Abstract
The family of NFAT (nuclear factor of activated T-cells) transcription factors plays an important role in cytokine gene regulation. In peripheral T-cells NFATc1 and -c2 are predominantly expressed. Because of different promoter and poly(A) site usage as well as alternative splicing events, NFATc1 is synthesized in multiple isoforms. The highly inducible NFATc1/A contains a relatively short C terminus, whereas the longer, constitutively expressed isoform NFATc1/C spans an extra C-terminal peptide of 246 amino acids. Interestingly, this NFATc1/C-specific terminus can be highly sumoylated. Upon sumoylation, NFATc1/C, but not the unsumoylated NFATc1/A, translocates to promyelocytic leukemia nuclear bodies. This leads to interaction with histone deacetylases followed by deacetylation of histones, which in turn induces transcriptionally inactive chromatin. As a consequence, expression of the NFATc1 target gene interleukin-2 is suppressed. These findings demonstrate that the modification by SUMO (small ubiquitin-like modifier) converts NFATc1 from an activator to a site-specific transcriptional repressor, revealing a novel regulatory mechanism for NFATc1 function.
Differentiation of peripheral T lymphocytes progresses from the naïve to effector and memory stages (1). The various subpopulations are defined by patterns of cytokines produced and functional abilities. Similar to other CD4+ T-cells, CD4+CD25+ regulatory T-cells are highly dependent on, but do not produce IL-25 themselves. In general, lymphokine expression in T-cells, including IL-2, is controlled to a substantial degree by the action of NFAT transcription factors (2-4).
NFAT proteins belong to a family of transcription factors whose genuine Ca2+-dependent members are designated as NFATc1-c4. NFATc1 and NFATc2 are highly expressed in peripheral T-cells and control, in particular, the expression of lymphokines. Additional targets controlled by NFATs are the promoters of p21Waf1 (CDKN1A), CD40 ligand (CD40LG), CD95 ligand (FASLG), and NFATC1 itself. Studies on NFAT-deficient mice suggest that NFATc1 and -c2 have divergent functions in lymphokine gene expression in vivo. Although NFATc2-/- mice suffer from lymphoproliferative disorder, peripheral T-cells from NFATc1-/- mice are rather defective in proliferation. This suggests contrasting roles in T-cell proliferation and activation for NFATc1 and -c2. The effects on IL-2, the lymphokine important for proliferation, are negligible in the case of NFATc2-/- and ambiguous for NFATc1-/-, i.e. one of two knock-out strains investigated synthesized more IL-2 after secondary stimulation (5), raising the possibility that under some circumstances NFATc1 could have a negative effect on IL-2 expression.
All NFATs share a DNA binding domain which is very similar in its conformation to the Rel DNA binding domain of Rel/NF-κB factors and, therefore, was designated as Rel similarity domain (2-4). The N terminus harbors a strong transactivation domain (TAD) (designated as TAD-A) and a regulatory domain. In T lymphocytes, NFATc1 is expressed in six isoforms (6). The NFATc1 isoforms c1/α and c1/β contain either the N-terminal α peptide spanning 42 amino acids (aa) or the β peptide spanning 29 aa, whereas c1/A, c1/B, and c1/C differ in the length of their C termini. The long isoforms c1/αC and βC harbor an extra C-terminal peptide of 246 aa constituting an additional transactivation domain, TAD-B (7). Interestingly, the long C terminus of NFATc1 is homologous to the C terminus of NFATc2 (7). Therefore, the question arises of whether NFATc1/C functionally resembles NFATc2 or has a function of its own, different from NFATc1/A and NFATc2.
Functions of proteins can be achieved by post-translational modification, for example with SUMO. Similar to ubiquitinylation, sumoylation is mediated by activating E1, conjugating E2, and ligating E3 enzymes. Sumoylation occurs mostly within the consensus core motif Ψ-Lys-X-Glu, where Ψ is a large hydrophobic aa (8, 9). Sumoylation often correlates with relocalization of the targeted proteins toward promyelocytic leukemia nuclear bodies (PML-nbs) (10) or heterochromatin (11). Accordingly, although sumoylation of transcription factors might occasionally enhance, most often it represses their activation capacity (12).
PML-nbs are specific nuclear matrix-associated subdomains (13), which act as storage sites for regulatory proteins (14). They house various kinds of protein factors involved in gene regulation, DNA repair, tumor suppression, and apoptosis. Accordingly, posttranslational modifications and protein-protein interactions with residual proteins occur. Established members of PML-nbs are histone acetylases and deacetylases (HDACs). (Hyper)-acetylation at lysines in N-terminal tails of core histones regulates transcription. Hypoacetylation on the other hand, poses the locus for subsequent changes such as methylation of the deacetylated lysines, leading to heterochromatinization and silencing (15).
Here we show that NFATc1 is sumoylated endogenously in CD4+ T-cells, because the long isoform of NFATc1, NFATc1/C, is targeted at two sites within its C-specific terminus by SUMO1. Sumoylation directs NFATc1/C into PML-nbs. Furthermore, sumoylated NFATc1/C recruits HDACs, leading to deacetylation of histones within the IL-2 promoter. This is in agreement with the localization of NFATc1/C to heterochromatin. Surprisingly, only some NFATc1 target genes, especially IL-2, are suppressed, whereas others, such as IFNγ and IL-13, are even enhanced. This indicates that sumoylation is necessary for the fine-tuning of NFATc1 function.
EXPERIMENTAL PROCEDURES
Cell Culture and Stimulations—Human lymphoid A3.01 and murine EL-4 thymoma cells as well as 293T human embryonic kidney (HEK) cells were cultured and stimulated as before (11).
Conventional CD4+ T-cells (GK1.5-FITC) and CD25+ Treg cells (7D4-PE) were isolated (16) from spleen cells by positive selection using high gradient MACS (Miltenyi Biotec, Bergisch-Gladbach, Germany) according to the manufacturer's instructions. The CD4-sort as well as the CD25-sort was performed twice. CD4+ T-cells were subsequently depleted from CD25+ Treg cells using monoclonal antibody PC61 and enriched >99%. They showed no proliferative response in the presence of Con A or soluble anti-CD3 monoclonal antibody, indicating negligible contamination with accessory cells. CD25+-enriched Treg cells were additionally depleted from CD8+ T-cells using anti-CD8 Dynabeads (Dynal Biotech, Hamburg, Germany), and the purity of the resulting CD25+ Treg cells was typically >95%. Preactivation of CD25+ Treg cells was performed using a combination of plate-bound anti-CD3 monoclonal antibody (mAb; 145-2C11, 3 μg/ml) and anti-CD28 mAb (37.51; 10 μg/ml) in the presence of murine recombinant interleukin-2 (100 ng/ml). After 72 h the cells were harvested and cultured again in the absence of any stimulus for additional 48 h.
DNA Constructs—Luciferase reporter gene plasmid pTATAluc+ (17) containing 293 bp of the proximal murine IL-2 promoter (18), retroviral vectors pEGZ/MCS (19) and pEGZ/MCS-HA, and pHA-hNFATc1/A (HA-c1/A) and pHA-hNFATc1/C (HA-c1/C) expression constructs have been described previously (20). In addition, NFATc1 isoforms, mutants, and C-terminal peptides were fused at their 3′ ends to the modified hormone binding domain of the estrogen receptor as described for a chimeric cMycER™ construct (21) or, accordingly, to enhanced yellow fluorescence protein (EYFP) (bicistronic vector pIZ-EYFP). In the case of the C-terminal peptides fused to the estrogen receptor (ER), a nuclear localization signal (5′-GATCTACCTTACGTTTTTTCTTTGGTG-3′) was added in between. SUMO1 was fused in-frame to the 5′ end of NFATc1. The mutations of lysines to arginines within the SUMO sites of NFATc1 were designed as K349R (5′-CAGTGGCGCTCCGGGTGGAGCCC-3′, K702R (5′-CGTTCCAATTATACGAACAGAACCCAC-3′), and K914R (5′-CTGTAACGGTCCGGCGAGAGCC-3′).
Transient transfection and reporter gene assay. 293T HEK cells were transfected by using the conventional calcium phosphate technique and EL-4 cells by a standard DEAE/Dextran protocol. 36 h post-transfection, luciferase activity was measured from the cells that were either left untreated or treated with TPA plus ionomycin (T/I) overnight. Relative light units were corrected for the transfection efficacy due to total protein concentrations (relative luciferase activity = 0.5/protein concentrations × absolute luciferase activity). Normalized mean values of at least three independent experiments are depicted in relative light units or as -fold activation over empty vector control in the cotransfection experiments. To check the equal expression pattern of the exogenously expressed proteins, Western blots were performed from the same protein extracts loading exactly the amount of lysate as revealed for calculation of luciferase activity. EL-4 cells were evaluated by green fluorescence as well as cotransfected with pCMV-GLuc (New England Biolabs) as a reference. For immunofluorescence (IF), 293T HEK cells were transfected by Superfect method according to the Qiagen protocol with various HA-NFATc1 and FLAG-SUMO-1 constructs. After 48 h of transfection, cells were stimulated with 10 ng of TPA, 1 μm ionomycin, and 2 mm CaCl2.
