CPT-induced signal, suggesting that multiple independent pathways regulate TAF1 alternative splicing. To understand how signals modulate the function of splicing factors, we characterized one of the CPT targets, Tra2 (Transformer-2). CPT was found to down-regulate Tra2 protein levels. CPT-induced Tra2 down-regulation was ATR-dependent and temporally paralleled the change in TAF1 alternative splicing, supporting the conclusion that Tra2 directly regulates TAF1 alternative splicing. Additionally, CPT-induced Tra2 down-regulation occurred independently of new protein synthesis, suggesting a post-translational mechanism. The proteasome inhibitor MG132 reduced CPT-induced Tra2 degradation and TAF1 alternative splicing, and mutation of evolutionarily conserved Tra2 lysine 81, a potential ubiquitin conjugation site, to arginine inhibited CPT-induced Tra2 degradation, supporting a proteasome-dependent alternative splicing mechanism. We conclude that CPT-induced TAF1 alternative splicing occurs through ATR-signaled degradation of a subset of splicing-regulatory proteins.
Alternative splicing is a fundamental mechanism operating in metazoan organisms to regulate protein expression (1–3). In humans and Drosophila, 30–95% of pre-mRNAs undergo alternative splicing, resulting in mature mRNAs that encode proteins with different sequences or that contain a premature stop codon and are subject to degradation (4–11). Alternative splicing plays key roles in developmental processes, such as Drosophila sex determination, and defects in alternative splicing are commonly associated with human disorders, such as spinal muscular atrophy (12, 13). Thus, understanding the molecular details of alternative splicing mechanisms has global implications for understanding both normal and disease states.
At its basic level, the choice of splicing pattern for a pre-mRNA involves the regulation of spliceosome assembly on introns to be excised. Spliceosomes contain five small nuclear ribonucleoproteins (snRNPs)3 (U1, U2, U4, U5, and U6) and numerous regulatory proteins, including the heterodimeric complex U2AF (U2 auxiliary factor) (14–16). Spliceosomes serve to define 5′ and 3′ splice sites at exon/intron boundaries and carry out the two transesterification reactions of splicing.
Splicing-regulatory proteins that control interactions between general splicing complexes and pre-mRNA splice sites are central to the process of alternative splicing. Serine/arginine-rich (SR) proteins constitute a major class of splicing-regulatory proteins. All SR proteins contain one or two RNA recognition motifs (RRMs) and at least one arginine/serine-rich (RS) dipeptide domain (17). RRMs possess sequence-specific RNA binding activity, and, thus, can determine pre-mRNA substrate specificity, and RS domains mediate protein-protein interactions (17, 18). To illustrate, Tra2 (Transformer-2) is an evolutionarily conserved SR protein that contains N- and C-terminal RS domains and a central RRM. In Drosophila, Tra2 directs sex-specific alternative splicing of the dsx (doublesex) pre-mRNA. In vitro studies indicate that, through interactions with other SR proteins, Tra2 facilitates recruitment of general splicing complexes to female-specific regulatory sequences in the dsx pre-mRNA, resulting in utilization of an alternative 3′ splice site (19, 20). In mammals, Tra2 orthologs direct tissue-specific alternative splicing of numerous pre-mRNAs, including those that encode proteins involved in breast cancer and sexual differentiation (21, 22).
To elucidate mechanisms of signal-dependent alternative splicing, we have been studying splicing of the Drosophila TAF1 (TBP-associated factor 1) pre-mRNA in response to the anti-cancer drug camptothecin (CPT), an inhibitor of topoisomerase I (23–25). CPT also affects the alternative splicing of other genes in Drosophila and genes in humans (26, 27).
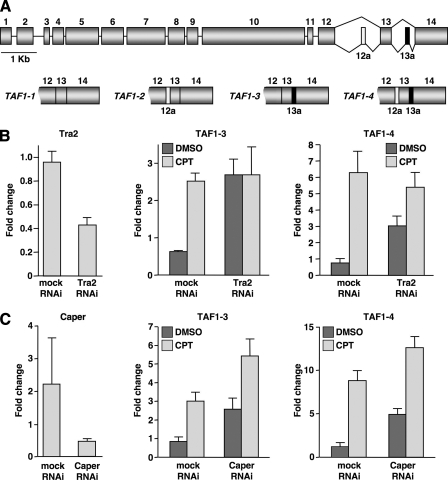
TAF1 encodes the largest subunit of the general transcription factor TFIID, which plays a central role in the transcription of most protein-coding genes (28–31). Alternative splicing of the Drosophila TAF1 pre-mRNA involves the regulated inclusion of cassette exons 12a and 13a and generates four TAF1 mRNA isoforms (TAF1-1, TAF1-2, TAF1-3, and TAF1-4) (Fig. 1A) (25). TAF1-2 and TAF1-4 include exon 12a, which encodes an AT-hook DNA binding motif conferring DNA binding activity to TAF1-2 and TAF1-4 protein isoforms (32). TAF1 mRNA isoforms are differentially expressed in Drosophila tissues, indicating the existence of tissue-specific mechanisms of alternative splicing (32, 33). Furthermore, TAF1-3 and TAF1-4 splicing is up-regulated in Drosophila S2 cells in response to CPT (25).
FIGURE 1.
Tra2 may be a targeted by the CPT-induced signaling pathway that activates TAF1-3 and TAF1-4 splicing. A, a schematic diagram of the Drosophila TAF1 pre-mRNA (top) and mature TAF1 mRNAs (bottom) generated by alternative splicing. Details are provided by Katzenberger et al. (25). Exons are depicted as boxes and introns as lines. Exons colored gray are constitutively included in the mature mRNA, and exons 12a and 13a (colored in white and black, respectively) are alternatively included in the mature mRNA to produce four distinct mRNAs, TAF1-1, TAF1-2, TAF1-3, and TAF1-4. For simplicity, only exons 12–14 are shown for the TAF1 mRNA isoforms. B, qPCR of Tra2 (n = 3), TAF1-3 (n = 6), and TAF1-4 (n = 6) levels in mock or Tra2 RNAi knockdown cells treated with DMSO or 20 μm CPT. C, qPCR of Caper (n = 3), TAF1-3 (n = 6), and TAF1-4 (n = 6) levels in mock or Caper RNAi knockdown cells treated with DMSO or 20 μm CPT. Error bars, S.E. values.
Up-regulation of TAF1-3 and TAF1-4 splicing by CPT is mediated by the DNA damage response kinase ATR (ATM-RAD3-related) and the effector kinase CHK1 (checkpoint kinase 1). In addition, as indicated by studies in which the strength of TAF1 splice sites was altered, up-regulation of TAF1-4 splicing involves altered recruitment of general splicing complexes to splice sites associated with exons 12a, 13, and 13a (26). Based on these findings, we hypothesized that splicing-regulatory proteins function downstream of the CPT-induced signaling pathway to mediate the change in TAF1 alternative splicing.
Here we present the results of a primary screen for TAF1 pre-mRNA alternative splicing-regulatory proteins and a secondary screen to discriminate targets of the CPT-induced signaling pathway. Further characterization of one of the CPT-targeted splicing-regulatory proteins, Tra2, revealed that the ATR pathway modulates TAF1 alternative splicing by signaling Tra2 degradation.
EXPERIMENTAL PROCEDURES
RNAi Screen and Quantitative Real Time PCR (qPCR)—Synthesis of dsRNA for the RNAi screen was carried out using plasmids and protocols described by Park et al. (34). Single-stranded RNAs were resuspended in buffer containing 5 mm KCl and 10 mm NaH2PO4 and were annealed by heating to 95 °C for 5 min and slow cooling overnight to generate dsRNA. RNAi was performed as described by Katzenberger et al. (25). For the primary RNAi screen, TAF1 isoform levels were determined by agarose gel electrophoresis of reverse transcription-PCR products, as described in Katzenberger et al. (25). A list of the 243 genes that were screened is available upon request.
