Abstract
Segregation Distorter (SD) is a selfish, coadapted gene complex on chromosome 2 of Drosophila melanogaster that strongly distorts Mendelian transmission; heterozygous SD/SD + males sire almost exclusively SD-bearing progeny. Fifty years of genetic, molecular, and theory work have made SD one of the best-characterized meiotic drive systems, but surprisingly the details of its evolutionary origins and population dynamics remain unclear. Earlier analyses suggested that the SD system arose recently in the Mediterranean basin and then spread to a low, stable equilibrium frequency (1–5%) in most natural populations worldwide. In this report, we show, first, that SD chromosomes occur in populations in sub-Saharan Africa, the ancestral range of D. melanogaster, at a similarly low frequency (∼2%), providing evidence for the robustness of its equilibrium frequency but raising doubts about the Mediterranean-origins hypothesis. Second, our genetic analyses reveal two kinds of SD chromosomes in Africa: inversion-free SD chromosomes with little or no transmission advantage; and an African-endemic inversion-bearing SD chromosome, SD-Mal, with a perfect transmission advantage. Third, our population genetic analyses show that SD-Mal chromosomes swept across the African continent very recently, causing linkage disequilibrium and an absence of variability over 39% of the length of the second chromosome. Thus, despite a seemingly stable equilibrium frequency, SD chromosomes continue to evolve, to compete with one another, or evade suppressors in the genome.
Author Summary
Mendel's first law of segregation holds that a heterozygous parent will transmit alternative alleles to offspring equally. Segregation Distorter (SD) is a naturally occurring selfish gene complex in D. melanogaster that subverts Mendel's first law. During spermatogenesis in heterozygous SD/SD + males, SD effectively kills SD +-bearing sperm, monopolizing fertilization. SD chromosomes carry a distorter gene and a complement of genetically linked enhancers, often held together by inversions. Thus, SD chromosomes are selfish, co-adapted gene complexes. Although SD is one of our best-characterized selfish gene systems, we still have a poor understanding of its evolutionary history and population dynamics. We therefore performed a large screen for SD chromosomes in African populations of D. melanogaster and studied their genetic properties and history. We found a new SD chromosome type, SD-Mal (endemic to Africa), that has a perfect transmission advantage and lacks recombination over much of the chromosome. This new SD chromosome rapidly swept across sub-Saharan Africa sometime within the last ∼3,000 years. These findings show that selfish gene complexes evolve continuously to evade suppression by other genes in the genome and to compete with one another for a place in the population.
Introduction
The Segregation Distorter (SD) system of the fruitfly, Drosophila melanogaster, is a naturally occurring meiotic drive complex—instead of fair Mendelian transmission, heterozygous SD/SD + males transmit SD chromosomes to most, if not all, progeny [1]–[8]. Full strength distortion is caused by three interacting loci clustered around the centromere of chromosome 2 (an autosome): the trans-acting Segregation distorter (Sd) locus; an upward modifier, Enhancer of SD (E(SD)); and a cis-acting distortion-insensitive allele at the target locus, Responder (Rsp i). (By convention, Sd refers to the locus whereas SD refers to chromosomes assumed to carry the full complex of loci.) SD chromosomes are thus Sd E(SD) Rsp i, whereas SD + chromosomes, which lack the distorting Sd locus and usually carry sensitive alleles of Rsp, are Sd + E(SD) + Rsp s (Figure 1A). During spermiogenesis in heterozygous SD/SD + males, the sperm-specific histone transition required for proper chromatin packaging is disrupted in Rsp s-bearing SD + sperm, leaving functional Rsp i-bearing SD sperm to monopolize fertilization [9]–[12]. For decades, the SD system has been a model in evolutionary genetics, not only for being selfish, propagating at the expense of its bearers, but as a coadapted gene complex whose fitness is determined by multiple epistatic interactors [5]–[7], [13]–[15].
Figure 1. A molecular screen for SD chromosomes.
(A) Part of chromosome arm 2L and all of 2R are shown with the approximate cytological locations of SD loci. The centromere occurs at the transition between cytological divisions 40 and 41. (B) A three-primer assay was used to screen isofemale lines for the presence of the Sd-RanGAP duplication. There are two potential primer pairs: the F-R1 primer pair, a positive control, amplifies a 463-bp product from RanGAP; the F-R2 primer pair amplifies a 353-bp product from the proximal breakpoint of the Sd-RanGAP duplicate gene, if present. Note that the R2 primer anneals to the 5′ region of both RanGAP and Sd-RanGAP; for Sd-RanGAP, however, there is no corresponding forward primer. An example gel is shown: flies carrying Sd-RanGAP yield two amplicons (from Sd-RanGAP and RanGAP), whereas those lacking Sd-RanGAP produce only one (from RanGAP only).
The evolution and persistence of the SD complex depend critically on genetic linkage. Multilocus drive systems can only invade a population when recombination is restricted among loci, as the transmission advantage of distorter chromosomes (Sd Rsp i) must not be offset by the formation of so-called ‘suicide’ chromosomes (Sd Rsp s) that distort against themselves [16]. The clustering of SD loci around the centromere of chromosome 2, where crossing over is reduced, is therefore unsurprising [15]. Epistatic selection further favors the evolution of secondary suppressors of recombination [15],[17],[18]. Many SD chromosomes, for instance, have recruited a pericentric inversion, In(2LR)39D-42A, that further reduces crossing over in the centromeric region, while some have recruited paracentric inversions on 2R (reviewed in [2],[5],[6],[17]). The paracentric inversions are thought to reduce crossing over between the centromeric SD elements and modifiers of distortion distributed across 2R, such as Modifier of SD (M(SD)), Stabilizer of SD (St(SD)), and possibly others [19]–[22]. Thus, SD chromosomes have evolved a complex of multiple, epistatically interacting loci with coadapted alleles whose linkage relationships are usually further tightened by one or more chromosomal inversions.
The geographic distribution of inversions on different SD chromosomes may shed light on the origins, and possibly the age, of the complex. SD can be found in nearly all populations of D. melanogaster at a frequency of ∼1–5% [23] (but see ref. [24]). In North America, Hawaii, Japan, and Australia, SD chromosomes invariably carry inversions (though not necessarily the same ones). In Italy and Spain, however, both inversion-bearing and presumably ancestral, inversion-free SD chromosomes occur. The presence of both derived and ancestral types has been taken as evidence that SD chromosomes originated in the Mediterranean basin [3],[4]. An origin in Mediterranean Europe further implies that the SD complex evolved recently, as D. melanogaster is a sub-Saharan African species whose range expanded to Europe only ∼15,000 years ago, probably via a single major out-of-Africa founder event [25]–[28]. The first population genetic analysis of SD found little divergence between four loci on SD versus SD + chromosomes, consistent with a recent origin for the complex [14],[29].
Much about the evolutionary history and population dynamics of SD in natural populations remains unclear. For one, a recent, Mediterranean origin for SD in the D. melanogaster lineage has important implications, explaining its absence from closely related species and suggesting that the multiple genetic components of the complex evolved very quickly. But the Mediterranean origins hypothesis hinges on few data—the presence of inversion-free SD chromosomes from collections in Italy and Spain and nowhere else. For another, what little is known about the population dynamics of SD comes from laborious, large-scale phenotypic assays to determine the frequency of Sd and Rsp in natural populations (e.g., [30],[31]). These have revealed that in natural populations of D. melanogaster worldwide, the frequency of SD is remarkably similar (1–5%) and thus presumably stable. The stability of SD occurs because its intrinsic transmission advantage is balanced by several forces: the sterility of many SD/SD males [32]; the reduced sperm numbers in SD-bearing flies [33]; the presence of suppressors of distortion [24],[30],[31],[34]; and selection against linked deleterious mutations that accumulate in the large non-recombining regions of SD chromosomes [1]. This apparent stability may however mask an underlying evolutionary turnover among competing SD chromosomes predicted by theory [15]. Using realistic parameters, Charlesworth and Hartl [15] showed that an inversion-free SD chromosome will invade a SD + population and spread to a low-frequency equilibrium; this mixed population, however, is susceptible to invasion by an inversion-bearing SD chromosome that will displace the inversion-free SD and spread to the same low-frequency equilibrium. It appears, then, that SD chromosomes may evolve continuously, as a small subpopulation of second chromosomes in D. melanogaster, competing with one another and evading suppressors.
Fifty years of SD work has produced a rich body of genetics and theory [2], [5]–[8], and recently the molecular basis of distortion has begun to emerge [8], [35]–[38]. The two main SD loci have been identified: Sd is a partial, tandem duplication of the gene RanGAP, called Sd-RanGAP [37]; and Rsp is a large array of 120-bp AT-rich repeats in the centric heterochromatin of 2R [38], where alleles with ≤300 repeats are “insensitive” (Rsp i), 700–1100 are “sensitive” (Rsp s), and ≥1100 are “super-sensitive” (Rsp ss; [3],[4],[38]). Sd-RanGAP encodes truncated RanGAP (Ran-GTPase Activating Protein; [37]), a protein with essential and evolutionarily conserved functions in nuclear transport, mitosis, and chromatin processing [39]–[41]. The truncated Sd-RanGAP protein is enzymatically active but mislocalizes to the nucleus (normally RanGAP is cytoplasmic) where, for reasons not yet clear, it causes segregation distortion in SD/SD + males [8],[35],[36].