Viral Infection, RNA Purification, RNase Protection Assay, Real-time PCR, and Enzyme-linked Immunosorbent Assay—Viral infection was carried out as described previously (22). RNA was purified using Trizol reagent (Invitrogen), and 5 μg of RNA was analyzed by RNase protection assays performed according to the Pharmingen Riboquant™ protocol. As multi-probe template set, the mCK1 or hCK1 set was used. cDNA was synthesized with RevertAid Moloney murine leukemia virus reverse transcriptase following the recommendations of the supplier (MBI Fermentas, St. Leon-Rot, Germany). Real-time PCR was performed using the following oligonucleotides: primers for NFATc1 E1+E3 and E2+E3 (qNFATc1Ex1F 5′-CGGGAGCGGAGAAACTTTGC-3′, qNFATc1Ex2F 5′-AGGACCCGGAGTTCGACTTC-3′, qNFATc1Ex3R 5′-CAGGGTCGAGGTGACACTAG-3′), hypoxanthine-guanine phosphoribosyl transferase (forward), GTT GGA TAC AGG CCA GAC TTT GTT G; HGPRT (reverse), GAG GGT AGG CTG GCC TAT AGG CT. For detection of NFATc2 mRNA, the Quanti-Tect primer assay Mm_NFATc2_SG (Qiagen) was used. Real-time PCR analyses to quantify the expression of NFATc1 E1+E3, NFATc1 E2+E3, NFATc2, and HGPRT mRNAs were performed in triplicate on an iCycler (Bio-Rad) using the Absolute PCR SYBR Green Mixes (Abgene). After normalization of the data according to the expression of HGPRT mRNA, the relative expression levels of NFATc1 E1+E3, NFATc1 E2+E3, and NFATc2 mRNAs were calculated. Enzyme-linked immunosorbent assays were performed for detection of IL-2 secretion using monoclonal antibodies according to the manufacturer's instructions (BD Biosciences).
Immunoprecipitations (IP) and Western or Immunoblot (IB) Assays—For IP, post-transfection 107 cells were resuspended in 1 ml of lysis buffer (50 mm Tris HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, protease inhibitor mixture (Roche Diagnostics), and 20 mm N-ethylmaleimide (Sigma-Aldrich). 50 μl were kept as a lysate control. Otherwise, the whole cell extract was incubated with 3 μg of anti-FLAG (M2, Sigma), 3 μg of anti-Myc (sc-40, Santa Cruz) or 3 μg of NFATc1 antibody (AB1-205, ImmunoGlobe), and 40 μl of protein A or G beads (Upstate Biotechnology, Inc.). The beads were washed 3 times with buffer (50 mm Tris HCl, pH 7.4, 150 mm NaCl, and protease inhibitor mixture). For Western blots, the precipitates and input control lysates were fractionated by 8-12% SDS-PAGE and electroblotted onto a nitrocellulose membrane. For detection, anti-HA (M-12CA, Roche Diagnostics), anti-FLAG, anti-SUMO (sc-5308, Santa Cruz) or anti-Myc and a peroxidase-coupled secondary antibody were used with a standard enhanced chemiluminescence system (Pierce). When endogenous sumoylation was tested, at first nuclei were prepared as for chromatin immunoprecipitation (ChIP) assays, a total of 15 μg of anti-NFATc1 (raised against the common N-terminal fragment of NFATc1, expressed as a fusion with glutathione S-transferase, Pineda Antibody Service, Berlin, Germany or 556602, mouse IgG1, BD Pharmingen) were used for IP, and the subsequent Western blot was analyzed by anti-SUMO (sc-5308, Santa Cruz) and finally detected with SuperSignal West Femto trial kit (Thermo Scientific/Pierce).
ChIP—ChIP analysis was carried out as described (11, 23) but with the addition of 20 mm N-ethylmaleimide (Sigma-Aldrich) within the lysis buffer. 1 × 107 cells were fixed for 5 min at room temperature with 1% formaldehyde. 30 units (A260) of chromatin were immunoprecipitated with 3 μg of rabbit polyclonal anti-acetylhistone 3 H3-Ac (#06-599, Upstate), anti-ERα (sc-542, Santa Cruz), or anti-HDAC2 (#39533, Active Motif) and subsequently by protein-A-Sepharose. DNA was purified from reverse-cross-linked chromatin followed by PCR amplification (1/10 of the immunoprecipitated DNA and input DNA) using primer pairs for either the human IL-2 promoter or the β-actin promoter: human IL-2_for (5′-CTT GCT CTT GTT CAC CA-3′) plus human IL-2_rev (5′-CTG TAC ATT GTG GCA GGA-3′). giving rise to a 493-bp fragment. and hβ-ac_for (5′-CCT CCT CCT CTT CCT CA-3′) plus hβ-ac_rev (5′-CGA GCC ATA AAA GGC AA-3′). giving rise to 193-bp DNA.
IF and Confocal Microscopy—For NFATc1, SUMO-1, and PML triple colocalization as well as for heterochromatin and polymerase II (pol II) colocalization studies, NFATc1-ER-infected A3.01 cells were harvested on slides by cytospins, whereas transiently transfected 293T HEK cells had been grown on coverslips. Cells were fixed in 4% formaldehyde and permeabilized with 0.2% Triton X-100 followed by blocking in 1:20 times diluted donkey serum for 20 min. For primary antibody staining, rabbit polyclonal NFATc1-NT (1:200), mouse monoclonal HA (1:500), mouse SUMO-1 (1:50), goat PML (1:50, sc-9862, Santa Cruz), rabbit polyclonal anti-trimethylhistone H3 (Lys-9, 1:500, 07-442, Upstate), and mouse monoclonal pol II (1:500, 05-623, Upstate) antibody were used. The secondary antibody combination used was anti-rabbit Cy3 (1:500), anti-mouse Cy5 (1:500), and anti-goat Cy2 (1:100) (Jackson ImmunoResearch). The slides were mounted with Fluoromount-G (Southern Biotechnology Associates) containing 4′,6-diamidino-2-phenylindole. Images were collected with a confocal microscope (Leica TCS SP2 equipment, objective lens: HEX PL APO, 40×/1.25-0.75) and analyzed by Leica LCS software. The images were digitally zoomed 6 or 7 times. For endogenous NFATc1 sumoylation, mouse CD4+ and CD4+CD25+ T-cells were collected on slides by cytospins (300 rpm, 2 min) and dried overnight at room temperature. Formaldehyde fixation, permeabilization and staining were performed as mentioned above. Quantitation was done as follows; cells clearly showing more than 50% (usually above 75%) merging of NFAT dots and SUMO/PML-nb dots were considered as a positive score. Roughly 100 cells were evaluated, and the percentage of positive cells was calculated.
RESULTS
NFATc1/C Interacts with the SUMO-conjugating Enzyme Ubc9—A yeast two-hybrid screen was performed using the NFATc1/C-specific C-terminal peptide as bait and a human spleen cDNA library as prey. Ubc9 (37x) and PIAS1 (4x) were detected as the predominant NFATc1/C-specific interaction partners. Ubc9 is the only known SUMO-conjugating enzyme, whereas PIAS1 acts as a substrate-specific SUMO ligase. Indeed, within NFATc1/αC (c1/αC), three SUMO consensus sites were identified at the position of aa 349, 702, and 914, whereas the shortest isoforms of NFATc1/A (c1/A) harbor only the site at aa 349 (Fig. 1A). In NFATc1/βC the corresponding sites are K349, K689, and K901. The N-terminal α- or β-peptides are free of SUMO-consensus sites. In silico analysis predicts a strong sumoylation for the C-terminal SUMO consensus sites at lysines 702 and 914 and a slightly weaker sumoylation for the common SUMO site at lysine 349.6 To create ΔSUMO mutants, the sumoylatable lysines were altered to arginines by site-directed mutagenesis, and the corresponding NFATc1 mutants are designated with K349R, K702R, or K914R for both lysines on the C terminus mutated, K702R/K914R, and in case of the triple mutation, K349R/K702R/K914R. For better comparison, all constructs contain the N-terminal α-peptide.
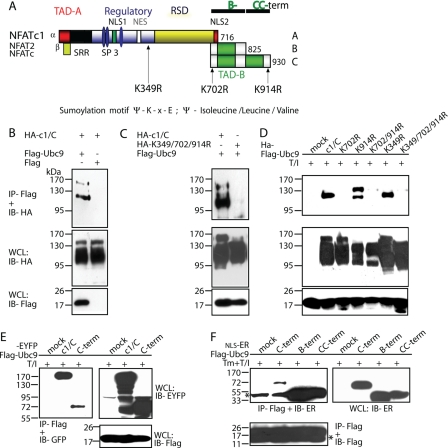
FIGURE 1.
