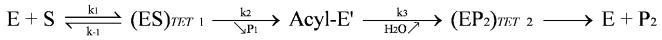Abstract
The effect of modulating the structural dynamics by incremental glycosylation on enzyme activity and thermostability has thus far only been investigated in detail for the serine protease α-chymotrypsin (for a recent review see Solá et al., Cell. Mol. Life Sci. 2007, 64(16):2133–2152). Herein, we extend this type of study to a structurally unrelated serine protease, specifically, subtilisin Carlsberg. The protease was glycosylated with activated lactose and various lactose-subtilisin conjugates were obtained and characterized. Near UV-CD spectroscopy revealed that the tertiary structure was unaffected by the procedure. H/D exchange FT-IR spectroscopy was performed to assess the structural dynamics of the enzyme. It was found that increasing the level of glycosylation caused a linearly dependent reduction in structural dynamics. This led to an increase in thermostability and a decrease in the catalytic turnover rate for both, acylation and de-acylation step. These data highlight the possibility that a structural dynamics – activity relationship might be a phenomenon generally found in serine proteases.
Keywords: chemical glycosylation, enzyme activity, protein dynamics, protein stability, subtilisin Carlsberg
INTRODUCTION
Subtilisin-like proteases (a.k.a. subtilases) constitute one of the principal structural classes of enzymes belonging to the serine protease family (Lindstrom-Lang and Ottesen, 1947; Siezen et al, 1991; Siezen and Leunissen, 1997). Subtilases are one of the most studied enzyme classes because its members are directly involved in the regulation of many important metabolic processes (e.g., processing of endocrine and neuronal peptide pro-hormones, embryonic and cellular homeostasis). Their ability to regulate important biological cascades through their catalytic function has brought broad interest to further the understanding of their catalytic power at the molecular level. Importantly, their de-regulation is involved in the development of various diseases (e.g., obesity, diabetes, high cholesterol levels, cancer, and viral infections) (Leung et al., 2000; Bontemps et al., 2007). Subtilases are also involved in the metabolism of many pathogenic organisms, e.g., the Malaria parasite (Withers-Martinez et al, 2004), and offer novel therapeutic targets. A deeper understanding of how subtilases’ function and are regulated will benefit further advancement of biomedical research.
The structure of subtilisin Carlsberg, the model enzyme investigated herein, has been determined by X-ray crystallography (Bode et al., 1986, 1987). It is a 275 residue monomeric protein with two alpha/beta domains which are composed of seven strands of parallel β-sheets surrounded by five α-helices. The active site residues of the catalytic triad are composed of His64 and Ser221 located at the N-terminus of adjacent α-helices and Asp32 situated at the C-terminal end of the central β-sheet strand. The general mechanism of serine protease catalysis (Figure 1) involves the formation of a substrate-enzyme complex (ES) followed by formation and breakdown of the first tetrahedral intermediate (ES)TET1 leading to the liberation of the first product and formation of enzyme-acyl intermediate. The catalytic cycle ends with the hydrolysis of this intermediate.
Figure 1.
Scheme of the general mechanism of serine protease catalysis.
The determination of the three-dimensional structure is the basis for understanding details of its catalytic mechanism (Hedstrom, 2002) and function (Shaw and Pal, 2007). However, information on the structure is not sufficient for understanding the detailed mechanism of action because enzymatic activity may also depend on conformational dynamics (Solá et al., 2007). Proteins are dynamic and not static systems and their internal structural motions are frequently crucial for their biological function (Bolton and Perutz, 1970; Bode et al., 2007; Zaccai, 2000). Enzymes make extensive use of conformational changes throughout their catalytic cycle that are believed to be essential to their function (Doring et al., 1999; Karplus and Kuriyan, 2005; Karplus et al., 2005). It is commonly assumed that in order to work properly, most enzymes must be stable enough to retain their native three-dimensional structures, but flexible enough to allow sufficient substrate binding, chemical reaction, and product release (Kempner, 1993).