Surprisingly, in the decade since its discovery [37], there have been no direct evolutionary analyses of Sd-RanGAP, the gene that actually causes distortion. In this paper, we study the molecular population genetics of the SD complex to investigate its evolutionary history and recent population dynamics. First, we perform the first screen for SD in populations from Africa, the ancestral range of D. melanogaster. Second, we study patterns of DNA sequence variation at the distorter, Sd-RanGAP, as well as its parent gene, RanGAP, and eight noncoding loci on chromosome 2. Finally, we characterize the strength of distortion, inversion status, and mutational load of SD chromosomes. We show that Sd-RanGAP is present in Africa and that a new SD chromosome type has spread very recently across the African continent, causing a large-scale selective sweep among SD chromosomes. These results call into question our current understanding of the timing and location of SD's origins and suggest that, despite its remarkably stable population frequency, SD evolution is not at equilibrium.
Results
A Screen for SD Chromosomes in African Populations
We used a three-primer PCR assay to screen 452 isofemale lines collected from 13 localities in Africa for the Sd-RanGAP duplication (Figure 1B; Table 1). We found 12 SD chromosomes from across the continent, including west (e.g., Benin, Gabon, Cameroon) and east Africa (e.g., Zimbabwe, Kenya; Table 2). Assuming that all isofemale lines are homozygous, the population frequency of SD is 12/452 = 0.027; and assuming that all isofemale lines are heterozygous, the population frequency of SD is 12/904 = 0.013. These estimates suggest that SD chromosomes occur in Africa at a frequency of 1.3–2.7%, similar to its frequency in other natural populations [23].
Table 1. Isofemale lines screened for SD chromosomes.
| Geographic origin | Lines screened | SD chromosomes |
| Benin | 7 | 1 |
| Cameroon | 132 | 3 |
| Congo | 15 | 0 |
| Eritrea | 26 | 0 |
| Gabon | 32 | 1 |
| Ghana | 2 | 0 |
| Kenya | 41 | 5 |
| Malawi | 19 | 0 |
| Niger | 44 | 0 |
| Nigeria | 10 | 0 |
| South Africa | 13 | 0 |
| Uganda | 37 | 0 |
| Zimbabwe | 74 | 2 |
| Total | 452 | 12 |
Table 2. SD chromosomes from Africa.
| SD chromosome | Geographic origin | Inversionsa |
| SD-GN09 | Gabon | In(2R)Mal |
| SD-KM87 | Malindi, Kenya | In(2L)t+In(2R)Mal |
| SD-KM92 | Malindi, Kenya | In(2R)Mal |
| SD-KN20 | Kenya | In(2R)Mal |
| SD-KY38 | Malindi, Kenya | In(2L)t+In(2R)Mal |
| SD-KY91 | Kenya | In(2R)Mal |
| SD-MD21 | Mbalang-Djalingo, Cameroon | In(2R)Mal |
| SD-NK04 | Nkouondja, Cameroon | In(2R)Mal |
| SD-ZK178 | Lake Kariba, Zimbabwe | In(2L)t+In(2R)Mal |
| SD-ZK216 | Lake Kariba, Zimbabwe | In(2L)t+In(2R)Mal |
| SD-BN19 | Benin | No Inversions |
| SD-MD31 | Mbalang-Djalingo, Cameroon | No Inversions |
In(2R)Mal has cytological breakpoints 44F3–12;54E3–10+51B6–11;55E3–12, and In(2L)t has breakpoints 22D3–22E1;34A8–34A9.
Low Divergence between Sd-RanGAP and RanGAP
After genetically extracting the SD chromosomes, we sequenced the ∼4.5-kb Sd-RanGAP sequence from all 12 as well as the homologous region of the parent gene, RanGAP, from 10 wildtype (non-SD) chromosomes sampled from Zimbabwe (see Methods). RanGAP and Sd-RanGAP show typical levels of silent divergence per site from the RanGAP homolog in the outgroup species, D. simulans, with K sil = 0.0471 and 0.0478, respectively. Silent divergence between the duplicate genes, RanGAP and Sd-RanGAP, within D. melanogaster is more than an order of magnitude lower, K sil = 0.0027 (see also ref. [37]). These findings confirm that Sd-RanGAP arose in D. melanogaster well after the split from D. simulans [29]. Using D. simulans RanGAP as an outgroup sequence, we polarized the substitutions between D. melanogaster RanGAP and Sd-RanGAP. Of five fixed differences between RanGAP and Sd-RanGAP, all were fixed in the common ancestor of the Sd-RanGAP sequences: three noncoding changes, one fixed 6-bp deletion, and a single nonsynonymous change (Figure 2). The first intron of RanGAP contains the gene, Hs2st, raising the possibility that some “silent” changes in one gene are not silent in the other. However, of the five fixed substitutions occurring in Sd-RanGAP, only four affect Hs2st: two are noncoding and two are synonymous.
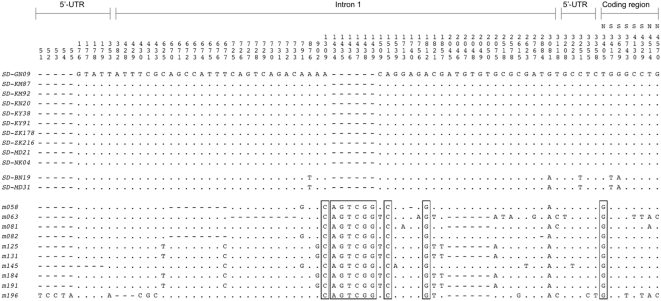
Figure 2. Variation in 12 Sd-RanGAP and 10 RanGAP sequences from African populations of D. melanogaster.
Sd-RanGAP and RanGAP show four nucleotide differences and one indel difference. (N = nonsynonymous; S = synonymous; all other changes are noncoding.)
DNA Sequence Variation at RanGAP and Sd-RanGAP
The amount and distribution of DNA sequence variability at RanGAP is not unusual for an autosomal locus sampled from African populations of D. melanogaster. First, among the 10 wildtype RanGAP sequences, we detect 29 segregating sites (Figure 2), with two measures of DNA sequence polymorphism per site, π = 0.0020 and θ = 0.0023. These values show that RanGAP harbors less variability than the average autosomal locus in African populations (π = 0.0104 and θ = 0.0114; ref. [27]), but this is not unexpected as RanGAP resides in a centromere-proximal region (37E) with a relatively low rate of crossing over and is thus especially susceptible to background selection and hitchhiking effects [42],[43]. Three polymorphisms are synonymous, two are nonsynonymous, and 23 are noncoding, with 66% falling in the large first intron (3.2 kb). The site frequency spectrum at RanGAP does not deviate significantly from standard neutral expectations (Tajima's D = −0.606, P = 0.297; Fay and Wu's H = −4.711, P = 0.127 [44],[45]), where significance was evaluated from 10,000 coalescent simulations conditioning on the observed θ and assuming no recombination). The moderately negative Tajima's D is consistent with recent expansion in the African D. melanogaster populations as inferred from other autosomal loci [27].
Sd-RanGAP is less variable than RanGAP: among the 12 Sd-RanGAP sequences, we detect only five segregating sites (Figure 2), with π = 0.0003 and θ = 0.0004, and a site frequency spectrum skewed towards a moderate excess of rare variants, although not significantly (Tajima's D = −0.313, P = 0.395; Fay and Wu's H = −0.242, P = 0.250). Of the five polymorphisms, two are synonymous and three are noncoding. The lower variability at Sd-RanGAP relative to RanGAP is, of course, expected as Sd-RanGAP is present on only ∼2% of second chromosomes. There are no shared polymorphisms between RanGAP and Sd-RanGAP and hence no evidence for recent gene conversion due to ectopic recombination [46]. The lack of recombination between the two loci implies that Sd-RanGAP evolves as an isolated subpopulation of sequences with a distinct genealogical history.
Unusual Haplotype Structure at Sd-RanGAP
We found six haplotypes among the ten wildtype RanGAP sequences, with levels of linkage disequilibrium (LD) typical of an autosomal locus in Africa (ZnS = 0.247; [27],[47]). In contrast, the spatial distribution of polymorphic sites in the 4.5-kb Sd-RanGAP sequences is unusual: mutations at five segregating sites are in perfect linkage disequilibrium (ZnS = 1.0), forming just two haplotypes (K = 2). The major haplotype occurs ten times in the sample (M = 10) and the minor haplotype twice. We used coalescent-based haplotype configuration tests to estimate the probability of observing such unusual haplotype structure under standard neutral model assumptions [48]. We performed 100,000 coalescent simulations without recombination (a conservative assumption), assuming a sample size of n = 12 and five segregating sites (S = 5). The cumulative probability that the observed haplotype configuration, or one more extreme, occurs by chance is P = 0.0378. Two features of the haplotype configuration, in particular, differ significantly from the expectations of a neutral genealogical process: the major haplotype is too common in the sample, P(M≥10|n = 12, S = 5) = 0.0313; and there are too few kinds of haplotypes, P(K≤2|n = 12, S = 5) = 0.0285. Both features of the data are consistent with an incomplete selective sweep in which the major haplotype has quickly and recently risen to high frequency, but not fixation, among SD chromosomes [49].