NFATc1/C harbors three SUMO consensus motifs which facilitate interaction with Ubc9. A, schematic representation of NFATc1 isoforms and the SUMO consensus motifs (Ψ-Lys-X-Glu; Ψ= isoleucine/leucine/valine) shown by arrows. The short isoforms harbor only the common Lys-349 site, whereas NFATc1/C contains sites at Lys-702 and -914 in addition. Lysines to arginine exchanges create ΔSUMO mutations. The mutation of both C-terminal lysines is designated as K702R/K914R and of all three sites as K349R/K702R/K914R. An indication for the B-term and CC-term peptides are given. TAD, transactivation domain; RSD, Rel similarity domain; NLS, nuclear localization signal; NES, nuclear export signal; SRR and SP, Ser/Thr phosphorylation sites, respectively. B, NFATc1/C physically interacts with Ubc9 in vivo. HA-tagged c1/C-coding vector was transfected into 293T HEK cells, either in combination with pcDNA expressing FLAG-tagged Ubc9 or FLAG only. After 6 h of stimulation with T/I and a total of 48 h, protein lysates were prepared and subjected to anti-FLAG IP followed by immunoblot detection with anti-HA. Whole cell lysates (WCL) were also directly analyzed by immunoblotting with anti-HA and anti-FLAG, respectively. C, the SUMO site-deficient mutant K349R/K702R/K914R cannot recruit Ubc9. Procedures were the same as in B, except that K349R/K702R/K914R was included. D, the SUMO site at Lys-702 is the most relevant. Procedures were the same as in C but including all different SUMO site deficient mutants. E, the C terminus of NFATc1/C is sufficient for binding Ubc9. Procedures were the same as before, but EYFP-tagged NFATc1/C or only the C terminus were analyzed. F, both the NFATc1/B specific (B-term) and the NFATc1/C only (CC-term) C termini interact with Ubc9. Procedures were as before, but by use of constructs with nuclear localization signal-estrogen receptor fusions (estrogen receptor α), which are released for nuclear localization by addition of 4-hydroxytamoxifen. *, heavy or light chain of the precipitation antibody.
The authenticity of the yeast two-hybrid screens was verified by cotransfecting 293T HEK cells with constructs encoding HA-tagged NFATc1, wild type, and mutants and FLAG-tagged Ubc9 followed by IP. c1/C could be precipitated with Ubc9 by anti-FLAG but not upon cotransfection with the empty vector pcFLAG (Fig. 1B). In the immunoprecipitations with wild type, at least two precipitated bands were detected. Interestingly, in immunoprecipitations with K349R/K702R/K914R, no bands appeared, indicating that the SUMO consensus sites in c1/C are necessary for proper Ubc9 interaction (Fig. 1C). When all different SUMO site-deficient mutants were analyzed for Ubc9 interaction, the first site within the C terminus, Lys-702, proved to be the most important one, whereas the mutation Lys-349 did not influence the binding of Ubc9 at all (Fig. 1D). Therefore, interaction of Ubc9 should be mainly directed to the C-terminal peptide. And indeed, precipitation of Ubc9 was able to pull down the full C-terminal peptide fused to enhanced yellow fluorescence protein (Fig. 1E) and even when divided into the B terminus-specific (B) and the remaining far C terminus-specific (CC) peptides (Fig. 1F; for fragments see Fig. 1A). We conclude that both NFATc1/C-specific SUMO sites are able to efficiently interact with Ubc9.
Lysines 702 and 914 within the C-terminal Peptide of NFATc1/C Are Potent Sumoylation Sites—We next sought to check if c1/C is indeed ligated to SUMO. To this end vectors expressing HA-tagged c1/C and FLAG-SUMO1 were cotransfected into 293T HEK cells. After immunoprecipitation, 3 (-4) SUMO-modified c1/C bands were observed (Fig. 2A). To see whether the predicted SUMO sites and the (de-)phosphorylation state of NFATc1/C are of importance, 293T HEK cells were transfected either with plasmids expressing c1/C or K349R/K702R/K914R along with FLAG-SUMO1 or pcFLAG (Fig. 2B). Cells were left untreated or activated by T/I, which mimics T-cell receptor signaling, thereby leading to dephosphorylation and nuclear translocation of NFATs. SUMO-modified c1/C bands were detected in both untreated and T/I-treated cells (Fig. 2B, first two lanes). More importantly, no SUMO-modified band could be detected in NFATc1/C-K349R/K702R/K914R protein (Fig. 2B). Because SENPs (SUMO-specific proteases) render sumoylation very transient, the necessity of N-ethylmaleimide (a SENP inhibitor) within the lysis buffer was checked. Accordingly, robust c1/C sumoylation could only be observed when N-ethylmaleimide was added, which points to the dynamic and transient nature of NFATc1 sumoylation (supplemental Fig. S1A).
FIGURE 2.
NFATc1/C is sumoylated in vivo. A, NFATc1/C was sumoylated. HA-tagged c1/C-coding vector was transfected into 293T HEK cells either with FLAG-tagged SUMO1- or FLAG-expressing pcDNA, and cells were treated with T/I for 6 h. Anti-FLAG immunoprecipitation was followed by IB with anti-HA (arrows, SUMO-modified NFATc1/C) and whole cell lysates (WCL) were analyzed for expression by anti-HA and anti-FLAG. B, the SUMO site-deficient mutant K349R/K702R/K914R cannot be sumoylated. Procedures were as in A, but K349R/K702R/K914R was included, and cells were left unstimulated or treated for 6 h as indicated. C, SUMO site mutants reveal site-specific sumoylation pattern of c1/C. HA-tagged c1/C and different SUMO site mutants of c1/C were subjected to IP as in A, and expression was compared by anti-HA (NFAT) and anti-FLAG (SUMO) IB in whole cell lysates. D, NFATc1 precipitation reveals sumoylated c1/C. Procedures were as in A, except NFATc1/C-ER-stimulated by Tm + T/I for 4 h, and either anti-ER or anti-NFATc1 was used for IP and anti-SUMO1 for IB. E, in CD4+ T-cells endogenous NFATc1 is sumoylated. Nuclei from CD4+ T-cells after 7 days restimulated or not by ionomycin or T/I for 5 h were subjected to IP and IB as in D; IP-anti-NFATc1 + IB-anti-SUMO1 (arrow, sumoylated NFATc1/C, fourth lane, no antibody for IP); reprobe of IB by anti-NFATc1, IB of nuclear lysates (NL) by anti-SUMO1 and anti-NFATc1.
To analyze the individual sumoylation sites in c1/C, immunoprecipitation experiments were performed after cotransfecting 293T HEK cells with HA-tagged NFATc1/C-encoding constructs, either NFATc1/C wild type or different ΔSUMO mutants (single Lys to Arg, K702R/K914R and K349R/K702R/K914R) and FLAG-tagged SUMO1 expression vector (Fig. 2C). Transfection of the construct encoding wild type resulted in three prominent and one uppermost weak SUMO-specific band (arrows). In the case of K702R, bands 1 and 3 persisted, whereas band 2 disappeared, suggesting this band as being specific for lysine 702 sumoylation. On the other hand, band 2 persisted in transfections with K914R, whereas bands 1 and 3 disappeared, indicating those two as specific for lysine 914 sumoylation. Furthermore, the overall sumoylation efficiency of K702R and K914R was drastically reduced compared with wild type c1/C. Intriguingly, the common SUMO site mutant K349R gave rise to the same, although weaker, banding pattern as wild type c1/C, implying that in the presence of strong SUMO sites at lysine 702 and 914, the internal site at lysine 349 is less important for sumoylation. To verify the dominance of the C-terminal sites, transfections were performed with K702R/K914R. As expected, no SUMO-specific bands could be observed. This proved that the lysine 349 is irrelevant or totally dependent on C-terminal sumoylation. No bands of sumoylated NFATc1/C could be detected for the triple ΔSUMO mutant K349R/K702R/K914R.
We further examined sumoylation of the short isoform NFATc1/A which only harbors the common SUMO site at lysine 349. Surprisingly, we detected weak sumoylation of c1/A which was abolished in the case of c1/AK-349R (supplemental Fig. S1B). This indicates that this site is a true, but extremely weak sumoylation site. The latter conclusion is supported by the fact that no function could be pinpointed to this particular site (see below). Taken together, the long isoform-specific sites at lysines 702 and 914 are the most important sites for NFATc1/C sumoylation, and mutation in any of those leads to a dramatic reduction in overall sumoylation pattern.
NFATc1/C Is Sumoylated in Vivo—Although typically only a small proportion of proteins can be identified as sumoylated (24) and detection of endogenous sumoylation is described as very difficult, we analyzed if sumoylation of NFATc1 could be verified in primary T-cells. First, antibodies were tested in transfection experiments. Reverse experiments as before were performed with precipitation of c1/C-ER by anti-ER and anti-NFATc1 and Western blot detection by anti-SUMO1 antibodies. Identical banding pattern proved the specificity of c1/C sumoylation as well as the usefulness of the anti-NFATc1 antibody for precipitation of endogenous NFATc1 (Fig. 2D). On the other hand, the experiment demonstrated the comparably low amount of sumoylated NFAT detectable, when no exogenous SUMO1 was present (Fig. 2D, third lane). For evaluation of in vivo sumoylation, CD4+ T-cells were isolated from lymph nodes and stimulated with plate-bound anti-CD3 and anti-CD28 plus IL-2 for 3 days and rested for a further 4 days in the presence of IL-2. Then NFATc1, mainly c1/C; see Fig. 4E (E2 + E3), was translocated to the nucleus upon treatment with the Ca2+ ionophor ionomycin, where the autoregulatory loop leads to the predominant reexpression of the short isoform (25); the addition of the diacylglycerol analogon TPA induces the transcription of NFATc1/A directly (Fig. 2E, nuclear lysates NL: IB-NFAT). Immunoprecipitation of nuclear lysates by anti-NFATc1 antibody enriched mainly the shorter isoforms of NFATc1 (Fig. 2E, IP-NFATc1 + IB-NFATc1), but immunoblot detection of those precipitates by anti-SUMO1 revealed the typical high molecular weight bands of sumoylated NFATc1/C (IP-NFATc1 + IB-SUMO1) proving that NFATc1 is endogenously sumoylated under physiological conditions. The high amount of sumoylated proteins, although detected by the more sensitive ECL system, might be indicative for the relevance of sumoylation in nuclei of primary CD4+ T-cells (Fig. 2E, NL: IB-SUMO1).
