Abstract
Reprogramming of somatic cells to pluripotency, thereby creating induced pluripotent stem (iPS) cells, promises to transform regenerative medicine. Most instances of direct reprogramming have been achieved by forced expression of defined factors using multiple viral vectors1-7. However, such iPS cells contain a large number of viral vector integrations1,8, any one of which could cause unpredictable genetic dysfunction. While c-Myc is dispensable for reprogramming9,10, complete elimination of the other exogenous factors is also desired since ectopic expression of either Oct4 or Klf4 can induce dysplasia11,12. Two transient transfection reprogramming methods have been published to address this issue13,14. However, the efficiency of either approach is extremely low, and neither has thus far been applied successfully to human cells. Here we show that non-viral transfection of a single multiprotein expression vector, which comprises the coding sequences of c-Myc, Klf4, Oct4 and Sox2 linked with 2A peptides, can reprogram both mouse and human fibroblasts. Moreover, the transgene can be removed once reprogramming has been achieved. iPS cells produced with this non-viral vector show robust expression of pluripotency markers, indicating a reprogrammed state confirmed functionally by in vitro differentiation assays and formation of adult chimeric mice. When the single vector reprogramming system was combined with a piggyBac transposon15,16 we succeeded in establishing reprogrammed human cell lines from embryonic fibroblasts with robust expression of pluripotency markers. This system minimizes genome modification in iPS cells and enables complete elimination of exogenous reprogramming factors, efficiently providing iPS cells that are applicable to regenerative medicine, drug screening and the establishment of disease models.
Keywords: iPS cells, reprogramming, pluripotency, 2A peptide, multiprotein expression vector, non-viral transfection
Efficient multiprotein expression has been reported in a variety of cell types, including human ES cells, using the 2A peptide sequence of foot-and-mouth disease virus (F2A), or 2A-like sequence from other viruses17,18. Recently, this multiprotein expression strategy has also been applied for reprogramming with transient transfection14 and doxycycline (dox)-inducible lentiviral vector19,20. Here we have taken advantage of the strategy to generate virus-free factor-removable iPS cells flanking a 2A-peptide-linked reprogramming cassette, c-Myc-Klf4-Oct4-Sox2 (MKOS)-ires-mOrange, by loxP sites (Supplementary Figure 1). Initially we investigated whether the 2A peptide-mediated multiprotein expression could achieve robust expression of c-Myc, Klf4, Oct4 and Sox2, when transcribed from the ubiquitously expressed synthetic CAG enhancer/promoter21. When the vector was transfected into HEK293 cells, expression of Klf4, Oct4 and Sox2 could be detected by immunoblotting (Supplementary Figure 2a). While high expression of endogenous c-Myc in HEK293 cells precluded clear identification of exogenous c-Myc, a phosphorylated form at Thr 58 that was subjected to subsequent ubiquitination22 was enriched in the transfectants, suggesting excess c-Myc was degraded (Supplementary Figure 2a, b). Appropriate nuclear localization of exogenous Oct4 and Sox2 was also observed in the transfected HEK 293 cells (Supplementary Figure 2c).
When the vector, pCAG2LMKOSimO, was introduced into MEFs, some mOrange positive cells converted to an ES cell-like morphology at day 5-6, and by day 9 colonies containing alkaline phosphatase positive cells appeared (data not shown). Moreover morphologically ES cell-like colonies picked between days 20-30 succeeded to grow maintaining an ES cell-like morphology on gelatin (Supplementary Figure 3a). We then went on to estimate the reprogramming efficiency using Nanog reactivation as a marker of reprogramming3,4. MEFs from TNG mice, which have a GFP reporter inserted at the Nanog start codon23, and MEFs from wild-type 129 mice were transfected with the pCAG2LMKOSimO plasmid and cultured on either γMEFs or gelatin. The number of transiently transfected mOrange positive cells was measured by flow cytometry at day 2. The number of reprogrammed colonies judged by GFP positivity (TNG MEFs) or anti-Nanog immunofluorescence (129 MEFs) (Supplementary Figure 3b) was scored at day 28 (Table 1). By comparing stable transfection efficiency with Nucleofection (3.6 % of transiently transfected cells, see Supplementary Figure 3c for details) and the number of reprogrammed colonies, we calculate overall reprogramming effiiency as average 2.5% (Supplementary Table 1). While the estimation method is different from that used in viral reprogramming systems (efficiency; ∼0.1% 2,3,7), this relatively high efficiency may depend on several factors in this non-viral method, including expression of the four reprogramming factors from a single transcript and use of the CAG enhancer/promoter, which may be less prone to silencing.
