Abstract
We report here the use of novel “sloppy” molecular beacon probes in homogeneous PCR screening assays in which thermal denaturation of the resulting probe-amplicon hybrids provides a characteristic set of amplicon melting temperature (Tm) values that identify which species is present in a sample. Sloppy molecular beacons possess relatively long probe sequences, enabling them to form hybrids with amplicons from many different species despite the presence of mismatched base pairs. By using four sloppy molecular beacons, each possessing a different probe sequence and each labeled with a differently colored fluorophore, four different Tm values can be determined simultaneously. We tested this technique with 27 different species of mycobacteria and found that each species generates a unique, highly reproducible signature that is unaffected by the initial bacterial DNA concentration. Utilizing this general paradigm, screening assays can be designed for the identification of a wide range of species.
A classic approach for determining the identity of a bacterial species is to employ PCR to exponentially amplify a selected segment of a 16S rRNA gene (26), utilizing a pair of “universal primers” that bind to highly conserved sequences at the ends of the target region, and to then use a sophisticated technique to identify a species-specific sequence in the middle of the resulting amplicons (12). These identification methods include nucleotide sequence analysis (27) and electrophoretic determination of the sizes of fragments produced by incubation of the amplicons with selected restriction endonucleases (17). However, these techniques are time consuming, costly, and labor intensive.
A more desirable approach is to use a set of fluorescently labeled, sequence-specific oligonucleotide hybridization probes, each of which hybridizes to a different species-specific sequence under conditions in which only perfectly complementary probe-target hybrids form.
In a screening assay, only one probe in the set will form a hybrid, thereby identifying the species that is present. This method is rapid and can be carried out in sealed reaction tubes, utilizing probes possessing interacting label moieties, such as TaqMan probes (10), LightCycler probes (24), or molecular beacons (19), all of which generate distinctive fluorescence signals when they hybridize to complementary target amplicons. Multiplex PCR screening assays that contain a number of different probes can also be prepared, each probe specific for a different species and each possessing a differently colored fluorophore (21, 22). However, a comprehensive screening procedure requires upwards of 100 different species-specific probes and has to be carried out in many PCR assay tubes. Although arrays of probes on hybridization chips have been utilized in screening assays to determine the identity of 16S rRNA amplicons (5), this approach is nonhomogeneous, thereby risking the cross contamination of untested samples, and has not been adopted by clinical laboratories.
A particularly attractive, homogeneous method for distinguishing one amplicon from another is to measure amplicon stability by heating the amplicons to determine the temperature at which each amplicon falls apart (9, 14, 25). This approach has been used for distinguishing different bacterial species (4) and different fungal species (7). However, the amplicon melting temperature (Tm) in and of itself does not provide sufficient resolution to distinguish hundreds of different species.
In this report, we introduce a novel screening technique for the identification of bacterial species that utilizes elements of these prior approaches. Our strategy requires only a single gene amplification assay containing a set of four differently colored molecular beacon probes of a unique design. After the completion of amplification, the molecular beacons in the reaction tube are hybridized to the amplicons at a relatively low temperature. The temperature is then slowly raised to determine, from the consequent changes in fluorescence intensity of each molecular beacon in the set, the temperature at which each of the four probe-target hybrids melts apart (Tm). The resulting set of four Tm values uniquely identifies the species present in the sample. These assays, despite utilizing only four different probes, have the potential to identify hundreds of different bacterial species.
The molecular beacon probes used in the assays reported here possess unusually long probe sequences. Unlike sequence-specific molecular beacons (18), which possess short probe sequences (18 to 26 nucleotides long) that form probe-target hybrids only with perfectly complementary, or nearly perfectly complementary, target sequences (23), these “sloppy” molecular beacon probes are designed to form probe-target hybrids with the amplicons generated from all of the species that we wish to identify. In order to enable this unusual permissive property, the probe sequences in these molecular beacons are about 40 nucleotides long. Consequently, they form probe-target hybrids even if the duplexes possess a substantial number of mismatched base pairs.
The principle underlying the use of sloppy molecular beacon probes in screening assays is that the Tm value of the probe-target hybrid reflects the degree to which the probe sequence is complementary to the target sequence in the amplicon. Although the Tm value of a hybrid formed by a sloppy molecular beacon probe does not provide sufficient information to identify the species from which the amplicon was generated, the simultaneous use of a set of sloppy molecular beacons, each possessing a different probe sequence and each labeled with a differently colored fluorophore, provides a set of Tm values that serves as a unique, species-specific signature.
To illustrate the principles underlying this method and to explore the conditions under which reliable results can be obtained, we prepared a model assay that simultaneously utilizes four different sloppy molecular beacon probes to distinguish 27 different species of mycobacteria. Unique signatures were obtained for each species and were not affected by the initial bacterial DNA concentration. In a blinded test of 55 different mycobacterial DNA samples, the signature from each sample (except two) matched one of the previously determined species-specific signatures. The amplicons from the two discrepant samples were sequenced and turned out to be from new species not in the database.
MATERIALS AND METHODS
Mycobacterial DNAs.