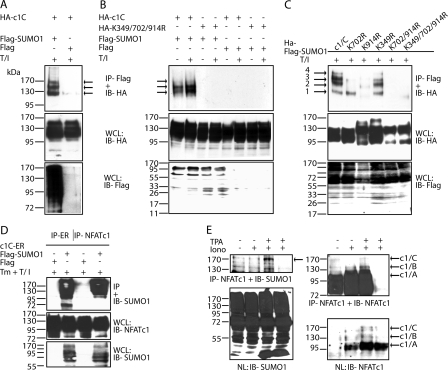
FIGURE 4.
Sumoylation represses NFATc1 transcriptional activity. A, IL-2 promoter activity is increased upon non-sumoylation of NFATc1/C. 293T HEK cells were transfected with 10 μg of pHA-NFATc1/C-EGZ or ΔSUMO mutants along with 1 μg of a luciferase reporter plasmid driven by the IL-2 promoter. After 36 h luciferase activity was measured from cells that were either left untreated or treated with T/I for 16 h. Data are represented as the mean ± S.E. To check for equal NFAT expression, Western blots were performed from these protein extracts. RLU, relative light units. B, fusion with SUMO1 impairs NFATc1 activity. Procedures are as in A, except SUMO1(S)-fused NFATc1 (-ER) clones were transfected in addition. Indicated is Fold difference in relation to basal activity (mock = 1). C, sumoylated NFATc1/C suppresses secretion of endogenous IL-2. Retrovirally infected A3.01 cells were stimulated by Tm+T/I for 48 h, and supernatants were analyzed by enzyme-linked immunosorbent assay. D, nTreg cells exhibit a predominant colocalization of NFATc1 with SUMO. CD4+ and CD4+CD25+ T-cells were isolated from mouse lymph nodes and stimulated with plate-bound anti-CD3 and anti-CD28 antibodies for 3 days. After further 2 days of resting cells were restimulated with T/I for 6 h. IF was performed with anti-NFATc1 and anti-SUMO1 followed by confocal microscopy. The scale bar represents 5 μm. DAPI, 4′,6-diamidino-2-phenylindole. E, nTregs mainly express the long isoform NFATc1/C (E2 + E3), initiated at the second promoter, P2, after restimulation. Real-time reverse transcription-PCR was performed from freshly isolated (0 h), stimulated as in D (5d), and restimulated with plate-bound anti-CD3 and anti-CD28 antibodies for 3 h (5d + 3 h) (no bars= not detectable). After normalization to the expression of HGPRT mRNA, relative expression levels of NFATc1 E1+E3, NFATc1 E2+E3, and NFATc2 mRNAs were calculated, whereas the value gained from unstimulated CD4+ (NFATc1 E2+E3 or NFATc2) was taken as 1.
Sumoylation Directs NFATc1/C to Nuclear Bodies—SUMO-modified proteins colocalize with nuclear bodies, in human cells, in particular, with PML-nbs. Therefore, we established A3.01 cells, a human lymphoid T-cell line by retroviral infection expressing c1/A, c1/C, and K349R/K702R/K914R. Equal expression was verified by confocal microscopy and immunoblot detection (supplemental Fig. S2).
To establish stable clones, an inducible system had to be chosen. NFAT proteins were fused to part of the ERα, which can be released from interaction with Hsp70 by the treatment with 4-hydroxytamoxifen (Tm), thereby providing an additional control for the nuclear translocation of NFATs. Cells were activated with 4-hydroxytamoxifen plus T/I for 4 h, and IF experiments were performed. Without treatment, all NFATc1-ER proteins remained in the cytosol (Fig. 3A), whereas upon stimulation all translocated to the nucleus. Tm and T/I were both needed for proper nuclear translocation and subnuclear appearance (supplemental Fig. S5A and below). Because sumoylation of NFATc2 is responsible for nuclear anchorage (26), the exact percentage of nuclear appearance was analyzed for NFATc1 wild type and all single, double, and triple ΔSUMO mutants, but no difference could be observed (supplemental Fig. S3A). Within the nucleus, the long isoform c1/C merged with SUMO1 as well as PML-nbs, whereas c1/A neither colocalized with SUMO1 nor with PML-nbs (Fig. 3A). More importantly, K349R/K702R/K914R behaves like c1/A, i.e. it neither colocalized with SUMO1 nor with PML-nbs. Therefore, only heavy sumoylation directs c1/C into PML-nbs. To consolidate these data, triple colocalization experiments were performed in 293T HEK cells, which were transfected transiently with NFATc1/A, c1/C, or K349R/K702R/K914R, this time without any estrogen receptor fusion part, together with SUMO1-expressing plasmids. As observed in A3.01 T-cells, upon sumoylation, NFATc1 was directed to PML-nbs (supplemental Fig. S3B). When all different ΔSUMO mutants were included for nuclear colocalization with SUMO bodies, it became obvious that the amount of sumoylation matters. Although the single Lys to Arg mutants of the C terminus reduced the colocalization to half, the double (or triple) mutant could only colocalize with about 10% (supplemental Fig. S3C).
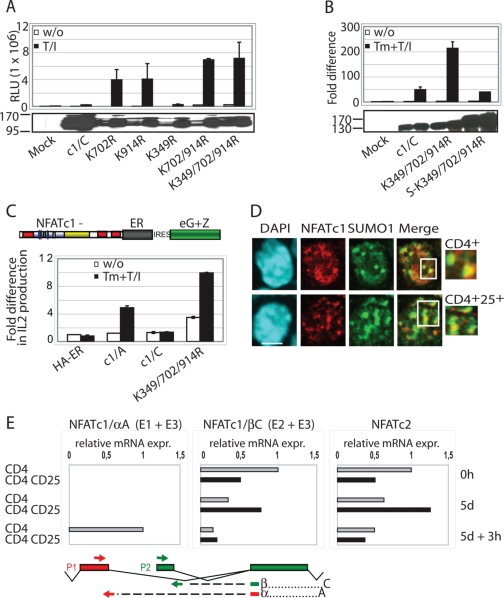
FIGURE 3.
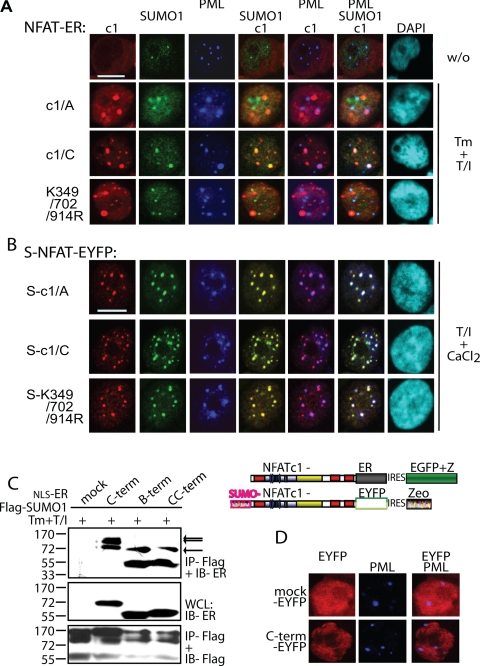
Sumoylation directs NFATc1 into PML-nbs. A, the sumoylatable long isoform colocalizes with PML-nbs. Human A3.01 cells retrovirally infected with constructs expressing c1/A-, c1/C-, and K349R/K702R/K914R-ER and selected by zeocin were left untreated or stimulated (w/o) with Tm+T/I for 4 h. IF was performed with anti-ERα (to detect exogenous NFATc1), anti-SUMO1, and anti-PML followed by laser scanning confocal microscopy. B, the fusion with SUMO directs any NFATc1 isoform to PML-nbs. 293T HEK cells were transfected with SUMO(S)-c1/A, -c1/C, and -K349R/K702R/K914R fused to EYFP at the C terminus and treated with T/I+CaCl2 for 1 h. IF with anti-SUMO1 and anti-PML was performed for triple localization analyzed by confocal microscopy. The scale bar represents 10 μm. C, the C-terminal peptide is sumoylated. 293T HEK cells were transfected with FLAG-SUMO1 and C terminus-specific peptides fused to ER with the addition of an nuclear localization signal. Analyzes were performed as for Fig. 1F. WCL, whole cell lysates. D, the sumoylated C terminus is not sufficient for recruitment to PML-nbs. Procedures were as in A but after infection of A3.01 with a retroviral vector expressing the C terminus-specific peptide fused to EYFP. DAPI, 4′,6-diamidino-2-phenylindole; IRES, internal ribosome entry site.
To provide further evidence that sumoylation is responsible for colocalizing with PML-nbs, SUMO-fused NFATc1 (S-NFATc1)-encoding constructs were generated and transfected into 293T HEK cells. Now all different NFATc1-proteins, irrespective whether they were short, long, or SUMO site-deficient, merged with PML-nbs, indicating that SUMO modification is a hallmark for sequestration of NFATc1 into PML-nbs (Fig. 3B).