Table 1. Nucleofection conditions and number of Nanog-GFP/Nanog positive colonies.
| Nucleofection condition1 | 2 days after nucleofection2 | No. of Nanog/Nanog-GFP +ve colonies at day 283 |
||||||
|---|---|---|---|---|---|---|---|---|
| Experiment No. |
passage No. |
DNA (μg) |
cell No / well |
mOrange +ve (%) |
+ve cell No / well |
on γMEFs | on gelatin | |
| TNG | 1 | 4 | 2 | 100,000 | 8.2 | 8,200 | - | 6 |
| 2 | 3 | 2 | 35,000 | 13.3 | 4,655 | - | 3.5* | |
| 129 | 1 | 3 | 2 | 30,000 | 21.0 | 6,300 | - | 3.3** |
| 2 | 4 | 2 | 20,000 | 18.6 | 3,720 | 6 | 2 | |
| 3 | 5 | 5 | 50,000 | 10.4 | 5,200 | 8 | 1 | |
| 4 | 5 | 10 | 50,000 | 12.0 | 6,000 | 7 | 6 | |
2×106 MEFs from TNG or 129 mice (passage number is shown) were transfected using indicated amount of linearised DNA in each experiment.
mOrange positive (+ve) cell number at day 2 was estimated from harvested cell number from one well and percentage of mOrange +ve cells.
Number of Nanog-GFP colonies from TNG MEFs or Nanog positive colonies (by immunofluorescence) from 129 MEFs at day 28 on irradiated MEFs (γMEFs) or on gelatin is shown. -; no data. *; average of two wells. **; average of three wells.
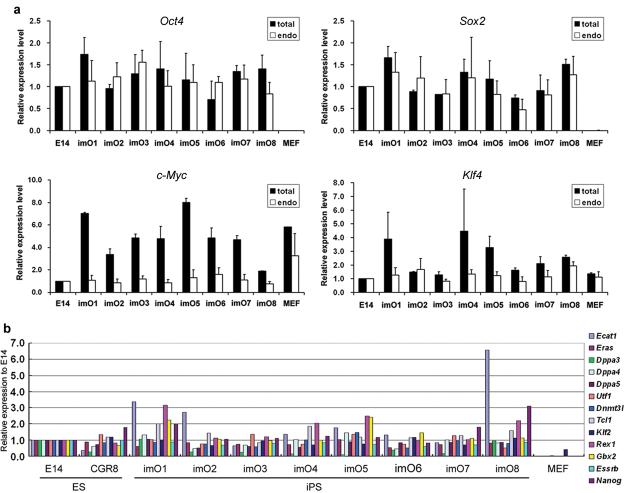
Stable cell lines were established by picking colonies derived from transfection of 129 MEFs (8/12 morphologically ES cell-like colonies picked) and TNG MEFs (5/9 GFP positive colonies picked) with pCAG2LMKOSimO. We examined the expression of the reprogramming factors in 8 cell lines, imO1-imO8. All cell lines showed robust reactivation of endogenous Oct4 and Sox2, comprising the majority of total Oct4 and Sox2 transcripts (Figure 1a). Endogenous c-Myc expression, which was higher in MEFs than ES cells, became similar to ES cells in all cell lines, while there was no large change in endogenous Klf4 expression levels (Figure 1a). Total c-Myc and Klf4 expression were high relative to ES cells, but total Oct4 and Sox2 expression were not, although the exogenous transcript encodes all four genes. This observation could be explained by the fact that the expression level of c-Myc and Klf4 is lower than that of Oct and Sox2 in ES cells (c-Myc<Klf4<Oct4≅Sox2, quantified by real-time PCR, data not shown). The total amount of the four reprogramming factor proteins was also comparable to ES cells in all cell lines examined (Supplementary Figure 4a). In conjunction with Oct4 and Sox2, 13 additional pluripotent markers, which are highly enriched in both ES cells and iPS cells1,4,24, were reactivated in all cell lines screened (Figure 1b). Bisulphite genomic sequencing analyses revealed that the promoter region of Oct4 and Nanog is highly unmethylated in the iPS cells (Supplementary Figure 4c). These results indicate that efficient reprogramming had occurred due to the non-viral single vector.
