Abstract
The zinc finger transcription factor Krox20 plays an essential role in the vertebrate hindbrain segmentation process. It positively or negatively controls a large variety of other regulatory genes, coordinating delimitation of segmental territories, specification of their identity, and maintenance of their integrity. We have investigated the molecular mechanisms of Krox20 transcriptional control by performing a detailed structure-function analysis of the protein in the developing chick hindbrain. This revealed an unsuspected diversity in the modes of action of a transcription factor in a single tissue, since regulation of each of the five tested target genes requires different parts of the protein and/or presumably different co-factors. The multiplicity of Krox20 functions might rely on this diversity. Investigation of known Krox20 co-factors was initiated in relation to this analysis. Nab was shown to act as a negative feedback modulator of the different Krox20 activating functions in the hindbrain. HCF-1 was found to bind to a Krox20 N-terminal region, which was shown to rely on multiple elements, including acidic domains, to convey Nab activation and Krox20 autoregulation.
The development of the vertebrate hindbrain involves a transient segmentation process, dividing it along the anterior-posterior axis into 7-8 metameric units called rhombomeres (r1-r8) (1-3). This subdivision underlies the differentiation of hindbrain neurons in stereotyped patterns and the repeated organization of branchiomotor cranial nerves (4, 5). It also participates in the specification of neural crest cells and in the establishment of their pathways of migration into the branchial arches (6-13), therefore playing a crucial role in the development of head and facial structures (14).
Hindbrain segmentation requires the delimitation of separate compartments, characterized by cell lineage restrictions (15) and specific patterns of gene expression related to the segment anterior-posterior identity (2). Numerous genes have been shown to play crucial roles at different levels of the segmentation process (16-31). Integrating this complex regulatory network constitutes a major challenge for reaching a comprehensive understanding of the segmentation process.
Krox20, also known as Egr2, is among these key regulatory genes and encodes a zinc-finger transcription factor (32, 33). It is expressed in two transverse stripes, that prefigure and subsequently correspond to r3 and r5 (34), and has been shown to be required for the formation and maintenance of these segments (28, 29, 35). More precisely, it was established that Krox20 plays multiple and complex roles in the initial formation and subsequent delimitation of the r3 and r5 territories (28, 36) in the specification of their identity and in particular of their odd-numbered character (29, 37) and in their stabilization by restriction of cell intermingling between adjacent rhombomeres (38, 39), therefore establishing that these processes are highly intertwined (35). To fulfill these different functions, Krox20 controls the expression, either positively or negatively, of a number of other regulatory genes. These include Krox20 itself (28, 40), its Nab co-factors, which modulate Krox20 activity (41, 42), several Hox genes, which participate in the specification of rhombomere identity (29, 37, 43-46), follistatin (29), which may be involved in signaling between rhombomeres, and Eph family members, which are involved in cell segregation between adjacent rhombomeres (38, 39, 47, 48). Many of the genes positively regulated by Krox20 constitute direct transcriptional targets of this factor, which binds to neighboring cis-acting sequences (39, 44-46), whereas the repression of one gene, Hoxb1, was shown not to involve direct binding of Krox20 to DNA (43). However, in most cases, the detailed molecular mechanisms underlying the various transcriptional activities of Krox20 have not been studied. Although several proteins have been shown to interact with Krox20 and/or to interfere with its activity, most of these studies were carried out in vitro and/or using artificial Krox20 targets, and it is therefore difficult to extrapolate their conclusions to any specific Krox20 target in an in vivo situation, including the hindbrain. Nab proteins have been shown to bind a motif called R1 present in Egr/Krox family members and were initially described as antagonists of Krox20 and Krox24 (also known as Egr1/Zif268) activating function in cultured cells (42, 49-51). However, this view was recently challenged by data suggesting that Nabs can also behave as positive modulators of Krox20 activity in cultured cells (52) and, like Krox20, promote myelination in Schwann cells (53, 54). HCF-1 and Ddx20 factors have been described as potential co-activator and co-repressor of Krox20, respectively, in cultured cells (55, 56). The only documented case in respect to hindbrain development is the interaction of Krox20 with PIASxβ, which is involved in the repression of Hoxb1 (43).
The understanding of Krox20 function in hindbrain development therefore justifies further efforts to decipher the mechanisms involved in the regulation of its various target genes in the embryo and in particular to identify which co-factors are involved. In the present work, using electroporation of the chick hindbrain, we have initiated such an in vivo analysis by localizing the domains of the Krox20 protein that are required for the regulation of five genes that are representatives of the various Krox20 transcriptional targets. This revealed an unexpected diversity in the modes of action of a transcription factor in a single tissue, since regulation of each of these targets appeared to require different parts of the protein and/or presumably different co-factors. This diversity is likely to constitute an important element for Krox20 to fulfill its multiple functions. Investigation of co-factors involved in Krox20 transcriptional function established that the Nab proteins act as general antagonists of transcriptional activation by Krox20 in the hindbrain, and HCF-1 was shown to bind to a Krox20 N-terminal region required for Krox20 autoregulation.
EXPERIMENTAL PROCEDURES
Expression Plasmid Constructions—The complete mouse Krox20-expressing plasmid, pAdRSVKrox20HA, is derived from pAdRSVKrox20 (37) by the addition of a sequence encoding an HA3 epitope (TACCCATACGACGTACCAGACTACGCATCG) just before the STOP codon. The N-terminal deletions were generated by the replacement of the AvrII-BglII fragment from the pAdRSVKrox20HA plasmid by 5′-deleted versions generated by PCR using the Expand High Fidelity PCR system (Roche Applied Science). The 5′ primers contained an AvrII restriction site followed by an ATG initiation codon and 21 nucleotides encoding the 7 N-terminal amino acids of the deleted constructs and the 3′ primer encoded the HA tag followed by the BglII restriction site. The Krox20 Δ159-189 construct was generated using the Exsite PCR-based site-directed mutagenesis kit (Stratagene), following the manufacturer's instructions. The pAdRSVKrox20I268FHA plasmid was obtained by introducing a point mutation changing ATC (isoleucine) into TTC (phenylalanine) at the level of codon 268 using the Quikchange multisite-directed mutagenesis kit (Stratagene). The VP16-Krox20 construct was generated by fusion of a sequence encoding VP16 activation domain (amino acids 413-490) (57) to the 3′ part of Krox20 (corresponding to amino acids 309-470) followed by cloning into the pAdRSVSp vector (58). The mouse Nab1- and Nab2-expressing plasmids, pAdRSVNab1HA and pAdRSVNab2HA, were constructed by cloning the coding sequences into the pAdRSVSp vector (58), with the addition of an HA epitope coding sequence just before the STOP codon. The coding sequences of Nab1 and Nab2 were obtained by PCR amplification from the IMAGE clones MRC 426565 and MRC 6306757, respectively, and using the Expand High Fidelity PCR system (Roche Applied Science). The mouse HCF-1 β-propeller-expressing plasmid was constructed by cloning the coding sequence corresponding to the first 380 amino acids of HCF-1 into the pCMVsport6 vector, with the insertion of a FLAG epitope sequence (GATTACAAGGATGACGACGATAAGCTA) just after the initiation codon. The insert was amplified by PCR from the RZPD IMAGE clone 6509535 using the Phusion PCR system (Finnzymes). For all constructs, cloning junctions and point mutations were verified by sequencing, and the sizes and levels of expression of the proteins were analyzed by immunoblotting performed on extracts from transiently transfected 293 cells or electroporated chick embryos, using anti-HA (anti-HA high affinity rat monoclonal antibody; Roche Applied Science; reference number 1867423, dilution 1:400) or anti-FLAG antibodies (anti-FLAG rabbit polyclonal IgG; Sigma; reference number F7425, dilution 1:400).