Still, detailed data on enzyme conformational motions and their relation to the catalytic function are available only for few systems because tools to systematically manipulate structural dynamics became available just recently (Solá and Griebenow, 2006a; Solá et al., 2007). One such method consists in the modification of surface residues (typically lysines) with chemically activated glycans (Solá and Griebenow, 2006a) or poly(ethylene glycol) (Castillo et al., 2008; Rodríguez-Martínez et al., 2008). In general, at increasing levels of modification enzyme structural dynamics is increasingly restricted and this leads to a loss in catalytic efficiency (Solá et al., 2006a). Glycosylation has in this instance the advantage over PEGylation that structural dynamics can be shifted over quite some range at increasing glycan molar content (Solá et al., 2007) in contrast to PEGylation where it was found that the effect levels of after few molecules are bound (Rodríguez-Martínez et al., 2008). Furthermore, in case of α-chymotrypsin PEGylation affected the substrate binding affinity making kinetic analysis more complex, while glycosylation did not. Still, the sole systematically investigated system thus far consists in α-chymotrypsin and therefore it is unclear whether the findings are representative for other enzymes, even though a recent theoretical study also came to this conclusion using another model protein (Shental-Bechor and Levy, 2008). Herein, we therefore investigated a structurally unrelated enzyme, subtilisin Carlsberg, to further test the relationship between glycosylation, protein stability, dynamics, and function. Some data on PEGylated subtilisin indicate a relationship between activity, enantioselectivity, and structural dynamics but these studies were performed in organic solvents and it is unclear how representative the data are for the enzyme in aqueous environments (Castillo et al., 2006, 2008).
MATERIALS AND METHODS
Materials
Subtilisin Carlsberg (EC 3.4.21.62) was from Sigma-Aldrich (St. Louis, MO); mono-(lactosylamido)-mono-(succinimidyl) suberate (SS-mLAC) and 2,4,6-trinitrobenzene sulfonic acid (TNBSA) were from Pierce (Rockford, IL) and Suc-Ala-Ala-Pro-Phe-pNA was purchased from Bachem (Bubendorf, Switzerland). All other chemicals were from various suppliers and of analytical quality.
Chemical Enzyme Modification
Covalent modification of subtilisin with lactose conjugates was performed by chemical glycosylation with SS-mLAC (500 Da) as described by Solá and Griebenow (2006a). To achieve protein conjugates with varying glycan molar content different amounts of lactose were added to subtilisin solutions (2 mg/mL) to achieve molar ratios of 3.5, 5.5, 7.5, and 9.5 mol of reagent per mol of protein in 0.1 M borate buffer, pH 9.0. Reaction mixtures were stirred at 4°C for 2 h followed by dialysis and lyophilization. Dried powders were stored at −20°C until use.
Determination of the Extent of Chemical Modification
Determination of the average lactose molar content of the conjugates was done via the TNBSA assay as described (Habeeb, 1966). Protein samples of subtilisin and of the different lactose-subtilisin conjugates were dissolved in 0.1 M sodium bicarbonate buffer (pH 8.5) to achieve concentrations of 0.20, 0.10, and 0.05 mg/mL. To 0.5 mL of these solutions and of buffer blanks was added 0.25 mL of a 0.01% (w/v) TNBSA solution. After incubation for 2 h at 37°C, 0.5 a 10% SDS solution and 0.25 mL of 1 N HCl were added. The absorbance of the solution was measured at 335 nm with Shimadzu 160 UV-Vis spectrophotometer using 10 mm path-length quartz cuvettes.
Circular Dichroism Spectroscopy
Near-UV CD measurements (260–310 nm) were performed using an Olis DSM-10 UV-Vis spectrophotometer. Protein concentration was 0.6 mg/mL in 10 mM potassium phosphate buffer at pH 7.8 and 25°C and spectra were recorded using a 1.0 cm path length quartz cell. Each spectrum was obtained by averaging 5 scans at 2 nm resolution. Solvent reference spectra were digitally subtracted from the protein CD spectra.
FTIR Amide H/D Exchange Kinetics
Protein Preparation for H/D Exchange Experiments
Prior to H/D experiments protein samples were dissolved in 10 mM potassium phosphate buffer (pH 7.8) to obtain a protein concentration of 0.5 mg/ml. Portions of 8 ml of these solutions and of their respective buffer blanks were flash frozen with liquid N2 and lyophilized. For FTIR measurements lyophilized buffer blanks and protein samples were dissolved in 200 μl of D2O yielding a protein concentration of 0.8 mM (20 mg/ml).
H/D Exchange Assay
Amide H/D exchange FTIR spectra were measured using a Nicolet NEXUS 470 infrared spectrometer equipped with a thermally controlled sample cell (Spectra-Tech Inc., Shelton, CT) using CaF2 windows and 25 μm Teflon spacers (Buck Scientific, East Norwalk, CT) at 25°C as previously described (Zavodszky et al., 1998; Solá and Griebenow, 2006a). Exchange time started at the moment of D2O addition. IR spectra (2,000-1,100 cm−1) were collected at 2, 4, 6, 8, 10, 20, 30, 40, 50, and 60 min and successively at 30 min intervals for a period of 6 h. For all spectra 56 scan at 4 cm−1 spectral resolution were collected and digitally averaged. Spectra of the buffer blanks were collected in the same cell setup prior to sample measurement and digitally subtracted from the sample spectra.
