Abstract
Legionella pneumophila is the causative agent of the severe and potentially fatal pneumonia Legionnaires' disease. L. pneumophila is able to replicate within macrophages and protozoa by establishing a replicative compartment in a process that requires the Icm/Dot type IVB secretion system. The signals and regulatory pathways required for Legionella infection and intracellular replication are poorly understood. Mutation of the rpoS gene, which encodes σS, does not affect growth in rich medium but severely decreases L. pneumophila intracellular multiplication within protozoan hosts. To gain insight into the intracellular multiplication defect of an rpoS mutant, we examined its pattern of gene expression during exponential and postexponential growth. We found that σS affects distinct groups of genes that contribute to Legionella intracellular multiplication. We demonstrate that rpoS mutants have a functional Icm/Dot system yet are defective for the expression of many genes encoding Icm/Dot-translocated substrates. We also show that σS affects the transcription of the cpxR and pmrA genes, which encode two-component response regulators that directly affect the transcription of Icm/Dot substrates. Our characterization of the L. pneumophila small RNA csrB homologs, rsmY and rsmZ, introduces a link between σS and the posttranscriptional regulator CsrA. We analyzed the network of σS-controlled genes by mutational analysis of transcriptional regulators affected by σS. One of these, encoding the L. pneumophila arginine repressor homolog gene, argR, is required for maximal intracellular growth in amoebae. These data show that σS is a key regulator of multiple pathways required for L. pneumophila intracellular multiplication.
Legionella pneumophila is a gram-negative opportunistic human pathogen that causes the severe and potentially fatal pneumonia Legionnaires' disease (30, 47, 67, 83). L. pneumophila's ability to replicate within human alveolar macrophages is essential for its capacity to cause disease (44-46). Transmission of L. pneumophila to the human lung occurs as a result of the inhalation of aerosolized contaminated water droplets (74), often from exposure to showers or whirlpool baths (96). Legionella species are ubiquitous in most naturally occurring and man-made aquatic systems, where the organism replicates within a variety of unicellular protozoan hosts (28, 38, 96). It has been suggested that the interaction of Legionella species with environmental protozoa has selected for the bacterium's evolutionary adaptation to intracellular life in mammalian cells (99).
Intracellular multiplication of L. pneumophila requires a series of ordered events that disrupt normal endocytic trafficking in both macrophage and protozoan host cells. These include preventing phagolysosome fusion and the acidification of the Legionella-containing vacuole (LCV), followed by the acquisition of membrane material derived from the Golgi and endoplasmic reticulum compartments of the host (71, 85, 93, 95). These events are dependent upon the Icm/Dot type IVB secretion system (TFBSS) (84, 90, 91). The Icm/Dot system is homologous to type IV conjugation systems and is able to translocate DNA (90, 105) and protein between bacteria as well as to translocate proteins from the bacteria into eukaryotic hosts (15, 18, 24, 25, 75). More than 150 putative Icm/Dot-translocated substrates have been identified (8, 14, 15, 18, 23, 25, 37, 52, 54, 61, 75, 78, 94, 103, 108), many based on the presence of eukaryotic-like protein domains (25), and these are predicted to modify endocytic trafficking and other host cell functions for the benefit of the bacterium (18, 48, 64, 75). However, the functions of the majority of Icm/Dot-translocated substrates are unknown, and most strains containing null mutations in the genes encoding Icm/Dot substrates are not defective for intracellular replication (77). The apparent functional redundancy of Icm/Dot-translocated substrates has resulted in the hypothesis that subsets of Icm/Dot substrates may target distinct protozoan hosts (77).