Cell Culture, Transfection, Co-immunoprecipitation, and Western Blotting Assays—COS-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum (Invitrogen). Transient transfection was performed with Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Cells were analyzed 48 h after transfection. For protein immunoprecipitation with the anti-HA antibody, transfected COS-7 cells were harvested in the following buffer: 100 mm Tris, pH 7.5, 250 mm KCl, 0.5% Triton X-100, 2 mm EDTA, 5 mm MgCl2, 5% glycerol, with protease inhibitor mixture (Roche Applied Science). Western blotting was performed using the ECL procedure according to the manufacturer's instructions (Amersham Biosciences) with a rabbit polyclonal anti-HA antibody (HA probe (Y-11); Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); reference number sc-805, dilution 1:2000) to quantify Krox20 protein or an anti-FLAG rabbit polyclonal antibody (Sigma; reference number F7425, dilution 1:2000) to detect the co-immunoprecipitated HCF β-propeller or a rabbit polyclonal anti-GFP (Invitrogen; reference number A11122, dilution 1:2000).
In Ovo Electroporation, Whole Mount Immunohistochemistry, and in Situ Hybridization—Electroporation and preparation of chick embryos for immunolabeling or in situ hybridization were performed as described previously (37). Briefly, fertilized hen eggs were incubated for 30 h up to stages HH8-HH10 before injection. DNA solution was injected at a concentration of 1 μg/μl into the neural tube of the embryo in ovo, and electroporation was performed with a BTX830 electroporator (Quantum) using the parameters previously described (37). 20-24 h after electroporation, embryos were recovered in phosphate-buffered saline, fixed in paraformaldehyde (4%), and processed for immunochemistry or in situ hybridization. Immunochemical detection of proteins was performed with the following antibodies: rat monoclonal against HA epitope (Roche Applied Science; 1:400) followed by biotin-coupled goat antibody anti-rat IgG (Jackson; 1:200) and Cy3-Streptavidin (Jackson; 1:200) or mouse monoclonal antibody against EphA4 (37) (1:20), followed by Alexa 488-coupled goat antibody anti-mouse IgG (Molecular Probes; 1:200). In situ hybridization was performed as described (59), using digoxigenin-labeled probes. The probes were as follows: chick Krox20 (37), chick EphA4 (60), chick Hoxb1 (61, 62), chick follistatin (61), chick Nab (nucleotides 927-1563 of Gallus gallus Nab mRNA, NM 204268), and chick Hcf (the probe was derived from HCF BBSRC chick expressed sequence tag ChEST953i24 and corresponds to the first 661 nucleotides of this sequence).
The activities of the mutant proteins were classified in different categories (activation (+), weak activation (+/0), no activity (0), and repression (-)) by evaluation of the number of positive or negative cells. The territories that were taken into account depend on the target gene, since the domains of competence are different for different targets. For the activation of Krox20 and Nab, we evaluated the total number of ectopic positive cells by embryo in r1, r2, r4, and r6 on the electroporated side and classified the embryos as follows: >200 cells (+); >20 and <50 cells (+/0); <10 cells (0). If >200 negative cells were found in r3 and r5, it was classified as (-). For the activation of EphA4, we have evaluated the number of ectopic positive cells in r2, r4, and r6 and classified the embryos as follows: >150 cells (+); >20 and <50 cells (+/0); <10 cells (0). For the repression of Hoxb1, if >100 negative cells were found in r4 on the electroporated side, the embryo was classified as (-). In the case of follistatin, the embryos in which a clear weaker density of positive cells was observed in r2 and r4 on the electroporated side were classified as (-).
RESULTS
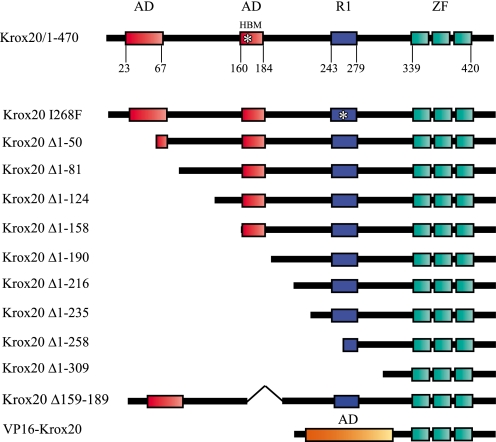
To analyze the mechanisms of the transcriptional regulation by Krox20 and to investigate the involvement of co-factors, we engaged into delineating the domains of the protein required for the control of different targets in the developing hindbrain. As a first step, the analysis of the evolutionary conservation of Krox20 amino acid sequence allowed the division of the protein into three regions (Fig. S1): (i) a large N-terminal domain corresponding approximately to two-thirds of the protein, which is relatively well conserved with nevertheless some small areas of divergence; (ii) an internal region (positions 312-425) that is extremely conserved and corresponds to the DNA binding domain (zinc fingers) flanked by upstream and downstream basic sequences; (iii) a short C-terminal region that is very poorly conserved and is therefore not likely to play an important functional role. This distribution led us to concentrate on the N-terminal domain that appeared more likely to harbor transcriptional activation and repression domains. We therefore generated a nested series of N-terminal external deletions carrying a C-terminal HA tag (Fig. 1; we have previously shown that the presence of a C-terminal tag does not affect the properties of the protein (37)), electroporated expression constructs in the chick embryo hindbrain, and compared their transcriptional activities with that of the wild type protein on five known Krox20 target genes.
FIGURE 1.
Presentation of the mutant Krox20 proteins. Shown is a schematic representation of wild type and mutant Krox20 proteins. Previously identified domains are indicated: two acidic domains (AD), the HBM site, likely to bind the HCF factor, the R1 domain, which binds the Nab cofactor, and the DNA binding domain consisting of three zinc fingers (ZF). The numbers below the line indicate amino acid positions. The name of each deletion mutant indicates the deleted region.
Transactivation of Different Targets Involves Distinct Mechanisms—We first focused on three target genes that are positively regulated by Krox20: Krox20 itself, Nab, and EphA4 (39-41). Krox20 and EphA4 have further been shown to constitute direct transcriptional targets of the Krox20 protein in the hindbrain, and the cis-acting elements mediating these regulations have been identified (39, 40). The three genes are ectopically activated by wild type Krox20 in chick in ovo electroporation experiments (37). This procedure allows unilateral introduction of DNA into the chick neural tube, the nonelectroporated side serving as a control to the experimental one. Mouse Nab genes (Nab1 and Nab2) have been shown to require Krox20 for their expression in r3 and r5 (41). In the chick, there is only one Nab gene known, and its regulation has not been investigated so far. We showed by in situ hybridization that chick Nab expression follows that of Krox20 in the hindbrain, with an initiation at around the 7 and 9 somite stages in r3 and r5, respectively (Fig. S2, A-D).