Processing of H/D Exchange Data
H/D exchange spectra from experiments were processed for quantitative analysis in form of hydrogen exchange decay plots. The fraction of unexchanged amide hydrogen atoms (X) was defined as:
| (1) |
where w(t) is the ratio of the amide II (1,550 cm−1) and amide I (1,637.5 cm−1) absorbencies corrected with the baseline absorbance (1,789 cm−1) at time t, w(0) is the amide II/amide I ratio of the undeuterated protein, and w(∞) is the amide II/amide I ratio for the fully deuterated protein. Quantitative analysis of the HX decay data was done by fitting a two-exponential model:
| (2) |
where A1 and A2 are the fraction of the fast and slow-exchanging amide protons and kHX,1 and kHX,2 are the apparent exchange rate constants. Results were interpreted thermodynamically under the EX2 exchange mechanism model (Krishna et al., 2004) where the Gibbs free energy of unfolding per mol of peptide hydrogen is based on the chemical exchange rate constant (k0) and the measured rate constant (k HX,1).
| (3) |
Determination of Protein Thermodynamic Stability by Differential Scanning Calorimetry (DSC)
Thermal denaturation experiments were performed on a CSC 6100 Nano II differential scanning calorimeter (Calorimetry Sciences Corp., London, UT). The protein concentration was adjusted to 1 mg/mL in 10 mM potassium phosphate buffer at pH 7.8. Heat capacity thermograms were determined at a 2°C/min scan rate. Thermodynamic unfolding parameters were determined by digital analysis of the heat capacity thermograms with the program Cp Calc by fitting to a two-state transition denaturation model (Kar et al., 2004).
Enzyme Activity Assays
Specific activities for the various subtilisin glycoconjugates were determined spectroscopically by following the formation of p-nitroanilide at 410 nm (ε410 = 8.8 mM−1·cm−1) on a Shimadzu UV 160 spectrophotometer with a 0.1 cm path length cuvette. Reactions were carried out in 10 mM potassium phosphate buffer, pH 7.8 (25°C) employing Suc-Ala-Ala-Pro-Phe-pNA as the substrate (DelMar et al., 1979). For determination of steady state kinetic parameters, initial velocities were determined at various substrate concentrations ranging from 0.01 to 0.3 mM. Eadie-Hofstee plot analysis was used to determine the Michaelis-Menten parameters kcat and KM. Determination of the individual rate constant (Ks, k2, and k3) was performed by kinetic dissection experiments employing Suc-Ala-Ala-Pro-Phe-SBzl as substrate (Hengge and Stein, 2004).
RESULTS AND oDISCUSSION
Chemical Glycosylation of Subtilisin and Characterization of the Glycoconjugates
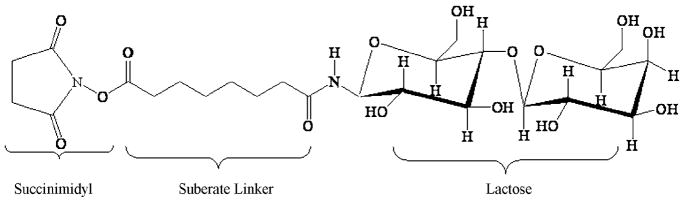
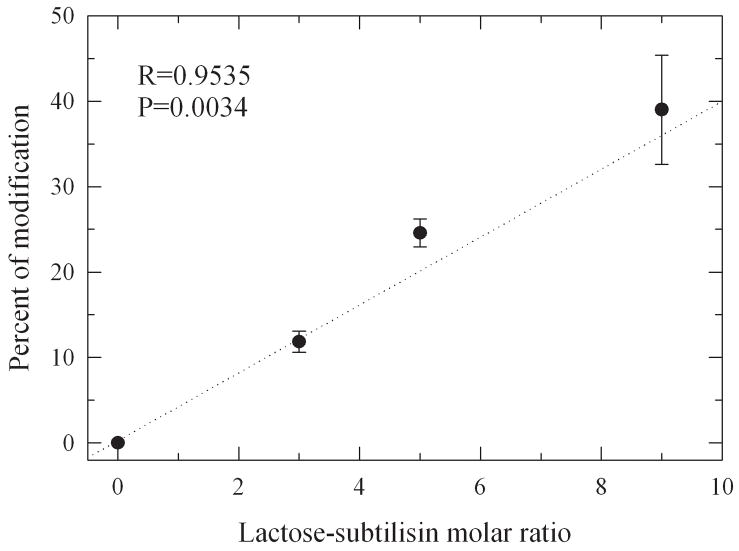
Herein we designed a series of differentially glycosylated subtilisin variants with sequentially reduced structural dynamics through chemical glycosylation with monofunctionally succinimidyl activated lactose (Figure 2). The succinimidyl functionality serves as leaving group during the glycosylation reaction which is selective for primary amines and allows coupling of the glycans to protein lysine residues and the amino terminus. Increasing the amount of activated lactose during synthesis leads to increased levels of glycosylation (Figure 3). The average number of lactose molecules bound to the protein was consecutively increased from 1 to around 4. Since the protein has 9 lysine residues this corresponds to 40% of the total glycan content that can theoretically be attached.