Lyophilized cultures of 27 different mycobacterial species were obtained from the American Type Culture Collection (ATCC; Manassas, VA) or from the collections of the Public Health Research Institute and the University of Medicine and Dentistry of New Jersey. The 27 species (and their ATCC numbers) were as follows: Mycobacterium abscessus (35752), Mycobacterium aichiense (27280), Mycobacterium asiaticum (25274), Mycobacterium aurum (23366), Mycobacterium avium (25291), Mycobacterium branderi (517888), Mycobacterium celatum (clinical isolate), Mycobacterium chubuense (27278), Mycobacterium diernhoferi (19340), Mycobacterium engbaekii (27354), Mycobacterium fortuitum (35931), Mycobacterium haemophilum (33206), Mycobacterium intracellulare (23434), Mycobacterium kansasii (12478), Mycobacterium lentiflavum (51985), Mycobacterium malmoense (29571), Mycobacterium marinum (927), Mycobacterium rhodesiae (27024), Mycobacterium senegalense (35796), Mycobacterium sherrisii (BAA-832), Mycobacterium shimoidei (27962), Mycobacterium szulgai (35799), Mycobacterium terrae (15755), Mycobacterium tokaiense (27282), Mycobacterium triviale (23290), Mycobacterium tuberculosis (25618), and Mycobacterium xenopi (19250). Bacterial colonies were grown on Löwenstein-Jensen slants (Becton Dickinson, Sparks, MD). Colonies were suspended in H2O and inactivated by incubation for 30 min at 80°C and then lysed by gentle shaking in 1% sodium dodecyl sulfate and 0.8 mg/ml proteinase K (Qiagen, Valencia, CA) for 60 min at 60°C. Genomic DNA was separated from the other components of the lysate by being incubated in 700 mM NaCl and 1.2% cetyltrimethylammonium bromide for 15 min at 60°C, followed by vigorous shaking with an equal volume of chloroform-isoamyl alcohol (24:1) and spinning in an Eppendorf centrifuge (VWR International, West Chester, PA) for 10 min to separate the aqueous phase. The DNA was then precipitated by mixing the aqueous phase with an equal volume of cold isopropanol, chilling the mixture for 30 min at −20°C, sedimenting the DNA by spinning it in an Eppendorf centrifuge for 10 min, washing the DNA in 80% ethanol, drying the DNA in a SpeedVac, and dissolving the DNA in 1 mM EDTA, 10 mM Tris-HCl (pH 8.3). All steps were carried out in a biosafety level 3 containment facility (16). The concentration of the DNA was then adjusted to approximately 15,000 genomes/μl based on its absorption (optical density at 260 nm).
Mycobacterial target sequences.
The 39-nucleotide-long target sequence that we used to distinguish 27 different mycobacterial species was located within 16S rRNA gene hypervariable region V2 (20), which is flanked by conserved sequences. The target sequence is different for each of the selected mycobacterial species (1, 3). Two universal PCR primers for the generation of 214-nucleotide-long mycobacterial DNA amplicon strands that include the target sequence were purchased from Integrated DNA Technologies (Coralville, IA). Each PCR primer was perfectly complementary to a conserved sequence within the 16S rRNA gene of every one of the 27 different mycobacterial species that we decided to test. The target primer was designed to be incorporated into the amplicon strand that serves as the target for the sloppy molecular beacon probes (the target strand), and the complement primer was designed to be incorporated into the complementary amplicon strand. The sequence of the target primer was 5′-ACACCCTCTCAGGCCGGCTACCCG-3′, and the sequence of the complement primer was 5′-CTCGAGTGGCGAACGGGTGAGTAACACG-3′. The identity of the target sequence in the 16S rRNA gene of each species was confirmed by nucleotide sequence analysis. For use in preliminary hybridization experiments, 27 different synthetic oligonucleotides each containing a different 39-nucleotide-long species-specific target sequence were purchased from Integrated DNA Technologies.
Sloppy molecular beacon probes.
Four molecular beacon probes were prepared by solid-phase synthesis on an Applied Biosystems 394 DNA synthesizer (Foster City, CA). During synthesis of the molecular beacons, controlled-pore glass columns (Biosearch Technologies, Novato, CA) were used to incorporate dabcyl or Black Hole Quencher 2 (BHQ2) at their 3′ ends, and 5′-amino-modifier C6 phosphoramidites (Glen Research, Sterling, VA) were incorporated at their 5′ ends. After being removed from the column, succinimidyl ester derivatives of fluorescein, Alexa Fluor 546, Alexa Fluor 594 (all from Invitrogen, Carlsbad, CA) or Cy5 (GE Life Sciences, Piscataway, NJ) were coupled to the 5′-amino groups. The molecular beacons were then purified by high-pressure liquid chromatography on a Beckman Coulter System Gold chromatograph (Fullerton, CA) through a C18 reverse-phase column (Waters Corporation, Milford, MA). A detailed protocol for molecular beacon probe synthesis can be found at the Molecular Beacons website (http://www.molecular-beacons.org).
The sequences of the sloppy molecular beacons were as follows: probe A (5′-fluorescein-CCGGCCGGATAGGACCACAGGATGCATGTCGTGTGGTGGAAAGCGCCGG-dabcyl-3′), probe B (5′-Alexa 546-CCGGGCGGATAGGACCACGGGATGCATGTGTTGTGGTGGAAAGCCCCGG-dabcyl-3′), probe C (5′-Alexa 594-CCGGGGGATAGGACCTCTAGGCGCATGCCTTTTGGTGGAAAGCCCCGG-BHQ2-3′), and probe D (5′-Cy5-CCGGCCGAATAGGACCACGCGCTTCATGGTGTGTGGTGGAAAGCGCCGG-BHQ2-3′), where underlining identifies the complementary arm sequences in each molecular beacon.
Thermal denaturation of hybrids formed with synthetic oligonucleotides.
Twenty-seven hybridization reaction mixtures, each containing 1,500 nM of a different species-specific oligonucleotide and a control reaction mixture containing no target oligonucleotides, were prepared in triplicate. Each 50-μl reaction mixture contained 35 nM probe A, 12 nM probe B, 25 nM probe C, 30 nM probe D, 50 mM KCl, 4 mM MgCl2, and 10 mM Tris-HCl (pH 8.3). The solutions were incubated for 20 min at 25°C to form hybrids. The stability of each of the four different types of hybrids formed in each reaction was determined automatically by a Bio-Rad iQ5 spectrofluorometric thermal cycler (Hercules, CA) which increased the temperature from 25°C to 95°C in 1°C steps, holding each temperature for 5 min. This multichannel fluorescence detection instrument recorded the fluorescence intensity from each probe at every temperature and from these data automatically calculated the Tm of each hybrid.