Legionella intracellular multiplication likely requires the sensing of, and response to, a myriad of signals, which suggests the need for a complex regulatory network (3, 31, 38-40, 62, 71, 72, 108). However, few regulators of intracellular multiplication have been identified, and their organization and interconnectivity are only partially understood. The two-component systems CpxRA and PmrAB directly regulate the transcription of subsets of Icm/Dot-translocated substrates (3, 108). An additional two-component system, LetAS (homologous to the GacAS system of pseudomonads [9, 32]), is proposed to respond to ppGpp by positively affecting the expression of postexponential growth phenotypes and is required for the efficient intracellular replication of L. pneumophila (5, 40, 71). The effect of LetAS has been linked to the global carbon storage regulator CsrA through shared phenotypes (72). Based on the GacAS system in Pseudomonas aeruginosa, LetAS is predicted to regulate the transcription of the small RNA csrB, which represses the CsrA protein (71). However, prior to this investigation no L. pneumophila csrB homolog had been characterized.
Null mutations in the rpoS gene, encoding the sigma factor σS, do not affect growth in rich medium but exhibit two interesting phenotypes: rpoS mutants retain significant stress resistance in postexponential phase, and they are not able to replicate in amoebae (38). The stress resistance phenotype of L. pneumophila is in contrast to Escherichia coli, in which rpoS mutants are much more sensitive to stress during postexponential growth than the wild type (43). In E. coli, σS is considered the general stress sigma factor because it senses and responds broadly to a variety of stress signals by regulating the transcription of a large number of target genes (42). In bacterial pathogens including Salmonella enterica serovar Typhimurium, Yersinia enterocolitica, and Vibrio cholerae, the σS regulon is involved not only in stress resistance but also in the regulation of virulence genes (49, 69, 76, 86). L. pneumophila rpoS mutants are unable to replicate in protozoa and primary macrophages (1) but are not defective for replication in human macrophage cell lines such as THP-1 and HL-60 (38). Because of its central role as a sigma factor and its altered host range compared to icm/dot TFBSS mutants, σS may be a key toward unlocking the regulatory requirements for Legionella intracellular multiplication.
In this study we show that although rpoS and icm/dot mutants share similar intracellular multiplication and trafficking defects, rpoS mutants are not defective in Icm/Dot-dependent protein translocation. To develop a comprehensive view of σS regulation in L. pneumophila and identify σS-regulated genes that might be required for intracellular multiplication, we compared the patterns of global gene expression of an rpoS mutant and wild-type L. pneumophila during exponential and postexponential growth in rich medium. The expression data were analyzed for global transcriptional effects of σS and interrogated for σS effects on the expression of genes known to be required for intracellular multiplication. We show that σS affects the expression of many genes encoding Icm/Dot substrates as well as genes encoding regulators required for intracellular multiplication. Through the mutational analysis of σS-affected transcription factor genes, we discovered that the L. pneumophila arginine repressor homolog, argR, is required for efficient intracellular multiplication. These data expand our understanding of the σS regulon and the requirements for L. pneumophila intracellular multiplication.
MATERIALS AND METHODS
Bacterial strains and mutants.