Figure 2.
Structure of succinimidyl activated lactose (SS-mLac) employed for chemical glycosylation.
Figure 3.
Percent of lactose-modification of subtilisin after chemical glycosylation.
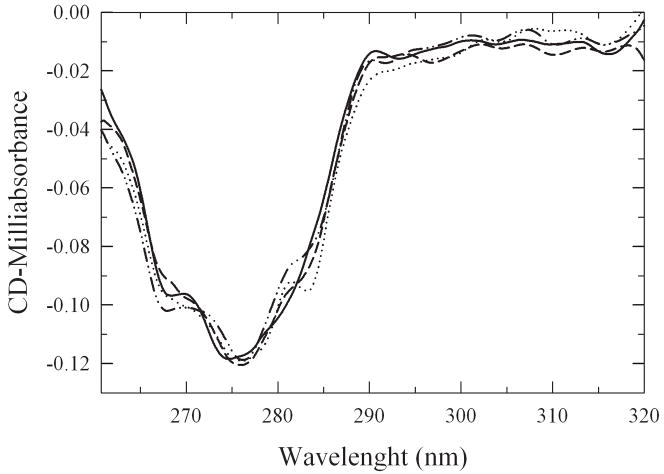
The structural impact of the glycosylation procedure was tested by measuring the near-UV circular dichroism spectra of the neo-glycoconjugates (Figure. 4). The significant signal centered around 275 nm disappears upon denaturation of the protein (Ganesan et al., 2006). Since there is no significant change in the CD spectra it can be concluded that glycosylation of subtilisin did not cause any significant tertiary structure changes, similar to our findings with α-chymotrypsin (Solá and Griebenow, 2006). Changes in enzyme properties as a result of glycosylation therefore must be due to other changes than structural.
Figure 4.
Near-UV CD spectra of subtilisin and subtilisin-lactose conjugates.
Changes in Structural Dynamics upon Chemical Glycosylation
Amide H/D exchange measurements have been used for over four decades to investigate protein structure and dynamics. Despite of the compact folded structure of most proteins, there are solvent exposed amide bonds that exchange relatively rapidly but the amide bonds that are inaccessible to the solvent or are participating in stable hydrogen bonds must undergo temporary structural distortion to allow for hydrogen exchange and thus have significantly slower exchange rates. This significant decrease in the exchange rates of these buried amide bonds in a folded protein makes H/D exchange an excellent and sensitive probe for monitoring protein conformational changes and dynamic processes (Ferraro et al., 2004).
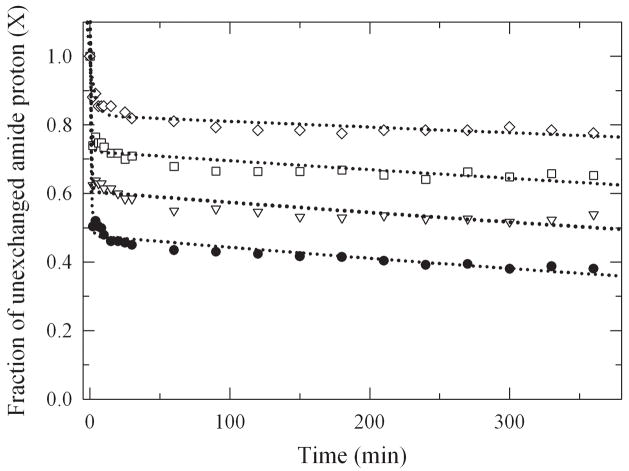
In order to study the global structural dynamics, H/D exchange kinetics were measured by FTIR spectroscopy by following changes in the amplitude of the amide II band (see Methods Section for details). Because of the technical limitations, the shortest interval of exchange monitored was after 2 min of incubation. The exchange kinetics of very fast exchanging amide bonds could therefore not be determined. The amount of amide hydrogen exchange (HX) was plotted in the form of HX decay plots where the fraction of unchanged amide hydrogen atoms (X) decreases over time due to the exchange process (Figure 5). It is evident that increasing the level of glycosylation led to an increase in the fraction of slowly exchanging amide bonds indicating increased structural rigidity of the enzyme. To obtain quantitative data, the decay plots were fitted with a bi-exponential model to obtain the individual exchange rates and amplitudes (Table 1). The amplitudes A1 and A2 represent the fraction of exchanging amide protons; kHX,1 and kHX,2 are the apparent exchange rate constants, respectively. The decay data reveal that upon chemical glycosylation the population and rate constants for the fast exchanging amide protons (A1, kHX,1) decreased with an increase in the slowly exchanging amide proton population (A2). There was no significant change in the rate of exchange for the slowly exchanging amide protons (kHX,2), but in the amount. The global Gibbs free-energy of unfolding (ΔGHX,1) for the various glycoconjugates was calculated by thermodynamic analysis of the H/D exchange kinetics (Solá and Griebenow, 2006a). This parameter represents the free energy of the global conformational dynamics of the protein (ΔGDyn ≈ ΔGHX,1). The results show reduced global structural dynamics as a function of the glycosylation level (Table 1). We also calculated the inverse of ΔGDyn to allow for correlation of other enzyme properties with the enzyme conformational mobility (ΔGHX,1−1) (Solá and Griebenow, 2006a).