For the secondary RNAi screen, TAF1 isoform levels were determined by qPCR, as described by Katzenberger et al. (25). Primer sets not previously described were as follows (oriented 5′ to 3′): Tra2, CAAGCCGCTGCATAGGAGTCTTTG and CTGGATGCGTTCGATAGGTC; Caper, TCCGCGTACCGAGACAAATCCTAC and GTTGTGCTCGGGCACTTGACATAC. The cDNA of interest was quantitated relative to actin by the formula, (EactinCt(actin))/(EtargetCt(target)) (35), where E is an empirically derived PCR efficiency factor, and Ct is the threshold value for amplification. cDNA levels were normalized to the level from an independent well of cells. Samples from separate wells of cells were used as independent replicates. Two-way analysis of variance and t tests were performed using Prism 4.0c (Graphpad). A Bonferroni correction was used when testing multiple hypotheses.
Plasmids—Plasmids that expressed influenza A virus hemagglutinin (HA) epitope-tagged Tra2 (HA-Tra2) or Rox8 (HA-Rox8) were constructed by cloning a PCR product into a copper-inducible expression vector, pRmHa-4 (36). PCR products were generated using the following primer sets (oriented 5′ to 3′): Rox8, ATGTACCCATACGATGTTCCAGATTACGCTGACGAGTCGCAACCGAAGAC and GGATCCTCATTGGGTCTGGTATTG; Tra2, ATGTACCCATACGATGTTCCAGATTACGCTTCTGACTACGATTACTGTGG and GGATTCTTAATAGCGCGATGAAGT. Construction of the pRmHa-4 plasmid that expressed HA-TAF1-1 was as described for HA-TAF1-4 (25). Site-directed mutagenesis of pRmHa-Tra2 was performed using the QuikChange II mutagenesis kit (Stratagene) and confirmed by sequencing. The green fluorescent protein (GFP) expression plasmid was generated by cloning an NcoI-SpeI fragment from pH-Stinger (Drosophila Genomics Resource Center; Barolo et al. (37)) into pBluescript II (Stratagene).
Cell Culture—Drosophila S2 cells were maintained at room temperature in Schneider's Drosophila medium containing 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Before transfection, cells were subcultured to a density of 1 × 106 cells/ml in 2 ml of medium in 6-well culture dishes. For all transfections, cells were transfected with 1 μg of experiment-specific plasmid DNA and 300 ng of GFP plasmid using Effectene (Qiagen). After 24 h, 200 μm CuSO4 was added to the cells, and after an additional 16 h, cells were treated using a 10 mm stock of CPT (Sigma) in DMSO (Sigma) to the indicated concentrations or, as a control, an equal volume of DMSO. Cells were maintained at room temperature for the indicated times. Where indicated, cells were simultaneously treated with CPT and 100 μg/ml cycloheximide (Calbiochem) in methanol, 20 mm caffeine (Sigma) in water, 1 μm wortmannin (Sigma) in DMSO, or 10 mm MG132 (Sigma) in DMSO. For ionizing radiation (IR) treatment, cells were irradiated with 40 grays of IR using a Mark 1 irradiator and allowed to recover at room temperature for the indicated times. Time course experiments were performed by two different protocols; either each time point was an independent sample (Figs. 2 (A and B), 4(A and B), and 6B), or each time point was an aliquot from a single sample (Figs. 2 (C and E) and 3A).
FIGURE 2.
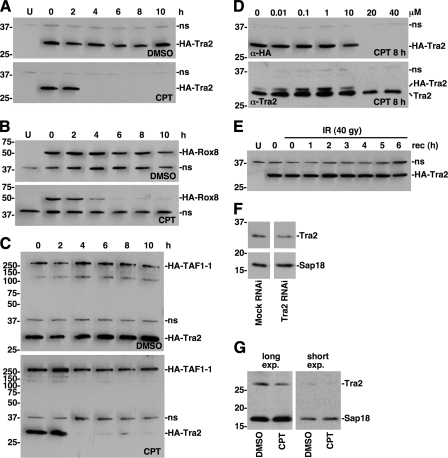
HA-Tra2 and HA-Rox8 expression is reduced by CPT treatment of S2 cells in a time- and dose-dependent manner. A, Western blot analysis for HA-Tra2 in S2 cells treated with DMSO or 20 μm CPT for the indicated times. Positions of protein molecular mass markers (in kDa) are indicated on the left. A nonspecific band (ns) detected by the secondary antibody was used as a loading control. Untransfected cells are indicated by U. Equal loading was confirmed by Coomassie Blue staining of total protein (data not shown). B, Western blot analysis of HA-Rox8 expression, as described for HA-Tra2 in A. C, Western blot analysis of HA-TAF1-1 and HA-Tra2 expression, as described for HA-Tra2 in A. D, Western blot analysis of HA-Tra2 expression in cells treated with the indicated concentration of CPT for 8 h. The same Western blot was probed sequentially with α-HA (top) and α-Tra2 (bottom) antibodies. E, Western blot analysis of HA-Tra2 expression in cells treated with 40 grays of IR and allowed to recover for the indicated times. F, Western blot analysis of endogenous Tra2 in mock and Tra2 RNAi cells. Analysis of the nuclear protein Sap18 served as a loading control. G, Western blot analysis of endogenous Tra2 in S2 cells treated with DMSO or 20 μm CPT for 8 h. Analysis of the nuclear protein Sap18 served as a loading control. Different length exposures of the same Western blot are shown.
FIGURE 4.
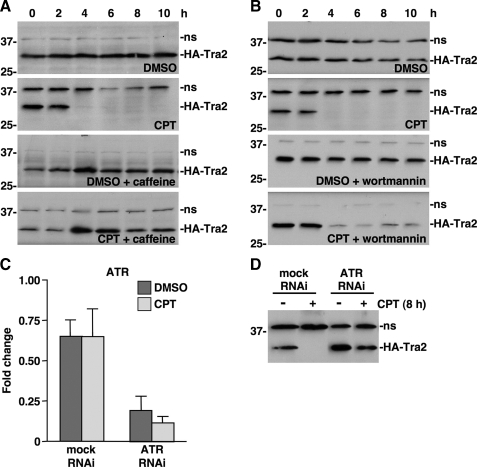
ATR kinase activity is required for CPT-induced down-regulation of HA-Tra2 expression. A, Western blot analysis of HA-Tra2 expression in cells treated with DMSO or 20 μm CPT in the presence or absence of caffeine for the indicated times. Positions of protein molecular mass markers (in kDa) are indicated on the left. A nonspecific band (ns) detected by the secondary antibody was used as a loading control. B, Western blot analysis of HA-Tra2 expression in the presence of wortmannin as described for caffeine in A. C, qPCR analysis of ATR levels in mock or ATR RNAi knockdown cells treated with DMSO or 20 μm CPT (n = 3). Error bars, S.E. values. D, Western blot analysis of HA-Tra2 expression in mock or ATR RNAi treated with DMSO (- lanes) or 20 μm CPT (+ lanes) for 8 h.
FIGURE 3.
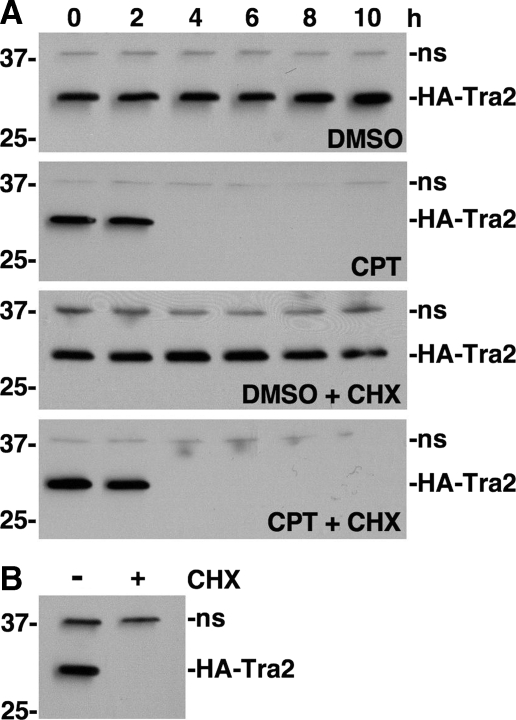
Protein synthesis is not required for CPT-induced down-regulation of HA-Tra2 expression. A, Western blot analysis of HA-Tra2 expression in cells treated with DMSO or 20 μm CPT in the presence or absence of cycloheximide (CHX) for the indicated times. Positions of protein molecular mass markers (in kDa) are indicated on the left. A nonspecific band (ns) detected by the secondary antibody was used as a loading control. B, Western blot analysis of HA-Tra2 expression in S2 cells treated with copper to induce HA-Tra2 expression in the absence or presence of CHX.