The bacterial strains used in this study are listed in Table 1. Media and antibiotics were used as previously described (18). L. pneumophila strains used in this study were L. pneumophila JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1 (87); LM1376 is an isogenic rpoS-null (lpg1284) derivative of JR32 (38); LELA3118 is an isogenic dotA-null (lpg2686) derivative of JR32 (87); KS79 is an isogenic ΔcomR (lpg2717) (24) derivative of JR32 that renders the bacteria genetically competent for DNA uptake (24, 92). Mutants made in this study were produced by creating allelic exchange fragments using long-flanking homology PCR as described previously (106), which consisted of approximately 1 kb of homology to each end of the target gene flanking a 1-kb kanamycin resistance cassette, based on previous results for Legionella natural transformation (92). Purified allelic exchange fragments were introduced into competent KS79 cells by natural transformation as described previously (92), selected on kanamycin, screened for gene replacement by PCR, and sequenced for verification of correct insertion as described previously (38). Mutants made by this method are listed in Table 2 and include GAH280 ΔargR (lpg0490) and GAH338 ΔletS (lpg1912), both of which are isogenic derivatives of KS79. The primers for long-flanking homology PCR are listed in Table S6 in the supplemental material. GAH199, a dotA-null strain, was made by the transformation of KS79 with LELA3118 genomic DNA prepared according to the manufacturer's protocol (Wizard Genomic DNA Purification Kit; Promega) and selection on kanamycin.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| L. pneumophila strains | ||
| JR32 | Am511 salt-sensitive isolate | 87 |
| KS79 | JR32 ΔcomR | 24 |
| LELA3118 | JR32 dotA::Tn903dIIlacZ | 87 |
| LM1376 | JR32 rpoS::Tn903dGent | 38 |
| GAH199 | KS79 dotA::Tn903dIIlacZ | This study |
| GAH235 | KS79 ΔrpoS::Kmr | This study |
| GAH280 | KS79 ΔargR::Kmr | This study |
| GAH338 | KS79 ΔletS::KmR | This study |
| GAH026 | JR32, pXDC31 | This study |
| GAH028 | LELA3118, pXDC31 | This study |
| GAH147 | LM1376, pXDC31 | This study |
| GAH281 | KS79, pXDC31 | This study |
| GAH282 | GAH199, pXDC31 | This study |
| GAH286 | GA280, pXDC31 | This study |
| GAH228 | KS79, pXDC61-FabI | This study |
| GAH214 | KS79, pXDC61-LepA | This study |
| GAH215 | GAH199, pXDC61-LepA | This study |
| GAH238 | GAH235, pXDC61-LepA | This study |
| SPF38 | JR32, pSF21 | This study |
| SPF39 | GAH338, pSF21 | This study |
| SPF40 | LM1376, pSF21 | This study |
| GAH301 | GAH199, pGAH123 | This study |
| GAH305 | KS79, pMMB207c, pGAH123 | This study |
| GAH306 | KS79, pGAH125, pGAH123 | This study |
| GAH312 | GAH280, pMMB207c, pGAH123 | This study |
| GAH314 | GAH280, pGAH125, pGAH123 | This study |
| Plasmids | ||
| pMMB207c | pMMB207 mobA | 91 |
| pBBR1MCS-2 | pBBR1MCS Kmr | 51 |
| pXDC31 | pMMB207c Ptac-GFP | This study |
| pXDC61-FabI | pMMB207c Ptac-TEM1-FabI | 24 |
| pXDC61-LepA | pMMB207c Ptac-TEM1-LepA | 24 |
| pSF21 | pMMB270c Ptac-letS | This study |
| pGAH125 | pMMB207c Ptac-argR | This study |
| pGAH123 | pBBR1MCS2 mobA, two copies of Ptac-GFP; Gentr | This study |
TABLE 2.
Transcriptional regulators affected by σS
| ORF | Protein description | Mutationa | Gene | Transcription level (log2 ratio of rpoS/Wt)b
|
|
|---|---|---|---|---|---|
| Exponential phase | Postexponential phase | ||||
| lpg0490 | Arginine repressor | Y | argR | 3.80 | −1.80 |
| lpg0586 | Transcriptional regulator | Y | −0.39 | −4.10 | |
| lpg0853 | Transcriptional regulator FleQ | N | fleQ | −1.75 | −2.81 |
| lpg1260 | Putative repressor protein of prophage | Y | prpA | 1.77 | 3.64 |
| lpg1292 | DNA-binding response regulator | N | pmrA | −0.94 | −2.14 |
| lpg1438 | DNA-binding response regulator | N | cpxR | −2.43 | −2.57 |
| lpg1446 | Transcriptional regulator | Y | −2.10 | −1.27 | |
| lpg1577 | RNA polymerase σE | Y | rpoE | −1.50 | −4.93 |
| lpg1782 | Flagellar biosynthesis sigma factor | N | fliA | −0.27 | −4.95 |
| lpg1796 | Transcriptional regulator, LysR family | Y | −1.86 | −3.19 | |
| lpg2138 | Transcriptional regulator, LysR family | Y | −0.41 | −2.75 | |
| lpg2376 | Transcriptional regulator, LysR family | Y | −0.76 | −3.02 | |
| lpg2723 | Transcriptional regulator, ArsR family | Y | −2.45 | −0.71 | |
Y, the indicated gene was deleted by allelic exchange in this study; N, the indicated gene was not deleted.