Figure 5.
HX decay plots of non-exchanged amide hydrogens at 25°C (pD 7.8) for subtilisin and the different lactose-subtilisin conjugates.
Table 1.
Kinetic and thermodynamic parameters derived from amide H/D exchange experiments for subtilisin and for various subtilisin-lactose conjugates.
| Subtilisin-Lactosea | A1b | kHX,1 (min−1) | A2b | kHX,2 (min−1) | ΔGHX,1c (kcal/mol) | (ΔGHX,1)−1 d (mol/kcal) |
|---|---|---|---|---|---|---|
| 0.0 ± 0.0 | 0.52 | 1.39 | 0.48 | 0.0008 | 2.43 | 0.411 |
| 1.0 ± 0.1 | 0.40 | 1.40 | 0.60 | 0.0005 | 2.43 | 0.412 |
| 2.2 ± 0.1 | 0.28 | 1.19 | 0.72 | 0.0004 | 2.52 | 0.397 |
| 3.5 ± 0.6 | 0.16 | 0.27 | 0.83 | 0.0002 | 3.32 | 0.301 |
Average moles of lactose bound per mol of subtilisin.
Ai is the fraction of amide protons that exchange with the rate constant kHX,i.
Gibbs free-energy of unfolding per mol of peptide hydrogen.
Protein mobility.
Thermodynamic Protein Stability
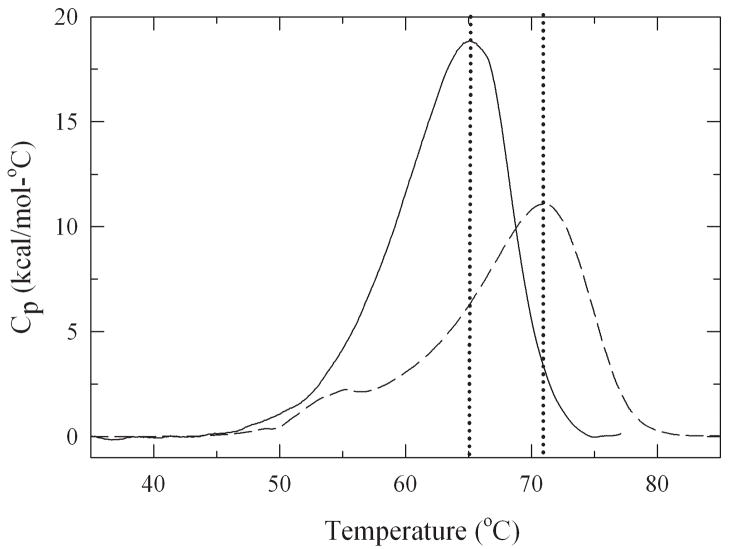
Thermal unfolding experiments were performed by differential scanning calorimetry to determine the effect of different degrees of glycosylation on protein stability (Figure 6). The unfolding temperature (Tm) rose linearly with increasing level of glycosylation and for the most modified glycoconjugate it increased by 7°C from 64°C to 71°C (Table 2).
Figure 6.
Thermograms obtained by differential scanning calorimetry for subtilisin (—) and the 2.2 glycan molar containing lactose-modified subtilisin (----) in 10 mM potassium phosphate buffer at pH 7.1.
Table 2.
Thermodynamic analysis of the data obtained by different scanning calorimetry.
| Lactose Contenta | Tm(°C) | ΔHm(kcal/mol K) | ΔCp (kcal/mol K) | ΔGu(25°C) (kcal/mol) |
|---|---|---|---|---|
| 0.0 ± 0.0 | 64 ± 1 | 283 ± 2 | 7.1 ± 0.9 | 16.5 ± 0.3 |
| 1.0 ± 0.1 | 62 ± 1 | 268 ± 5 | 5.5 ± 0.5 | 17.6 ± 0.1 |
| 2.2 ± 0.1 | 68 ± 1 | 256 ± 4 | 4.4 ± 0.5 | 18.6 ± 0.4 |
| 3.5 ± 0.6 | 71 ± 1 | 206 ± 5 | 3.3 ± 0.2 | 19.1 ± 0.4 |
Average moles of lactose bound per mol of subtilisin.