Thermal denaturation of hybrids formed with amplicons.
Twenty-seven PCR assays, each initiated with approximately 50,000 copies of genomic DNA from a different mycobacterial species and a control reaction mixture containing no bacterial DNA, were prepared in triplicate. Prior to setting up the individual reaction mixtures, a master mix was prepared of such a nature that when the components of the master mix were added to the reaction mixtures, the final concentration of the components provided by the master mix was 50 U/ml AmpliTaq Gold DNA polymerase (Applied Biosystems), 100 U/ml restriction endonuclease AluI (Invitrogen, Carlsbad, CA), 1,000 nM target primer, 35 nM complementary primer, 35 nM probe A, 12 nM probe B, 25 nM probe C, 30 nM probe D, 250 μM dATP, 250 μM dCTP, 250 μM dGTP, 250 μM dTTP, 50 mM KCl, 4 mM MgCl2, and 10 mM Tris-HCl (pH 8.3). The master mix was incubated for 90 min at 37°C to enable the restriction endonuclease to digest small traces of Escherichia coli DNA that contaminate the DNA polymerase (2), followed by incubation for 20 min at 65°C to inactivate the endonuclease. Individual reaction mixtures were then prepared by mixing 3 μl of genomic DNA with 47 μl of the AluI-digested master mix. The reaction mixtures were then sealed and incubated in the Bio-Rad iQ5 spectrofluorometric thermal cycler for 10 min at 95°C to activate the DNA polymerase, followed by 55 cycles of 20 s at 95°C, 30 s at 63°C, and 30 s at 72°C. The reaction mixtures were then incubated for 20 min at 25°C to form hybrids, and the stability of each of the four different types of hybrids formed in each reaction was determined, using the same procedure described above for the hybrids formed with target oligonucleotides.
Data processing.
The Tm values listed in the tables and used for data shown in one figure (see Tables 1 and 2 and Fig. 3) were obtained directly from the output of the computer program controlling the spectrofluorometric thermal cycler. However, in order to prepare figures that compare the denaturation profiles of different hybrids, we utilized the fluorescence intensity data obtained by the spectrofluorometric thermal cycler for each hybrid and corrected these data for the effect of temperature on the intrinsic fluorescence of the particular fluorophore used to label the probe. In addition, the fluorescence intensity value at every temperature was smoothed (to reduce the effects of random fluctuations) by taking a rolling average of every three consecutive readings. Finally, to correct for small differences in volume and for intrinsic differences in each of the 96 wells of the spectrofluorometric thermal cycler reaction plates, the data for probe A (see Fig. 2 and 4) were normalized to a common value at 80°C (where it is assumed that all probes have dissociated from their targets and exist in a random-coil configuration).
TABLE 1.
Stability of hybrids formed by the binding of sloppy molecular beacon probes to mycobacterial 16S rRNA gene amplicons
| Species |
Tm (°C)
|
|||
|---|---|---|---|---|
| Probe A | Probe B | Probe C | Probe D | |
| M. abscessus | 57 | 55 | 64 | |
| M. aichiense | 48 | 55 | 60 | |
| M. asiaticum | 71 | 76 | 52 | 56 |
| M. aurum | 64 | 62 | 63 | |
| M. avium | 65 | 61 | 64 | |
| M. branderi | 70 | 67 | 51 | 66 |
| M. celatum | 70 | 74 | 53 | 51 |
| M. chubuense | 68 | 73 | 53 | 61 |
| M. diernhoferi | 52 | 54 | 66 | |
| M. engbaekii | 61 | 63 | 73 | |
| M. fortuitum | 58 | 60 | 75 | |
| M. haemophilum | 64 | 60 | 71 | 51 |
| M. intracellulare | 62 | 58 | 64 | |
| M. kansasii | 67 | 65 | 66 | 56 |
| M. lentiflavum | 60 | 57 | 71 | |
| M. malmoense | 58 | 57 | 62 | 50 |
| M. marinum | 69 | 73 | 59 | |
| M. rhodesiae | 54 | 59 | 56 | |
| M. senegalense | 64 | 66 | 77 | |
| M. sherrisii | 65 | 63 | 63 | |
| M. shimoidei | 68 | 66 | 55 | 64 |
| M. szulgai | 62 | 60 | 65 | |
| M. terrae | 67 | 72 | 48 | 52 |
| M. tokaiense | 61 | 73 | 66 | |
| M. triviale | 63 | 63 | 51 | 66 |
| M. tuberculosis | 73 | 77 | 53 | 58 |
| M. xenopi | 57 | 58 | 52 | 49 |
TABLE 2.
Identification of mycobacterial species from blinded DNA samples
| Identification by mycobacterial culture (no. of samples) | Identification by species-specific signature (no. of samples) |
|---|---|
| M. avium complex (25) | M. avium (25) |
| M. avium complex (10) | M. intracellulare (10) |
| M. abscessus (6) | M. abscessus (6) |
| M. haemophilum (6) | M. haemophilum (6) |
| M. fortuitum (3) | M. fortuitum (3) |
| M. xenopi (3) | M. xenopi (2) + new signaturea (1) |
| M. kansasii (1) | M. kansasii (1) |
| M. gordonae (1) | New signatureb (1) |
| No-target control (1) | No-target control (1) |
Upon sequence analysis of the amplicon, this sample was identified as an M. xenopi variant that possessed a single-nucleotide substitution in the target sequence.
Upon sequence analysis of the amplicon, this sample was identified as M. gordonae, which was not one of the 27 mycobacterial species in the original database.
FIG. 3.