Western Blot Analysis—To prepare whole cell extracts, S2 cells were pelleted by centrifugation and lysed in Laemmli sample buffer (Bio-Rad). Equal volumes of extract were fractionated by 12% SDS-PAGE; transferred to Immobilon P polyvinylidene difluoride membrane (Millipore); probed with affinity-purified rabbit α-HA antibody (1:1000; Sigma), α-Pan(598)-Tra2β1 antibody (1:10,000; kindly provided by S. Stamm, University of Kentucky (38)), or α-Sap18 (1:2000) (39); and detected by the ECL Plus Western blotting System (GE Healthcare). The 22-amino acid peptide from human Tra2β1 used to generate α-Pan-Tra2 antibody is identical in sequence to Drosophila Tra2 at 13 positions, six of which are contiguous.
Immunofluorescence Microscopy—Immunofluorescence analysis of S2 cells was carried out as described by Pile et al. (39). A single sample of cells expressing HA-Tra2 and GFP was divided into two aliquots. One aliquot was treated with DMSO for 8 h, and the other was treated with 20 μm CPT for 8 h. The primary antibody was an affinity-purified rabbit α-HA (1:100; Sigma), and the secondary antibody was Alexa Fluor 594 goat α-rabbit IgG (1:400; Invitrogen). Cells were stained with 4′,6-diamidino-2-phenylindole (5 μg/ml), coverslips were mounted with Vectashield mounting medium (Vector Laboratories), and cells were imaged at the same magnification using an Axiovert 200M microscope (Zeiss).
RESULTS
A Screen for Regulators of TAF1 Alternative Splicing—To identify proteins that control endogenous TAF1-3 and TAF1-4 alternative splicing in Drosophila S2 cells, we employed an RNAi screen developed by Park et al. (34). dsRNA synthesized in vitro from plasmids was used to individually knock down expression of 243 genes encoding components of the splicing machinery and predicted RNA binding proteins. For each gene, S2 cells were incubated with dsRNA for 3 days, total RNA was extracted from the cells, and TAF1 mRNA isoform levels were analyzed. Initially, reverse transcription-PCR and agarose gel electrophoresis were used to compare TAF1 mRNA isoform levels between mock-treated and dsRNA-treated cells (data not shown). Genes that had a detectable effect on TAF1 mRNA isoform levels were reanalyzed in independent experiments by qPCR. qPCR was used to determine TAF1-3 and TAF1-4 levels.
The screen identified 15 genes whose knockdown phenotype in S2 cells was up-regulation of TAF1-3 and TAF1-4 splicing, the same change in TAF1 alternative splicing that occurs in response to CPT (Table 1) (25). Where examined, which was for 10 of the 15 genes, total TAF1 levels were not affected by RNAi knockdown, indicating that the increase in TAF1-3 and TAF1-4 levels was due to a change in the relative amounts of TAF1 mRNA isoforms (data not shown). Representative examples of the qPCR analysis are provided for Tra2 and Caper in Figs. 1, B and C, respectively, and analysis of U2AF38 was previously reported (25).
TABLE 1.
Regulators of TAF1-3 and TAF1-4 splicing
|
Genea
|
CG number (isoform)
|
Target
nucleotidesb
|
-Fold
increasec
|
Additive with
CPTd
|
|
|---|---|---|---|---|---|
| TAF1–3 | TAF1–4 | ||||
| Tra2 | CD10128 (Tra2-RD) | 503–812 | 3.7 | 2.7 | No |
| 140–472 | 2.7 | 2.3 | NDe | ||
| 480–806 | 2.7 | 2.2 | ND | ||
| Rox8/dTIAR | CG5422 (Rox8-RD) | 1575–2005 | 2.2 | 2.7 | No |
| 980–1612 | 2.3 | 2.4 | ND | ||
| 1683–2377 | 2.3 | 2.3 | ND | ||
| Tia1-like | CG34362 | 1559–1953 | 3.5 | 3.9 | No |
| (Tia1-like-RA) | 653–1405 | 3.5 | 3.6 | ND | |
| 1455–2155 | 3.9 | 4.1 | ND | ||
| Crn | CG3193 (Crn-RA) | 119–2227 | 2.8 | 2.9 | No |
| 289–1138 | 2.6 | 2.0 | ND | ||
| 1245–2107 | 2.4 | 2.2 | ND | ||
| PUF60/hfp | CG12085 (PUF60-RA) | 921–1170 | 3.5 | 2.8 | No |
| 1291–2093 | 2.8 | 2.6 | ND | ||
| U2AF38 | CG3582 (U2AF38-RA) | 196–990 | 2.5 | 3.0 | No |
| 196–592 | 3.5 | 3.0 | ND | ||
| 622–988 | 2.0 | 2.0 | ND | ||
| KEP1 | CG3584 | 268–720 | 2.0 | 2.3 | Yes |
| U1C | CG5454 | 109–546 | 1.8 | 2.5 | Yes |
| Brr2 | CG5931 | 5791–6172 | 3.3 | 3.0 | Yes |
| UAP56/Hel25E | CG7269 | 411–863 | 2.3 | 1.9 | Yes |
| eIF-4A | CG9075 | 831–1163 | 2.5 | 1.8 | Yes |
| Caper | CG11266 | 1120–1459 | 3.0 | 3.3 | Yes |
| Prp39 | CG1646 | 1076–1562 | 2.5 | 2.0 | Yes |
| LSM4 | CG31990 | 1304–1516 | 2.5 | 2.5 | Yes |
| Prp16 | CG32604 | 432–786 | 1.9 | 2.8 | Yes |
Naming of Tia1-like and Prp16 is based on sequence similarity
Nucleotides complementary to the dsRNA
Average -fold increase relative to mock RNAi-treated cells (n = 3)
Additivity of RNAi and CPT effects on TAF1-3 and TAF1-4 splicing
ND, not determined
Potential Downstream Regulators of CPT-induced TAF1-3 and TAF1-4 Splicing—To determine which of the 15 genes encode targets of the CPT-induced signaling pathway, we used qPCR to analyze TAF1-3 and TAF1-4 levels in S2 cells that were doubly treated with dsRNA and CPT. We reasoned that if a protein functioned downstream of CPT, then the double treatment would increase TAF1-3 and TAF1-4 to levels comparable with that of dsRNA treatment alone or CPT treatment alone. In contrast, if a protein functioned independently of CPT, then the double treatment would increase TAF1-3 and TAF1-4 to levels comparable with the additive effects of dsRNA treatment and CPT treatment. RNAi-treated cells were incubated with 20 μm CPT for 8 h, conditions previously shown to elicit a maximal increase in TAF1-3 and TAF1-4 levels (25).
RNAi knockdown reduced Tra2 mRNA levels to ∼40% of mock RNAi cells (Fig. 1B). Reducing the level of Tra2 resulted in a significant increase in TAF1-3 and TAF1-4 levels. However, CPT treatment of Tra2 RNAi cells did not significantly increase TAF1-3 or TAF1-4 levels relative to Tra2 RNAi alone (p > 0.05 and p > 0.05, respectively). Similar results were observed for Rox8/dTIAR (Drosophila Tia1-related protein), Crn (Crooked neck), Tia1-like (T-cell intracellular antigen 1-like), Hfp (PUF60/Half-pint), and U2AF38 (data not shown) (Table 1) (25). These results place Tra2 and the other five proteins in the CPT-induced signaling pathway and suggest that the pathway inhibits the function of these proteins. Therefore, we collectively refer to these proteins as CPT-targeted splicing-regulatory proteins.