Wt, wild type.
Growth of bacterial strains and medium preparation.
Media and antibiotics for the growth of L. pneumophila were used as described previously (18). Isolation of exponentially and postexponentially growing L. pneumophila was performed in the complex medium AYE [N-(2-acetamido)-2-aminoethanesulfonic acid-buffered yeast extract] in which cultures were started with an inoculum with an optical density (OD) of approximately 0.05 and grown with agitation at 37°C. Exponential phase cultures were collected at an OD of 0.70 to 0.80. Postexponential phase cultures were collected approximately 4 h following the cessation of growth, which occurred at an approximate OD of 3.0 to 3.5. For most experiments bacteria were grown in triplicate; however, six independent bacterial cultures for each of the strains under any growth condition were tested for gene expression profiling in this study.
Plasmid construction.
Plasmids pXDC31, pGAH125, and pSF21 were constructed from the restriction digest of green fluorescent protein (GFP), ArgR, and LetS PCR products, respectively, bearing the restriction sites EcoRI/HindIII (GFP) or KpnI/XbaI (ArgR and LetS) cloned into digested pMMB207c vector. Construction of pGAH123 consisted of the mutational inactivation of mobA, addition of two copies of ptac-GFP derived from pXDC31, and the addition of a gentamicin resistance cassette to pBBR1MCS2 in successive cloning steps.
TEM translocation assays.
Measurement of Icm/Dot-dependent substrate translocation was performed as previously described using published TEM fusion plasmids (24). Translocation experiments were performed three times, and the data shown are from one representative experiment.
Fluorescence microscopy of D. discoideum.
Dictyostelium discoideum AX2 cells were prepared and infected as described previously (18, 19) using bacteria expressing cytoplasmic DsRed and D. discoideum amoebae expressing a GFP-tagged subunit of the V-ATPase, VatM-GFP (22). The cells were observed by microscopy at 1 h and 4 h following infection, as previously described (18).
RNA isolation, cDNA preparation, and real-time qPCR experiments.
RNA for microarray and real-time quantitative PCR (qPCR) was prepared using an RNeasy Mini Kit following manufacturer's protocols (Qiagen). In qPCR experiments, RNA was treated with DNase I according to the manufacturer's instructions (Invitrogen) prior to cDNA preparation. For microarray and qPCR samples, cDNA was prepared using Superscript II reverse transcriptase as described by the manufacturer (Invitrogen). Real-time qPCR experiments were performed using an Applied Biosystems StepOne Plus 96-well reverse transcription-PCR system with Power SYBR Green PCR Master Mix following the manufacturer's instructions (Applied Biosystems). 16S RNA was used as the reference sample in all comparative threshold cycle (ΔΔCT) experiments. All qPCR primers were tested for amplification efficiency (see Table S7 in the supplemental material). Real-time qPCR data were analyzed using StepOne System software and Microsoft Excel. Primers used in qPCR experiments are shown in Table S7 in the supplemental material.
Microarray analysis.
The L. pneumophila strain Philadelphia-1 microarray consisting of 2,997 unique 70-mer oligonucleotides representing all of the L. pneumophila predicted open reading frames (ORFs) presented in duplicate was previously published (16). Oligonucleotides were resuspended to a concentration of 30 μM in 50% dimethyl sulfoxide and printed on UltraGaps aminosilane-coated slides (Corning Life Sciences) using a SpotArray 72 Microarray Printing System spotter (Perkin Elmer). After being spotted, the slides were stored in a vacuum at room temperature until further use.