Large-Scale Selective Sweep among African SD Chromosomes
If the major African SD haplotype has indeed risen to high frequency due to positive selection or superior segregation distortion, then the haplotype structure may extend beyond the Sd-RanGAP region. To test this possibility, we sequenced eight noncoding regions across chromosome 2 from all 12 African SD chromosomes and from 10 wildtype chromosomes (Figure 3; Table 3). The amount and distribution of DNA sequence variation among wildtype chromosomes was typical for African D. melanogaster populations [27], with θ ranging from 0.0053 to 0.0137 and Tajima's D ranging from −1.630 to 0.445 (P = 0.045 for locus G, but ≥0.05 for other loci; Table 3). In addition, there is ample evidence for recombination in all but one of the loci (region E has an unusual lack of variability and a small number of haplotypes, though not significantly so; Table 3; Figure 3).
Figure 3. Distribution of DNA sequence variation at Sd-RanGAP and eight non-coding regions in Africa.
Sequences were sampled from 12 SD and 10 wildtype second chromosomes from African populations of D. melanogaster. The positions of the two overlapping inversions, In(2R)Mal, and the sequenced loci are shown on chromosome 2 (only part of 2L is shown). Sequence variants are arbitrarily coded: gray matches SD-GN09, white does not. The red box highlights the long, mutation-free haplotype that spans from Sd-RanGAP on 2L to locus G (55B) on 2R.
Table 3. Summary of molecular population genetic analyses at Sd-RanGAP and eight non-coding regions on chromosome 2.
| locus | M | SD | J | K | E | F | G | H | I | |
| gene | tup | Sd-RanGAP | CG30497 | Myd88 | off-track | scab | staufen | plexus | CG34372 | |
| region | p_IGR | gene | intron | d_IGR | p_IGR | p_IGR | p_IGR | intron | d_IGR | |
| cytol. location | 37B | 37E | 43E | 45C | 48D6 | 51E | 55B | 58E | 59E | |
| Wildtype (non-SD) | n | 10 | - | 10 | 10 | 10 | 10 | 10 | 10 | 8 |
| L | 707 | - | 834 | 824 | 715 | 612 | 567 | 728 | 626 | |
| S | 14 (15) | - | 18 | 31 (32) | 13 | 12 | 12 | 11 | 20 | |
| π | 0.0092 | - | 0.0062 | 0.0124 | 0.0073 | 0.0076 | 0.0048 | 0.0042 | 0.0110 | |
| θ | 0.0075 | - | 0.0076 | 0.0137 | 0.0066 | 0.0069 | 0.0075 | 0.0053 | 0.0123 | |
| Tajima's D | 1.050 | - | −0.853 | −0.483 | 0.445 | 0.405 | −1.630 * | −0.997 | −0.554 | |
| Fay & Wu's H | −1.067 | - | 1.156 | 4.356 | 0.800 | −0.978 | −3.022 | −1.689 | −0.786 | |
| ZnS | 0.299 | - | 0.109 | 0.143 | 0.280 | 0.269 | 0.142 | 0.096 | 0.147 | |
| K s | 0.042 | - | 0.063 | 0.085 | 0.056 | 0.020 | 0.039 | 0.024 | 0.034 | |
| SD | n | 12 | 12 | 12 | 12 | 12 | 12 | 11 ** | 12 | 12 |
| L | 708 | 4515 | 865 | 825 | 700 | 612 | 567 | 734 | 626 | |
| S | 17 | 5 | 11 | 10 | 12 | 7 | 0 | 13 (14) | 24 | |
| π | 0.0075 | 0.0003 | 0.0024 | 0.0024 | 0.0042 | 0.0021 | 0.0000 | 0.0051 | 0.0114 | |
| θ | 0.0080 | 0.0004 | 0.0042 | 0.0040 | 0.0057 | 0.0038 | 0.0000 | 0.0063 | 0.0127 | |
| Tajima's D | −0.263 | −0.313 | −1.755 * | −1.705 * | −1.077 | −1.713 * | - | −0.794 | −0.462 | |
| Fay & Wu's H | −0.818 | −0.242 | −4.697 * | −7.424 * | −7.242 * | −4.091 * | - | 0.061 | 0.485 | |
| ZnS | 0.147 | 1.000 | 0.497 | 0.652 | 0.651 | 0.466 | - | 0.088 | 0.162 | |
| K s | 0.042 | 0.048 | 0.064 | 0.084 | 0.057 | 0.018 | 0.037 | 0.025 | 0.034 | |
| SD-Mal only | n | 10 | 10 | 10 | 10 | 10 | 10 | 9 ** | 10 | 10 |
| L | 708 | 4515 | 874 | 825 | 709 | 614 | 573 | 737 | 626 | |
| S | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 13 (14) | 23 | |
| π | 0.0071 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0058 | 0.0121 | |
| θ | 0.0070 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0067 | 0.0130 | |
| Tajima's D | 0.047 | - | - | - | - | - | - | −0.631 | −0.335 | |
| Fay & Wu's H | −1.244 | - | - | - | - | - | - | 0.267 | 0.711 | |
| ZnS | 0.199 | - | - | - | - | - | - | 0.099 | 0.184 | |
| K s | 0.042 | 0.048 | 0.064 | 0.085 | 0.057 | 0.019 | 0.037 | 0.025 | 0.034 |
*: P<0.05, determined by 10,000 coalescent simulations conditional on observed θ and assuming no recombination.
**: The SD-KY38 chromosome has a lethal mutation in the staufen region (G) and thus could not be sequenced from Df(2R)Pcl7B/SD-KY38 or Df(2R)Pcl11B/SD-KY38 flies.
p_IGR and d_IGR = proximal and distal intergenic region, respectively.
n = number of chromosomes sampled.
L = gapless length of sequenced region.
S = number of segregating sites (mutations) in the sample.
π = average number of pairwise differences per site.
θ = Watterson's measure of variability based on the number of segregating sites.
Z nS = average linkage disequilibrium among all pairwise combinations of S segregating sites in sequenced region.
K s = Jukes-Cantor corrected silent divergence per silent site between sample and D. simulans.
The distribution of variation among SD chromosomes differs strikingly from wildtype chromosomes in two ways. First, the frequency spectra at several loci show patterns consistent with a recent selective sweep. Of the eight noncoding regions surveyed, three loci (J, K, and F) show significant excesses of rare variants (Tajima's D; Table 3), four contiguous loci (J, K, E and F) show significant excesses of high-frequency derived variants (Fay and Wu's H; Table 3), and a fifth contiguous locus (G) possesses no variability at all. The three loci whose frequency spectra do not deviate from neutral expectations include the most distal locus on 2L (M at 37B) and the two most distal loci on 2R (H and I at 58E and 59E, respectively). Second, and more striking, the haplotype structure seen at Sd-RanGAP extends across most of chromosome arm 2R: the 10 major Sd-RanGAP chromosomes possess a single identical haplotype that extends from cytological region 37E on 2L (Sd-RanGAP) to region 55B on 2R (locus G; Table 3; Figure 3). The long distance LD does not extend to the most distal locus on 2L (locus M) or the two most distal loci on 2R (H and I; Table 3; Figure 3). Among all 12 SD chromosomes, forty-five segregating sites occur at Sd-RanGAP and the six regions extending to cytological subdivision 55B. Remarkably, all are differences between the major and minor SD chromosomes (17) or between the two minor SD chromosomes (28). The 10 major SD haplotypes are identical—there is not a single polymorphism in >8.1 kb of sequence. A haplotype configuration test assuming n = 12, S = 45, and no recombination confirms that this haplotype configuration (M = 10, K = 3) is highly unusual under a standard neutral genealogical process (P = 0.00002): the major haplotype is too common in the sample (P[M≥10|n = 12, S = 45]≤0.00001) and there are too few kinds of haplotypes (P[K≤3|n = 12, S = 34] = 0.00002). The chromosomal region between Sd-RanGAP and region 55B (G) spans ≥14 Mb and ∼30 cM, comprising more than 39% of the euchromatic length of chromosome 2. Taken together, the significantly skewed frequency spectra and the existence of an extraordinarily long, high-frequency, mutation-free haplotype suggest a large-scale selective sweep in progress among SD chromosomes [49].