RNAi knockdown reduced Caper mRNA levels to ∼25% of mock RNAi cells (Fig. 1C). In contrast to CPT treatment of Tra2 RNAi cells, CPT treatment of Caper RNAi cells significantly increased TAF1-3 and TAF1-4 levels relative to Caper RNAi alone (p < 0.01 and p < 0.001, respectively). Similar results were observed for the remaining eight proteins (data not shown). Thus, Caper and the remaining eight proteins may function independently of CPT-targeted splicing-regulatory proteins to regulate TAF1 alternative splicing.
For all of the CPT-targeted splicing-regulatory proteins, RNAi reduced the mRNA level of the targeted mRNA (Fig. 1, B and C) (data not shown). However, due to the nature of the RNAi mechanism with long dsRNAs, it was possible that the increase in TAF1-3 and TAF1-4 splicing was due to off-target effects. To address this possibility, we used qPCR to analyze TAF1-3 and TAF1-4 levels in S2 cells treated with nonoverlapping dsRNAs for each of the CPT-targeted splicing-regulatory proteins. In all cases, the nonoverlapping dsRNAs increased TAF1-3 and TAF1-4 levels, suggesting that off-target effects are not responsible for the change in TAF1 alternative splicing (Table 1) (data not shown).
CPT Treatment Reduces Expression of HA-Tra2 and HA-Rox8—Based on studies of other splicing-regulatory proteins, a CPT-induced signal could affect TAF1 alternative splicing by modulating the cellular localization, expression, or function of CPT-targeted splicing-regulatory proteins. For example, in the case of SR proteins, nucleocytoplasmic shuttling is subject to regulation by extracellular cues, relatively small changes in protein concentration affect alternative splicing patterns, and phosphorylation of the RS domains affects protein-protein interactions (17, 19, 22).
To elucidate the CPT-induced regulatory mechanism, we first examined the cellular localization of Tra2. S2 cells were transiently transfected with plasmids encoding N-terminally HA-tagged Tra2 and, as a transfection control, GFP. HA-Tra2 expression was induced by the addition of copper to the medium, DMSO (the solvent for CPT) or CPT was added to the medium, and, after incubation for 8 h, HA-Tra2 localization was determined by immunofluorescence microscopy. Detection of HA-Tra2 using an α-HA primary antibody and a fluorescently tagged secondary antibody revealed that HA-Tra2 predominantly localized to the nucleus of GFP-positive, control DMSO-treated cells (supplemental Fig. 1A). In contrast, when identical detection conditions were used, HA-Tra2 was not detected in GFP-positive, CPT-treated cells (supplemental Fig. 1B). When a longer image exposure time was used, the α-HA signal was detected in the nucleus and cytoplasm of both GFP-positive and GFP-negative CPT-treated cells, indicative of background staining (supplemental Fig. 1C). These data suggest that a CPT-induced signal affects HA-Tra2 expression.
To examine this conclusion, Western blot analysis of whole cell lysates was used to determine HA-Tra2 and HA-Rox8 expression levels. In support of the immunofluorescence results, this assay revealed that HA-Tra2 and HA-Rox8 expression was dramatically reduced after 4–6 h of CPT treatment (Fig. 2, A and B). There was some variability in the extent of HA-Tra2 loss at 4–6 h, but close to complete loss was consistently observed at 8 h (Figs. 3, 4, 5, 6). In contrast, expression of another HA-tagged protein, HA-TAF1-1, alone or in combination with HA-Tra2 was not substantially reduced by CPT treatment, even after 10 h, indicating that down-regulation of HA-Tra2 or HA-Rox8 was not due to the HA tag or inhibition of transcription from the expression plasmid (Fig. 2C) (data not shown). Additionally, Coomassie Blue staining of total cellular proteins fractionated by SDS-PAGE revealed a similar protein abundance pattern in DMSO- and CPT-treated cells over the 10-h time course, indicating some level of specificity to the CPT-induced protein down-regulation mechanism (data not shown).
FIGURE 5.
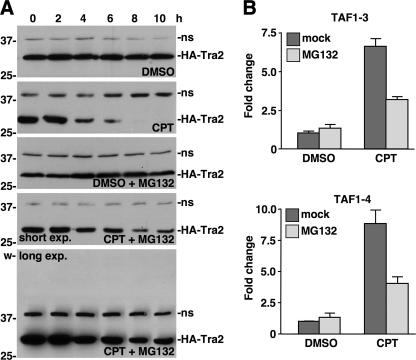
The proteasome is required for CPT-induced down-regulation of HA-Tra2 expression and activation of TAF1-3 and TAF1-4 splicing. A, Western blot analysis of HA-Tra2 expression in S2 cells treated with DMSO or 20 μm CPT in the presence or absence of MG132 over a 10-h time course. Short and long exposures are provided to assess the presence of more slowly migrating, polyubiquitinated forms of HA-Tra2. Positions of protein molecular mass markers (in kDa) and the well (w) are indicated on the left. A nonspecific band (ns) detected by the secondary antibody was used as a loading control. B, qPCR analysis of TAF1-3 and TAF1-4 levels in S2 cells treated with DMSO or 20 μm CPT in the presence or absence of MG132 (n = 3). Error bars, S.E. values.
FIGURE 6.
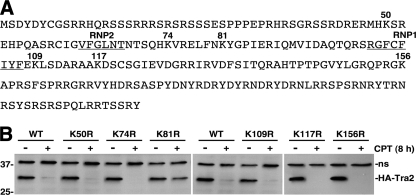
Lysine 81 is important for CPT-induced down-regulation of HA-Tra2 expression. A, the amino acid sequence of the Tra2 isoform (Tra2-RC) used in the studies. Lysine residues that were mutated to arginine are indicated by amino acid number. Lysine 81 is located within the RRM and between the RNP1 and RNP2 motifs, indicated by underlined amino acids. B, Western blot analysis of wild type or lysine mutant HA-Tra2 in S2 cells treated with DMSO (- lanes) or 20 μm CPT (+ lanes) for 8 h. A nonspecific band (ns) detected by the secondary antibody was used as a loading control. Positions of protein molecular mass markers (in kDa) are indicated on the left.
Consistent with the proposal that Tra2 directly regulates TAF1 alternative splicing, the timing and concentration dependence of CPT-induced down-regulation of HA-Tra2 expression was similar to the timing and concentration dependence of CPT-induced up-regulation of TAF1-3 and TAF1-4 splicing (Fig. 2, A and D) (25). Collectively, these data suggest that a CPT-induced signal specifically targets HA-Tra2 and HA-Rox8 for down-regulation.
HA-Tra2 Expression Is Not Affected by Ionizing Radiation—To assess the generality of signal-induced down-regulation of HA-Tra2, S2 cells expressing HA-Tra2 were exposed to IR. We previously found that IR activates the ATM (ataxia telangiectasia-mutated) pathway to signal the same change in TAF1 alternative splicing as CPT (25). However, HA-Tra2 expression was not affected after 0–6 h of recovery from 40 grays of IR, although TAF1-4 splicing was up-regulated after 2 h of recovery (Fig. 3E) (data not shown). Since TAF1 alternative splicing is mediated by the ATR signaling pathway in response to CPT and the ATM signaling pathway in response to IR, these data indicate that different signaling pathways that produce a given change in TAF1 alternative splicing target different splicing-regulatory proteins. This conclusion is supported by the finding that only a subset of genes identified in the RNAi screen function downstream of CPT (Table 1).
CPT Treatment Reduces Expression of Endogenous Tra2—To determine the extent to which endogenous Tra2 expression is affected by CPT-induced signals, we used an antibody raised against human Tra2β1 to detect Drosophila Tra2 (38). On Western blots of S2 whole cell extracts, the Tra2 antibody detected a single protein of the expected size (∼28 kDa), and RNAi knockdown of Tra2 mRNA caused a reduction in the level of the protein to a similar extent as the reduction in the level of the mRNA (Figs. 1B and 2F, and supplemental Fig. 2). These data support the specificity of the antibody.