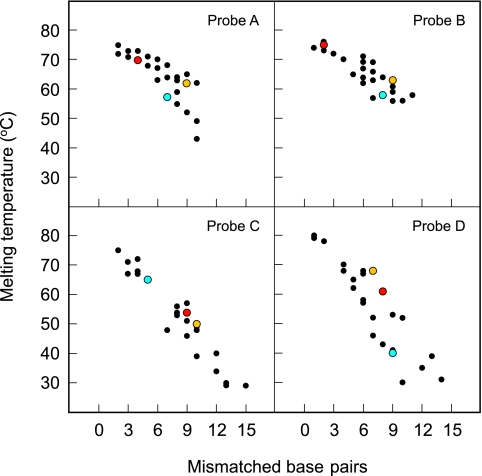
Dependence of hybrid stability on the relatedness of the probe sequence to the target sequence. The Tm of the hybrids formed by each of the four sloppy molecular beacon probes with 27 different species-specific oligonucleotide targets is plotted as a function of the number of mismatched base pairs in the hybrid. In general, the more mismatches present, the lower the Tm. Since each of the four molecular beacons possessed a different probe sequence, the set of four Tm values obtained for a particular species-specific target serves as a unique signature that identifies the species that is present. Results for M. chubuense (red), M. malmoense (blue), and M. triviale (orange) provide examples of unique species-specific signatures.
FIG. 2.
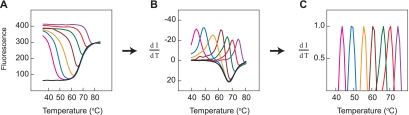
Determination of the stability of probe-target hybrids formed between sloppy molecular beacon probe A and seven different mycobacterial target oligonucleotides. (A) The fluorescence intensity of the molecular beacon in each hybrid was measured as a function of temperature. A drop in fluorescence intensity occurs at those temperatures where each hybrid dissociates. The increase in fluorescence intensity observed at higher temperatures with each species, and with the no-target control (black line), is due to the melting apart of the probe's hairpin stem. Each of the seven target oligonucleotides formed hybrids of differing stability (from weakest to strongest: M. aichiense, M. diernhoferi, M. abscessus, M. lentiflavum, M. senegalense, M. terrae, and M. asiaticum). (B) Utilizing the data from the left-hand panel, the derivative of fluorescence intensity with respect to temperature (dI/dT) is plotted as a function of temperature. (C) The data within approximately 2°C of each peak in the middle panel were normalized to the same height.
FIG. 4.
Determination of the stability of hybrids formed by sloppy molecular beacon probe A with seven different mycobacterial target amplicons. The fluorescence intensity of the molecular beacon in each hybrid is plotted as a function of temperature. A drop in fluorescence intensity is observed at those temperatures where each hybrid dissociates. The black line shows the fluorescence intensity of probe A in the no-target control reaction mixture. The hybrids formed from these seven mycobacterial amplicons gave the following Tm values: M. aichiense, 48°C; M. rhodesiae, 54°C; M. abscessus, 57°C; M. lentiflavum, 60°C; M. szulgai, 62°C; M. avium, 65°C; and M. terrae, 67°C.
In order to visually compare the Tm values of different hybrids (and to obtain an easily discernible representation of the multiprobe fluorescence signature generated by each mycobacterial species), the data showing the derivative of fluorescence intensity with respect to temperature (see Fig. 2C, 5, and 6B) were normalized so that the derivatives within approximately 2°C of each peak were plotted at values between 0 and 1.
FIG. 5.
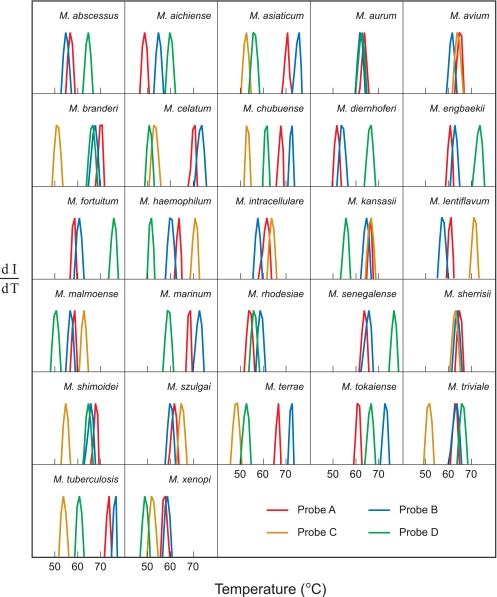
Species-specific signatures for 27 different mycobacteria. Each PCR assay was initiated with genomic DNA from a different mycobacterial species. The normalized derivatives of the fluorescence intensity of the hybrids formed by the binding of the resulting amplicons to the differently colored sloppy molecular beacon probes present in each reaction mixture (dI/dT) are plotted as a function of temperature. The set of three or four Tm values determined in each reaction by melting apart these hybrids serves as a unique signature that identifies the mycobacterial species whose genomic DNA was used to initiate amplification.
FIG. 6.
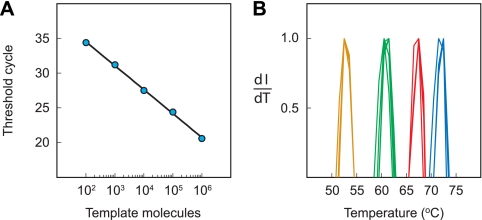
Effect of initial genomic DNA concentration on the species-specific signature. (A) Utilizing the fluorescence of probe A, the threshold cycle of each of the five PCRs is plotted as a function of the logarithm of the number of molecules of M. chubuense genomic DNA used to initiate the reaction. (B) The normalized derivative of fluorescence intensity with respect to temperature is plotted for each probe (dI/dT). The species-specific signatures obtained from each of the five reactions are virtually superimposable.
Blinded mycobacterial DNA samples.