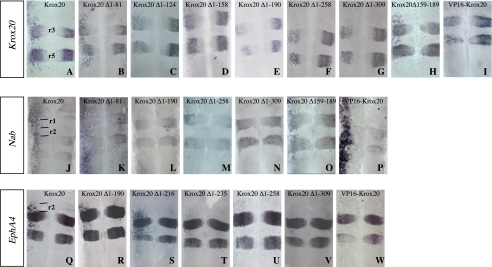
In ovo electroporation of the HA-tagged complete Krox20 construct efficiently induced Krox20, Nab, and EphA4 expression in the neural tube (Fig. 2, A, J, and Q). Interestingly, the rostral limits of efficient ectopic expression appeared different, corresponding to the r1/r2 boundaries for EphA4, whereas no rostral restriction was observed for Krox20 or Nab. Furthermore, ectopic activation of EphA4 was strong in r2 and progressively decreased in r4 and r6 (Fig. 2Q), whereas activation of Krox20 and Nab was relatively uniform in the hindbrain (Fig. 2, A and J).
FIGURE 2.
The various Krox20 transcriptional activities are mediated by different domains. Chick embryo neural tubes were electroporated on the left side with wild type or truncated Krox20 expression constructs, as indicated, in situ hybridized with the probes indicated in the left panel, and flat mounted. The effects of the ectopic expression of the different constructs are determined by comparison of the in situ hybridization patterns observed in the left (experimental) and right (control) sides of the neural tube. A-I, analysis of endogenous Krox20 expression indicates in particular that deletion of the 190 N-terminal amino acids abolishes ectopic activation and even leads to repression of Krox20 in r3 and r5. J-P, Krox20 electroporation leads to ectopic activation of Nab with no rostral restriction, which is also lost upon deletion of the 190 N-terminal amino acids. Grafting of the VP16 acidic domain on an N-terminally deleted Krox20 restores the ability to activate Nab (P), but not Krox20 (I). Q-W, analysis of EphA4 expression indicates that ectopic activation is restricted to r2, r4, and r6 with a level decreasing rostro-caudally. Deletion of the 235 N-terminal amino acids eliminates most of this activity. Anterior is up. r, rhombomere.
In conclusion, these data indicate that Krox20 can activate its own transcription as well as that of EphA4 and Nab in the hindbrain but that different regional restrictions apply to different targets. This suggests that different limiting and unevenly distributed co-factors are required for the transcriptional activation of these genes and therefore that the molecular mechanisms involved are distinct.
The Various Krox20 Transcriptional Activities Are Mediated by Different Domains—The next step was to compare the transcriptional activities of the different Krox20 N-terminal deletions on the three positive targets. Functional analysis of the deletions showed that transcriptional activations of Krox20 and Nab on one side and EphA4 on the other require different parts of Krox20. Whereas the deletion of the first 190 N-terminal amino acids (Krox20 Δ1-190) completely abolished the ability of Krox20 to activate endogenous Krox20 or Nab (Table 1 and Fig. 2, A, E, J, and L), this construct was still able to efficiently induce EphA4 expression (Table 1 and Fig. 2, Q and R). It is interesting to note that the deletion mutants that were not able to ectopically activate Krox20 actually partially repressed its expression in r3 and r5 (Table 1 and Fig. 2, E-G), therefore presenting a possible dominant-negative behavior that was not observed with the Nab target (Fig. 2, L and N).
TABLE 1.
Transcriptional activities of the different Krox20 mutants
The table summarizes the transcriptional activities of the various Krox20 mutants tested on five different target genes (Krox20, Nab, EphA4, follistatin, and Hoxb1) and evaluated as defined under “Experimental Procedures.” +, activation; +/0, very weak activation; −, repression; 0, no activity. ND, not determined. For each construct, the ratio in parenthesis indicates the number of embryos showing the indicated transcriptional pattern over the total number of observed embryos.
| Krox20 | Krox20 Δ1-50 | Krox20 Δ1-81 | Krox20 Δ1-124 | Krox20 Δ1-158 | Krox20 Δ1-190 | Krox20 Δ1-216 | Krox20 Δ1-235 | Krox20 Δ1-258 | Krox20 Δ1-309 | Krox20 Δ159-189 | Krox20 I268F | VP16 Krox20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Krox20 | + (27/32) | + (5/5) | + (4/4) | +/0 (11/11) | +/0 (10/10) | − (14/14) | ND | ND | − (6/6) | − (4/5) | + (3/4) | + (7/7) | 0 (6/6) |
| Nab | + (21/22) | ND | + (4/4) | + (4/5) | + (3/4) | 0 (17/17) | ND | ND | 0 (4/4) | 0 (4/4) | + (5/6) | + (16/17) | + (6/6) |
| KphA4 | + (36/38) | ND | + (4/4) | + (4/4) | + (7/7) | + (18/20) | + (10/12) | +/0 (7/7) | +/0 (8/8) | +/0 (8/8) | + (8/10) | + (12/13) | 0 (14/14) |
| follistatin | − (7/10) | ND | ND | − (3/4) | ND | − (5/12) | − (4/5) | 0 (5/5) | 0 (8/8) | 0 (5/5) | ND | ND | ND |
| Hoxb1 | − (20/26) | ND | ND | ND | ND | − (6/6) | ND | ND | − (3/4) | − (4/7) | ND | − (3/4) | ND |
Further deletions indicated that an element essential for EphA4 induction was located between positions 216 and 235 (Table 1 and Fig. 2, S and T). However, a very low level of EphA4 activation was maintained with Krox20 Δ1-235 as well as with further deletions eliminating the entire N-terminal region (Table 1 and Fig. 2, T-V).
The N-terminal part of Krox20 contains two acidic domains (Fig. 1) whose deletion was previously shown to affect transcriptional activation of an artificial reporter construct containing multimerized Krox20 binding sites in cultured cells (63). We therefore analyzed the possible involvement of these acidic domains in Krox20 activation function. For this purpose, we generated additional deletions covering the 1-190 region (Fig. 1). Deletion of the 81 N-terminal amino acids, eliminating the first acidic region, did not affect the capacity of Krox20 to activate endogenous Krox20 (Table 1 and Fig. 2B). However, further external deletions progressively reduced the activity, with significant drops when sequences 82-124 and 159-190 were eliminated (Table 1 and Fig. 2, B-E). Successive deletions of the N-terminal region also led to progressive decrease of the ability of Krox20 to activate Nab, with a complete loss when sequences 159-190 were eliminated (Table 1 and Fig. 2, K and L; data not shown). These data suggest that several regions located between positions 1 and 190 participate in Krox20 and Nab transcriptional activation. Since the second acid region is located between positions 159 and 190, these results are also consistent with a possible redundant role of the acidic regions.