Western blot analysis of whole cell extracts from S2 cells treated with DMSO or CPT for 8 h revealed that the level of endogenous Tra2 was reduced by CPT treatment (Fig. 2G). Furthermore, the level of CPT-induced Tra2 reduction was similar to the level of RNAi-induced Tra2 knockdown, consistent with the proposal that reducing Tra2 levels is sufficient to up-regulate TAF1-3 and TAF1-4 splicing. Additionally, the concentration dependence of the effect on endogenous Tra2 expression was similar to that of HA-Tra2 (Fig. 2D). Thus, although HA-Tra2 levels were more sensitive to the CPT signal than endogenous Tra2 levels, HA-Tra2 is a reasonable reporter of endogenous Tra2 expression.
Down-regulation of HA-Tra2 Expression Does Not Require New Protein Synthesis—Although CPT causes changes in S2 cell mRNA levels in less than 1 h, down-regulation of HA-Tra2 and HA-Rox8 expression occurred several hours after the addition of CPT, suggesting that the regulatory mechanism requires new protein synthesis (data not shown). To examine this hypothesis, S2 cells were treated with the protein synthesis inhibitor cycloheximide (CHX), and HA-Tra2 levels were assayed by Western blot. In the absence of CPT, CHX treatment had no detectable affect on HA-Tra2 levels until 10 h of treatment, indicating that the HA-Tra2 turnover rate is slow relative to CPT-induced HA-Tra2 down-regulation (Fig. 3A). Co-treatment of S2 cells with CPT and CHX inhibited new protein synthesis but did not affect CPT-induced down-regulation of HA-Tra2 expression (Fig. 3). These findings indicate that HA-Tra2 down-regulation does not require new protein synthesis, which is consistent with our prior finding that CPT-induced up-regulation of TAF1-3 and TAF1-4 splicing does not require new protein synthesis (25).
ATR Is Required for CPT-induced Down-regulation of HA-Tra2 Expression—To determine the involvement of the ATR signaling pathway in CPT-induced down-regulation of HA-Tra2, we employed small molecule kinase inhibitors. Caffeine inhibits the kinase activity of mammalian phosphatidylinositol 3-kinase family members, such as ATM and ATR, at low millimolar concentrations, whereas wortmannin inhibits the kinase activity of mammalian ATM but not ATR at low micromolar concentrations (40–42). S2 cells that expressed HA-Tra2 were treated with 20 μm CPT, 20 mm caffeine, or both CPT and caffeine and HA-Tra2 levels were determined by Western blot analysis. This experiment revealed that caffeine inhibited CPT-induced down-regulation of HA-Tra2 expression, indicating that phosphatidylinositol 3-kinase family kinase activity is involved in the HA-Tra2 regulatory mechanism (Fig. 4A). Similarly, S2 cells were treated with 20 μm CPT, 1 μm wortmannin, or both CPT and wortmannin. However, unlike caffeine, wortmannin did not inhibit CPT-induced down-regulation of HA-Tra2, suggesting that ATR-like kinase activity is involved in the mechanism (Fig. 4B).
To directly examine the requirement for ATR in CPT-induced down-regulation of HA-Tra2 expression, RNAi was used to knockdown ATR expression in S2 cells. On the first day of the experiment, S2 cells were incubated with ATR dsRNA, on the second day the cells were transfected with HA-Tra2 plasmid, on the third day HA-Tra2 expression was induced with copper, and on the fourth day the cells were treated with 20 μm CPT for 8 h.
qPCR and Western blot analyses, respectively, revealed that RNAi significantly reduced ATR mRNA levels in both the absence and presence of CPT (p = 0.0029) and inhibited CPT-induced down-regulation of HA-Tra2 expression (Fig. 4, C and D). Collectively, the kinase inhibitor and RNAi data indicate that ATR kinase activity is necessary for CPT-induced down-regulation of HA-Tra2 expression. Since the same signaling pathway regulates HA-Tra2 expression and TAF1 alternative splicing, it is likely that Tra2 directly regulates TAF1 alternative splicing.
Proteasome Inhibition Blocks CPT-induced Down-regulation of HA-Tra2 Expression—CPT-induced down-regulation of HA-Tra2 expression could occur by mechanisms that affect transcription, translation, or post-translation events. Transcription effects are unlikely, since the same promoter was used to drive transcription of both HA-Tra2 and HA-TAF1-1, but only HA-Tra2 expression was strongly affected by CPT treatment (Fig. 2, A and C). Translation effects are unlikely, since inhibition of new protein synthesis by CHX was not sufficient to down-regulate HA-Tra2 expression in 4–6 h (Fig. 3). Thus, we hypothesized that CPT affects post-translational events. More specifically, we hypothesized that CPT causes polyubiquitination and 26 S proteasome-dependent degradation of CPT-targeted splicing-regulatory proteins (43, 44). The ubiquitin-proteasome system is responsible for the vast majority of protein degradation in eukaryotes and has been implicated in the response to DNA damage induced by CPT (43, 45–47).
To examine the extent to which the proteasome is involved in CPT-induced down-regulation of HA-Tra2 expression, S2 cells were treated with the proteasome inhibitor MG132 (44). Western blot analysis revealed that MG132 inhibited CPT-induced down-regulation of HA-Tra2 expression (Fig. 5A) (data not shown). Unexpectedly, S2 cells co-treated with CPT and MG132 did not accumulate larger, polyubiquitinated forms of HA-Tra2. Taken together, these data indicate that HA-Tra2 is degraded by the proteasome in response to a CPT-induced signal but bring into question the involvement of ubiquitination in the degradation mechanism.
If proteasome-dependent degradation of splicing-regulatory proteins is a necessary component of the CPT signaling pathway that activates TAF1-3 and TAF1-4 splicing, then proteasome inhibition would be predicted to block CPT-induced TAF1-3 and TAF1-4 splicing. To test this prediction, TAF1 mRNA isoform levels were determined in S2 cells treated with CPT or MG132 relative to cells co-treated with CPT and MG132. qPCR analysis revealed that MG132 alone did not affect TAF1 alternative splicing, but MG132 significantly inhibited CPT-induced up-regulation of TAF1-3 and TAF1-4 splicing (p < 0.05 and p < 0.05, respectively) (Fig. 5B). These data indicate that up-regulation of TAF1-3 and TAF1-4 splicing results from CPT-induced, proteasome-dependent degradation of splicing-regulatory proteins.
Mutation of a Particular Lysine Residue in HA-Tra2 Inhibits CPT-induced HA-Tra2 Degradation—Based on involvement of the proteasome in CPT-induced HA-Tra2 degradation, we hypothesized that ubiquitination of a lysine residue in HA-Tra2 is part of the degradation mechanism (43). To determine the extent to which the six lysine residues in Drosophila Tra2 (Lys50, Lys74, Lys81, Lys109, Lys117, and Lys156) contribute to the degradation mechanism, we examined the expression of HA-Tra2 proteins in which individual lysines were mutated to arginine, which is structurally similar to lysine but is not a substrate for ubiquitination (Fig. 6A). S2 cells expressing HA-Tra2 proteins were either treated with DMSO, as a control, or 20 μm CPT for 8 h, and HA-Tra2 levels were assayed by Western blot.
Similar to wild type HA-Tra2, expression of the K50R, K74R, K109R, K117R, and K156R mutants was almost undetectable in cells treated with CPT for 8 h (Fig. 6B). In contrast, the K81R mutant was readily detected in cells treated with CPT for 8 h, albeit at lower levels than in cells treated with DMSO for 8 h. These data indicate that post-translational modification, most likely ubiquitination, of Lys81 in HA-Tra2 occurs in response to CPT and signals proteasome-dependent HA-Tra2 degradation.