Thermodynamic parameters were extracted from the thermograms (Table 2). Data obtained for the non-glycosylated protein were found to be in good agreement with published data (Bakhtiar et al., 2003). The heat capacity and the enthalpy of protein unfolding for the different glycoconjugates decreased at increasing glycosylation level compared to the non-modified enzyme. The changes in Gibbs free energy of unfolding for the lactose modified conjugates (ΔG (25°C)) increased (from 16.5 to 19.1 kcal/mol) at increasing degree of glycosylation (Table 2). Similar to our studies with α-chymotrypsin, glycosylation of subtilisin caused a decrease in protein dynamics. This rigidification of the protein caused an increase in thermodynamic protein stability. A statistically significant correlation was found between the Tm-values and the protein mobility (R=0.89, p=0.009). The same result was found for α-chymotrypsin where the Tm-values correlated with the changes in conformational dynamics upon glycosylation (Solá et al., 2006). The increase in protein stability is largely due to shielding of the protein surface from water which leads to an increase in bonding strength of those groups involved in stabilizing the protein tertiary structure (Solá and Griebenow, 2006b). In contrast, it was found that ΔGU(25°C) showed a more complex dependence on glycosylation levels due to the fact that glycosylation strongly affected the stability of the unfolded state (Solá et al., 2007). The Tm-value seems to largely only monitor the stability of the folded (native) state of proteins. This stability is related to the structural dynamics and thus the measurement of Tm-values which is quite simple could replace technically more complex H/D exchange measurements in routine stability experiments.
Impact of Conformational Dynamics on the Catalytic Activity of Subtilisin
It is well known that proteins typically are very dynamical systems and it has long been speculated that enzyme conformational motions are crucial for optimum activity. We have recently demonstrated that incremental glycosylation caused a decrease in structural dynamics in α-chymotrypsin and a decrease in catalytic activity (Solá and Griebenow, 2006a). For subtilisin, activity and dynamical data are only available for the PEG-modified enzyme in 1,4-dioxane, but the data obtained support the view that also for this enzyme structural dynamics and activity are linked (Castillo et al., 2008). To investigate this in detail, the enzymatic activity of subtilisin was determined from the hydrolysis of the peptide substrate N-succinyl-Ala-Ala-Pro-Phe p-nitroanilide (Suc-Ala-Ala-Pro-Phe-pNA). Michaelis-Menten parameters were derived from initial velocities measured at various substrate concentrations. It was found that increasing the level of glycosylation did not alter the substrate binding affinity (KM) but caused a significant reduction in the turn over rate (kcat) (Table 3). The results are similar to those obtained with a structurally unrelated protease, α-chymotrypsin (Solá and Griebenow, 2006a).
Table 3.
Michaelis-Menten kinetic parameters for subtilisin and the different glycoconjugates.
| Lactose Contenta | kcat (s−1) | Km (μM) | Ks (μM) | k2 (s−1) | k3 (s−1) | ΔG k2(kcal/mol) | ΔG k3(kcal/mol) |
|---|---|---|---|---|---|---|---|
| 0.0 ± 0.0 | 200 ± 2 | 3.7 ± 0.1 | 15 ± 2 | 725 ± 6 | 270 ± 3 | 12.32 ± 0.08 | 12.89 ± 0.03 |
| 1.0 ± 0.1 | 166 ± 3 | 2.7 ± 0.3 | 9 ± 2 | 540 ± 4 | 232 ± 5 | 12.47 ± 0.09 | 12.45 ± 0.09 |
| 2.2 ± 0.1 | 132 ± 3 | 3.6 ± 0.3 | 9 ± 5 | 340 ± 2 | 216 ± 7 | 12.77 ± 0.03 | 13.01 ± 0.02 |
| 3.5 ± 0.6 | 90 ± 1 | 3.7 ± 0.1 | 12 ± 3 | 291 ± 6 | 133 ± 2 | 12.86 ± 0.1 | 13.28 ± 0.05 |
Average moles of lactose bound per mol of subtilisin.
A kinetic dissection was performed using Suc-Ala-Ala-Pro-Phe-SBzl as the substrate and the individual rate constants (KS, k2, k3) for the different glycoconjugates were obtained (Table 3). This experiment revealed that the kinetics of enzyme acylation (k2) and deacylation (k3) are both reduced by chemical glycosylation. In contrast, the substrate binding step (KS) was unaffected by chemical glycosylation. It is interesting to note that it has recently been reported that substrate binding seems not to be influenced by protein dynamics and also does not influence protein dynamics (Liu and Konermann, 2008).