The Major SD Haplotype Recombines Less and Distorts More
The magnitude of a selective sweep is determined by two key parameters: the local rate of recombination and the strength of selection driving the major haplotype to high frequency. The SD chromosomes carrying the major haplotype are unique in both respects. First, recombination is suppressed along much of the major SD haplotype. We cytogenetically characterized the 12 African SD chromosomes by crossing SD/SD or SD/CyO males to virgin cn bw females (which are homozygous for standard-arrangement second chromosomes) and examined polytene chromosome squashes from larval salivary glands. None of the African SD chromosomes possess the In(2R)NS inversion (52A2–52B1;56F9–56F13) found on most non-African SD chromosomes [2]. Indeed, SD-BN19 and SD-MD31 are inversion-free chromosomes (Table 2). The other ten SD chromosomes, however, possess a complex chromosomal arrangement on 2R (Table 2). The cytological order (40–44F|54E–55E|51BC–44F|54E–51BC|55E–60) shows that these SD chromosomes have recruited two overlapping inversions: In(2R)51BC;55E first, followed by In(2R)44F;54E (Figure 4). These inversion breakpoints match a previously identified, but rare, African endemic chromosomal arrangement found in Malawi [50], hereafter called In(2R)Mal. In addition to In(2R)Mal, four SD chromosomes (SD-KN20, SD-KY87, SD-ZK178, SD-ZK216) carry the cosmopolitan In(2L)t inversion (22D3–D6;34A8–A9; Table 2).
Figure 4. Polytene chromosome squashes reveal two overlapping inversions on arm 2R of ten SD chromosomes.
The In(2R)Mal arrangement is endemic in African populations and involves two inversions, In(2R)51B6–11;55E3–12 and In(2R)44F3–12;54E3–10.
The association between major haplotype and the In(2R)Mal arrangement is perfect: all major haplotype SD chromosomes carry In(2R)Mal, whereas both minor haplotype SD chromosomes (SD-BN19 and SD-MD31) lack In(2R)Mal (Fisher's Exact P = 0.015). Hereafter, we refer to this new class of In(2R)Mal-bearing, major haplotype SD chromosomes as SD-Mal. To test the effect of the In(2R)Mal arrangement on crossing over, we crossed heterozygous SD-NK04/cn bw females (SD-NK04 is a SD-Mal chromosome) to cn bw males and recorded the frequency of recombination between cn (43E16) and bw (59E2). As negative controls, we crossed heterozygous SD-MD31/cn bw females to cn bw males (SD-MD31 is inversion free). Among progeny from the SD-MD31 control crosses with a standard arrangement second chromosome, 34.8% carried recombinant chromosomes (n = 2,155 progeny). In contrast, the In(2R)Mal arrangement almost entirely eliminates crossing over between cn and bw: among progeny from crosses with the In(2R)Mal-bearing SD-NK04 chromosome, only 0.2% carried recombinant chromosomes (n = 1,564). By restricting recombination with wildtype chromosomes, In(2R)Mal sequesters a large piece of chromosome arm 2R as an effectively non-recombining region. The lack of recombination helps to explain the long-range LD produced by the selective sweep (Figure 3) as well as the strong population differentiation at loci between SD-Mal and inversion free chromosomes (S nn = 1.0, P≤0.0001 [51], for the Sd-RanGAP to 55B regions concatenated; Table 4).
Table 4. Genetic differentiation between SD and non-SD chromosomes.
| Population comparison | M | J | K | E | F | G | H | I | |
| 37B | 43E | 45C | 48D6 | 51E | 55B | 58E | 59E | ||
| SD (12) vs. wildtype (10) | S nn | 0.655 | 0.841** | 0.864* | 0.848** | 0.777** | 0.541* | 0.314 | 0.462 |
| F st | 0.119 | 0.406 | 0.423 | 0.599 | 0.088 | 0.032 | −0.024 | −0.019 | |
| SD-Mal (10) vs. wildtype (10) | S nn | 0.620 | 1.000** | 0.950** | 1.000** | 0.855** | 0.514 | 0.379 | 0.373 |
| F st | 0.081 | 0.581 | 0.529 | 0.771 | 0.230 | 0.032 | −0.012 | −0.043 |
P<0.001 and ** P<0.0001, where probabilities determined by 10,000 permutations of the data.
We next assayed the strength of segregation distortion by estimating k, the proportion of progeny inheriting SD chromosomes from heterozygous SD/Rsp s males. In preliminary work, we found that the dominantly marked balancer chromosome, In(2LR)Gla (hereafter, Gla), carries a sensitive Responder (Rsp s). We therefore measured transmission from heterozygous SD/Gla flies (see Methods). Surprisingly, the two SD chromosomes bearing the minor haplotype showed no detectable distortion: k* = 0.538±0.025 for SD-BN19 and k* = 0.415±0.012 for SD-MD31 (mean k*±s.e. are corrected for viability; Table 5). SD-BN19 and SD-MD31 chromosomes also failed to cause distortion when heterozygous against the super-sensitive Rsp ss allele of the lt pk cn bw chromosome (not shown). In contrast, males heterozygous for SD-Mal chromosomes collectively sired 10,664 progeny and failed to produce a single Rsp s-bearing offspring (k* = 1.0; Table 5). The genetic and phenotypic data on recombination and distortion thus provide a clear explanation for the rise of the major haplotype-bearing SD-Mal chromosomes in Africa: they recombine less and distort more.
Table 5. Strength of segregation distortion for African SD chromosomes.
| Chromosome | Male transmission | Female transmission | ||||||
| k m | ±s.e. | k m* | ±s.e. | Total progeny | k f | ±s.e. | Total progeny | |
| SD-GN09 | 1.000 | 0.000 | 1.000 | 0.000 | 682 | 0.539 | 0.021 | 487 |
| SD-KM87 | 1.000 | 0.000 | 1.000 | 0.000 | 1124 | 0.561 | 0.037 | 431 |
| SD-KM92 | 1.000 | 0.000 | 1.000 | 0.000 | 1092 | 0.616 | 0.087 | 799 |
| SD-KN20 | 1.000 | 0.000 | 1.000 | 0.000 | 1072 | 0.595 | 0.024 | 1020 |
| SD-KY38 | 1.000 | 0.000 | 1.000 | 0.000 | 837 | 0.519 | 0.046 | 664 |
| SD-KY91 | 1.000 | 0.000 | 1.000 | 0.000 | 1544 | 0.579 | 0.064 | 230 |
| SD-MD21 | 1.000 | 0.000 | 1.000 | 0.000 | 997 | 0.486 | 0.05 | 752 |
| SD-NK04 | 1.000 | 0.000 | 1.000 | 0.000 | 1167 | 0.525 | 0.037 | 457 |
| SD-ZK178 | 1.000 | 0.000 | 1.000 | 0.000 | 842 | 0.533 | 0.023 | 967 |
| SD-ZK216 | 1.000 | 0.000 | 1.000 | 0.000 | 1307 | 0.555 | 0.034 | 813 |
| SD-BN19 | 0.569 | 0.025 | 0.538 | 0.025 | 1179 | 0.613 | 0.074 | 561 |
| SD-MD31 | 0.531 | 0.012 | 0.415 | 0.012 | 1531 | 0.621 | 0.035 | 1167 |
| cn bw | 0.602 | 0.030 | 0.504 | 0.033 | 580 | 0.615 | 0.036 | 912 |
| OreR | 0.654 | 0.024 | 0.558 | 0.026 | 912 | 0.615 | 0.013 | 876 |
k m and k f = proportion of progeny inheriting SD when transmitted by males and females, respectively.
k m* = viability-corrected estimate of proportion of progeny inheriting SD transmitted by males.
The Age of the SD Sweep
The complete absence of even low frequency polymorphisms in ∼8.1 kb of sequence distributed from Sd-RanGAP on 2L to cytological subdivision 55B on 2R (G) suggests that SD-Mal rose to high frequency among SD chromosomes quickly and recently. To obtain estimates of the upper 95% confidence limit for the age of the sweep, we assumed that the genealogy relating SD-Mal haplotypes is star-shaped, as expected for a selective sweep, and then estimated the time back to their most recent common ancestor [52]. The expected number of segregating sites in such a sample is E(S) = ntu, where n = number of lineages, t = time in the past when the lineages coalesce into a single common ancestor, and u = the total mutation rate of the sequenced regions. Assuming that the number of mutations on the ten lineages is Poisson distributed, we numerically solved for the probability of observing zero polymorphisms, P(S = 0) = e−ntu, for different times to the common ancestor, t. We used two different estimates of the sequence-specific mutation rate. First, we estimated the mutation rate per generation from θ, which equals 4N e u under standard neutral assumptions, estimated from the wildtype sequences and assuming that N e = 106 for D. melanogaster. Second, we estimated the mutation rate per year based on the number of fixed differences between D. melanogaster and D. simulans, assuming a divergence time of 3 Mya [53]. The two mutation rates yield qualitatively similar limits for the age of the sweep. Using the polymorphism-based estimate of u, the 95% upper confidence limit for the age of the sweep is 1,875 years. Using the divergence-based estimate of u, the 95% upper confidence limit for the age of the sweep is 3,360 years. Both estimates suggest that the major SD-Mal haplotype expanded across Africa very recently, within the last few thousand years.