Isolates of mycobacterial species obtained from patient samples during a 2-year period at the Memorial Sloan-Kettering Cancer Center were grown in culture medium, identified by standard biochemical tests (11) and by isotopically labeled DNA probes designed to selectively hybridize to the rRNA of particular mycobacterial species (Gen-Probe AccuProbes, San Diego, CA), and stored on Löwenstein-Jensen slants. For this study, a single colony from each isolate was streaked onto a 7H11 Middlebrook agar plate (Becton Dickinson) to ensure the viability and purity of the culture. Cells from three or four of the resulting colonies were lysed by being boiled in 1 ml H2O for 20 min to release their DNA (6) and then spun in an Eppendorf centrifuge for 5 min at 14,000 × g to remove cell debris. Three-microliter aliquots of each lysate (the identity of which was unknown to the group at the New Jersey Medical School) were used to initiate each 50-μl PCR assay.
RESULTS
Sloppy molecular beacon probes.
We designed and synthesized four different sloppy molecular beacons, each possessing a different probe sequence and each labeled with a differently colored fluorophore (Fig. 1). The four fluorophores (fluorescein on probe A, Alexa Fluor 546 on probe B, Alexa Fluor 594 on probe C, and Cy5 on probe D) were chosen because their fluorescence signals are readily distinguishable by the spectrofluorometric thermal cycler in which the PCR assays were carried out. Although each of the four probes was designed to hybridize to the target sequences in the amplicons of all of the 27 mycobacterial species, none of the four probes was perfectly complementary to any of the target sequences. Instead, the probe sequence in each of the four molecular beacons was selected so that its complementarity with the 27 different target sequences varied from species to species.
FIG. 1.
Schematic representation of the four sloppy molecular beacon probes. Each probe possessed a different sequence and a differently colored fluorophore, enabling the four probes to be used simultaneously in the same reaction mixture and to be distinguished from each other by the spectrofluorometric thermal cycler used to perform the assays. Probe A was labeled with fluorescein (red) and dabcyl (gray), probe B was labeled with Alexa Fluor 546 (blue) and dabcyl, probe C was labeled with Alexa Fluor 594 (orange) and BHQ2 (black), and probe D was labeled with Cy5 (green) and BHQ2.
Ideally, the probe sequence of each sloppy molecular beacon should be chosen so that its degree of complementarity with the target sequence from each species is different. However, the number of mismatched base pairs should not be so great that the hybrid cannot form. For the experiments reported here, we selected the sequences of probes A and B to be similar to each other, differing by only three nucleotides and forming hybrids that contained between 1 and 11 mismatched base pairs depending on which mycobacterial target sequence was present in the amplicon. On the other hand, we selected the sequences of probes C and D to differ from each other by 11 nucleotides and form hybrids that contained between 1 and 15 mismatched base pairs. The positions of the nucleotides at which the sequences of the four probes differed from one another were chosen to correspond to the nucleotides in the set of 27 target sequences that showed the most variation in the hope that each probe would respond differently to each target.
Preliminary hybridization experiments.
We prepared 27 hybridization reaction mixtures, each containing an excess of 1 of the 27 different target oligonucleotides; we also prepared a control reaction mixture that contained no targets. Each reaction mixture also contained the four differently colored molecular beacons, the concentration of each being chosen so that they would all produce fluorescence signals of approximately the same intensity. Hybrids were formed by being incubated at 25°C in the same buffer that we use for PCR assays. The Tm values of the four different hybrids formed in each reaction were determined automatically by a Bio-Rad iQ5 spectrofluorometric thermal cycler, which slowly increased the temperature from 25°C to 95°C in 1°C steps.
Figure 2A shows the probe-target hybrid denaturation profiles that were obtained for the hybrids formed by probe A with oligonucleotide target sequences from seven different mycobacteria. At low temperatures, a strong fluorescence signal was produced in each tube, indicating that the molecular beacon probe was hybridized to all seven species-specific targets. However, as the temperature was slowly increased, the intensity of the fluorescence signal produced by each species eventually dropped, indicating that the molecular beacon probe had dissociated from its target and formed a nonfluorescent conformation. The key observation here is that the temperature at which each species-specific probe-target hybrid dissociates is different, depending on the number of mismatched base pairs and on the location of those mismatched base pairs in the hybrid.
It is common to think of fluorescent hybridization probes as tools to determine whether a particular target sequence is present or absent in a sample by observing whether a fluorescence signal occurs or does not occur after incubation of the sample with the probe. However, these results demonstrate that sloppy molecular beacon probes form a fluorescent hybrid with many different target sequences even though they are not perfectly complementary to the targets.
The beauty of sloppy molecular beacons is that the stability of the probe-target hybrids that they form provides a characteristic Tm value that depends on the identity of the target.
Utilizing the data shown in Fig. 2A, the derivative of fluorescence intensity with respect to temperature was plotted as a function of temperature, with negative values plotted above the x axis (Fig. 2B). In this plot, the decrease in fluorescence intensity that occurs when a hybrid dissociates is seen as a peak that rises and then falls, with the highest rate of dissociation (the top of the peak) occurring at the hybrid's Tm. In order to easily compare the results obtained with different hybrids, the data that occur within approximately 2°C of each peak in Fig. 2B were normalized so that the values of the derivatives were plotted between 0 and 1, as shown in Fig. 2C.
The results of these preliminary hybridization experiments are graphically summarized in the four panels of Fig. 3. For each sloppy molecular beacon probe, the stability (Tm) of the hybrid that it forms with each of the 27 species-specific oligonucleotides is plotted as a function of the number of mismatched base pairs that occur in that hybrid. The results highlight the dependence of Tm on the relatedness of the nucleotide sequence of the probe to the nucleotide sequence of the target. In general, the more mismatched base pairs that occur in a hybrid, the lower its Tm value. These results demonstrate that the Tm obtained from the hybrid formed by any one of the probes provides information that helps to identify the target species to which the probe is hybridized but that Tm, in and of itself, is not sufficient to uniquely identify the target. However, the combination of the four Tm values obtained for the same target with four different probes provides much more information about the identity of the target (see, for example, the species-specific results highlighted by the colored dots in Fig. 3). These preliminary results illustrate the underlying principle of obtaining a species-specific signature through the use of a series of independent measurements. For example, if each of the sloppy molecular beacon probes was capable of providing only 10 distinguishable Tm values, then the results from four different probes, taken together, would provide 10,000 different species-specific signatures (10 × 10 × 10 × 10).