Conclusions
In this work we demonstrate that chemical glycosylation of a structurally completely unrelated serine protease, subtilisin Carlsberg, led to similar results than previously reported for α-chymotrypsin by us. Specifically, increasing the level of glycosylation caused a reduction in structural dynamics, an increase in thermostability, and a decrease in catalytic activity. This demonstrates that coupling of enzyme activity and structural dynamics also occurs in enzymes which do not have such a pronounced domain organization as found in α-chymotrypsin.
Acknowledgments
This publication was made possible by grant number S06 GM08102 from the National Institute for General Medical Sciences (NIGMS) at the National Institutes of Health (NIH) through the Support of Competitive Research (SCORE) Program. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS.
References
- Bakhtiar S, Andersson MM, Gessesse A, Mattiasson B, Hatti-Kaul R. Stability characteristics of a calcium-independent alkaline protease from Nesterenkonia sp. Enz Microb Technol. 2003;32(5):525–531. [Google Scholar]
- Bode C, Kovacs AI, Szalay MS, Palotai R, Korcsmaros T, Csermely P. Network analysis of protein dynamics. FEBS Lett. 2007;581:2776–2782. doi: 10.1016/j.febslet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Bode W, Papamokos E, Musil D, Seemueller U, Fritz H. Refined 1.2 Å crystal structure of the complex formed between subtilisin Carlsberg and the inhibitor eglin c. Molecular structure of eglin and its detailed interaction with subtilisin. EMBO J. 1986;5(4):813–818. doi: 10.1002/j.1460-2075.1986.tb04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W, Papamokos E, Musil D. The high-resolution X-ray crystal structure of the complex formed between subtilisin Carlsberg and eglin c, an elastase inhibitor from the leech Hirudo medicinalis. Structural analysis, subtilisin structure and interface geometry. Eur J Biochem. 1987;166(3):673–692. doi: 10.1111/j.1432-1033.1987.tb13566.x. [DOI] [PubMed] [Google Scholar]
- Bolton W, Perutz MF. Three dimensional Fourier synthesis of horse deoxyhaemoglobin at 2.8 Angstrom units resolution. Nature. 1970;228(5271):551–552. doi: 10.1038/228551a0. [DOI] [PubMed] [Google Scholar]
- Bontemps Y, Scamuffa N, Calvo F, Khatib AM. Potential opportunity in the development of new therapeutic agents based on endogenous and exogenous inhibitors of the proprotein convertases. Med Res Rev. 2008;27(5):631–648. doi: 10.1002/med.20072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo B, Mendez J, Al-Azzam W, Barletta G, Griebenow K. On the relationship between the activity and structure of PEG-alpha-chymotrypsin conjugates in organic solvents. Biotechnol Bioeng. 2006;94(3):565–574. doi: 10.1002/bit.20863. [DOI] [PubMed] [Google Scholar]
- Castillo B, Sola RJ, Ferrer A, Barletta G, Griebenow K. Effect of PEG-modification on subtilisin Carlsberg activity, enantioselectivity, and structural dynamics in 1,4-dioxane. Biotechnol Bioeng. 2008;99(1):9–17. doi: 10.1002/bit.21510. [DOI] [PubMed] [Google Scholar]
- DelMar EG, Largman C, Brodrick JW, Geokas MC. A sensitive new substrate for chymotrypsin. Anal Biochem. 1979;99(2):316–320. doi: 10.1016/s0003-2697(79)80013-5. [DOI] [PubMed] [Google Scholar]
- Döring K, Surrey T, Nollert P, Jähnig F. Effects of ligand binding on the internal dynamics of maltose-binding protein. Eur J Biochem. 1999;266(2):477–483. doi: 10.1046/j.1432-1327.1999.00880.x. [DOI] [PubMed] [Google Scholar]
- Ferraro DM, Lazo ND, Robertson AD. EX1 hydrogen exchange and protein folding. Biochemistry. 2004;43(3):587–594. doi: 10.1021/bi035943y. [DOI] [PubMed] [Google Scholar]
- Ganesan A, Price NC, Kelly SM, Petry I, Moore BD, Halling PJ. Circular dichroism studies of subtilisin Carlsberg immobilised on micron sized silica particles. Biochim Biophys Acta Proteins and Proteomics. 2006;1764(6):1119–1125. doi: 10.1016/j.bbapap.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Habeeb AF. Determination of free amino groups in proteins by trinitrobenzenesulfonic acid. Anal Biochem. 1966;14(3):328–336. doi: 10.1016/0003-2697(66)90275-2. [DOI] [PubMed] [Google Scholar]
- Hutcheon GA, Parker MC, Moore BD. Measuring enzyme motility in organic media using novel H-D exchange methodology. Biotechnol Bioeng. 2000;70(3):262–269. [PubMed] [Google Scholar]
- Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102(12):4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Hengge AC, Stein RL. Role of protein conformational mobility in enzyme catalysis: acylation of alpha-chymotrypsin by specific peptide substrates. Biochemistry. 2004;43(3):742–747. doi: 10.1021/bi030222k. [DOI] [PubMed] [Google Scholar]
- Kar K, Alex B, Kishore N. Thermodynamics of the interactions of calcium chloride with alpha-chymotrypsin. J Chem Thermodyn. 2002;34:319–336. [Google Scholar]
- Karplus M, Gao YQ, Ma J, van der Vaart A, Yang W. Protein structural transitions and their functional role. Philos Transact A Math Phys Eng Sci. 2005;363(1827):331–355. doi: 10.1098/rsta.2004.1496. [DOI] [PubMed] [Google Scholar]
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci USA. 2005;102(19):6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempner ES. Movable lobes and flexible loops in proteins. Structural deformations that control biochemical activity. FEBS Lett. 1993;326(1–3):4–10. doi: 10.1016/0014-5793(93)81749-p. [DOI] [PubMed] [Google Scholar]
- Krishna MMG, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34(1):51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Leung D, Abbenante G, Fairlie DP. Protease inhibitors: current status and future prospects. J Med Chem. 2000;43(3):305–341. doi: 10.1021/jm990412m. [DOI] [PubMed] [Google Scholar]
- Linderstrom-Lang K, Ottesen M. A new protein from ovalbumin. Nature. 1947;1947:159. doi: 10.1038/159807a0. [DOI] [PubMed] [Google Scholar]
- Liu YH, Konermann L. Conformational dynamics of free and catalytically active thermolysin are indistinguishable by H/D exchange mass spectrometry. Biochemistry. 2008;47:6342–6351. doi: 10.1021/bi800463q. [DOI] [PubMed] [Google Scholar]
- Privalov PL, Tsalkova TN. Micro- and macro-stabilities of globular proteins. Nature. 1979;280(5724):693–696. doi: 10.1038/280693a0. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Martínez JA, Solá RJ, Castillo B, Cintrón-Colón HR, Rivera-Rivera I, Barletta G, Griebenow K. Stabilization of α-chymotrypsin upon PEGylation correlates with reduced structural dynamics. Biotechnol Bioeng. 2008 doi: 10.1002/bit.22014. Accepted Article Available Online: Jun 11 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AK, Pal SK. Activity of subtilisin Carlsberg in macromolecular crowding. J Photochem Photobiol B. 2007;86(3):199–206. doi: 10.1016/j.jphotobiol.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: A close look at thermodynamic stabilization. Proc Natl Acad Sci USA. 2008;105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Leunissen JAM. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997;3:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, de Vos WM, Leunissen JAM, Dijkstra BW. Homology modeling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases. Protein Eng. 1991;4:719–737. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- Solá RJ, Griebenow K. Chemical glycosylation: new insights on the interrelation between protein structural mobility, thermodynamic stability, and catalysis. FEBS Lett. 2006a;580(6):1685–1690. doi: 10.1016/j.febslet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Solá RJ, Griebenow K. Influence of modulated structural dynamics on the kinetics of alpha-chymotrypsin catalysis. Insights through chemical glycosylation, molecular dynamics and domain motion analysis. FEBS J. 2006b;273(23):5303–5319. doi: 10.1111/j.1742-4658.2006.05524.x. [DOI] [PubMed] [Google Scholar]
- Solá RJ, Al-Azzam W, Griebenow K. Engineering of protein thermodynamic, kinetic, and colloidal stability: chemical glycosylation with monofunctionally activated glycans. Biotechnol Bioeng. 2006;94(6):1072–1079. doi: 10.1002/bit.20933. [DOI] [PubMed] [Google Scholar]
- Solá RJ, Rodríguez-Martínez JA, Griebenow K. Modulation of protein biophysical properties by chemical glycosylation: biochemical insights and biomedical implications. Cell Mol Life Sci. 2007;64(16):2133–2152. doi: 10.1007/s00018-007-6551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers-Martinez Ch, Létitia J, Blackman MJ. Subtilisin-like proteases of the malaria parasite. Mol Microbiol. 2004;53(1):55–63. doi: 10.1111/j.1365-2958.2004.04144.x. [DOI] [PubMed] [Google Scholar]
- Zaccai G. How soft is a protein? A protein dynamics force constant measured by Neutron scattering. Science. 2000;288(5471):1604–1607. doi: 10.1126/science.288.5471.1604. [DOI] [PubMed] [Google Scholar]
- Závodszky P, Kardos J, Svingor Á, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci USA. 1998;95(13):7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]