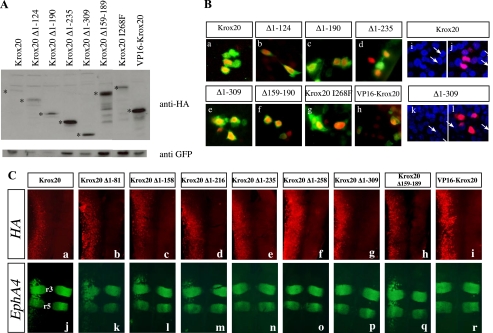
To establish that the observed modifications in target gene expression indeed reflected the transcriptional activity of each deleted protein, we carried out several control experiments. First, to eliminate a contribution of possible variations in the relative stability of the mutant proteins, we performed Western blotting analyses with an antibody directed against the HA epitope on extracts prepared from electroporated embryos. The N-terminally deleted proteins were all present in higher amounts than the wild type Krox20 protein (Fig. 3A). Previously, we demonstrated that the N-terminal part of Krox20 does not contribute to DNA binding activity in vitro (63). We have now verified that the present deletions do not affect the subcellular localization of the proteins by performing immunolabeling experiments that confirmed that all the deleted proteins localized within the cell nucleus (Fig. 3B). Finally, we performed double-immunolabeling analyses to detect both endogenous EphA4 and exogenous HA-labeled Krox20. Analysis of wild type and deletion mutants confirmed that loss of transcriptional activation was not due to inefficient expression of the mutant proteins and that the restriction in ectopic EphA4 expression did not reflect a lack of homogeneity in the distribution of the electroporated Krox20 (Fig. 3C).
FIGURE 3.
Deleted Krox20 proteins are efficiently expressed and localized in the nucleus. A, estimation of the relative amounts of Krox20 mutant proteins following electroporation. Chick embryo neural tubes were co-electroporated with each of the HA-tagged Krox20 deletion mutant plasmids and a GFP expression vector (pEGFP-N1) (43). Western blotting was then performed using an anti-HA antibody and an anti-GFP antibody for normalization. The bona fide deleted Krox20 proteins are marked with asterisks. Their relative amounts are higher than or equivalent to that of the wild type protein. B, subcellular localization of the deleted Krox20 proteins. Chick embryo neural tubes were co-electroporated with pEGFP-N1 and constructs encoding wild type or mutant HA-tagged Krox20, as indicated. Double immunolabeling against HA and GFP was performed and the nuclei were counterstained with Hoechst 33342. Comparisons of anti-HA (red) and anti-GFP (green) staining (a-h) or of anti-HA (red) and Hoechst (blue) staining (i-l) indicate that the various Krox20 mutants are essentially nuclear. i and k, Hoechst staining only. a, i, and j correspond to the same field as e, k, and l. The arrows point to the nuclei positive for Krox20 in these latter cases. C, ectopic target gene expression reflects mutant Krox20 protein transcriptional activity. Chick embryo neural tubes were electroporated on the left side with wild type or truncated HA-tagged Krox20 expression constructs, as indicated, double-immunolabeled for the HA epitope (a-i; red) and the EphA4 protein (j-r; green), and flat mounted. These data establish that loss of transactivation by constructs having lost the 235 N-terminal amino acids is not due to lack of expression. It also shows that the anterior-posterior restriction of the EphA4 ectopic expression domain is not accounted for by the area of efficient electroporation.
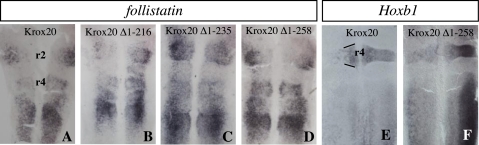
We then investigated the cases of two other target genes, follistatin and Hoxb1, previously shown to be repressed by Krox20 (29, 37). Deletion Krox20 Δ1-216 repressed follistatin like the complete protein, whereas further deletions up to positions 235 and 258 did not (Table 1 and Fig. 4, A-D). Regulation of Hoxb1 showed different requirements, since deletion of the 258 N-terminal amino acids did not prevent repression (Table 1 and Fig. 4, E and F). The limited role of the N-terminal part of the protein in Hoxb1 repression is consistent with previous work, in which a Hoxb1 repression domain was mapped to the zinc finger region (43).
FIGURE 4.
Localization of a follistatin repression domain. A-D, chick embryo neural tubes were electroporated on the left side with wild type or truncated Krox20 expression constructs, as indicated, in situ hybridized with the follistatin probe, and flat mounted. This reveals a partial repression of follistatin gene expression, which is lost upon deletion of the 235 N-terminal amino acids. E and F, similar analysis of Hoxb1 expression shows a repression by Krox20, which is maintained upon elimination of most of the N-terminal region, with, however, a slight reduction. This latter effect might be an indirect consequence of the lack of induction of endogenous Krox20 by Krox20 Δ1-258.
In conclusion, this analysis indicates that at least three different Krox20 domains are required for transcriptional regulation of the tested targets; region 1-190 is necessary for the activation of Krox20 and Nab, and region 217-235 is necessary for the activation of EphA4 and the repression of follistatin, whereas the zinc finger domain is sufficient for the repression of Hoxb1. These data raise the possibility that each of these domains might contain interaction interfaces for different co-factors that mediate transcriptional specificity.
Nab Acts as a Krox20 Antagonist for Transcriptional Activation in the Hindbrain—Among Krox20 interactors, Nab proteins were shown to bind to the R1 domain of Krox20 and to behave either as antagonist (42, 49-51) or as positive modulators (52) of Krox20-activating function in cultured cells. Therefore, we performed experiments designed to clarify the role of Krox20-Nab interactions during hindbrain segmentation. We first performed gain-of-function experiments involving in ovo electroporation of mouse Nab1 or Nab2 expression plasmids, followed by the analysis of the expression of Krox20 targets. Both Nab1 and Nab2 repressed Krox20 and EphA4 genes (Fig. 5, A and E). Repression is mainly observed in the dorsal region, since electroporation is more efficient in this part of the neural tube. Nab was also repressed by Nab1 but not significantly by Nab2 (Fig. 5, B and F). In contrast, EphA4 was efficiently repressed by Nab2 but weakly repressed by Nab1 (Fig. 5, C and G). As expected, Hoxb1, which is not expressed in the Krox20-positive territory, was not affected by ectopic Nab expression (Fig. 5, D and H). Altogether, these data support the view that Nabs antagonizes Krox20 activation function in the hindbrain, with some specificity in the activities of Nab1 and Nab2.
FIGURE 5.