Support for the biological importance of Tra2 Lys81 was provided by sequence comparison of the Drosophila Tra2 protein with Tra2 proteins from other organisms, including those from plants, fish, amphibians, and mammals. This analysis revealed that Lys81 and Lys117, but not the other four lysine residues, are evolutionarily conserved (data not shown). It is interesting to note that some organisms, such as mice, contain two Tra2 genes; one gene encodes a protein with lysine residues at positions equivalent to 81 and 117, and the other gene does not. This suggests that in addition to Lys81, Lys117 is important for Tra2 regulation, possibly as a site for post-translational modification, and that in some organisms, expression of Tra2 isoforms that lack Lys81 can bypass signal-induced degradation.
DISCUSSION
Common Targets for Alternative Splicing Regulation—To date, the RNAi screen used to identify regulators of TAF1 alternative splicing has been used to identify regulators of six other alternative splicing events (34, 48). In accord with these studies, genes that affected TAF1 alternative splicing encode core components of the spliceosome (U2AF subunits (U2AF38, PUF60, and Caper) and helicases (UAP56 and a U5 snRNP subunit (Brr2)) and an RNA-binding protein (Rox8) (34). Thus, there are two basic classes of alternative splicing proteins. Members of the first class, which are enriched for core spliceosomal components and exemplified by U2AF subunits, function combinatorially to regulate different types of alternative splicing. This is consistent with the finding that core spliceosomal components in yeast perform transcript-specific roles in splicing and that knockdown of U2AF35 isoforms in human cells alters the alternative splicing pattern of a subset of genes (49, 50). In contrast, members of the second class, exemplified by Tra2, regulate relatively few alternative splicing events. Members of both classes are targets of the CPT-induced signal, suggesting that the two classes function coordinately to regulate alternative splicing.
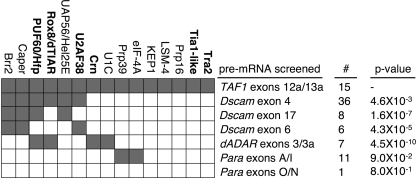
Comparison of the data sets from RNAi splicing screens, using the TAF1 screen as the exemplar, revealed that no single gene affected all of the alternative splicing events, and some genes only affected a single alternative splicing event, providing evidence that RNAi knockdown did not have global effects on alternative splicing (Fig. 7). Moreover, comparison of the data sets revealed that the identified sets of genes were not independent between screens. To illustrate, there is statistically significant overlap between genes that regulated TAF1 alternative cassette exon splicing and genes that regulate dADAR alternative 5′ splice site utilization (p = 4.5 × 10-10) or genes that regulate Dscam mutually exclusive inclusion of exon 17 (p = 1.6 × 10-7). Thus, although different types of alternative splicing appear mechanistically distinct, they involve a common set of regulatory proteins.
FIGURE 7.
A comparison of the proteins identified in Drosophila RNAi screens for regulators of alternative splicing. Proteins that affected TAF1 alternative splicing are indicated above the grid. Those characterized as functioning downstream of the CPT-induced signal are indicated in boldface type. Pre-mRNAs whose alternative splicing was examined in an RNAi screen are indicated to the right of the grid (34, 48). Gray boxes indicate proteins that affected alternative splicing of these pre-mRNAs when knocked down by RNAi. The total number of proteins identified in the RNAi screens is indicated on the right (#). All screens were assumed to test 243 genes. p values are derived from a χ2 test in which the null hypothesis was that the compared screens are independent. Because six χ2 tests were performed, a significant result was defined as below p = 0.0083 (i.e. 0.05/6). Note that the p value for Para exons A/I and Para exons O/N are not significant (i.e. the TAF1 screen is independent of these screens).
Models for TAF1 Alternative Splicing Regulation by CPT-targeted Splicing-regulatory Proteins—A priori, the expectation was that the RNAi screen would identify canonical negative regulators of alternative exon inclusion, such as heterogeneous ribonucleoproteins (hnRNPs), that typically function antagonistically to SR proteins through binding exonic splicing silencers (14, 17, 51, 52). Loss of hnRNPs would permit assembly of splicing complexes at alternative exon splice sites and thereby promote alternative exon inclusion. However, TAF1-3 and TAF1-4 splicing were not affected by knockdown of hnRNPs. The screen contained seven of the nine Drosophila hnRNPs, three of which (hnRNP A1 (hrb87F/hrp36), hnRNP K (hrb57A/bl), and hnRNP A2/B1 (sqd)) affect Dscam exon 4 alternative splicing when knocked down by RNAi in S2 cells (34).
Instead, all of the CPT-targeted splicing-regulatory proteins identified in the screen are primarily characterized as positive regulators of exon inclusion. Tra2 is an SR protein that facilitates sex-specific recognition of alternative splice sites in Drosophila by binding exonic splicing enhancers and, through association with the RS domains, recruiting U2AF or the U2 snRNP to nearby splicing elements (20, 53). Crn is an unusual protein almost entirely composed of half-tetratricopeptide repeat protein-protein interaction modules. In yeast, Crn facilitates productive U4/U6.U5 tri-snRNP interactions with the assembling spliceosome (54). Tia1-like and Rox8 are related proteins that contain RRMs (55). In mammalian cells, these proteins bind uridine-rich sequences and activate splicing of proapoptotic genes that contain alternative exons with weak 5′ splice sites followed by uridine-rich stretches (56). Finally, U2AF38 and PUF60 are core components of the spliceosome that function cooperatively in 3′ splice site recognition (57, 58). U2AF38 is a U2AF35 homolog that binds the 3′ splice site AG dinucleotide, and PUF60 is a U2AF65 family member that binds the polypyrimidine tract.
Two scenarios are envisioned for how reducing the level of Tra2 or other CPT-targeted splicing-regulatory proteins causes increased inclusion of TAF1 alternative exons 12a and 13a. First, Tra2 may bind intronic splicing silencers to suppress alternative exon inclusion. In support of this mechanism, Tra2 represses splicing of the M1 intron from its own pre-mRNA in a concentration-dependent manner. In Drosophila primary spermatocytes, Tra2 represses M1 intron removal by binding an intronic splicing silencer and interfering with spliceosome assembly (59). Moreover, in Drosophila somatic cells, where Tra2 levels are lower than primary spermatocytes and the M1 intron is normally removed, raising the level of Tra2 above endogenous levels is sufficient to repress M1 removal. Thus, in the case of TAF1 alternative splicing, Tra2 may bind intronic splicing silencers in introns flanking exons 12a and 13a to repress inclusion of these exons, and the CPT-induced reduction in Tra2 level relieves this repression. Although binding sites for Tra2 in the TAF1 pre-mRNA have not been identified, an evolutionarily conserved intronic splicing silencer, termed IE-A, is located in the intron between exons 12 and 12a (26).
Alternatively, Tra2 may bind an exonic splicing enhancer in exon 13 to promote splicing between constitutive exons 12, 13, and 14, resulting in alternative exon 12a and 13a exclusion. Tra2 knockdown would reduce splicing complex assembly at exon 13 splice sites, allowing complexes at exon 12a and 13a splice sites to compete for pairing with exon 12 and 14 splice sites, respectively. The U2AF subunits U2AF38 and PUF60 may be integral to this mechanism, since alternative splicing can be controlled by signal-dependent regulation of U2AF occupancy on the pre-mRNA (57, 60). Consistent with this scenario, mutating the exon 13 3′ splice site to improve the match to the 3′ splice site consensus sequence increases exclusion of exon 12a (26). In this scenario, the six CPT-targeted splicing-regulatory proteins may function coordinately to define exon 13 splice sites, explaining why loss of any one of the proteins by RNAi is sufficient to activate exon 12a and 13a inclusion. Overall, our results indicate that the precise balance of splicing-regulatory proteins in a cell can profoundly affect pre-mRNA alternative splicing patterns.
Proteasome-dependent Degradation of Splicing-regulatory Proteins Modulates Alternative Splicing—Our data indicate that TAF1 alternative splicing in response to CPT results from ATR signal-dependent proteasome degradation of specific splicing-regulatory proteins. In addition to the observed proteasome-dependent degradation of Tra2 and Rox8 in response to CPT, support for a role for the proteasome in alternative splicing comes from finding that a proteasome inhibitor abrogated CPT-induced TAF1 alternative splicing in wild type S2 cells (Fig. 5). These findings in Drosophila cells contrast with the situation in yeast, where proteasome inhibitors do not appear to affect splicing (61). The lack of a role for the proteasome in splicing in yeast may simply reflect the fact that alternative splicing is rare in yeast.