We had demonstrated interaction of Ubc9 with the C-terminal peptides (Fig. 1, E and F). Accordingly, those peptides are also sumoylated (Fig. 3C). Nonsumoylated peptides are also pulled down, most likely because they are within the complex, but the pattern revealed one band for the peptides containing only one SUMO site (B and CC) and two for the entire C terminus (Fig. 3C, arrows). Surprisingly, although the C-terminal peptide localizes to the nucleus if fused to nuclear localization signal-ER or EYFP, it is not recruited to PML-nbs (Fig. 3D). This indicates that sumoylation is a requisite but not sufficient for extended interaction with the proteins in PML-nbs.
The presence of exogenous Ubc9 did not enhance sumoylation of NFATc1/C (supplemental Fig. S4A) and had no influence on relocalization of NFATc1/A or K349R/K702R/K914R within the nucleus (data not shown). In contrast but in line with published data (27), the presence of exogenous PIAS1, which was the E3 ligase identified by the yeast two-hybrid screen, supported PML-nb recruitment of K349R/K702R/K914R without sumoylating it (supplemental Fig. S4B). Differently, PIAS1 had no influence on NFATc1/A. Together, this indicates that only the long isoform NFATc1/C recruits a complex mediated by the sumoylation machinery which allows stable recruitment to PML-nbs.
Sumoylation Represses the Transcriptional Activity of NFATc1—To pursue whether sumoylation affects NFATc1 function, a luciferase assay was performed with the NFATc1-driven IL-2 promoter. A strong up-regulation of reporter gene activity was observed by both single mutants, K702R and K914R, compared with wild type NFATc1/C (Fig. 4A). This is in line with immunofluorescence, revealing a significant reduction in colocalization between K702R or K914R and SUMO1 compared with wild type c1/C (supplemental Fig. S3C). When either the C-terminal or all three SUMO sites were mutated, as in K702R/K914R or K349R/K702R/K914R, the transactivation potential increased even further. In contrast to the C-terminal sumoylation mutants, the activity of K349R appeared to be similar to that of wild type c1/C (Fig. 4A). This agrees with the observation that the extent of K349R-sumoylation is similar to wild type c1/C, establishing this site as dispensable for NFATc1/C function. Furthermore, no difference was detected in transactivation potential of c1/A and c1/A-K349R mutant, confirming that weak sumoylation of NFATc1/A does not affect its activity (supplemental Fig. S1C).
Moreover, fusing SUMO to transcriptionally hyperactive K349R/K702R/K914R led to a dramatic down-regulation of its transactivation potential (Fig. 4B), identifying this functional effect on c1/C as sumoylation-mediated. Taken together, sumoylation of the C terminus, specific for the long isoform, exerts a negative effect on NFATc1 function.
Sumoylation Renders NFATc1/C Less Potent on Endogenous IL-2 Expression—To test the effect of NFATc1 sumoylation on endogenously expressed IL-2, retrovirally transduced A3.01 cells were assayed in RNase protection (supplemental Fig. S5B) and enzyme-linked immunosorbent assay (Fig. 4C). Although TPA/ionomycin treatment alone leads to incomplete and improper nuclear translocation (supplemental Fig. S5A), IL-2 expression is influenced artificially (supplemental Fig. S5B, third lane in each panel). This might be due to the remaining interaction between the estrogen receptor part and Hsp70, for example, masking or preventing sumoylation. Nevertheless, in accordance with nuclear appearance and RNA data from fully stimulated cells, i.e. T/I plus 4-hydroxytamoxifen, a marked reduction (4×) of IL-2 secretion could be detected in the presence of the sumoylatable long isoform and a sound 8-fold increase upon the eradication of SUMO sites (K349R/K702R/K914R).
Because NFATc1/C sumoylation leads to suppression of IL-2 gene activity and thereby IL-2 production, nTregs (CD4+CD25+), which do not produce IL-2, were investigated. Thus, we performed immunofluorescence on in vitro restimulated CD4+ effector T- and nTreg-cells by using an anti-NFATc1 antibody recognizing all isoforms. Both types of CD4+ T-cells expressed NFATc1 as well as SUMO1, and in both cell types some of the endogenous NFATc1 colocalized with SUMO1 bodies (Fig. 4D). However, in nTregs NFATc1 signals were always found to be reduced. More excitingly, in nTregs the extent of colocalized SUMO1 with NFATc1 appeared to be more pronounced. Thus, the maintenance of anergy-like phenotype of nTreg cells could to be supported by the higher sumoylation level of c1/C, which in turn leads to IL-2 suppression. To evaluate the expression of the short and long isoforms of NFATc1 in both CD4+ T-cell types, RNA was isolated at certain time points and subjected to real-time PCR experiments (Fig. 4E). Primer pairs for the highly inducible short, α-peptide-containing (exon 1-encoded, Fig. 1A) and the more constitutive long, β-peptide-containing (exon2-encoded) were compared. The data revealed that high induction occurs in conventional CD4+ T-cells after restimulation but that nTreg cells predominantly express NFATc1/C, giving a good explanation for the more pronounced colocalization of NFATc1 with SUMO1 in CD4+CD25+ nTreg cells.
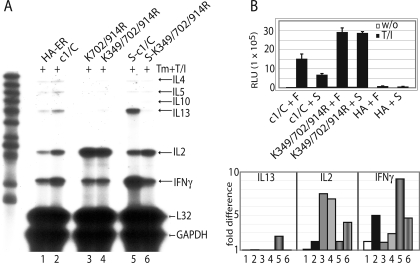
Sumoylation of NFATc1 Can Both Suppress and Enhance Lymphokine Induction—In addition to IL-2, numerous other lymphokine genes are controlled by NFAT, and we asked whether those are also suppressed by sumoylation. Therefore, the murine thymoma cell line EL-4, which express Th1 as well as Th2 type lymphokines upon stimulation, were retrovirally infected with NFATc1-ER constructs. After treatment with 4-hydroxytamoxifen plus TPA/ionomycin for 24 h, RNA was isolated and assayed by RNase protection assay. Again, K349R/K702R/K914R (and K702R/K914R) exerted a much stronger transactivation on IL-2 mRNA than did wild type (Fig. 5A, lanes 2-4). Intriguingly, all other lymphokines tested, i.e. of Th1- (IFNγ) and Th2- (IL4, -5, -10, and -13) type exhibited reduced transcript levels under the control of non-sumoylatable NFATc1/C. Accordingly, when SUMO was fused to wild type (S-c1/C), the amount of IFNγ and IL-13 transcripts was robustly increased above wild type levels (lanes 2 and 5). This suggests a dependence of NFATc1 on sumoylation for full-fledged IFNγ and IL-13 gene expression, whereas IL-2 expression is suppressed.
FIGURE 5.
Sumoylation of NFATc1 can both suppress and enhance lymphokine induction. A, IL-2 mRNA is repressed, but RNA levels of IL-13 and IFNγ are further up-regulated upon sumoylation of NFATc1. 5 μg RNA of retrovirally infected EL-4 cells (NFATc1/C-ER, its ΔSUMO and SUMO fusion mutants) were subjected to RNase protection assay after stimulating the cells with Tm+T/I for 24 h using the mCK1 template set. Also a graph of phosphor-imaging values is given of three independent experiments where 1-6 correspond to the lanes of the autoradiography. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, the presence of SUMO suppresses the transactivation potential of NFATc1. As in Fig. 4A, except murine EL-4 cells were transfected and plasmids expressing FLAG (F) or FLAG-SUMO1 (S) were cotransfected with those encoding NFATc1/C, K349R/K702R/K914R, or empty vector (HA) along with an IL-2-luciferase plasmid.
Because all SUMO-fused NFATc1 proteins, irrespective of intact endogenous SUMO sites, are localized to PML-nbs (Fig. 3B), S-c1/C and S-K349R/K702R/K914R were expected to induce a similar lymphokine profile. Surprisingly, although the IL-2 mRNA level was reduced in the case of S-K349R/K702R/K914R compared with K349R/K702R/K914R (Fig. 5A, fourth and sixth lanes), it was still enhanced in comparison to S-c1/C (Fig. 5A, fifth and sixth lanes). Furthermore, in the case of IFNγ and IL-13 RNA expression, the loss of the SUMO sites in c1/C could not be compensated by the presence of the N-terminal-fused SUMO. This indicates that the positions of sumoylation and/or amount of SUMO-moieties on NFATc1 are determining factors in controlling SUMO-mediated function.
To elicit whether the abundant presence of SUMO1 would have the same effect as the fusions to NFATc1, EL-4 cells were transiently transfected with the IL-2 promoter-driven reporter construct, in conjunction with plasmids expressing c1/C, wild type, or K349R/K702R/K914R as well as SUMO1. Again, K349R/K702R/K914R was able to exert a more pronounced transactivation potential than wild type, and more importantly, the cotransfection with SUMO1 was able to reduce wild type but not K349R/K702R/K914R-mediated activity (Fig. 5B).