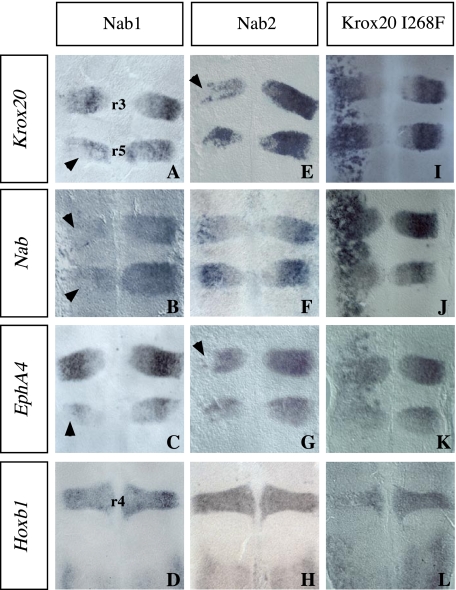
Nab factors antagonize Krox20-activating functions. Chick embryo neural tubes were electroporated on the left side with the expression constructs indicated above each column, in situ hybridized with the probes indicated on the left, and flat mounted. A-H, Krox20 is repressed following both mouse Nab1 and Nab2 overexpression (A and E; the arrowheads point to regions where repression is the strongest). Nab is also repressed by Nab1 (B) but not significantly by Nab2 (F). In contrast, EphA4 is only weakly repressed by Nab1 (C, arrowhead) but more efficiently by Nab2 (G, arrowhead). Hoxb1 expression is not affected by Nab1 or Nab2 (D and H). Note that the repression is mostly observed in the dorsal part of the neural tube, reflecting a higher efficiency of electroporation, as observed previously (37). Similar effects are observed in cases of activation (see I, for instance. I-L, the Krox20 I268F protein very efficiently activates Krox20, Nab, and EphA4 and represses Hoxb1 like the wild type protein. Anterior is up. r, rhombomere.
Our external deletions were not informative about the role of the Krox20-Nab interaction, since the R1 domain, which is required for this interaction, is located downstream to the two regions required for transcriptional activation (Fig. 1 and Fig. S1). Therefore, to further investigate the implication of the Krox20-Nab interaction in the regulation of Krox20 targets, we introduced a mutation changing an isoleucine into phenylalanine at position 268 in the R1 domain (I268F), which was previously shown to abolish the interaction between Nab and Krox20 or Krox24 (42). In ovo electroporation of a construct encoding a complete Krox20 carrying this mutation (Krox20I268F) led to very efficient activation of endogenous Krox20, Nab, and EphA4 and to repression of Hoxb1 (Fig. 5, I-L). Since Krox20 is capable of activating Nab along the entire neural tube, our data are consistent with the following interpretation: wild type Krox20 activates Nab, which partially antagonizes its up-regulatory activity and therefore limits transcriptional activation of its targets, whereas Krox20I268F escapes this limitation. In conclusion, altogether our data suggest that Nab acts as a negative modulator of the different Krox20-positive transcriptional activities in the hindbrain.
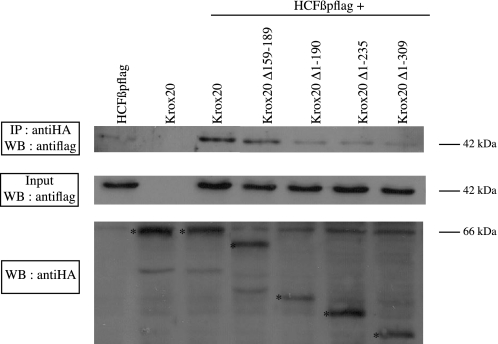
Analysis of Krox20-HCF Interaction—We have shown that the 190 N-terminal amino acids of Krox20 are required for transactivation of Krox20 and Nab. In addition, it has been reported that in cultured cells, HCF-1 stimulates activation by Krox20 of a co-transfected reporter and that a putative HCF binding motif (HBM) is located at amino acid positions 162-165, within the second acidic region of Krox20 (55). To investigate whether HCF could act as a Krox20 co-factor during hindbrain segmentation, we first analyzed HCF expression in the chick developing neural tube. Two HCF (HCF-1 and HCF-2) genes were described in mammals, but only one HCF family member is known in the chick. In situ hybridization revealed that the chick gene is expressed in a decreasing rostro-caudal gradient in the neural tube, overlapping with Krox20 expression domains (Fig. S2, E-J) and thus may be available for cooperation with Krox20. We then tested the requirement of the HBM in Krox20 activation function. For this purpose, we generated an internal deletion, Δ159-189, eliminating the HBM as well as the second acidic region (Fig. 1). Surprisingly, this internal deletion was still able to activate Krox20 and Nab expression (Fig. 2, H and O), indicating that the previously identified HBM was not required for Krox20 and Nab activation. To clarify this issue, we investigated HCF-Krox20 interaction using co-immunoprecipitation experiments. Expression constructs for wild type or mutant Krox20 were co-transfected into COS-7 cells with a plasmid encoding a FLAG-tagged version of the β-propeller domain of HCF-1, which is known to be responsible for the interaction with the HBM (64, 65). Co-immunoprecipitations confirmed that the HCF-1 β-propeller specifically interacted with wild type Krox20 (Fig. 6). Elimination of 190 or more N-terminal amino acids, covering the region that is required for Krox20 and Nab activation, reduced co-immunoprecipitation of HCF-1 to background levels, indicating a significant reduction or elimination of the interaction (Fig. 6). In contrast, the internal deletion (Δ159-189), which does not prevent transcriptional activation, only marginally affected the interaction with HCF-1 (Fig. 6). These data indicate that HCF-1 binding correlates with transcriptional activation and that, in contrast to what was proposed previously (55), the HBM at positions 162-165 is not required for binding to Krox20, suggesting that the N-terminal part of Krox20 contains an additional site(s) for interaction with HCF-1. Since no other canonical HCF binding motif ((D/E)HXY) is present in the 1-190 N-terminal region of Krox20, this interaction must involve another type of motif.
FIGURE 6.
Analysis of Krox20-HCF interaction. This was investigated by co-immunoprecipitation of HA-tagged Krox20 with FLAG-tagged HCF-1 β-propeller. COS-7 cells were transfected with the expression plasmids indicated above each line. Cell lysates were subjected to immunoprecipitation (IP) with anti-HA antibody, and the precipitates were analyzed by Western blotting (WB) using anti-FLAG antibody. A 42 kDa band corresponding to the HCF-1 β-propeller is observed after co-transfection with wild type Krox20. Some nonspecific precipitation of the HCF-1 β-propeller occurs in the absence of Krox20 (first lane). Elimination of the N-terminal 190 amino acids of Krox20 reduces co-immunoprecipitation to background levels, whereas the internal deletion (Δ159-189) preserves the interaction. Analysis of the input (10%) indicates that the levels of HCF-1 β-propeller are identical in all cases. The Western blotting analysis of the Krox20 mutant proteins (the bands corresponding to the bona fide proteins are marked with asterisks) is shown underneath. Consistent results have been obtained in three independent experiments.