Figure 1. Efficient reactivation of pluripotent markers in iPS cells generated by a non-viral multiprotein expression vector.
a. Quantitative PCR for total and endogenous c-Myc, Klf4, Oct4 and Sox2 expression. Data is shown as relative expression to an ES cell line, E14Tg2a (E14). Error bars indicate the s.d. generated from triplicates. b. Quantitative PCR for pluripotent markers. Two independent ES cell lines, E14Tg2a (E14) and CGR8, were analyzed together with iPS cell lines. Data is shown as relative expression to E14Tg2a, and represents one of two independent experiments.
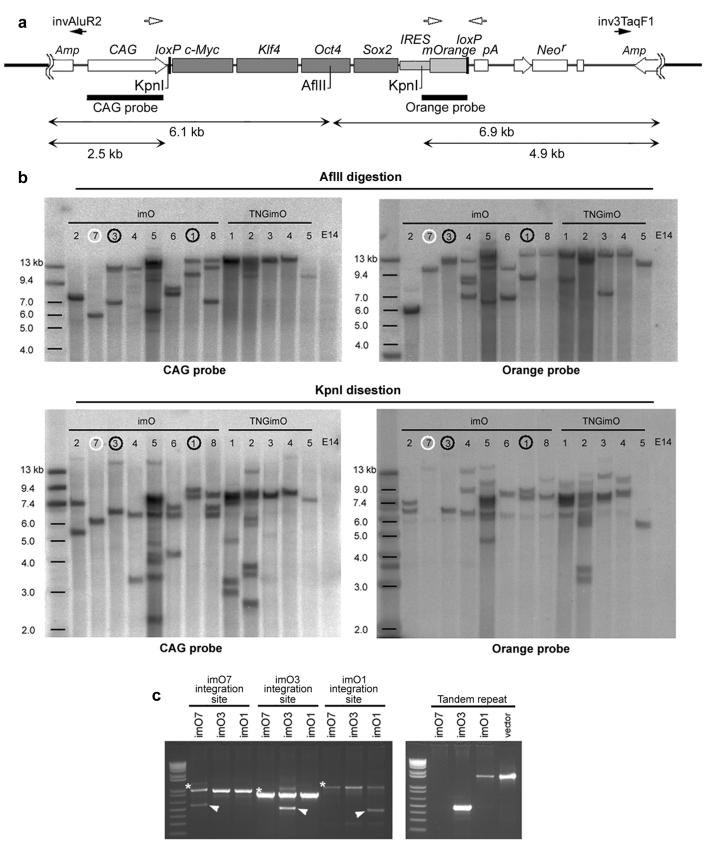
The number of vector integration sites was analyzed by Southern blotting in cell lines imO1-imO8, as well as 5 TNG iPS cell lines, TNGimO1-imO5 (strategy shown in Figure 2a). Of the 13 cell lines, imO7 and TNGimO5 were found to have a single vector integration in one site, indicating that a stable single-copy insertion of the non-viral expression vector can achieve direct reprogramming, effectively minimizing genome modification in iPS cells (Figure 2b). We did not find reprogrammed cell lines without vector integration. The integration sites of imO1, imO3 and imO7, as well as the tandem repeats in imO1 and imO3, were identified by inverse PCR and confirmed by genomic PCR (Supplementary Figure S5 and Figure 2c). All integration sites of the individual cell lines are different, supporting the notion that reprogramming does not depend upon vector integration at specific loci8. Expression of the genes at the integration sites was detectable in MEFs, ES cells, and all examined iPS cell lines, without significant influence imposed by vector integration (Supplementary Figure 5d). Adult cells (footpad fibroblasts) were also succesfully reprogrammed via definite single-copy vector integration (Supplementary Figure 6).