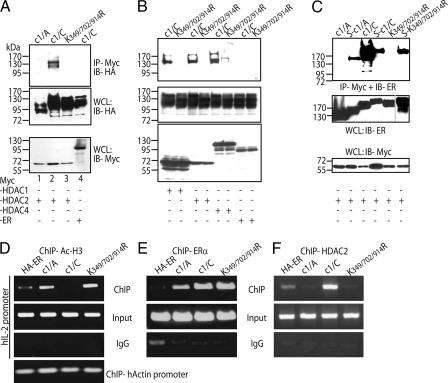
SUMO1 Promotes NFATc1 Interaction with HDACs and Reduces Histone Acetylation at the IL-2 Promoter—Earlier studies described that SUMO1-modified transcription factors can associate with the transcriptional repressor HDAC2 (28). Therefore, immunoprecipitation experiments were performed after transfection of 293T HEK cells with plasmids encoding Myc-tagged HDAC2 and HA-tagged NFATc1. Indeed, HDAC2 interacts with c1/C (Fig. 6A), whereas no or a poor interaction was detected with c1/A and K349R/K702R/K914R, indicating that the SUMO1 modification on NFATc1 serves as a tag for the interaction with HDAC2. Furthermore, c1/C, but not K349R/K702R/K914R, can also be efficiently precipitated by HDAC1 and HDAC4, which shows that class-I as well as class-II HDACs can interact with sumoylated NFATc1/C (Fig. 6B). When SUMO-fused NFATc1 was subjected to coimmunoprecipitation, it revealed that sumoylation mediates interaction with HDAC2, as SUMO-NFATc1/A and SUMO-K349R/K702R/K914R were precipitated (Fig. 6C). Intriguingly, however, the efficiency of interaction of HDAC2 with NFATc1/C is much better than with SUMO-NFATc1/C.
FIGURE 6.
Sumoylation promotes NFATc1 association with class I and II HDACs, which mediates histone deacetylation at the IL-2 promoter. A, HDAC2 interacts with NFATc1/C but not K349R/K702R/K914R. 293T HEK cells transfected with constructs coding for Myc-tagged HDAC 2 and different forms of HA-tagged NFATc1 were treated with T/I for 6 h. Anti-Myc IP was followed by anti-HA antibody for IB, and the expression levels were analyzed directly in whole cell lysates by anti-HA and anti-Myc. In lane 4 specificity was checked by the use of full-length c-Myc-ER construct in place of Myc-tagged HDAC2. WCL, whole cell lysates. B, also, HDAC1 and 4 interact with NFATc1/C. Procedures were as in A, except HDAC1 and HDAC4 constructs were included. C, SUMO fusion recruits HDAC2 to NFATc1. Procedures were as in A, but NFATc1-ER constructs were transfected, and IB was performed by anti-ER. D, sumoylatable NFATc1/C deacetylates histone H3 at the IL-2 promoter. ChIP assay of retrovirally infected A3.01 cells (Figs. 3 and 4), stimulated with Tm+T/I for 6 h, was performed with anti-Ac-H3. DNA from precipitated chromatin was PCR-amplified by using human IL-2 or actin promoter-specific primers. This is one of three independent experiments, and the result was also confirmed in murine EL-4 cells. E, NFATc1 isoforms bind equally to the IL-2 promoter. Procedures were as in C, except anti-ERα was (subsequently) applied to the same cell lysates. F, NFATc1/C expression mediates HDAC2 binding to the IL-2 promoter. Procedures were as in C, except cells were only stimulated for 3 h, and anti-HDA2 was applied.
As sumoylation promotes c1/C and HDAC interaction, this event could lead to deacetylation of histones at the IL-2 promoter, rendering chromatin less accessible for transcription. To test this, ChIP assays with an antibody raised against acetylated histone H3 (a mark for active chromatin) and chromatin from those retrovirally transduced A3.01 cells were performed. To validate the relative amount of immunoprecipitated IL-2 chromatin, primers specific for the human IL-2 promoter were used in PCR assays. In c1/C-expressing cells, a sound reduction in the level of histone acetylation on the IL-2 promoter was detected, whereas it was comparably high in c1/A- and K349R/K702R/K914R-expressing cells (Fig. 6D). While assaying the IFNγ or IL-13 promoters, no changes could be observed (data not shown). When, subsequently, an anti-ERα antibody was applied to precipitate exogenous NFATs, an equal binding of c1/C and K349R/K702R/K914R could be detected to the IL-2 promoter (Fig. 6E). This is in line with the results of electrophoretic mobility shift assays which elucidated the same binding capacity of NFATc1 in vitro irrespective of isoform and state of sumoylation (data not shown). Taken together, these data proved that by sumoylated NFATc1/C, HDACs are recruited to the IL-2 promoter (Fig. 6F), thereby clearing marks of active chromatin.
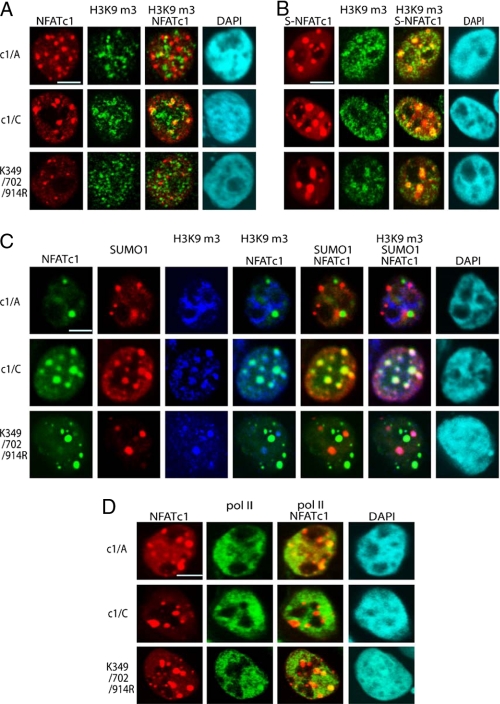
Sumoylation Withdraws NFATc1 from Transcriptional Hotspots and Relocalizes It to Condensed Chromatin—One of the best marks for condensed chromatin is trimethylated histone 3 lysine 9 (H3K9m3). Therefore, we examined if c1/C would colocalize with H3K9m3 upon sumoylation. In three independent immunofluorescence experiments, a pronounced colocalization (56%) of c1/C was observed within condensed chromatin but only 29% of c1/A and 31% of K349R/K702R/K914R (Fig. 7A). Confirming the role of SUMO as a key factor for directing NFATc1 to repressed chromatin, all SUMO fusion constructs were detected exclusively at H3K9m3-positive sites (Fig. 7B). Moreover, to question the presence of SUMO1 at condensed chromatin, NFATc1 colocalization studies were performed with anti-H3K9m3 as well as anti-SUMO antibodies. As expected, in a substantial amount of nuclear territories, wild type NFATc1/C was found to colocalize with SUMO1 and H3K9m3, suggesting SUMO1 as an important tag for subnuclear orientation to sites of repression (Fig. 7C).
FIGURE 7.
Sumoylation directs NFATc1 into transcriptionally inactive chromatin. A, 293T HEK cells were transfected with plasmids encoding pHA-NFATc1/A, -c1/C, and -K349R/K702R/K914R and stimulated with T/I+CaCl2 for 1 h. IF was performed to detect NFATc1 with anti-HA and heterochromatin with anti-H3K9m3. Colocalization was analyzed by confocal microscopy. The scale bar represents 10 μm. B, 293T HEK cells were transfected with plasmids encoding SUMO1(S)-c1/A, -c1/C, and - K349R/K702R/K914R, which were C-terminally fused to EYFP. Cells were stimulated with T/I+CaCl2 for 1 h. IF was performed with anti-H3K9m3, and colocalization of SUMO-fused NFATc1 (S-NFATc1) and heterochromatin were analyzed by confocal microscopy. C, FLAG-SUMO1 was transfected along with c1/A, c1/C, and K349R/K702R/K914R plasmids, C-terminally fused to EYFP into 293T HEK cells, and stimulated with T/I+CaCl2 for 1 h followed by IF with anti-SUMO and anti-H3K9m3. Tri-colocalization of NFATc1, SUMO1, and H3K9m3 was analyzed by confocal microscopy. D, without sumoylation, NFATc1 colocalizes to hotspots of transcription. Transfection and stimulation was as in C. To reveal the colocalization pattern of NFATc1 and pol II-bodies, IF was performed with anti-pol II antibodies. The scale bar represents 10 μm. DAPI, 4′,6-diamidino-2-phenylindole.
Strikingly, also NFATc1 in its active forms (c1/A and K349R/K702R/K914R) established a punctuated distribution within the nucleus. Previously, it has been shown that numerous transcriptional hotspots including RNA pol II bodies (29) exist within the nucleus. Intriguingly, by colocalization experiments with antibodies raised against NFATc1 and pol II, we found that c1/A and K349R/K702R/K914R, but not c1/C, partially overlays with these pol II bodies (Fig. 7D). The appearance of the pol II bodies is not very distinct, but it might indicate that the transcription factor NFATc1 associates with the highly active transcription machinery, whereas sumoylation can withdraw NFATc1 from those transcriptional factories.
DISCUSSION
NFAT transcription factors have strongly been implicated in T-cell activation including regulation of numerous cytokine genes. Although the role of individual NFATc members has been addressed by NFAT-deficient mice, their isoform-specific signaling capacities have been less well studied. Hence, we addressed the functional difference between the shortest and longest isoforms of NFATc1, namely NFATc1/A and NFATc1/C. We observed that these two isoforms exert a differential effect on IL-2 expression; although both NFATc1/A and c1/C show a high transactivation potential, the long isoform can negatively be regulated by sumoylation. As a consequence, NFATc1/C becomes suppressive for IL-2 expression.