Involvement of Krox20 Acidic Domains in Transcriptional Activation—The results summarized in Table 1 are consistent with an implication of the two Krox20 acidic domains in the transcriptional activation of Krox20 and Nab, although they indicate that elimination of only one of them does not significantly affect activity. To investigate whether an acidic domain is sufficient for Krox20 activation function, we grafted the acidic domain of VP16 to a version of Krox20 deleted of most of the N-terminal region (Krox20 Δ1-309) to obtain VP16-Krox20 (Fig. 1). This portion of the VP16 acidic domain does not contain any HCF binding motif, and consistently we found that VP16-Krox20 did not efficiently interact with HCF-1 (data not shown). Functional analysis in electroporated chick embryos indicated that VP16-Krox20 was able to activate Nab expression very efficiently (Fig. 2P), whereas it was completely inactive on Krox20 and EphA4 (Fig. 2, I and W).
In conclusion, our data indicate that, in the case of Nab, a strong acidic domain appears to circumvent the deletion of the N-terminal region, supporting the implication of the endogenous acidic domains in Nab regulation. In the case of Krox20, the involvement of the acid regions cannot be excluded, although they are not sufficient. This suggests the requirement of another motif, which might be the HCF binding site(s) or still another interaction domain(s) located in the 1-190 region. In any case, the striking differences observed with the Krox20 and Nab targets indicate that their regulation involves molecular mechanisms that are at least partly distinct, despite the requirement of the same region of the Krox20 protein.
DISCUSSION
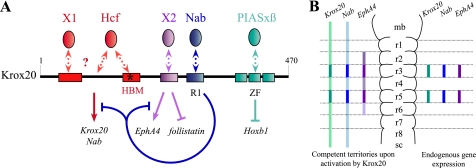
Transcriptional Regulation by Krox20 Involves Various Molecular Mechanisms—In this work, we have performed the first in vivo and systematic structure-function analysis of the Krox20 protein, focusing on its activity in the developing hindbrain. This analysis revealed an unexpected diversity in the modes of action of a transcription factor for the regulation of its transcriptional targets in a single tissue. Differences have been detected at three levels. (i) Three of the target genes (Krox20, Nab, and EphA4) are activated by Krox20 and two (follistatin and Hoxb1) are repressed. (ii) The regulation involves different domains of the Krox20 protein and presumably different interactors, as schematized in Fig. 7A. Hence, Krox20 and Nab activation requires elements located in the N-terminal 1-190 region, whereas this region is not necessary for EphA4 induction, which necessitates a short domain located between positions 216 and 235. Repression of follistatin requires the same 216-235 domain, whereas only the DNA binding domain appears necessary for Hoxb1 repression. Furthermore, although Krox20 and Nab depend on the same Krox20 region, they are likely to rely on partially different sets of cofactors. (iii) The competence for target gene activation shows regional variations in the neural tube, as summarized in the legend to Fig. 7B. Krox20 cannot activate EphA4 rostrally to the r2/r1 boundary, whereas there is no rostral limit to the activation of Krox20 and Nab.
FIGURE 7.
Schematic representation of Krox20 functional organization and of its competence territories in the hindbrain. A, several distinct domains are involved in the different Krox20 transcriptional activities and constitute established or putative binding sites for co-factors. The N-terminal part of the protein, which contains two acidic regions (red boxes), interacts with HCF-1, and at least another unknown cofactor (X1) is required for Krox20 and Nab activation. A domain located between positions 216 and 235 (purple box) is required for EphA4 activation and follistatin repression and may bind one or several unknown factors (X2). The previously described R1 domain (blue box) binds Nab factors that antagonize Krox20 activation functions and establish a negative feedback loop. The zinc finger (ZF) DNA binding domain is sufficient for Hoxb1 repression via the interaction with PIASxβ. B, endogenous Krox20, Nab, and EphA4 expression is restricted to r3 and r5 (right). Upon exogenous Krox20 expression, ectopic activation of the three targets is observed (left) with distinct territories of competence; Krox20 and Nab expression shows no restriction, and EphA4 expression is confined to the r2-r4-r6 region with a decreasing rostro-caudal gradient. mb, midbrain; r, rhombomere; sc, spinal cord.
These different properties in the transcriptional regulation of target genes suggest the involvement of distinct molecular mechanisms and of specific co-factors. In the present work, we delineate the precise role of the Nab co-factor in the hindbrain, provide evidence that HCF-1 interacts with the N-terminal region of Krox20, and discuss its role as well as those of other co-factors (see below).
Concerning the different regulatory domains identified within the Krox20 protein, there is an intriguing coincidence; the same region, the 216-235 domain, is required for both activation of EphA4 and repression of follistatin (Fig. 7A). The small size of this domain raises the possibility that the same motif might be involved in the two processes, mediated by the binding of a common interactor. Since Krox20 is a direct transcriptional activator of EphA4 (39), it is possible that Krox20 also directly activates another factor according to the same mechanism and that this factor is involved in the repression of follistatin.
Nab Acts as a Negative Feedback Modulator of Krox20 in the Hindbrain—Previous studies performed in cultured cells have shown that Nab can act either as an antagonist of Krox20 (42, 50, 51) or as a co-activator (52), generating some confusion on its activity in vivo. In the present work, we have established for the first time that Nab antagonizes Krox20-activating functions in an in vivo situation, during hindbrain development. Indeed, ectopic expression of Nab led to repression of the three up-regulated targets tested in this study (Fig. 5). Moreover, we have shown that in ovo electroporation of a dominant-negative version of Nab (66) also leads to overexpression of endogenous Nab in r3 and r5 (data not shown). Since Nab is itself activated by Krox20 in the hindbrain (41) (this work) and binding of Krox20 to the Nab promoter has been revealed by chromatin immunoprecipitation analysis (67), these data suggest that it belongs to a direct, negative feedback loop controlling Krox20 activation function.
The present work is consistent with previous observations indicating that Nab misexpression in the zebrafish embryo results in alterations in hindbrain patterning (41), and it provides a molecular explanation for the previously observed patterning defects. Despite the conclusions derived from the gain-of-function experiments, the double Nab knock-out (54) and the Krox20-I268F knock-in mice (53) do not show any major hindbrain phenotypes. This suggests the existence of redundant mechanisms, in addition to the Nabs, to prevent Krox20 overexpression, presumably because this factor is so critical for the development of the hindbrain.
Complexity of the Krox20 N-terminal Region Required for Nab and Krox20 Activation—Our study investigated the possible implication of another co-factor, HCF-1, in Krox20 function. HCF-1 was identified as an interactor of herpes simplex virus VP16 (68) and contains a transactivation domain (69). The factors interacting with the β-propeller region of HCF-1 usually contain an HBM (70-74). An HBM was previously identified in Krox20, and its disruption was shown to reduce the activation by Krox20 of a co-transfected target reporter in cultured cells, although the consequences for Krox20-HCF interactions were not investigated (55). In the present work, we show that HCF-1 is expressed in the developing hindbrain and that it binds the N-terminal 1-190 region of Krox20, which is required for transcriptional activation of Krox20 and Nab. However, the HBM does not appear to be required for this interaction or for transcriptional activation of these targets. These data therefore suggest that either another Krox20-HCF interaction site is present within the 1-190 domain or HCF interacts indirectly with this region. This domain also contains two acidic regions, neither of which is required for Nab or Krox20 activation. Nevertheless, we have shown that a strong acidic domain can rescue a deleted Krox20 for Nab activation, suggesting that the acidic regions play an important, redundant role in the regulation of the latter target. In the case of Krox20 activation, the acidic regions may also be required, but factors binding to other domains, possibly HCF-1, are likely to be involved in addition (Fig. 7A).