Figure 2. Non-viral iPS cells with a single vctor integration site.
a. Schematic diagram of restriction enzyme sites, AflII and KpnI in pCAG2LMKOSimO and two probes, CAG probe and Orange probe, used for Southern blotting (bars). Black arrows (invAluR2, inv3TaqF1) indicate position of primers used to detect tandem repeat integration in c. b. Southern blotting analysis for AflII (top two panels) and KpnI (bottom two panels) digested genome of imO1-8 and TNGimO1-5, using CAG probe (left panels) and Orange probe (right panels). c. Validation of the integration site and tandem repeat integration. A single integration site of imO7 (white circle in b), and a single integration site with tandem integration (same orientation) of imO1 and imO3 (black circles in b) was identified by inverse PCR (data not shown) and validated by genomic PCR. Asterisk; band from wild-type allele. Arrowhead; integration site-specific band. Detail of the integration site is shown in Supplementary Figure 5. Note different band size of tandem repeats is caused by vector degradation accompanied by random integration.
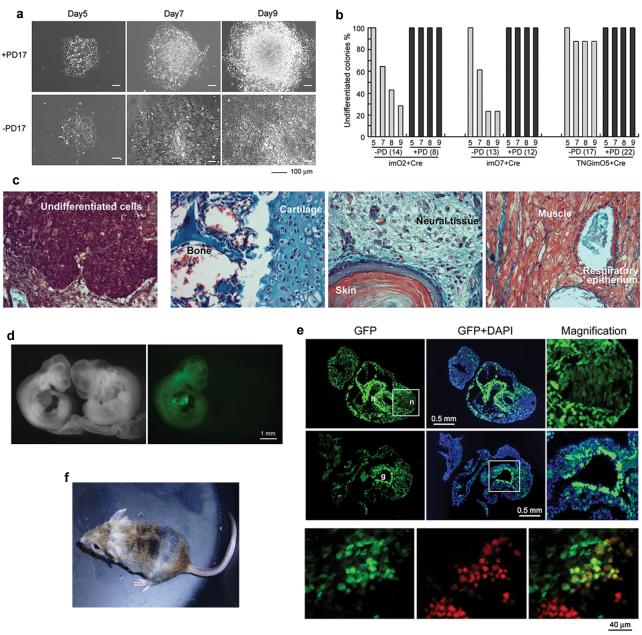
To remove the exogenous reprogramming factors, we performed transient Cre transfection and tracked the loss of fluorescent reporter expression. Surprisingly, many of the mOrange negative colonies started to differentiate after Cre transfection (Supplementary Figure 7a). This excision-mediated differentiation was prevented by culturing the cells in the presence of the Fgf receptor inhibitor, PD173074, which inhibits mouse ES cell differentiation25 (Figure 3a). The percentage of differentiated colonies after Cre-mediated excision differed among clones, possibly reflecting variable stability of the reprogrammed state (Figure 3b). All tested reprogramming cassette-excised clones, which were generated from imO2, imO3, imO7 and TNGimO5 and expanded in the presence of PD173074, could subsequently maintain an undifferentiated morphology for at least 5 passages after removal of PD173074 (data not shown). The presence of PD173074 may allow cells to adapt upon rapid loss of exogenous reprogramming factors in single cells at colony density. Whereas Cre-excised cell lines from imO7 and TNGimO5 could be maintained over 20 passages while maintaining an undifferentiated morphology, cell lines derived from imO2 and imO3 were prone to generate more differentiated cells at later passages in the absence of PD173074, suggesting a different reprogramming level for each clone (data not shown). The endogenous gene expression of c-Myc, Klf4, Sox2, and Oct4 was maintained in the Cre-excised cell lines, imO2Ec3, imO3c8 and imO7c8, which are derived from imO2 with additional constitutive EGFP expression imO2E1, imO3 and imO7, respectively (Supplementary Figure 8a). Expression of other pluripotency genes was sustained (Supplementary Figure 8b), indicating that our single-vector system has enabled complete elimination of exogenous genes without disturbing maintenance of the iPS cell state.