Accumulation of Linked Lethal Mutations
We performed complementation tests among all pairwise combinations of the 12 SD chromosomes, producing 12 SD i/SD i and 66 SD i/SD j genotypes. Both minor SD chromosomes (SD-BN19 and SD-MD31) are homozygous viable, but all ten SD-Mal chromosomes are homozygous lethal (Table 6). Crosses among SD-Mal chromosomes, however, show that all ten fall into unique complementation groups—none of the lethal mutations is shared among major SD-Mal chromosomes (Table 6). This distribution of lethal mutations supports a star-shaped genealogy: all of the lethal mutations must have arisen on the external branches of the genealogical history of the SD-Mal chromosomes in our sample. These complementation data also reveal that lethal mutations are significantly over-represented on SD-Mal chromosomes relative to wildtype chromosomes: 29% of wildtype second chromosomes are lethal or semi-lethal [54] versus 100% of SD-Mal chromosomes (Fisher's exact P = 0.0015). The large In(2R)Mal rearrangement on SD-Mal chromosomes provides a large non-recombining target for lethal mutations that can persist by hitchhiking with the SD system.
Table 6. Complemenation tests for all SD i/SD j combinations.
| SD-GN09 | SD-KM87 | SD-KM92 | SD-KN20 | SD-KY38 | SD-KY91 | SD-MD21 | SD-NK04 | SD-ZK178 | SD-ZK216 | SD-BN19 | SD-MD31 | |
| SD-GN09 | Lethal | MF | MF | MF | MF | MF | MF | MF | MF | MF | MF | MF |
| SD-KM87 | FF | Lethal | MF | MS | MF | MF | MF | MF | MF | MF | MF | MF |
| SD-KM92 | FF | FF | Lethal | MF | MF | MF | MF | MF | MS | MF | MF | MF |
| SD-KN20 | FF | FF | FF | Lethal | MS | MF | MS | MS | MF | MF | MF | MF |
| SD-KY38 | FF | FF | FF | FF | Lethal | MF | MF | MF | MS | MF | MF | MF |
| SD-KY91 | FF | FF | FF | FF | FF | Lethal | MF | MF | MS | MF | MF | MF |
| SD-MD21 | FF | FF | FF | FF | FF | FF | Lethal | MS | MS | MF | MF | MF |
| SD-NK04 | FF | FF | FF | FF | FF | FF | FF | Lethal | MF | MF | MF | MF |
| SD-ZK178 | FF | FF | FF | FF | FF | FF | FF | FF | Lethal | MS | MF | MF |
| SD-ZK216 | FF | FF | FF | FF | FF | FF | FF | FF | FF | Lethal | MF | MF |
| SD-BN19 | FF | FF | FF | FF | FF | FF | FF | FF | FF | FF | Viable, MF, FF | MF |
| SD-MD31 | FF | FF | FF | FF | FF | FF | FF | FF | FF | FF | FF | Viable, MF, FF |
Above the diagonal, MS = male sterile and MF = male fertile; below the diagonal FF = female fertile.
Fertility in SD i/SD j Flies
For the 66 viable SD i/SD j and 2 viable SD i/SD i genotypes, we tested the fertility of both sexes. None of the 68 genotypes were female-sterile, but 10 were male-sterile (Table 6). SD-ZK178 is male-sterile in combination with five other SD chromosomes; SD-KN20 is male sterile in combination with four others; and SD-NK04 is male-sterile in combination with SD-ZK216. The patterns of complementation for male fertility are complex. For instance, SD-KY38 and SD-MD21 complement one another and yet both fail to complement SD-ZK178. Similarly, SD-ZK178 and SD-NK04 complement one another and yet both fail to complement SD-ZK216. Assuming that male sterility results from male-sterile mutations on chromosome 2, the data in Table 6 require a circular complementation map with at least 10 unique lesions. A more plausible hypothesis, however, is that male sterility results not from linked male-sterile mutations but from interactions among different alleles at SD complex loci [32],[55]. Indeed, previous work has shown that deletion of one copy of Sd rescues sterility in otherwise male-sterile SD i/SD j combinations, supporting a connection between distortion and sterility [55]. The complex patterns of fertility complementation in SD i/SD j males cannot, however, be explained by intragenic complementation at the Sd locus, as the Sd-RanGAP sequences among SD-Mal chromosomes are identical, suggesting that interactions involving other SD loci must be involved.
Discussion
Two major findings emerge from our analysis of the SD system. First, SD occurs in ancestral, African populations of D. melanogaster at a frequency similar to that of other populations worldwide. This discovery raises doubts about the Mediterranean-origins hypothesis. Second, the evolution and rapid spread of a newer, stronger SD chromosome has left a dramatic population genetic signature: a remarkably long haplotype, spanning more than 39% of chromosome 2—roughly 30 cM—that is both free of polymorphisms (Table 3, Figure 3) and differentiated from other chromosomes in the population (Table 4). These findings suggest that a new SD chromosome type endemic to Africa, SD-Mal, has swept across the continent sometime within the last few thousand years.
SD in Africa
The Mediterranean-origins hypothesis is based on the geographic distribution of inversions on SD chromosomes: inversion-bearing SD chromosomes occur throughout the world; but both inversion-bearing and inversion-free, presumably ancestral, SD chromosomes occur in Spain and Italy [3],[4]. The presence of ancestral SD chromosomes suggests that the complex may have arisen in Spain or Italy or nearby. Our discovery of SD chromosomes in African populations of D. melanogaster raises questions about the Mediterranean-origins hypothesis. Did SD originate in the Mediterranean and subsequently invade sub-Saharan Africa via back-migration? Or did SD originate in Africa and then make its way to Europe (and the rest of the world) as part of the D. melanogaster out-of-Africa event, ∼15,000 years ago [25]–[28]? The presence of inversion-free SD chromosomes in Benin and Cameroon (SD-BN19 and SD-MD31, respectively) would seem to make a sub-Saharan African origin as likely as a Mediterranean one. In either case, the fact that inversion-free SD chromosomes occur in both Africa and the Mediterranean suggests that Sd-RanGAP dispersed from one location to the other shortly after it originated and then subsequently acquired different inversions on different continents.
The relative youth of the Sd-RanGAP duplication makes distinguishing between sub-Saharan African and Mediterranean origins with the present data difficult. We cannot, for instance, precisely date the origin of Sd-RanGAP from RanGAP based on the five fixed differences (1 indel, 4 nucleotide changes) by assuming a simple neutral molecular clock for two reasons. First, we cannot exclude the rapid, non-neutral fixation of changes in Sd-RanGAP. Second, some (or all) of the five fixed differences may have been segregating as the ancestral RanGAP sequence that ultimately gave rise to Sd-RanGAP. This putative ancestral RanGAP haplotype may be missing from our population sample by chance, or because it was lost from the population, or because it does not occur in African populations. Determining the time and place of origin for the SD system will therefore require deeper resequencing of Sd-RanGAP and RanGAP from both Europe and Africa.
Evolutionary Turnover of SD Chromosomes
The population genetic analyses revealed six striking patterns among SD chromosomes (Table 3; Figure 3): significant excesses of rare variants; significant excesses of high frequency derived variants; an unusual distribution of haplotype frequencies (10+2 or 10+1+1; Figure 3); exceedingly long-range LD; a complete absence of polymorphism in >8.1 kb spanning >39% of the length of SD-Mal chromosomes; and significant population genetic differentiation between SD-Mal and other chromosomes (Table 4). Together these observations suggest that SD-Mal has spread to high frequency among SD chromosomes in Africa sometime within the last 3,000 years. Why might one type of SD chromosome rise in frequency so quickly, apparently displacing other SD chromosomes? The answer seems straightforward: SD-Mal chromosomes distort more than SD-BN19 and SD-MD31 and recombine less over the length of 2R, perhaps preserving a favorable distortion-enhancing combination of alleles in the In(2R)Mal region. Similar displacement of one SD type (SD-5) by another (SD-72) appears to have occurred during a 30-year period in populations in Wisconsin [31]. Thus, the apparently stable equilibrium frequency of SD chromosomes in D. melanogaster populations worldwide (1–5%) appears to mask a dynamic turnover among competing SD chromosome types.
There are at least two, non-exclusive explanations for the turnover of SD chromosomes. First, the SD system may be sufficiently new that it has not yet reached a stable evolutionary equilibrium: older Sd-RanGAP bearing chromosomes are still being displaced by new ones, like SD-Mal in Africa or SD-72 in North America [31], as predicted by theory [15]. Second, an ultimately stable evolutionary equilibrium for SD chromosomes may not exist: SD may be engaged in a perpetual coevolutionary conflict with the rest of the genome [17]. Indeed, there is considerable variation among populations in the frequency of insensitive Rsp i alleles [30],[31] and other unlinked genetic variants that affect distortion (e.g., [24],[34]). Under this scenario, the rise of SD-Mal and decline of SD-BN19 and SD-MD31 could reflect a transitional phase in the genetic conflict in Africa: SD-BN19 and SD-MD31 may no longer cause distortion because they have come under the effective control of unlinked suppressors in the genome, whereas adaptive changes specific to SD-Mal chromosomes allow them to escape suppression.