Use of LATE-PCR.
The simultaneous use of sloppy molecular beacon probes in PCR assays generates hybrids that are sometimes relatively weak because they possess many mismatched base pairs. This raises a number of concerns for the selection of an effective design for the PCR assays, all of which could cause a decrease in the intensity of the fluorescence from weaker hybrids, including the following: (i) competition between probes that form strong hybrids and probes that form weak hybrids might diminish the number of weaker hybrids that form; (ii) competition between the probes and the complementary amplicon strands for binding to the target strands might diminish the number of weaker hybrids that form; and (iii) secondary and tertiary structures that are present in the target strands might diminish the formation of weaker hybrids. We therefore decided to utilize linear-after-the-exponential (LATE)-PCR, which is an efficient asymmetric PCR format (13, 15), in which so many target strands are synthesized that their number exceeds the number of sloppy molecular beacon probes present in the assay and in which many more target strands are synthesized than complementary strands. By eliminating sources of competition for the binding of probes to target strands, weaker hybrids are more abundant, and their fluorescence is therefore more likely to be sufficiently intense for their Tm to be measured.
Determination of species-specific signatures.
We carried out 27 LATE-PCR assays, each containing the four sloppy molecular beacon probes and genomic DNA from 1 of the 27 different mycobacterial species, and we carried out control assays that did not contain any mycobacterial DNA. After the completion of amplification, the Tm values of the hybrids formed by the four differently colored sloppy molecular beacon probes with each species-specific target amplicon were determined automatically by the Bio-Rad iQ5 spectrofluorometric thermal cycler.
To illustrate the nature of the results that were obtained, the denaturation profiles of the hybrids formed by probe A with seven different mycobacterial target amplicons are shown in Fig. 4. As can be seen, the lower the stability of the hybrid (as evidenced by locations where a drop in intensity occurred), the weaker the overall intensity of the fluorescence signal. The lower fluorescence intensity of the less-stable hybrids was not due to competition among the four probes, since the same results were obtained in preliminary experiments in which only probe A was present. Moreover, the reduction in fluorescence intensity seen in weaker hybrids formed from amplicon strands was not seen in corresponding hybrids formed from oligonucleotides, implying that the lower fluorescence of the less-stable probe-amplicon hybrids is due to the presence of secondary and tertiary structures in the amplicon strands that restrict the access of the probes to the target sequence. Despite the lowering of the fluorescence intensity from the less-stable hybrids, the results demonstrate that the fluorescence signal from all seven hybrids was sufficiently intense to enable each hybrid's characteristic Tm value to be determined.
See Tables SA, SB, SC, and SD in the supplemental material for a comprehensive listing of the results that were obtained with amplicons. In the supplemental material, there is a different table for each of the four sloppy molecular beacon probes, and each table shows the nucleotide sequence of each species-specific target and highlights in black letters those nucleotides that are not complementary to the corresponding nucleotide in the probe sequence. The Tm value obtained for each of the hybrids is listed in the right-hand column, and the results are shown in the order of hybrid stability, with the most stable hybrids listed at the top of each table. These results show that, in addition to there being a roughly inverse correlation between the number of mismatched base pairs in a hybrid and its stability, Tm values are affected by the identity of the mismatches, the identity of neighboring base pairs, the location of the mismatches within the probe-target hybrid, and whether or not the mismatches occur in a run of adjacent mismatches. The results also show that the fluorescence intensity of probe-target hybrids that possess more than 10 mismatched base pairs was too low to obtain useful Tm values. Despite this observation, the set of three or four Tm values that was obtained for each species is sufficiently unique to constitute a species-specific signature that identifies the species that was present in each sample. Table 1 summarizes the results obtained for each of the 27 mycobacterial species that were tested.
Figure 5 shows, for each of the 27 mycobacterial species, the normalized derivatives obtained by melting the hybrids formed by the PCR amplicons with each of the differently colored sloppy molecular beacon probes. An examination of these results shows that the genomic DNA from each species produces a combination of Tm values that distinguishes that species from all of the other species tested. Even mycobacterial species whose 39-nucleotide-long target sequences are extraordinarily similar produce reproducibly distinguishable species-specific signatures.
For example, the target sequences from M. asiaticum and M. tuberculosis differ from each other only by the substitution of a single adenosine for a guanosine, yet the Tm values for the hybrids formed by M. tuberculosis amplicons with probes B and C are consistently 1°C higher than the Tm values for the corresponding hybrids formed by M. asiaticum, and the Tm values for the hybrids formed by M. tuberculosis amplicons with probes A and D are consistently 2°C higher than the Tm values for the corresponding hybrids formed by M. asiaticum. In order to illustrate the precision of the Tm measurements, every PCR assay was repeated three times, and all three multiprobe fluorescence signatures obtained for each mycobacterial species were virtually superimposable.
Effect of initial mycobacterial DNA concentration on the species-specific signature.
A particularly attractive aspect of exponential gene amplification techniques, such as PCR, is that over a wide range of amounts of template DNA initially present, the final amounts of amplified DNA do not vary very much. This is a very useful attribute when the amplicons are used to form probe-target hybrids for the measurement of hybrid stability, since the Tm of the hybrid is affected by target concentration. To illustrate this desirable feature, we carried out five different LATE-PCR assays, each initiated with a different number of M. chubuense genomic DNA molecules. The reactions were initiated with as little as 100 molecules of genomic DNA and as many as 1,000,000 molecules of genomic DNA. After amplification, the denaturation profiles of the four differently colored probe-target hybrids that were present in each reaction mixture were determined.