Conclusion—This study reveals a unique diversity and complexity in the mechanisms of transcriptional regulation by Krox20 in the developing hindbrain, raising the possibility that this might constitute an important feature for Krox20 to fulfill its multiple and complex functions. The existence of several types of transcriptional mechanisms mediated by Krox20 raises the possibility that target genes controlled by the same mechanisms are more strictly co-regulated and that their classification according to this basis might be of functional significance. This work illustrates the interest of conducting similar in vivo analyses on other master regulatory genes involved in the control of numerous targets.
Supplementary Material
Acknowledgments
We are grateful to Christine Vesque-Kishtoo, Charlotte Labalette, and Julien Ghislain for critical reading of the manuscript.
This work was supported by grants from the Institut National de la Santéet de la Recherche Médicale, Ministère de l'Education Nationale de la Recherche et de la Technologie (MENRT), Agence Nationale pour la Recherche, Association pour la Recherche sur le Cancer, and Association Française contre les Myopathies.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: HA, hemagglutinin; HBM, HCF binding motif.
References
- 1.Lumsden, A. (1990) Trends Neurosci. 13 329-335 [DOI] [PubMed] [Google Scholar]
- 2.Lumsden, A., and Krumlauf, R. (1996) Science 274 1109-1115 [DOI] [PubMed] [Google Scholar]
- 3.Wingate, R. J., and Lumsden, A. (1996) Development 122 2143-2152 [DOI] [PubMed] [Google Scholar]
- 4.Lumsden, A., and Keynes, R. (1989) Nature 337 424-428 [DOI] [PubMed] [Google Scholar]
- 5.Clarke, J. D., Erskine, L., and Lumsden, A. (1998) Dev. Dyn. 212 14-26 [DOI] [PubMed] [Google Scholar]
- 6.Birgbauer, E., Sechrist, J., Bronner-Fraser, M., and Fraser, S. (1995) Development 121 935-945 [DOI] [PubMed] [Google Scholar]
- 7.Bronner-Fraser, M. (1995) Perspect. Dev. Neurobiol. 3 53-62 [PubMed] [Google Scholar]
- 8.Ghislain, J., Desmarquet-Trin-Dinh, C., Gilardi-Hebenstreit, P., Charnay, P., and Frain, M. (2003) Development 130 941-953 [DOI] [PubMed] [Google Scholar]
- 9.Kulesa, P. M., and Fraser, S. E. (2000) Development 127 1161-1172 [DOI] [PubMed] [Google Scholar]
- 10.Lumsden, A., Sprawson, N., and Graham, A. (1991) Development 113 1281-1291 [DOI] [PubMed] [Google Scholar]
- 11.Serbedzija, G. N., Bronner-Fraser, M., and Fraser, S. E. (1992) Development 116 297-307 [DOI] [PubMed] [Google Scholar]
- 12.Trainor, P. A., and Krumlauf, R. (2000) Nat. Rev. Neurosci. 1 116-124 [DOI] [PubMed] [Google Scholar]
- 13.Trainor, P. A., Sobieszczuk, D., Wilkinson, D., and Krumlauf, R. (2002) Development 129 433-442 [DOI] [PubMed] [Google Scholar]
- 14.Kontges, G., and Lumsden, A. (1996) Development 122 3229-3242 [DOI] [PubMed] [Google Scholar]
- 15.Fraser, S., Keynes, R., and Lumsden, A. (1990) Nature 344 431-435 [DOI] [PubMed] [Google Scholar]
- 16.Barrow, J. R., Stadler, H. S., and Capecchi, M. R. (2000) Development 127 933-944 [DOI] [PubMed] [Google Scholar]
- 17.Bell, E., Wingate, R. J., and Lumsden, A. (1999) Science 284 2168-2171 [DOI] [PubMed] [Google Scholar]
- 18.Chisaka, O., Musci, T. S., and Capecchi, M. R. (1992) Nature 355 516-520 [DOI] [PubMed] [Google Scholar]
- 19.Choe, S. K., and Sagerstrom, C. G. (2004) Dev. Biol. 271 350-361 [DOI] [PubMed] [Google Scholar]
- 20.Deflorian, G., Tiso, N., Ferretti, E., Meyer, D., Blasi, F., Bortolussi, M., and Argenton, F. (2004) Development 131 613-627 [DOI] [PubMed] [Google Scholar]
- 21.Frohman, M. A., Martin, G. R., Cordes, S. P., Halamek, L. P., and Barsh, G. S. (1993) Development 117 925-936 [DOI] [PubMed] [Google Scholar]
- 22.Lufkin, T., Dierich, A., LeMeur, M., Mark, M., and Chambon, P. (1991) Cell 66 1105-1119 [DOI] [PubMed] [Google Scholar]
- 23.McKay, I. J., Muchamore, I., Krumlauf, R., Maden, M., Lumsden, A., and Lewis, J. (1994) Development 120 2199-2211 [DOI] [PubMed] [Google Scholar]
- 24.McNulty, C. L., Peres, J. N., Bardine, N., van den Akker, W. M., and Durston, A. J. (2005) Development 132 2861-2871 [DOI] [PubMed] [Google Scholar]
- 25.Pasini, A., and Wilkinson, D. G. (2002) BioEssays 24 427-438 [DOI] [PubMed] [Google Scholar]
- 26.Rijli, F. M., Mark, M., Lakkaraju, S., Dierich, A., Dolle, P., and Chambon, P. (1993) Cell 75 1333-1349 [DOI] [PubMed] [Google Scholar]
- 27.Rossel, M., and Capecchi, M. R. (1999) Development 126 5027-5040 [DOI] [PubMed] [Google Scholar]
- 28.Schneider-Maunoury, S., Topilko, P., Seitandou, T., Levi, G., Cohen-Tannoudji, M., Pournin, S., Babinet, C., and Charnay, P. (1993) Cell 75 1199-1214 [DOI] [PubMed] [Google Scholar]
- 29.Seitanidou, T., Schneider-Maunoury, S., Desmarquet, C., Wilkinson, D. G., and Charnay, P. (1997) Mech. Dev 65 31-42 [DOI] [PubMed] [Google Scholar]
- 30.Studer, M., Lumsden, A., Ariza-McNaughton, L., Bradley, A., and Krumlauf, R. (1996) Nature 384 630-634 [DOI] [PubMed] [Google Scholar]
- 31.Waskiewicz, A. J., Rikhof, H. A., Hernandez, R. E., and Moens, C. B. (2001) Development 128 4139-4151 [DOI] [PubMed] [Google Scholar]
- 32.Chavrier, P., Zerial, M., Lemaire, P., Almendral, J., Bravo, R., and Charnay, P. (1988) EMBO J. 7 29-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph, L. J., Le Beau, M. M., Jamieson, G. A., Jr., Acharya, S., Shows, T. B., Rowley, J. D., and Sukhatme, V. P. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 7164-7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson, D. G., Bhatt, S., Chavrier, P., Bravo, R., and Charnay, P. (1989) Nature 337 461-464 [DOI] [PubMed] [Google Scholar]
- 35.Voiculescu, O., Taillebourg, E., Pujades, C., Kress, C., Buart, S., Charnay, P., and Schneider-Maunoury, S. (2001) Development 128 4967-4978 [DOI] [PubMed] [Google Scholar]