The discovery of two Sd-RanGAP bearing chromosomes that fail to cause distortion is surprising—indeed, classical phenotypic screens for segregation distortion undoubtedly would have misclassified SD-BN19 and SD-MD31 as wildtype chromosomes. While these chromosomes may now be suppressed, there are four other possibilities. One is that SD-BN19 and SD-MD31 have experienced mutations causing a loss of distortion. Mutational disruption of the Sd-RanGAP sequence seems unlikely, however, as all five differences that distinguish SD-BN19 and SD-MD31 from SD-Mal are silent. A second possibility is that recombination has stripped SD-BN19 and SD-MD31 chromosomes of essential modifiers required for distortion. Wildtype chromosomes that carry Sd-RanGAP transgenes but lack upward modifiers cause either very weak or even no distortion [37]. However, both of these scenarios—disruption by mutation or recombination—require that we explain the seemingly improbable coincidental loss of distortion by two identical, and relatively rare, Sd-RanGAP haplotypes. A third possibility is that SD-BN19 and SD-MD31 are not “SD chromosomes” but rather ancestral Sd-RanGAP-bearing chromosomes that never caused drive. This scenario would imply that SD chromosomes evolved from a neutral, non-driving ancestral haplotype: Sd-RanGAP arose as new duplication, drifted to sufficiently high frequency to become established via migration in Europe and in Africa, and then subsequently recruited genetic modifiers that conferred distortion. This history, if true, implies that African and non-African SD chromosomes independently acquired convergent distorting gene complexes. A final possibility is that SD-BN19 and SD-MD31 may cause distortion but not in the particular genetic backgrounds used in our assay. Further genetic analyses are required to distinguish these possibilities.
Epistatic Selection Shapes Variation on SD-Mal Chromosomes
The long SD-Mal haplotype spans Sd-RanGAP, region 43E (locus J), and the In(2R)Mal inversions (K, E, F, and G; Figure 3) but does not extend distal to Sd-RanGAP on 2L or distal to In(2R)Mal on 2R. The structure of the SD-Mal haplotype probably reflects the hitchhiking effects of epistatic selection. First, consistent with the lack of loci known to affect distortion distal to Sd-RanGAP, SD and SD + chromosomes are free to recombine without consequence on the distal part of 2L, preventing LD there [4],[29]. Second, although In(2R)Mal suppresses recombination within the inverted regions, there is opportunity for crossing over in the interval between the SD complex loci (Sd, E(SD), and Rsp) and the proximal breakpoint of the In(2R)Mal. The perfect LD across this interval suggests that strong epistatic selection maintains the association between the SD loci and the In(2R)Mal inversions. In principle, double-recombinants in the interval between centromeric SD loci and In(2R)Mal could preserve their association, but these may be rare events relative to the strength of epistatic selection favoring SD-Mal. Thus, positive epistatic selection on the SD-In(2R)Mal genotype may have caused hitchhiking effects to dominate the intervening sequence between them, explaining the skewed frequency spectrum, LD and lack of variability on SD-Mal chromosomes in region 43E (locus J). It is also possible that epistatic selection directly preserves an association with a M(SD) allele in the SD-In(2R)Mal interval [20], but we do not yet know if SD-Mal carries M(SD). Third, inversions on 2R have been interpreted as tightening the association between SD and St(SD), a modifier (or region of polygenic modifiers; ref. [56]) that increases the strength of distortion, putatively located near the tip of 2R [21],[22]. The fact that we fail to detect LD between SD and loci in cytological regions 58–59 (H and I; Figure 3) suggests that either no St(SD) loci reside in (or distal to) regions 58–59 as previously reported [22] or that no such St(SD) loci enhance distortion on SD-Mal chromosomes. It is important to note that St(SD), like M(SD), was characterized from non-African SD chromosomes; African SD chromosomes may carry a distinct set of linked modifiers.
Explaining the Global SD Equilibrium
Although there appears to be competition among SD chromosomes, the overall frequency of SD in populations throughout the world is remarkably similar (1–5%; but see ref. [24]). Considering that different populations have experienced different environments, genetic backgrounds, and demographic histories, the seemingly stable frequency of SD suggests that its equilibrium is the result of strong deterministic forces. What prevents SD from reaching higher frequencies or even fixation? Three factors limit the spread of SD. First, as SD frequency increases, so does selection for insensitive Rsp i alleles and other genetic suppressors. Second, as SD frequency increases, intrinsically male-sterile SD i/SD j genotypes become more common, placing an upper-limit on the spread of SD (Table 6; ref. [32]). Third, SD/SD + males have been shown to suffer reduced male fertility, as might be expected when 50% of sperm are destroyed [9]. Finally, many SD chromosomes worldwide, including the new SD-Mal chromosomes, carry linked recessive lethal and other deleterious mutations (Table 6). The large non-recombining, inverted blocks of chromosome that become associated with SD present a large mutational target. Without recombination, linked recessive lethal and other deleterious mutations are able to persist by hitchhiking with SD. It remains unclear if these factors are sufficient to explain the distortion-selection balance that causes the frequency of SD to settle at 1–5% in D. melanogaster populations worldwide.
Conclusions
The hitchhiking effects of selfish meiotic drive gene complexes have shaped patterns of DNA sequence variability in at least five other cases: four selfish X chromosome systems (one in Drosophila pseudoobscura [57], two in Drosophila simulans [58],[59], and one in Drosophila recens [60]) that drive in the male germline and a selfish autosomal centromere that drives in the female germline of the monkeyflower, Mimulus guttatus [61]. Like SD, all five of these drive systems are associated with haplotypes of reduced variability and three show long-range LD—the signatures of partial selective sweeps. Notably, all five are balanced polymorphisms in which the drive elements are prevented from going to fixation by modifiers or countervailing selection. It is important to note that these well characterized drive systems may not be representative, as there is a clear detection bias: to be discovered and characterized, drive systems must be conspicuous (e.g., causing strong drive or distorting sex ratios) and segregate within populations (i.e., balanced) [7]. But what about those drive elements that are not balanced and thus able to spread to fixation? These would also invade when concentrated in the centromeric regions of autosomes or on sex chromosomes (little or no crossing over occurs between the X and Y) and then sweep through populations, causing complete rather than partial selective sweeps. The extent to which hitchhiking effects of selfish meiotic drive systems contribute to overall patterns of DNA sequence variation, reducing variability around centromeres and on sex chromosomes (e.g., ref. [62]), remains to be determined.
Methods
PCR-Screen for SD Chromosomes
We used a molecular assay to screen for SD chromosomes in a collection of 452 isofemale lines from across sub-Saharan Africa, kindly provided by Drs. John Pool, Charles Aquadro and Andy Clark (Cornell University). We used a single-reaction PCR assay involving three primers, a forward primer (F) and two reverse primers (R1 and R2): F = TTTGGAGACTGCCTGATCAAAACTAATG; R1 = CAACGTCGCGGAGGAGACTGCCTATGT; R2 = CGTGTTCTGAGCGTTTCGCACAGTGTAT. One primer pair (F-R1) amplifies a 463-bp fragment from the parent gene, RanGAP (a positive control), and the other (F-R2) amplifies a 353-bp SD-specific fragment that spans the breakpoint of the Sd-RanGAP-RanGAP junction (Figure 1B). Only one amplicon results from flies that lack SD chromosomes and two result from flies that carry SD (Figure 1B).
Extracting SD Chromosomes
Isofemale lines found to be SD-positive by PCR assay could be homozygous SD/SD or heterozygous SD/SD+. We therefore extracted SD chromosomes onto a common genetic background, then maintained homozygous viable SD chromosomes as homozygous stocks, and maintained homozygous lethal SD chromosomes over the CyO balancer chromosome. To extract SD chromosomes, we crossed 3–5 w 118; In(2LR)Gla, wg Gla-1 Bc 1/CyO (hereafter, w 118; Gla/CyO) virgin females to 3–5 males from the SD-positive isofemale lines. We then collected 5 white-eyed CyO sons and individually backcrossed them to 5–10 w 118; Gla/CyO females. Once larvae appeared in the backcross vials, we PCR-tested the 5 white-eyed CyO sons for SD (see above) and retained progeny from a single SD-positive male. We then crossed w 118/w 118; SD/CyO virgin daughters to w 118; SD/CyO sons. If the SD chromosome was homozygous viable, we used the progeny to establish a w 118; SD/SD stock; if the SD chromosome was homozygous lethal, we maintained a w 118; SD/CyO stock. Last, we confirmed that all of the final stocks carried the SD chromosome by PCR assay.
Inversion-Typing SD Chromosomes
Many SD chromosomes possess one or more inversions on chromosome 2 (reviewed in ref. [2]). To determine the inversion types of SD chromosomes, we examined polytene chromosomes from larval salivary gland squashes. We crossed virgin cn bw females to SD males to generate larvae; cn bw chromosomes have standard arrangement second chromosomes. Salivary glands were dissected from F1 larvae in 1% Na-citrate hypotonic solution on siliconized slides and then transferred and fixed for 10–15 seconds in 45% acetic acid. The dissections were stained with 1% lacto-aceto-orcein for 25–35 minutes. We determined inversion breakpoints by comparing photographs with the standard maps of chromosome 2.
Complementation Tests among SD Chromosomes
We performed complementation tests between all pairwise combinations of SD chromosomes. For all homozygous lethal SD chromosomes, we tested the viability of all SD i/SD j combinations by crossing five SD i/CyO virgin females to 3–5 SD j/CyO males. If CyO + progeny appear, then the lethality of SD i and SD j chromosomes must map to different complementation groups. We also tested the male and female fertility of viable SD i/SD j combinations. At least two replicates each of 3–5 SD i/SD j males and 3–5 virgin SD i/SD j females were crossed to OreR virgin females and males, respectively. SD i/SD j flies that produced larvae were considered fertile, whereas those that failed to produce any progeny over multiple replicates were considered sterile.