The results (Fig. 6A) demonstrate that the number of thermal cycles required to complete the symmetric phase of each LATE-PCR was, as expected, an inverse linear function of the logarithm of the number of molecules of genomic DNA initially present. Significantly, the normalized derivatives obtained from the five PCRs were virtually superimposable, despite the wide range of initial DNA concentrations tested (Fig. 6B), confirming that the Tm values of the hybrids formed by sloppy molecular beacon probes with the amplified DNA of a bacterial species yield a characteristic species-specific signature irrespective of the initial DNA concentration.
Test of blinded mycobacterial DNA samples.
To see whether species-specific signatures can be used to reliably identify mycobacterial species, we utilized the four sloppy molecular beacon probes in LATE-PCR assays to test 55 blinded DNA samples that were prepared from mycobacteria isolated from patients over a 2-year period at the Memorial Sloan-Kettering Cancer Center. The set of Tm values obtained from each of the 55 samples was compared to the 27 previously determined species-specific signatures listed in Table 1. The signatures obtained from 53 of the 55 samples correctly matched the signature of 1 of the 27 species already in our database. However, the signatures obtained from two of the samples did not match any of the 27 previously determined signatures. Sequence analysis of the amplicons present in these two discrepant samples indicated that one sample (probe A Tm, 65°C; probe B Tm, 58°C; and probe D Tm, 55°C) was Mycobacterium gordonae, which was not in our database, and the other sample (probe A Tm, 61°C; probe B Tm, 59°C; probe C Tm, 54°C; and probe D Tm, 54°C) was a variant of M. xenopi that possessed a single-nucleotide substitution in its target sequence, also not in our database. The two new species-specific signatures can now be added to our database, illustrating how the scope of these assays can be enhanced as additional species are tested. These results are summarized in Table 2.
DISCUSSION
The methods presented here are an extension of the pioneering work carried out by Carl Wittwer and his colleagues on the information that can be obtained from the melting of amplicons (9, 14, 25). What distinguishes our own work is the realization that the simultaneous use of a set of differently colored sloppy molecular beacon probes in a homogeneous nucleic acid amplification assay, in combination with an analysis of the melting curves obtained from the resulting probe-target hybrids, provides a unique set of Tm values that serves as a species-specific signature that identifies the species present in a sample.
Underlying this approach is the recognition that reliable Tm values can be obtained despite the presence of factors that lower the magnitude of the fluorescence signal generated by the probe-target hybrids. These factors include the effect of competition between the probes and the complementary amplicon strands, the effect of competition among the probes for binding to the available target strands, and the effect of secondary and tertiary structures in the target strands that partially restrict access of the probes to the amplicon. Although all of these factors alter the abundance of the hybrids, they do not significantly alter the stability of the hybrids, and that is the property that is measured.
In general, it is desirable to utilize smaller amplicons, if possible, as this is likely to limit the formation of secondary and tertiary structures in the target strand that restrict the access of the probes. It may also be worthwhile to explore the use of sloppy molecular beacon probes in assay formats that generate single-stranded RNA amplicons (8) rather than DNA amplicons, such as assays that employ transcription-mediated amplification or nucleic acid sequence-based amplification. These target amplification techniques do not generate cRNA strands. Moreover, they might be more sensitive because they can directly amplify a segment of the abundant 16S rRNA rather than a segment of the 16S rRNA gene, which is present only in one or two copies per cell.
The results of the experiments reported here provide guidance as to how sloppy molecular beacon probes should best be designed. In order to ensure that the magnitude of the fluorescence signal generated by the probe-target hybrids is sufficiently intense to enable their Tm values to be determined, the probe sequence of each molecular beacon should be selected in such a manner that all of the species-specific hybrids will contain less than 10 mismatched base pairs (and have Tm values above 45°C), assuming that the target sequence is about 40 nucleotides in length. In addition, in order to ensure a diverse set of species-specific signatures, the sequence of each of the probes should differ from the sequence of each of the other probes by at least four nucleotide substitutions. It would be helpful to have a computer program for the selection of an optimal set of probe sequences. However, the selection of a useful set of probe sequences is not a difficult task, since we obtained good results by simply examining the set of target sequences and designing probes that varied from one another in those regions of the target sequences that contained the most sequence variations.
Although these screening assays are designed to detect the presence of a single species in a sample, the melting curves that are obtained will often enable the identification of two different species when they are both present. In these situations, there will usually be two separate drops in fluorescence intensity in the melting curve for each of the sloppy molecular beacon probes, with each drop occurring at a Tm value characteristic of one of the two species.
Real-world screening assays can be designed to identify species that occur in different genera and to differentiate species under conditions where the identification of the species that is present in a sample has clinical significance. In this regard, we have been developing an assay that utilizes sloppy molecular beacon probes to identify sepsis-causing bacteria of differing genera in normally sterile human blood samples (S. Chakravorty et al., unpublished data).
It seems entirely feasible to design assays that distinguish any related set of species, irrespective of whether they are, for example, fungi, protozoa, nematodes, or fish. The key to developing single-tube assays that are able to distinguish a long list of species is the selection of a target sequence region that is shared by all of the species of interest, that can be amplified by a pair of universal primers, and that possesses just the right degree of sequence variability so that the presence of each species results in the generation of a unique species-specific signature. Since all living organisms are related to each other, sharing a common genetic code and sharing highly conserved, multielement genetic instruction sets for the control and achievement of gene replication, gene expression, and protein synthesis, there are many common genetic sites to choose from, and algorithms can be developed to select an optimal target site when provided with a set of relevant sequences for all the species that need to be distinguished.