Proteasome-dependent degradation of splicing-regulatory proteins is not unprecedented. Lai et al. (62) showed in mammalian cells that in response to inhibition of RNA polymerase II transcription by 5,6-dichloro-1β-d-ribofuranosyl-benzimidazole, SRp55 is hyperphosphorylated at the RS domain, relocalized to nuclear speckles, and degraded by the proteasome upon overexpression of the SR protein kinase Clk/Sty. In this case, Clk/Sty causes proteasome-dependent degradation of SRp55 but not other SR proteins, and the degradation signal involves a C-terminal RS domain and an adjacent proline-rich domain. Venables et al. (63) showed in mammalian cells that T-STAR, a SAM68 splicing-regulatory protein family member, is targeted for proteasome-dependent degradation by the E3 ubiquitin ligase SIAH1 and that T-STAR-dependent alternative splicing is modulated by SIAH1. Finally, Saeki et al. (64) showed that insulin treatment of human hematopoietic cells rapidly causes proteasome-dependent degradation of SRp20. Thus, it appears that reducing the level of specific splicing-regulatory proteins by proteasome-dependent degradation may be a common mechanism for regulating alternative splicing in response to intracellular or extracellular signals.
Signal-dependent Degradation of Splicing-regulatory Proteins—Collectively, our data indicate that activation of the ATR signaling pathway by CPT treatment of S2 cells causes proteasome-dependent degradation of Tra2, resulting in up-regulation of TAF1-3 and TAF1-4 splicing. The events that occur between ATR activation and Tra2 degradation remain to be resolved. The finding that protein synthesis is not required for CPT-induced Tra2 degradation indicates the involvement of post-translational modifications in the degradation mechanism. Frequently, signal-dependent protein degradation by the proteasome involves protein phosphorylation followed by polyubiquitination (65).
In terms of phosphorylation, there is no direct evidence that Tra2 is phosphorylated prior to CPT-induced degradation. Tra2 function in alternative splicing is regulated by phosphorylation of the RS domains and by a direct interaction with protein phosphatase 1 (17, 66). However, Rox8 is also degraded in response to CPT, yet it does not contain an RS domain or a protein phosphatase 1 binding site. One possibility is that Tra2, Rox8, and other CPT-targeted splicing-regulatory proteins are in a complex, and signal-dependent degradation of Tra2 results in destabilization of the complex and degradation of the constituent proteins.
In terms of polyubiquitination, proteasome inhibition by MG132 did not lead to the accumulation of larger, polyubiquitinated forms of Tra2 in CPT-treated cells, bringing into question the involvement of ubiquitination in the degradation mechanism (Fig. 5A). One possibility is that MG132 also inhibits an event that occurs prior to and is required for Tra2 ubiquitination, such as ATR activation. In fact, proteasome inhibitors affect phosphorylation of ATM and ATR substrates in response to DNA-damaging agents (45–47). On the other hand, ubiquitination is supported by the analysis of HA-Tra2 lysine mutants. HA-Tra2(K81R), but not other single lysine mutants, was partially resistant to CPT-induced degradation, suggesting that ubiquitination of Lys81 is necessary for degradation (Fig. 6). The partial effect of K81R may be due to ubiquitination of an alternative lysine residue or to a ubiquitin-independent mechanism (67).
In summary, signal-dependent degradation of splicing-regulatory proteins may be a general mechanism to regulate alternative splicing. With assays in place, it should now be possible to identify signaling events and enzymes involved in post-translational modification of splicing-regulatory proteins that mediate signal-induced TAF1 alternative splicing.
Supplementary Material
Acknowledgments
We are extremely grateful to J. Park and B. Graveley for generously providing the plasmids and protocols for conducting the RNAi screen and to C. Sumanasekera and S. Stamm for providing the Tra2 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant T32 GM08688 (to M. S. M.). This work was also supported by National Science Foundation Grants MCB-0614059 and MCB-0743403 (to D. A. W.) and by a predoctoral fellowship from the PhRMA Foundation (to M. S. M.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: snRNP, small nuclear ribonucleoprotein; CHX, cycloheximide; CPT, camptothecin; dsRNA, double-stranded RNA; GFP, green fluorescent protein; HA, hemagglutinin; hnRNP, heterogeneous ribonucleoprotein; IR, ionizing radiation; qPCR, quantitative real-time PCR; RNAi, RNA interference; RRM, RNA recognition motif; RS, arginine/serine-rich; SR, serine/arginine-rich; E3, ubiquitin-protein isopeptide ligase.
References
- 1.Lynch, K. W. (2007) Adv. Exp. Med. Biol. 623 161-174 [DOI] [PubMed] [Google Scholar]
- 2.Shin, C., and Manley, J. L. (2004) Nat. Rev. Mol. Cell Biol. 5 727-738 [DOI] [PubMed] [Google Scholar]
- 3.Stamm, S. (2002) Hum. Mol. Genet. 11 2409-2416 [DOI] [PubMed] [Google Scholar]
- 4.Black, D. L. (2000) Cell 103 367-370 [DOI] [PubMed] [Google Scholar]
- 5.Brett, D., Hanke, J., Lehmann, G., Hasse, S., Delbruck, S., Krueger, S., Reich, J., and Bork, P. (2000) FEBS Lett. 474 83-86 [DOI] [PubMed] [Google Scholar]
- 6.Graveley, B. R. (2001) Trends Genet. 17 100-107 [DOI] [PubMed] [Google Scholar]
- 7.Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Amour, C. D., Santos, R., Schadt, E. E., Stoughton, R., and Shoemaker, D. D. (2003) Science 302 2141-2144 [DOI] [PubMed] [Google Scholar]
- 8.Kan, Z., States, D., and Gish, W. (2002) Genome Res. 12 1837-1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mironov, A. A., Fickett, J. W., and Gelfand, M. S. (1999) Genome Res. 9 1288-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan, Q., Shai, O., Lee, L. J., Frey, B. J., and Blencowe, B. J. (2008) Nat. Genet. 40 1413-1415 [DOI] [PubMed] [Google Scholar]
- 11.Wang, E. T., Sandberg, R., Luo, S., Khrebtukova, I., Zhang, L., Mayr, C., Kingsmore, S. F., Schroth, G. P., and Burge, C. B. (2008) Nature 456 470-476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Förch, P., and Valcárcel, J. (2003) Prog. Mol. Subcell. Biol. 31 127-151 [DOI] [PubMed] [Google Scholar]
- 13.Singh, N. N., Androphy, E. J., and Smith, R. N. (2004) Crit. Rev. Eukaryotic Gene Expression 14 271-285 [DOI] [PubMed] [Google Scholar]
- 14.Black, D. L. (2003) Annu. Rev. Biochem. 72 291-336 [DOI] [PubMed] [Google Scholar]
- 15.Brow, D. A. (2002) Annu. Rev. Genet. 36 333-360 [DOI] [PubMed] [Google Scholar]
- 16.Patel, A. A., and Steitz, J. A. (2003) Nat. Rev. Mol. Cell Biol. 4 960-970 [DOI] [PubMed] [Google Scholar]