Sumoylated NFATc1/C Suppresses IL-2 Expression—Numerous findings showed that the induction of the IL-2 promoter in T-cells depends critically on the activity of NFATc factors (2-4), although the loss of NFATc2 had no effect on IL-2 expression, and NFATc1-/- T-cells exhibited only modestly reduced or normal levels of IL-2 in response to primary anti-CD3 antibody stimulation (5, 30). In contrast, NFATc1/c2 double-deficient T-cells were found to be devoid in their production of lymphokines, including IL-2 (31). Therefore, the activities of NFATc1 and -c2 are essential but are largely redundant with respect to activation-induced IL-2 expression. The modification with SUMO marks NFATc1/C for interaction with HDACs, most likely within PML-nbs, which in turn heterochromatizes areas where sumoylated NFATc1/C binds to DNA; for example, to the IL-2 promoter.
Here, we demonstrate that down-regulation of IL-2 is specifically achieved by NFATc1/C. In contrast to the NFATc1/A, the long isoform NFATc1/C is constitutively expressed in peripheral T-cells (Fig. 4E) and Chuvpilo et al. (20). Therefore, the long isoform might keep IL-2 production in check before full activation and also terminate it when differentiation proceeds. It was recently revealed that in memory (like in effector cells), but not in naive CD4+ T-cells, NFATc1 was strongly expressed (32). Strikingly, in naive cells which show predominant expression of the long, sumoylatable isoform, the induction level of IL-2 is low, whereas it is high in memory cells. Also, nTreg cells, which do not express IL-2, synthesize no NFATc1/αA after restimulation, unlike conventional CD4+ cells, which do synthesize NFATc1/αA (Fig. 4E). This is in line with our observation that NFATc1 colocalization with SUMO is enhanced in nTregs. Moreover, the increase in IL-2 after restimulation of NFATc1-/- T-cells (5) might be due to the concomitant loss of NFATc1/C.
Recently, also sumoylation of NFATc2 was demonstrated (26). The two sites within the NFATc1/C-specific terminus are the homologous recognition sites within NFATc2, but in the latter sumoylation supports nuclear anchorage and subnuclear localization to active sites. After all, it seems that only NFATc1/C can act as a repressor on IL-2 when sumoylated and recruiting HDACs.
Expression of IFNγ and IL-13 Is Enhanced by Sumoylated NFATc1/C—When SUMO was fused to c1/C, IL-13 and IFNγ were strongly transactivated. On both promoters the interaction of NFATc1 with other transcription factors might be influenced by sumoylation. The residues involved in contact to AP1, the common partner in T-cells, are located largely in the N-terminal part of the Rel similarity domain of NFAT (3), clearly distinct from the C-terminal SUMO sites. However, heavy sumoylation might enhance interaction with other positive factors or sterically hamper the interaction with negatively acting partners. Interestingly, some basic-leucine-zipper as well as helix-turn-helix domain proteins shown to interact with NFAT act as repressors (3). For the IFNγ and IL-13 promoters, no such NFAT partners have yet been identified, but their release by sumoylation of NFATc1/C would explain the elevated expression level of IFNγ and IL-13.
The Amount and the Site of Sumoylation Matters—There are two consensus sites within the C terminus (Lys-702 and -914) of NFATc1/C which can be sumoylated independently of each other. This seems to be in contrast to NFATc2 for which sumoylation of the homologous first site is a requisite for the modification of second site (26). Although the authors claim this site in NFATc2 (Lys-897 within IKQE) to be unique, it corresponds to Lys-914 within VKRE in NFATc1/C (7). In line with a dependence of SUMO sites on each other, the overall efficiency of sumoylation was reduced by either single site mutation in Lys-702 or -914. Additionally, weak sumoylation of the common D site was dependent on A and B sites. Moreover, the interaction with Ubc9 only occurred in the presence of intact SUMO sites, indicating that the affinity is drastically reduced upon lysine to arginine exchange, which is in line with published data (33). By the predominant dependence of Ubc9 interaction with Lys-702, it can be concluded that within intact NFATc1/C this site is the primary docking site, whereas the data gained with the C-terminal peptides also reveal independent interaction potential of both sumoylation sites. Taken together, it can be concluded that (i) weak SUMO sites depend on the presence of strong ones for modification, (ii) the common D site is very weak, (iii) the site at Lys-897 within NFATc2 exhibits a much weaker affinity to Ubc9 than K914 of NFATc1/C which might be due to the two varied aa of the SUMO motif, and (iv) even sumoylation of homologous sites can result in a completely different functional outcome.
Furthermore, fusing SUMO to c1/C wild type or to K349R/K702R/K914R was not comparable. Although SUMO wild type, exhibiting now four SUMO-moieties, achieved a strong positive effect on IFNγ and IL-13, the activity of SUMO-K349R/K702R/K914R resembled K349R/K702R/K914R. Also, although the interaction with HDAC2 is clearly mediated by the SUMO moiety, the addition of N-terminal SUMO to c1/C wild type leads to reduced interaction with HDAC2. These data revealed that either the amount and/or the site of sumoylation matters. It is obvious that protein-protein interactions mediated by protruding SUMO moieties depend on the location within a ternary structure. Therefore, we envisage a SUMO tag as sufficient for PML-nb localization (as demonstrated for all SUMO fusion NFATc1 constructs) but not in all cases sufficient for full function mediated by site-specific sumoylation of NFATc1/C. Surprisingly, the C-terminal peptides are efficiently sumoylated but not detectable at PML-nbs. Most likely, sumoylation has the potential of recruitment to PML-nbs, but the nuclear distribution is only stable if other parts of NFATc1/C interact with residual proteins of the nuclear bodies.
The SUMO-NFATc1/C-recruited Complex Remodels Chromatin at the IL-2 Locus—Chromatin remodeling at the IL-2 locus has recently been studied. AlthoughCD28 costimulation led to hyperacetylation (34), anergy induction by Ikaros induced histone deacetylation (35). Here we show that upon sumoylation NFATc1/C recruits HDACs to the IL-2 promoter. Therefore, the SUMO tag possibly serves as a platform on NFATc1 to build a repressor complex with mSin3a, NuRD, or SMART/Ncor (36). As HDACs leave an imprint for transcriptionally inactive chromatin, sumoylated NFATc1/C colocalized with condensed chromatin indicated by H3K9m3. Surprisingly, we did not observe any significant colocalization of PML-nbs with H3K9m3 (data not shown). Therefore, we propose that sumoylated NFATc1 is first directed to PML-nbs, where it recruits the repressor complex, and then transverses to the periphery or outside of PML-nbs to promote heterochromatinization. This view is supported by previous observations that HP1, HDACs, DAXX, and PML itself shuttle in and out of PML-nbs to associate with heterochromatin (37, 38). Non-sumoylated NFATc1 associates with pol II. Because IL-2 was efficiently transactivated by c1/A and K349R/K702R/K914R, we hypothesized that the nuclear dots formed by them represent areas of transcriptional activity. Intranuclear foci have been identified containing multimolecular complexes for different processes including mRNA synthesis. They contain pol II entailing a higher degree of active transcriptional processes (38, 39). Although transcriptionally active c1/A and K349R/K702R/K914R associated with such transcription factories, SUMO modification excluded NFATc1/C from those transcriptional hotspots. Because sumoylation is a transient modification, it is possible that desumoylated NFATc1/C can then take part in transactivation. In such a scenario the second transactivation domain found within the c1/C-specific C terminus (7) would be unmasked, leading to the observed high transactivation potential of the NFATc1/C-ΔSUMO mutant. Accordingly, although H3K9m3 is the best characterized mark for inactive chromatin, new data established that H3K9m3 can also be associated with transcriptional elongation through mammalian chromatin (40). In relation to their differentiation stage-dependent activity, genes are regarded to shuttle toward or from pericentromeric heterochromatin (41). Therefore, regulated and reversible sumoylation of NFATc1/C may support finetuned T-cell differentiation. A not mutually excluding, but different scenario comes from the observation that although sumoylation is a transient modification (which explains the low fraction of sumoylated NFATc1 detected in primary CD4+ T-cells), formerly sumoylated transcription factors are kept in stable repressor complexes (24). Here, the highly (upon CD4+ T-cell activation) induced expression of the short NFATc1/A isoform (20, 25) would replace the repressive NFATc1/C, leading to sufficient transactivation of IL-2 for proliferation, differentiation, or effector function.
Supplementary Material
Acknowledgments
We are indebted to Melanie Schott and Ursula Sauer for excellent technical assistance and to Doris Michel and Ilona Pietrowski for further support. We thank I. Berberich for cloning and providing the pIZ-EYFP vector, H. Hofmann for pcFLAG-SUMO and pcFLAG-Ubc9, T. Kouzarides for pcDNA-HDAC1, -2, -4-Myc-His constructs, and R. Rost for purification and providence of the anti-NFATc1 antibody (Pineda Antibody Service). We are very grateful to A. Schimpl and S. Klein-Hessling for discussion and reading the manuscript.
This work was further by supported by Deutsche Forschungsgemeinschaft Grants SFB 466/B3, BE 2309/2-1, and SPP1365, by the Wilhelm-Sander-Foundation, and the Mildred-Scheel-Foundation for Cancer Research.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S5.