Estimating the Strength of Segregation Distortion
We estimated the strength of distortion for each SD chromosome by measuring the rate of transmission, k, of the SD chromosome through heterozygous SD/Gla males. In preliminary work, we screened a series of balancer chromosomes (Bal) for sensitivity to distortion by assaying transmission from SD-5/Bal males. SD-5 is a well-characterized, non-African SD chromosome. These crosses revealed that the In(2LR)Gla chromosome (hereafter, Gla) carries a sensitive Rsp s allele. Gla is an effective balancer of most of the second chromosome and carries a dominant eye-phenotype marker. We estimated k by individually crossing five SD/Gla males of each SD chromosome to five 3–5 day old cn bw virgin females each. After four days, each cross was transferred to a fresh food vial every fourth day. We then scored all progeny emerging until 20 days after the parents were removed from each of the four vials.
The rate of transmission of SD to progeny depends both on the strength of distortion and on the relative viability of the SD chromosome. Therefore, to distinguish the strength of distortion from relative viability, we measured the rate of transmission of SD chromosomes through heterozygous SD/Gla females. As distortion is male-specific, the rate of transmission of SD through females allows estimation of SD relative viability. By using the Gla balancer to minimize recombination on the second chromosome in females, we could estimate the viability of intact SD chromosomes like those transmitted through males (which lack recombination in D. melanogaster). For each SD chromosome we set up three replicate crosses of five 3–5 day old SD/Gla virgin females with three 3–5 day old cn bw males. After four days, each cross was transferred to fresh vial every fourth day. We used our estimates of relatively viability to estimate a corrected strength of distortion, k*, following ref. [63].
Sequencing of Sd-RanGAP
To sequence the new Sd-RanGAP duplicate gene, we first isolated SD chromosomes in heterozygous state over a chromosomal deficiency, Df(2L)Sd77, which deletes the 37D1–37D2;38C1–38C2 region including the RanGAP locus. After isolating genomic DNA from SD/Df(2L)Sd77 flies, we PCR amplified two fragments from the Sd-RanGAP region with two sets of primers. All PCR products therefore come from the SD chromosome. The first set amplifies a 2,994-bp fragment from the 5′-half of Sd-RanGAP. The forward primer (F4) binds the distal intergenic region between Sd-RanGAP and the neighboring gene CG10237; the reverse primer (R4) binds in intron 1 of Sd-RanGAP (which, on the reverse strand, is exon 2 of Hs2st). The second primer set amplifies a 2,410-bp fragment from the 3′-half of Sd-RanGAP with a 280-bp overlap with the first fragment. The forward primer (F6) binds in the first intron of Sd-RanGAP (which, on the reverse strand, is intron 2 of Hs2st); the reverse primer (R6) binds the intergenic region between Sd-RanGAP and RanGAP. Both the R4 and F6 primers bind two genomic locations in flies with SD chromosomes. First, R4 binds the first intron of Sd-RanGAP and the homologous sequence of the parent gene RanGAP. However, when the F4-R4 primer pair is used and PCR extension times are constrained, only product from the first R4 binding location results. Second, F6 binds the first intron of Sd-RanGAP and the homologous sequence of RanGAP. However, when the F6-R6 primer pair is used, only the 3′-half of Sd-RanGAP is amplified. We used Exo-SAP to clean PCR products and then sequenced both strands of the PCR products using internal sequencing primers (Table S1), BigDye Terminator chemistry, and standard cycle sequencing protocols. All sequences were manually edited using Sequencher v. 4.5 (Gene Codes). We obtained outgroup sequences via BLAST searches of the D. simulans genome [62].
Sequencing Non-Coding Regions on Chromosome 2
In addition to Sd-RanGAP, we sequenced the parent gene and eight non-coding regions across chromosome 2 from a collection of SD chromosomes and from 10 wildtype chromosomes from Zimbabwe. As many SD chromosomes, and some wildtype ones, are homozygous lethal (see RESULTS), we could not make homozygous lines for sequencing for all stocks. Instead, for homozygous lethal lines, we used deficiencies to produce flies hemizygous for the focal chromosomal regions. The eight regions ranged in size from 567–874 bp long (Table 3). We sequenced fragments from the proximal intergenic region of tup (cytological position = 37B; deficiency used for hemizygous flies = Df(2L)Exel7073); a large intron from CG30947 (43E; Df(2R)Exel6054); the distal intergenic region of Myd88 (45C; Df(2R)Np3); the proximal intergenic region of off-track (48D6; Df(2R)BSC39); the proximal intergenic region of scab (51E; Df(2R)Jp1); the proximal intergenic region of staufen (55B; Df(2R)Pcl7B); a large intron of plexus (58E4–8; Df(2R)Exel7173); a large intron of CG34372 (59E1; Df(2R)bw-S46). To sequence the parent gene, RanGAP, from the 10 wildtype chromosomes, we used the Df(2L)Sd77.
Population Genetic Analyses
We performed most population genetic analyses using DnaSP [64]. Probability values for Tajima's D and Fay and Wu's H were obtained from 10,000 coalescent simulations with no recombination, conditioning on the observed θ. For coalescent-based haplotype configuration tests we used the haploconfig software [48].
Supporting Information
Primer sequences for sequencing Sd-RanGAP.
(0.06 MB DOC)
Acknowledgments
We thank Mary Kate Gaetani and Seema Thomas for technical assistance; the Bloomington Stock Center and Harvard Medical School for providing fly stocks, including those from the Exelixis collection. Dan Garrigan, Sarah Kingan, Todd Schlenke, Shanwu Tang and four anonymous reviewers provided helpful advice and/or comments on the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by funds to DCP from a NIH Ruth L. Kirschstein NRSA postdoctoral fellowship, the Radcliffe Institute for Advanced Study at Harvard University, and the University of Rochester. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sandler L, Hiraizumi Y, Sandler I. Meiotic drive in natural populations of Drosophila melanogaster. I. The cytogenetic basis of segregation distortion. Genetics. 1959;44:232–250. doi: 10.1093/genetics/44.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartl DL, Hiraizumi Y. Segregation distortion. In: Novitski E, Ashburner M, editors. The Genetics and Biology of Drosophila melanogaster. New York: Academic Press; 1976. [Google Scholar]
- 3.Temin RG, Ganetzky B, Powers PA, Lyttle TW, Pimpinelli S, et al. Segregation Distorter (SD) in Drosophila melanogaster: genetic and molecular analysis. American Naturalist. 1990;137:287–331. [Google Scholar]
- 4.Wu C-I, Hammer MF. Molecular evolution of ultraselfish genes of meiotic drive systems. In: Selander RK, Clark AG, Whittam TS, editors. Evolution at the Molecular Level. Sunderland, Massachusetts: Sinauer Associates; 1991. [Google Scholar]
- 5.Lyttle TW. Segregation Distorters. Annual Review of Genetics. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- 6.Lyttle TW. Cheaters sometimes prosper: distortion of mendelian segregation by meiotic drive. Trends in Genetics. 1993;9:205–210. doi: 10.1016/0168-9525(93)90120-7. [DOI] [PubMed] [Google Scholar]
- 7.Presgraves DC. Drive and sperm: evolution and genetics of male meiotic drive. In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Elsevier Press; 2008. [Google Scholar]
- 8.Kusano A, Staber C, Chan HYE, Ganetzky B. Closing the (Ran)GAP on segregation distortion in Drosophila. BioEssays. 2003;25:108–115. doi: 10.1002/bies.10222. [DOI] [PubMed] [Google Scholar]
- 9.Hartl DL, Hiraizumi Y, Crow JF. Evidence for sperm dysfunction as the mechanism of segregation distortion in Drosophila melanogaster. Proceedings of the National Acadamy of Sciences. 1967;58:2240–2245. doi: 10.1073/pnas.58.6.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauschteck-Jungen E, Hartl DL. Defective histone transition during spermiogenesis in heterozygous Segregation Distorter males of Drosophila melanogaster. Genetics. 1982;101:57–69. doi: 10.1093/genetics/101.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kettaneh NP, Hartl DL. Histone transition during spermiogenesis is absent in segregation distorter males of Drosophila melanogaster. Science. 1976;193:1020–1021. doi: 10.1126/science.821147. [DOI] [PubMed] [Google Scholar]
- 12.Tokuyasu KT, Peacock WJ, Hardy RW. Dynamics of spermiogenesis in Drosophila melanogaster. VII. Effects of Segregation Distorter (SD) chromosome. Journal of Ultrastructure Research. 1977;58 doi: 10.1016/s0022-5320(77)80011-7. [DOI] [PubMed] [Google Scholar]
- 13.Burt A, Trivers RA. Genes in conflict. Harvard University Press; 2006. [Google Scholar]
- 14.Palopoli MF. Genetic partners in crime: Evolution of ultraselfish supergene that specializes in sperm sabotage. In: Wolf JB, Brodie ED, Wade MJ, editors. Epistasis and the evolutionary process. New York: Oxford University Press; 2000. pp. 113–128. [Google Scholar]