Supplementary Material
Acknowledgments
We thank Soumitesh Chakravorty for suggesting the use of restriction endonuclease AluI, Barry Kreiswirth for providing mycobacterial species, Arjun Raj for stimulating discussions, Larry Wangh and Aquiles Sanchez for guidance on the design of LATE-PCR assays, Susan Massarella for expert assistance in clinical mycobacteriology, and Amy Piatek for carrying out early experiments.
This research was supported by grants AI-056689 and EB-000277 from the National Institutes of Health.
Footnotes
Published ahead of print on 26 January 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Böddinghaus, B., T. Rogall, T. Flohr, H. Blöcker, and E. C. Böttger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 281751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll, N. M., P. Adamson, and N. Okhravi. 1999. Elimination of bacterial DNA from Taq DNA polymerases by restriction endonuclease digestion. J. Clin. Microbiol. 373402-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravorty, S., D. Helb, M. Burday, N. Connell, and D. Alland. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69330-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, J.-C., C.-L. Huang, C.-C. Lin, C.-C. Chen, Y.-C. Chang, S.-S. Chang, and C.-P. Tseng. 2006. Rapid detection and identification of clinically important bacteria by high-resolution melting analysis after broad-range ribosomal RNA real-time PCR. Clin. Chem. 521997-2004. [DOI] [PubMed] [Google Scholar]
- 5.Couzinet, S., C. Jay, C. Barras, R. Vachon, G. Vernet, B. Ninet, I. Jan, M.-A. Minazio, P. Francois, D. Lew, A. Troesch, and J. Schrenzel. 2005. High-density DNA probe arrays for identification of staphylococci to the species level. J. Microbiol. Methods 61201-208. [DOI] [PubMed] [Google Scholar]
- 6.El-Hajj, H. H., S. A. E. Marras, S. Tyagi, F. R. Kramer, and D. Alland. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 394131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erali, M., J. I. Pounder, G. L. Woods, C. A. Petti, and C. T. Wittwer. 2006. Multiplex single-color PCR with amplicon melting analysis for identification of Aspergillus species. Clin. Chem. 521443-1445. [DOI] [PubMed] [Google Scholar]
- 8.Guatelli, J. C., K. M. Whitfield, D. Y. Kwoh, K. J. Barringer, D. D. Richman, and T. R. Gingeras. 1990. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc. Natl. Acad. Sci. USA 871874-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gundry, C. N., J. G. Vandersteen, G. H. Reed, R. J. Pryor, J. Chen, and C. T. Wittwer. 2003. Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes. Clin. Chem. 49396-406. [DOI] [PubMed] [Google Scholar]
- 10.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6986-994. [DOI] [PubMed] [Google Scholar]
- 11.Kent, T. P., and G. P. Kubica. 1985. Public health mycobacteriology. A guide for the level III laboratory. Centers for Disease Control and Prevention, Atlanta, GA.
- 12.McCabe, K. M., Y. H. Zhang, B. L. Huang, E. A. Wagar, and E. R. McCabe. 1999. Bacterial species identification after DNA amplification with a universal primer pair. Mol. Genet. Metab. 66205-211. [DOI] [PubMed] [Google Scholar]
- 13.Pierce, K. E., J. A. Sanchez, J. E. Rice, and L. J. Wangh. 2005. Linear-after-the-exponential (LATE)-PCR: primer design criteria for high yields of specific single-stranded DNA and improved real-time detection. Proc. Natl. Acad. Sci. USA 1028609-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1997. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245154-160. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez, J. A., K. E. Pierce, J. E. Rice, and L. J. Wangh. 2004. Linear-after-the-exponential (LATE)-PCR: an advanced method of asymmetric PCR and its uses in quantitative real-time analysis. Proc. Natl. Acad. Sci. USA 1011933-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somerville, W., L. Thibert, K. Schwartzman, and M. A. Behr. 2005. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J. Clin. Microbiol. 432996-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor, T. B., C. Patterson, Y. Hale, and W. W. Safranek. 1997. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J. Clin. Microbiol. 3579-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1649-53. [DOI] [PubMed] [Google Scholar]
- 19.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14303-308. [DOI] [PubMed] [Google Scholar]
- 20.Van de Peer, Y., S. Chapelle, and R. De Wachter. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 243381-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varma-Basil, M., H. El-Hajj, S. A. E. Marras, M. H. Hazbon, J. M. Mann, N. D. Connell, F. R. Kramer, and D. Alland. 2004. Molecular beacons for multiplex detection of four bacterial bioterrorism agents. Clin. Chem. 501060-1062. [DOI] [PubMed] [Google Scholar]
- 22.Vet, J. A. M., A. R. Majithia, S. A. E. Marras, S. Tyagi, S. Dube, B. J. Poiesz, and F. R. Kramer. 1999. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. USA 966394-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vet, J. A. M., B. J. Van der Rijt, and H. J. Blom. 2002. Molecular beacons: colorful analysis of nucleic acids. Expert Rev. Mol. Diagn. 277-86. [DOI] [PubMed] [Google Scholar]
- 24.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22130-131. [DOI] [PubMed] [Google Scholar]
- 25.Wittwer, C. T., G. H. Reed, C. N. Gundry, J. G. Vandersteen, and R. J. Pryor. 2003. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 49853-860. [DOI] [PubMed] [Google Scholar]
- 26.Woese, C. R., L. J. Magrum, R. Gupta, R. B. Siegel, D. A. Stahl, J. Kop, N. Crawford, J. Brosius, R. Gutell, J. J. Hogan, and H. F. Noller. 1980. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 82275-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zucol, F., R. A. Ammann, C. Berger, C. Aebi, M. Altwegg, F. K. Niggli, and D. Nadal. 2006. Real-time quantitative broad-range PCR assay for detection of the 16S rRNA gene followed by sequencing for species identification. J. Clin. Microbiol. 442750-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.