Footnotes
The abbreviations used are: IL, interleukin; ER, estrogen receptor; H3K9m3, histone 3 lysine 9 trimethylated; HDAC, histone deacetylase; IB, immunoblot; IF, immunofluorescence; IFN, interferon; IP, immunoprecipitation; NFAT, nuclear factor of activated T-cells; PML-nb, promyelocytic leukemianuclear body; SUMO, small ubiquitin-like modifier; Tm, 4-hydroxytamoxifen; T/I, TPA plus ionomycin; aa, amino acids; TPA, 12-O-tetradecanoylphorbol-13-acetate; ChIP, chromatin immunoprecipitation; HEK cells, human embryonic kidney cells; EYFP, enhanced yellow fluorescence protein; HA, hemagglutinin; pol, polymerase; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase.
A. Nayak, J. Glöckner-Pagel, M. Vaeth, J. E. Schumann, M. Buttmann, T. Bopp, E. Schmitt, E. Serfling, and F. Berberich-Siebelt, unpublished data.
References
- 1.Weaver, C. T., Harrington, L. E., Mangan, P. R., Gavrieli, M., and Murphy, K. M. (2006) Immunity 24 677-688 [DOI] [PubMed] [Google Scholar]
- 2.Serfling, E., Berberich-Siebelt, F., Chuvpilo, S., Jankevics, E., Klein-Hessling, S., Twardzik, T., and Avots, A. (2000) Biochim. Biophys. Acta 1498 1-18 [DOI] [PubMed] [Google Scholar]
- 3.Hogan, P. G., Chen, L., Nardone, J., and Rao, A. (2003) Genes Dev. 17 2205-2232 [DOI] [PubMed] [Google Scholar]
- 4.Macian, F. (2005) Nat. Rev. Immunol. 5 472-484 [DOI] [PubMed] [Google Scholar]
- 5.Ranger, A. M., Hodge, M. R., Gravallese, E. M., Oukka, M., Davidson, L., Alt, F. W., de la Brousse, F. C., Hoey, T., Grusby, M., and Glimcher, L. H. (1998) Immunity 8 125-134 [DOI] [PubMed] [Google Scholar]
- 6.Serfling, E., Chuvpilo, S., Liu, J., Hofer, T., and Palmetshofer, A. (2006) Trends Immunol. 27 461-469 [DOI] [PubMed] [Google Scholar]
- 7.Chuvpilo, S., Avots, A., Berberich-Siebelt, F., Glockner, J., Fischer, C., Kerstan, A., Escher, C., Inashkina, I., Hlubek, F., Jankevics, E., Brabletz, T., and Serfling, E. (1999) J. Immunol. 162 7294-7301 [PubMed] [Google Scholar]
- 8.Iniguez-Lluhi, J. A., and Pearce, D. (2000) Mol. Cell. Biol. 20 6040-6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill, G. (2004) Genes Dev. 18 2046-2059 [DOI] [PubMed] [Google Scholar]
- 10.Verger, A., Perdomo, J., and Crossley, M. (2003) EMBO Rep. 4 137-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berberich-Siebelt, F., Berberich, I., Andrulis, M., Santner-Nanan, B., Jha, M. K., Klein-Hessling, S., Schimpl, A., and Serfling, E. (2006) J. Immunol. 176 4843-4851 [DOI] [PubMed] [Google Scholar]
- 12.Gill, G. (2005) Curr. Opin. Genet. Dev. 15 536-541 [DOI] [PubMed] [Google Scholar]
- 13.Seeler, J. S., and Dejean, A. (1999) Curr. Opin. Genet. Dev. 9 362-367 [DOI] [PubMed] [Google Scholar]
- 14.Negorev, D., and Maul, G. G. (2001) Oncogene 20 7234-7242 [DOI] [PubMed] [Google Scholar]
- 15.Jenuwein, T., and Allis, C. D. (2001) Science 293 1074-1080 [DOI] [PubMed] [Google Scholar]
- 16.Bopp, T., Becker, C., Klein, M., Klein-Hessling, S., Palmetshofer, A., Serfling, E., Heib, V., Becker, M., Kubach, J., Schmitt, S., Stoll, S., Schild, H., Staege, M. S., Stassen, M., Jonuleit, H., and Schmitt, E. (2007) J. Exp. Med. 204 1303-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschmied, J., and Duschl, J. (1997) Biotechniques 23 436-438 [DOI] [PubMed] [Google Scholar]
- 18.Serfling, E., Avots, A., and Neumann, M. (1995) Biochim. Biophys. Acta 1263 181-200 [DOI] [PubMed] [Google Scholar]
- 19.Kuss, A. W., Knodel, M., Berberich-Siebelt, F., Lindemann, D., Schimpl, A., and Berberich, I. (1999) Eur. J. Immunol. 29 3077-3088 [DOI] [PubMed] [Google Scholar]
- 20.Chuvpilo, S., Zimmer, M., Kerstan, A., Glockner, J., Avots, A., Escher, C., Fischer, C., Inashkina, I., Jankevics, E., Berberich-Siebelt, F., Schmitt, E., and Serfling, E. (1999) Immunity 10 261-269 [DOI] [PubMed] [Google Scholar]
- 21.Littlewood, T. D., Hancock, D. C., Danielian, P. S., Parker, M. G., and Evan, G. I. (1995) Nucleic Acids Res. 23 1686-1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berberich-Siebelt, F., Klein-Hessling, S., Hepping, N., Santner-Nanan, B., Lindemann, D., Schimpl, A., Berberich, I., and Serfling, E. (2000) Eur. J. Immunol. 30 2576-2585 [DOI] [PubMed] [Google Scholar]
- 23.Soutoglou, E., and Talianidis, I. (2002) Science 295 1901-1904 [DOI] [PubMed] [Google Scholar]
- 24.Hay, R. T. (2005) Mol. Cell 18 1-12 [DOI] [PubMed] [Google Scholar]
- 25.Chuvpilo, S., Jankevics, E., Tyrsin, D., Akimzhanov, A., Moroz, D., Jha, M. K., Schulze-Luehrmann, J., Santner-Nanan, B., Feoktistova, E., Konig, T., Avots, A., Schmitt, E., Berberich-Siebelt, F., Schimpl, A., and Serfling, E. (2002) Immunity 16 881-895 [DOI] [PubMed] [Google Scholar]
- 26.Terui, Y., Saad, N., Jia, S., McKeon, F., and Yuan, J. (2004) J. Biol. Chem. 279 28257-28265 [DOI] [PubMed] [Google Scholar]
- 27.Sachdev, S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F., and Grosschedl, R. (2001) Genes Dev. 15 3088-3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, S. H., and Sharrocks, A. D. (2004) Mol. Cell 13 611-617 [DOI] [PubMed] [Google Scholar]
- 29.Chakalova, L., Debrand, E., Mitchell, J. A., Osborne, C. S., and Fraser, P. (2005) Nat. Rev. Genet. 6 669-677 [DOI] [PubMed] [Google Scholar]
- 30.Yoshida, H., Nishina, H., Takimoto, H., Marengere, L. E., Wakeham, A. C., Bouchard, D., Kong, Y. Y., Ohteki, T., Shahinian, A., Bachmann, M., Ohashi, P. S., Penninger, J. M., Crabtree, G. R., and Mak, T. W. (1998) Immunity 8 115-124 [DOI] [PubMed] [Google Scholar]
- 31.Peng, S. L., Gerth, A. J., Ranger, A. M., and Glimcher, L. H. (2001) Immunity 14 13-20 [DOI] [PubMed] [Google Scholar]
- 32.Dienz, O., Eaton, S. M., Krahl, T. J., Diehl, S., Charland, C., Dodge, J., Swain, S. L., Budd, R. C., Haynes, L., and Rincon, M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7175-7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampson, D. A., Wang, M., and Matunis, M. J. (2001) J. Biol. Chem. 276 21664-21669 [DOI] [PubMed] [Google Scholar]
- 34.Thomas, R. M., Gao, L., and Wells, A. D. (2005) J. Immunol. 174 4639-4646 [DOI] [PubMed] [Google Scholar]
- 35.Bandyopadhyay, S., Dure, M., Paroder, M., Soto-Nieves, N., Puga, I., and Macian, F. (2007) Blood 109 2878-2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Downes, M., Ordentlich, P., Kao, H. Y., Alvarez, J. G., and Evans, R. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10330-10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luciani, J. J., Depetris, D., Usson, Y., Metzler-Guillemain, C., Mignon-Ravix, C., Mitchell, M. J., Megarbane, A., Sarda, P., Sirma, H., Moncla, A., Feunteun, J., and Mattei, M. G. (2006) J. Cell Sci. 119 2518-2531 [DOI] [PubMed] [Google Scholar]
- 38.Zimber, A., Nguyen, Q. D., and Gespach, C. (2004) Cell. Signal. 16 1085-1104 [DOI] [PubMed] [Google Scholar]
- 39.Gall, J. G. (2001) FEBS Lett. 498 164-167 [DOI] [PubMed] [Google Scholar]
- 40.Vakoc, C. R., Sachdeva, M. M., Wang, H., and Blobel, G. A. (2006) Mol. Cell. Biol. 26 9185-9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baxter, J., Merkenschlager, M., and Fisher, A. G. (2002) Curr. Opin. Cell Biol. 14 372-376 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.