- 15.Charlesworth B, Hartl DL. Population dynamics of the segregation distorter polymorphism of Drosophila melanogaster. Genetics. 1978;89:171–192. doi: 10.1093/genetics/89.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartl DL. Genetic dissection of segregation distortion. I. Suicide combinations of SD genes. Genetics. 1974;76:477–486. doi: 10.1093/genetics/76.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crow JF. Why is Mendelian segregation so exact? BioEssays. 1991;13:305–312. doi: 10.1002/bies.950130609. [DOI] [PubMed] [Google Scholar]
- 18.Thomson GJ, Feldman MW. Population genetics of modifiers of meiotic drive. II. Linkage modification in the Segregation Distorter system. Theoretical Population Biology. 1974;5:155–162. doi: 10.1016/0040-5809(74)90038-0. [DOI] [PubMed] [Google Scholar]
- 19.Miklos GLG. The genetic structure of chromosomes carrying Segregation Distorter. Canadian Journal of Genetics and Cytology. 1972;14:235–243. doi: 10.1139/g72-030. [DOI] [PubMed] [Google Scholar]
- 20.Hiraizumi Y, Martin DW, Eckstrand IA. A modified model of segregation distortion in Drosophila melanogaster. Genetics. 1980;95:693–706. doi: 10.1093/genetics/95.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandler L, Hiraizumi Y. Meiotic drive in natural populations of Drosophila melanogaster. IV. Instability at the Segregation-Distorter locus. Genetics. 1960;45:1269–1287. doi: 10.1093/genetics/45.9.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waddle F, Moore C, Hadley L, Hildebrand R. Stabilizer of Segregation Distortion. Drosophila Information Service. 1994;75:96. [Google Scholar]
- 23.Hartl DL. Segregation distortion in natural and artificial populations of Drosophila melanogaster. In: Mulcahy DL, editor. Gamete competition in plants and animals. Amsterdam: North-Holland Publishing; 1975. [Google Scholar]
- 24.Hiraizumi Y, Thomas AM. Suppressor systems of segregation distorter (SD) chromosomes in natural populations of Drosophila melanogaster. Genetics. 1984;106:279–292. doi: 10.1093/genetics/106.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baudry E, Viginier B, Veuille M. Non-African populations of Drosophila melanogaster have a unique origin. Molecular Biology and Evolution. 2004;21:1482–1491. doi: 10.1093/molbev/msh089. [DOI] [PubMed] [Google Scholar]
- 26.Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutter S, Li H, Beisswanger S, DeLorenzo D, Stephan W. Distinctly different sex ratios in African and European populations of Drosophila melanogaster inferred from chromosomewide single nucleotide polymorphism data. Genetics. 2007;177:469–480. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. Public Library of Science Biology. 2006;2:e166. doi: 10.1371/journal.pgen.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palopoli MF, Wu C-I. Rapid evolution of a coadapted gene complex: evidence from the Segregation Distorter (SD) system of meiotic drive in Drosophila melanogaster. Genetics. 1996;143:1675–1688. doi: 10.1093/genetics/143.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartl DL, Hartung N. High frequency of one element of Segregation Distorter in natural populations of Drosophila melanogaster. Evolution. 1975;29:512–518. doi: 10.1111/j.1558-5646.1975.tb00840.x. [DOI] [PubMed] [Google Scholar]
- 31.Temin RG, Marthas M. Factors influencing the effect of segregation distortion in natural populations of Drosophila melanogaster. Genetics. 1984;107:375–393. doi: 10.1093/genetics/107.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartl DL. Complementation analysis of male fertility among the segregation distorter chromosomes of Drosophila melanogaster. Genetics. 1973;73:613–629. doi: 10.1093/genetics/73.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartl DL. Dysfuntional sperm production in Drosophila melanogaster males homozygous for the segregation distorter elements. Proceedings of the National Acadamy of Sciences. 1969;63:782–789. doi: 10.1073/pnas.63.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cicchetti R, Argentin G, Nicoletti B. The segregation distortion (SD) phenomenon in wild populations of Drosophila melanogaster: interaction between chromosomes 3 and SD chromosomes 2. Genetica. 1990;81:77–84. doi: 10.1007/BF00226445. [DOI] [PubMed] [Google Scholar]
- 35.Kusano A, Staber C, Ganetzky B. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Developmental Cell. 2001;1:351–361. doi: 10.1016/s1534-5807(01)00042-9. [DOI] [PubMed] [Google Scholar]
- 36.Kusano A, Staber C, Ganetzky B. Segregation distortion induced by wild-type RanGAP in Drosophila. Proceedings of the National Acadamy of Sciences. 2002;99:6866–6870. doi: 10.1073/pnas.102165099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science. 1999;283:1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- 38.Wu C-I, Lyttle TW, Wu M-L, Lin GF. Association between DNA satellite sequences and the Responder of Segregation Distortion in D. melanogaster. Cell. 1988;54:179–189. doi: 10.1016/0092-8674(88)90550-8. [DOI] [PubMed] [Google Scholar]
- 39.Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 40.Minakhina S, Myers R, Druzhinina M, Steward R. Crosstalk between the actin cytoskeleton and Ran-mediated nuclear transport. BMC Cell Biology. 2005;6:32. doi: 10.1186/1471-2121-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 42.Charlesworth B. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genetical Research. 1996;68:131–149. doi: 10.1017/s0016672300034029. [DOI] [PubMed] [Google Scholar]
- 43.Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 44.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fay JC, Wu C-I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Betran E, Rozas J, Navarro A, Barbadilla A. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics. 1997;146:89–99. doi: 10.1093/genetics/146.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Innan H, Zhang K, Marjoram P, Tavare S, Rosenberg NA. Statistical tests of the coalescent model based on the haplotype frequency distribution and the number of segregating sites. Genetics. 2005;169:1763–1777. doi: 10.1534/genetics.104.032219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Depaulis F, Mousset S, Veuille M. Detecting selective sweeps with haplotype tests: Hitchhiking and haplotype tests. In: Nurminsky D, editor. Selective Sweep. New York: Kluwer Academic/Plenum; 2005. [Google Scholar]
- 50.Aulard S, David JR, Lemeunier F. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genetical Research. 2002;79:49–63. doi: 10.1017/s0016672301005407. [DOI] [PubMed] [Google Scholar]
- 51.Hudson RR. A new statistic for detecting genetic differentiation. Genetics. 2000;155:2011–2014. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rozas J, Segarra C, Ribo G, Aguade M. Molecular population genetics of the rp49 gene region in different chromosomal inversions of Drosophila subobscura. Genetics. 1999;151:189–202. doi: 10.1093/genetics/151.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hey J, Kliman RM. Population genetics and phylogenetics of DNA sequence variation at multiple loci within the Drosophila melanogaster species complex. Molecular Biology and Evolution. 1993;10:804–822. doi: 10.1093/oxfordjournals.molbev.a040044. [DOI] [PubMed] [Google Scholar]
- 54.Hiraizumi Y, Crow JF. Heterozygous effects on viability, fertility, rate of development, and longevity of Drosophila chromosomes that are lethal when homozygous. Genetics. 1960;45:1071–1083. doi: 10.1093/genetics/45.8.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brittnacher JG, Ganetzky B. On the components of segregation distortion in Drosophila melanogaster. II. Deletion mapping and dosage analysis of the SD locus. Genetics. 1983;103:659–673. doi: 10.1093/genetics/103.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miklos GLG. An investigation of the components of segregation distorter systems in Drosophila melanogaster. Genetics. 1972;70:405–418. doi: 10.1093/genetics/70.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babcock CS, Anderson WW. Molecular evolution of the sex-ratio inversion complex in Drosophila pseudoobscura: analysis of the esterase-5 gene region. Molecular Biology and Evolution. 1996;13:287–308. doi: 10.1093/oxfordjournals.molbev.a025589. [DOI] [PubMed] [Google Scholar]
- 58.Derome N, Baudry E, Ogereau D, Veuille M, Montchamp-Moreau C. Selective sweeps in a 2-locus model for sex-ratio meiotic drive in Drosophila simulans. Molecular Biology and Evolution. 2008;25:409–416. doi: 10.1093/molbev/msm269. [DOI] [PubMed] [Google Scholar]
- 59.Kingan SB, Hartl DL. Signatures of selection on the genes of the Winters sex-ratio in Drosophila simulans. Molecular Biology and Evolution submitted [Google Scholar]
- 60.Dyer K, Charlesworth B, Jaenike J. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proceedings of the National Academy of Sciences. 2007;104:1587–1592. doi: 10.1073/pnas.0605578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 62.Begun DJ, Holloway AK, S K, Hillier LW, Poh YP, et al. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans. Public Library of Science Biology. 2007;5:2534–2559. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Powers PA, Ganetzky B. On the components of segregation distortion in Drosophila melanogaster. V. Molecular analysis of the Sd locus. Genetics. 1991;129:133–144. doi: 10.1093/genetics/129.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences for sequencing Sd-RanGAP.
(0.06 MB DOC)