- 36.Swiatek, P. J., and Gridley, T. (1993) Genes Dev. 7 2071-2084 [DOI] [PubMed] [Google Scholar]
- 37.Giudicelli, F., Taillebourg, E., Charnay, P., and Gilardi-Hebenstreit, P. (2001) Genes Dev. 15 567-580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellitzer, G., Xu, Q., and Wilkinson, D. G. (2000) Curr. Opin. Neurobiol. 10 400-408 [DOI] [PubMed] [Google Scholar]
- 39.Theil, T., Frain, M., Gilardi-Hebenstreit, P., Flenniken, A., Charnay, P., and Wilkinson, D. G. (1998) Development 125 443-452 [DOI] [PubMed] [Google Scholar]
- 40.Chomette, D., Frain, M., Cereghini, S., Charnay, P., and Ghislain, J. (2006) Development 133 1253-1262 [DOI] [PubMed] [Google Scholar]
- 41.Mechta-Grigoriou, F., Garel, S., and Charnay, P. (2000) Development 127 119-128 [DOI] [PubMed] [Google Scholar]
- 42.Russo, M. W., Sevetson, B. R., and Milbrandt, J. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 6873-6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Dominguez, M., Gilardi-Hebenstreit, P., and Charnay, P. (2006) EMBO J. 25 2432-2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manzanares, M., Nardelli, J., Gilardi-Hebenstreit, P., Marshall, H., Giudicelli, F., Martinez-Pastor, M. T., Krumlauf, R., and Charnay, P. (2002) EMBO J. 21 365-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nonchev, S., Vesque, C., Maconochie, M., Seitanidou, T., Ariza-Mc-Naughton, L., Frain, M., Marshall, H., Sham, M. H., Krumlauf, R., and Charnay, P. (1996) Development 122 543-554 [DOI] [PubMed] [Google Scholar]
- 46.Sham, M. H., Vesque, C., Nonchev, S., Marshall, H., Frain, M., Gupta, R. D., Whiting, J., Wilkinson, D., Charnay, P., and Krumlauf, R. (1993) Cell 72 183-196 [DOI] [PubMed] [Google Scholar]
- 47.Gilardi-Hebenstreit, P., Nieto, M. A., Frain, M., Mattei, M. G., Chestier, A., Wilkinson, D. G., and Charnay, P. (1992) Oncogene 7 2499-2506 [PubMed] [Google Scholar]
- 48.Taneja, R., Thisse, B., Rijli, F. M., Thisse, C., Bouillet, P., Dolle, P., and Chambon, P. (1996) Dev. Biol. 177 397-412 [DOI] [PubMed] [Google Scholar]
- 49.LeBlanc, S. E., Jang, S. W., Ward, R. M., Wrabetz, L., and Svaren, J. (2006) J. Biol. Chem. 281 5453-5460 [DOI] [PubMed] [Google Scholar]
- 50.Svaren, J., Sevetson, B. R., Apel, E. D., Zimonjic, D. B., Popescu, N. C., and Milbrandt, J. (1996) Mol. Cell. Biol. 16 3545-3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swirnoff, A. H., Apel, E. D., Svaren, J., Sevetson, B. R., Zimonjic, D. B., Popescu, N. C., and Milbrandt, J. (1998) Mol. Cell. Biol. 18 512-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sevetson, B. R., Svaren, J., and Milbrandt, J. (2000) J. Biol. Chem. 275 9749-9757 [DOI] [PubMed] [Google Scholar]
- 53.Desmazières, A., Decker, L., Vallat, J. M., Charnay, P., and Gilardi-Hebenstreit, P. (2008) J. Neurosci. 28 5891-5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le, N., Nagarajan, R., Wang, J. Y., Svaren, J., LaPash, C., Araki, T., Schmidt, R. E., and Milbrandt, J. (2005) Nat. Neurosci. 8 932-940 [DOI] [PubMed] [Google Scholar]
- 55.Luciano, R. L., and Wilson, A. C. (2003) J. Biol. Chem. 278 51116-51124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gillian, A. L., and Svaren, J. (2004) J. Biol. Chem. 279 9056-9063 [DOI] [PubMed] [Google Scholar]
- 57.Das, G., Hinkley, C. S., and Herr, W. (1995) Nature 374 657-660 [DOI] [PubMed] [Google Scholar]
- 58.Giudicelli, F., Gilardi-Hebenstreit, P., Mechta-Grigoriou, F., Poquet, C., and Charnay, P. (2003) Dev. Biol. 253 150-162 [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson, D. G., and Nieto, M. A. (1993) Methods Enzymol. 225 361-373 [DOI] [PubMed] [Google Scholar]
- 60.Sajjadi, F. G., and Pasquale, E. B. (1993) Oncogene 8 1807-1813 [PubMed] [Google Scholar]
- 61.Graham, A., and Lumsden, A. (1996) Development 122 473-480 [DOI] [PubMed] [Google Scholar]
- 62.Guthrie, S., Muchamore, I., Kuroiwa, A., Marshall, H., Krumlauf, R., and Lumsden, A. (1992) Nature 356 157-159 [DOI] [PubMed] [Google Scholar]
- 63.Vesque, C., and Charnay, P. (1992) Nucleic Acids Res. 20 2485-2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.LaBoissiere, S., Walker, S., and O'Hare, P. (1997) Mol. Cell. Biol. 17 7108-7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson, A. C., Freiman, R. N., Goto, H., Nishimoto, T., and Herr, W. (1997) Mol. Cell. Biol. 17 6139-6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Svaren, J., Sevetson, B. R., Golda, T., Stanton, J. J., Swirnoff, A. H., and Milbrandt, J. (1998) EMBO J. 17 6010-6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srinivasan, R., Jang, S. W., Ward, R. M., Sachdev, S., Ezashi, T., and Svaren, J. (2007) BMC Mol. Biol. 8 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerster, T., and Roeder, R. G. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 6347-6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luciano, R. L., and Wilson, A. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13403-13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freiman, R. N., and Herr, W. (1997) Genes Dev. 11 3122-3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin, J., Puigserver, P., Donovan, J., Tarr, P., and Spiegelman, B. M. (2002) J. Biol. Chem. 277 1645-1648 [DOI] [PubMed] [Google Scholar]
- 72.Lu, R., Yang, P., Padmakumar, S., and Misra, V. (1998) J. Virol. 72 6291-6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahajan, S. S., Little, M. M., Vazquez, R., and Wilson, A. C. (2002) J. Biol. Chem. 277 44292-44299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wysocka, J., Myers, M. P., Laherty, C. D., Eisenman, R. N., and Herr, W. (2003) Genes Dev. 17 896-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.