Figure 3. Reprogramming cassette excision and pluripotency of the non-viral iPS cells.
a. Many of the undifferentiated colonies at 5 days post Cre-transfection differentiated by day 9 in the absence of an Fgf receptor inhibitor, PD173074 (bottom). b. Percentage of reprogramming cassette-free undifferentiated colonies in the absence (−PD) and presence (+PD) of PD173074. Numbers of monitored reprogramming cassette-excised colonies are indicated in parentheses. Experiments were performed in three cell lines, imO2, imO7 and TNGimO5. c. Undifferentiated cells in imO7 teratoma (left) and various tissues in imO7c8 teratoma (the other three panels). d. imO3Ec5 derived chimeric embryos (green) at 10.5 dpc (left panels). e. Transversal sections at 9.5 dpc (upper six panels) and genital ridge at 12.5 dpc (bottom panels). imO3Ec5 contributed to ectoderm (neural tissue; n with magnification), mesoderm (heart; h), endoderm (gut; g with magnification) and germ cells (red staining with anti-Oct4 antibody). f. An adult imO7c8 derived chimeric mouse.
We next examined the differentiation ability of the cell lines before and after reprogramming cassette excision. Cre-excised cell lines imO3c8 and imO7c8 downregulated the pluripotency markers Oct4, Nanog and Rex1, and up-regulated markers of all three germ layers in embryoid bodies, thus behaving in the same way as ES cells26 (Supplementary Figure 9a). Furthermore, β-tubulin positive neurons were generated efficiently in a monolayer neural differentiation protocol27 (Supplementary Figure 9b). Differentiation of the factor-retained parental cell lines imO3 and imO7 was less efficient (Supplementary Figure 9a, b). Both imO7 and imO7c8 produced teratocarcinomas when transplanted to the kidney capsule, although imO7 tumours contained more undifferentiated cells than those from imO7c8 tumours (Figure 3c). Finally we injected the reprogramming vector-free iPS cells into C57BL/6 blastocysts (results summarized in Supplementary Table 2). To identify chimeric embryos two cell lines with constitutive EGFP expression, imO3Ec5 and imO7Ec3, were used, and high contribution chimeric embryos were generated at high frequency from both cell lines (Figure 3d and Supplementary Table 2). Contribution to all three germ layers - ectoderm, mesoderm and endoderm - was observed, in addition to Oct4 positive germ cells in the 12.5dpc genital ridge (Figure 3e). Three Cre-excised iPS cell lines, imO1c5, imO7c8 and TNGimO3cC5, gave rise to live chimeras, indicating iPS cells derived with the non-viral multiprotein expression vector are genuinely pluripotent (Figure 3f).
To address the reprogramming ability of the non-viral single-vector system in human cells, we enhanced stable transfection efficiencies using a piggyBac (PB) transposon gene delivery system15. Co-transfection of two PB transposons carrying a doxycycline (dox) inducible MKOS-ires-geo cassette and a constitutively active CAG-rtTA transactivator construct16 was applied to human embryonic fibroblasts (Supplementary Figure 10a). In this design, upon genomic integration, the two transposons allow dox-inducible activation of MKOS expression in wild-type cells. We observed iPS-like colony formation 14 days post transfection (dpt) when the cells were maintained in hES cell culture conditions supplemented with dox. In total, fifteen colonies were picked from 14-25dpt from four wells of 6-well plates, initially containing either 3.2×104 or 6.4×104 fibroblasts/well. When dox was withdrawn on 32dpt, three clones successfully propagated as stable cell lines, while the exogenous factor expression was uninduced as indicated by negative staining for lacZ activity (Supplementary Figure 10b). All clonal lines displayed human ES cell morphology (Supplementary Figure 10b) and were positive for alkaline phosphatase (not shown). Robust expression of endogenous pluripotency markers, SSEA4, NANOG, TRA-160 and TRA-181 was confirmed in all the three cell lines (Supplementary Figure 10c). These results demonstrate that the non-viral single-vector system can reprogram human fibroblasts and strongly suggests that it can also be applied to the production of exogenous factor-free, non-viral human iPS cells.