- 17.Lin, S., and Fu, X. D. (2007) Adv. Exp. Med. Biol. 623 107-122 [DOI] [PubMed] [Google Scholar]
- 18.Maris, C., Dominquez, C., and Allain, F. H. (2005) FEBS J. 272 2118-2131 [DOI] [PubMed] [Google Scholar]
- 19.Qi, J., Su, S., McGuffin, M. E., and Mattox, W. (2006) Nucleic Acids Res. 34 6256-6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sciabica, K. S., and Hertel, K. J. (2006) Nucleic Acids Res. 34 6612-6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venables, J. P., Dalgliesh, C., Paronetto, M. P., Skitt, L., Thorton, J. K., Saunders, P. T., Sette, C., Jones, K. T., and Elliot, D. J. (2004) Hum. Mol. Genet. 13 1525-1534 [DOI] [PubMed] [Google Scholar]
- 22.Watermann, D. O., Tang, Y., Zur Hausen, A., Jäger, H., Stamm, S., and Stickler, E. (2006) Cancer Res. 66 4774-4780 [DOI] [PubMed] [Google Scholar]
- 23.Pommier, Y. (2006) Nat. Rev. Cancer 6 789-802 [DOI] [PubMed] [Google Scholar]
- 24.Hertzberg, R. P., Caranfa, M. J., and Hecht, S. M. (1989) Biochemistry 28 4629-4638 [DOI] [PubMed] [Google Scholar]
- 25.Katzenberger, R. J., Marengo, M. S., and Wassarman, D. A. (2006) Mol. Cell. Biol. 26 9256-9267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marengo, M. S., and Wassarman, D. A. (2008) RNA 14 1681-1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solier, S., Lansiaux, A., Logette, E., Wu, J., Soret, J., Tazi, J., Bailly, C., Dosoche, L., Solary, E., and Corcos, L. (2004) Mol. Cancer Res. 2 53-61 [PubMed] [Google Scholar]
- 28.Chen, B. S., and Hampsey, M. (2002) Curr. Biol. 12 R620-R622 [DOI] [PubMed] [Google Scholar]
- 29.Kim, T. H., Barrera, L. O., Zheng, M., Qu, C., Singer, M. A., Richmond, T. A., Wu, Y., Green, R. D., and Ren, B. (2005) Nature 436 876-880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matangkasombut, O., Auty, R., and Buratowski, S. (2004) Adv. Protein Chem. 67 67-69 [DOI] [PubMed] [Google Scholar]
- 31.Weinzierl, R. O., Dynlacht, B. D., and Tjian, R. (1993) Nature 362 511-517 [DOI] [PubMed] [Google Scholar]
- 32.Metcalf, C. E., and Wassarman, D. A. (2006) J. Biol. Chem. 281 30015-30023 [DOI] [PubMed] [Google Scholar]
- 33.Metcalf, C. E., and Wassarman, D. A. (2007) Dev. Dyn. 236 2836-2843 [DOI] [PubMed] [Google Scholar]
- 34.Park, J. W., Parisky, K., Celotto, A. M., Reenan, R. A., and Graveley, B. R. (2004) Proc. Natl. Acad. Sci. 101 15974-15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaffl, M. W. (2001) Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunch, T. A., Grinblat, Y., and Goldstein, L. S. (1988) Nucleic Acids Res. 16 1043-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barolo, S., Carver, L. A., and Posakony, J. W. (2000) BioTechniques 29 726-732 [DOI] [PubMed] [Google Scholar]
- 38.Stoilov, P., Daoud, R., Nayler, O., and Stamm, S. (2004) Hum. Mol. Genet. 13 509-524 [DOI] [PubMed] [Google Scholar]
- 39.Pile, L. A., Schlag, E. M., and Wassarman, D. A. (2002) Mol. Cell. Biol. 22 4965-4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham, R. T. (2004) DNA Repair 3 883-887 [DOI] [PubMed] [Google Scholar]
- 41.Sarkaria, J. N., Busby, E. C., Tibbetts, R. S., Roos, P., Taya, Y., Karnitz, L. M., and Abraham, R. T. (1999) Cancer Res. 59 4375-4382 [PubMed] [Google Scholar]
- 42.Sarkaria, J. N., Tibbetts, R. S., Busby, E. C., Kennedy, A. P., Hill, D. E., and Abraham, R. T. (1998) Cancer Res. 58 4375-4382 [PubMed] [Google Scholar]
- 43.Ciechanover, A., and Ben-Saadon, R. (2004) Trends Cell Biol. 14 103-106 [DOI] [PubMed] [Google Scholar]
- 44.Lee, D. H., and Goldberg, A. L. (1998) Trends Cell Biol. 8 397-403 [DOI] [PubMed] [Google Scholar]
- 45.Jacquemont, C., and Taniguchi, T. (2007) Cancer Res. 67 7395-7405 [DOI] [PubMed] [Google Scholar]
- 46.Mu, J. J., Wang, Y., Luo, H., Leng, M., Zhang, J., Yang, T., Besusso, D., Jung, S. Y., and Qin, J. (2007) J. Biol. Chem. 282 17330-17334 [DOI] [PubMed] [Google Scholar]
- 47.Sakasai, R., and Tibbetts, R. S. (2008) J. Biol. Chem. 283 13549-13555 [DOI] [PubMed] [Google Scholar]
- 48.Olson, S., Blanchette, M., Park, J., Savva, Y., Yeo, G. W., Yeakley, J. M., Rio, D. C., and Graveley, B. R. (2007) Nat. Struct. Mol. Biol. 14 1134-1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pacheco, T. R., Moita, L. F., Gomes, A. Q., Hacohen, N., and Carmo-Fonseca, M. (2006) Mol. Cell. Biol. 17 4187-4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pleiss, J. A., Whitworth, G. B., Bergkessel, M., and Guthrie, C. (2007) PLoS Biol. 5 e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, C. W., and Valcárcel, J. (2000) Trends Biochem. Sci. 25 381-388 [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Contreras, R., Cloutier, P., Shkreta, L., Fisette, J.-F., Revil, T., and Chabot, B. (2007) Adv. Exp. Med. Biol. 623 123-147 [DOI] [PubMed] [Google Scholar]
- 53.Wu, J. Y., and Maniatis, T. (1993) Cell 75 1061-1070 [DOI] [PubMed] [Google Scholar]
- 54.Chung, S., McLean, M. R., and Rymond, B. C. (1999) RNA 5 1042-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brand, S. F., Pichoff, S., Noselli, S., and Bourbon, H. M. (1995) Gene (Amst.) 154 187-192 [DOI] [PubMed] [Google Scholar]
- 56.Förch, P., and Valcárcel, J. (2001) Apoptosis 6 463-468 [DOI] [PubMed] [Google Scholar]
- 57.Hastings, M. L., Allemand, E., Duelli, D. M., Myers, M. P., and Krainer, A. R. (2007) PLoS ONE 2 e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mollet, I., Barbosa-Morais, N. L., Andrade, J., and Carmo-Fonseca, M. (2006) FEBS J. 273 4807-4816 [DOI] [PubMed] [Google Scholar]
- 59.Chandler, D. S., Qi, J., and Mattox, W. (2003) Mol. Cell. Biol. 23 5174-5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tisserant, A., and König, H. (2008) PLoS ONE 3 e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dembla-Rajpal, N., Seipeit, R., Wang, Q., and Rymond, B. C. (2004) Biochim. Biophys. Acta. 1680 34-45 [DOI] [PubMed] [Google Scholar]
- 62.Lai, M.-C., Lin, R.-U. and Tarn, W.-Y. (2003) Biochem. J. 371 937-945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venables, J. P., Bourgeois, C. F., Dalgliesh, C., Kister, L., Stevenin, J., and Elliot, D. J. (2005) Hum. Mol. Genet. 14 2289-2303 [DOI] [PubMed] [Google Scholar]
- 64.Saeki, K., Yasugi, E., Okuma, E., Breit, S. N., Nakamura, M., Toda, T., Kaburagi, Y., and You, A. (2005) Am. J. Physiol. 289 E419-E428 [DOI] [PubMed] [Google Scholar]
- 65.Hunter, T. (2007) Mol. Cell 28 730-738 [DOI] [PubMed] [Google Scholar]
- 66.Novoyatieva, T., Heinrich, B., Tang, Y., Benderska, N., Butchbach, M. E., Lorson, C. L., Lorson, M. A., Ben-Dov, C., Fehlbaum, P., Bracco, L., Burghes, A. H., Bollen, M., and Stamm, S. (2008) Hum. Mol. Genet. 17 52-70 [DOI] [PubMed] [Google Scholar]
- 67.Orlowski, M. O., and Wilk, S. (2003) Arch. Biochem. Biophys. 415 1-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.