We demonstrate that single non-viral vector with 2A peptide-linked reprogramming factors can achieve reprogramming efficiently and the exogenous reprogramming factors can be completely removable from the iPS cells using subsequent Cre-transfection. Absolute avoidance of unpredictable exogenous reprogramming factor reactivation is important not only for clinical applications but also for drug screening, since some small molecules affect epigenetic genome modification28 which could cause unexpected reactivation and unreliable screening results. Cre recombinase boasts the most efficient in vivo and in vitro recombination system currently known, and a part of the vector backbone remained in the integration site following Cre-mediated factor deletion would be a tolerable remnant for in vitro iPS applications. We also demonstrated the single-vector reprogramming system combined with PB transposon delivery system for human cell reprogramming. PB transposons are completely removable from their integration site without any residual change in the original DNA sequence15,16. This PB-based single-vector reprogramming system will enable the generation of non-genetically modified human iPS cells as shown in the mouse16, which is ideal for regenerative medicine.
Methods Summary
A vector, pCAG2LMKOSimO, which have c-Myc, Klf4, Oct4 and Sox2 coding regions linked with 2A peptide sequences driven by CAG enhancer/promoter18,21, were constructed as described in the Methods. The vectors were introduced into MEFs using Nucleofector II (Amaxa) and cells were cultured in ES cell culture condition for up to 4 weeks. Colonies showing ES cell-like morphology were picked and cultured either on irradiated MEFs (γMEFs) or gelatin after trypsinization. Gene expression, integration number/sites of the vector in the established cell lines were analyzed by quantitative PCR, immunoblotting, Southern blotting and inverse PCR, respectively. The reprogramming cassette was excised by Cre transient transfection in the presence or absence of an Fgf receptor inhibitor PD173074 (100 ng/ml), and pluripotency of the cell lines were examined in vitro (embryoid body formation, neural differentiation) and in vivo (teratoma formation, blastocyst injection).
Full Methods accompany this paper.
Supplementary Material
Acknowledgements
We thank A. Tsakiridis for advice on using 2A peptide sequence and A. Nagy for providing data from his laboratory on generating human reprogrammed cell lines by combining the PB transposon system and the 2A sequences joined reprogramming factors and his comments on the manuscript. We also thank I. Chambers for providing TNG MEFs, discussion and comments on the manuscript, V. Wilson for advice on teratoma analysis, T. Kunath, S. Lowell, C. Blackburn and K. Vintersten for discussions and comments on the manuscript, B. Hendrich for permission for starting preliminary experiments of this work in his laboratory. We also thank J. Ure, L. Robertson, R. McLay, and R. Wilkie for technical assistance, Biomed unit staff for mouse husbandry. K.K. is the recipient of MRC career development fellowship in stem cell research. A.P. is the recipient of a BBSRC CASE PhD studentship. M.M., P.M. and K.W. were supported by grants from the Canadian Stem Cell Network and JDRF.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Reprints and permission information is available at npg.nature.com/reprintsandpermissions.
The authors declare no competing financial interests.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 3.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 6.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 7.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 8.Aoi T, et al. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science. 2008 doi: 10.1126/science.1154884. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 10.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Foster KW, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced Pluripotent Stem Cells Generated Without Viral Integration. Science. 2008 doi: 10.1126/science.1162494. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of Mouse Induced Pluripotent Stem Cells Without Viral Vectors. Science. 2008 doi: 10.1126/science.1164270. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woltjen K, et al. Nature. 2009 [Google Scholar]
- 17.Hasegawa K, Cowan AB, Nakatsuji N, Suemori H. Efficient multicistronic expression of a transgene in human embryonic stem cells. Stem Cells. 2007;25:1707–1712. doi: 10.1634/stemcells.2006-0813. [DOI] [PubMed] [Google Scholar]
- 18.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 19.Sommer CA, et al. iPS Cell Generation Using a Single Lentiviral Stem Cell Cassette. Stem Cells. 2008 doi: 10.1634/stemcells.2008-1075. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carey BW, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 22.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 23.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008 doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunath T, et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 26.Kaji K, et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 27.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 28.Brueckner B, Kuck D, Lyko F. DNA methyltransferase inhibitors for cancer therapy. Cancer J. 2007;13:17–22. doi: 10.1097/PPO.0b013e31803c7245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.