To prepare the samples for microarray hybridization, 20 μg of total RNA from each of the samples was converted to cDNA by reverse transcription in the presence of aminoallyl-dUTP and fluorescently labeled by coupling the resulting cDNA with the succinimidyl ester fluorescent dyes Alexa Fluor 546 and Alexa Fluor 647 (Invitrogen), following the manufacturer's protocols. Labeled cDNA was hybridized to Corning UltraGAPs coated slides and washed following Corning, Inc., protocols. Hybridized arrays were scanned using a ScanArray Express (PerkinElmer) instrument at 5-μm resolution, and the resulting hybridization intensities for all probes from both channels on each array were exported for further analysis. Raw signal intensities were corrected for dye labeling effects within and between all slides using the normalize.lowess R-function implemented in the Bioconductor affy microarray analysis package (33). Genes with a P value of ≤0.005 and for which the ratio of the log2 spot intensity of the mutant (LM1376) over the wild type (JR32) was less than or equal to −2 or greater than or equal to +2 were considered for further analysis. The resulting sets of differentially expressed genes were further analyzed using hierarchical clustering algorithms implemented within the Spotfire DecisionSite software suite (Tibco Spotfire, Inc.).
Eukaryotic cell line maintenance and kinetic measurement of intracellular multiplication.
Acanthamoeba castellanii was maintained in peptone-yeast-glucose medium as described previously (38). Preparation for measurement of L. pneumophila intracellular multiplication was similar to previously described methods (38) with minor adaptations for 96-well plates. Monolayers of A. castellanii cells were formed in 96-well plates at 1 × 105 cells per well. L. pneumophila strains harboring GFP-positive plasmids (pXDC31 or pGAH123) were induced overnight on charcoal-yeast extract plates containing 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and then resuspended and diluted to the desired multiplicity of infection (MOI) in the appropriate medium for infection, which included an antibiotic for plasmid maintenance and 0.5 mM IPTG for constitutive expression of GFP. Infection of A. castellanii was carried out in Ac buffer as described previously (38) containing 0.5 mM IPTG and 5 μM chloramphenicol (Gibco). Prior to infection, cell attachment medium was removed and replaced with infection medium containing L. pneumophila strains at the desired MOIs. Plates were centrifuged for 10 min at 2,000 rpm. Intracellular multiplication was monitored automatically by measuring GFP fluorescence at an excitation of 485 nm and emission of 520 nm in a Tecan Infinite M200 plate reader every hour for 72 h. Fluorescence data, expressed in relative fluorescence units, was collected using Magellan software and exported to Microsoft Excel for background subtractions, time x/time zero calculations (to produce normalized relative fluorescence values), and graphical analysis. All intracellular multiplication experiments were performed at least three times, and the data shown are from one representative experiment performed in triplicate and averaged.
Northern blot analysis.
RNA was isolated by using TRIzol reagents as described by the manufacturer (Invitrogen). One microgram of RNA was separated on a 6% Tris-borate-EDTA-urea polyacrylamide gel (Invitrogen) and transferred to a positively charged nylon membrane (Micropore) using a semidry gel blotting system (Bio-Rad) for 20 min at 200 mA. Blots were prehybridized in Ultrahyb-Oligo (Ambion) for at least 1 h before overnight hybridization with 5′ biotin-labeled oligonucleotide probes (rsmY probe, 5′-biotin-GCAGCGAAGTACATCCTTTGTACTGGTCCCTTAGTTGACTTCCTGTCAGACATATCC; rsmZ probe, 5′-biotin-CGCAGTCATCCGTATAAGAACTTGCGTTCTTATTGTCATCCTGACAAATC). Blots were then washed two times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.5% sodium dodecyl sulfate, and the biotin probes were detected by using a Chemiluminescent Nucleic Acid Detection Module (Pierce) as directed by the manufacturer.
Microarray data accession number.
The microarray data developed in this study have been deposited in the NCBI Gene Expression Omnibus database under accession number GSE14830.
RESULTS
rpoS mutant intracellular multiplication phenotypes are not due to a defective Icm/Dot TFBSS.
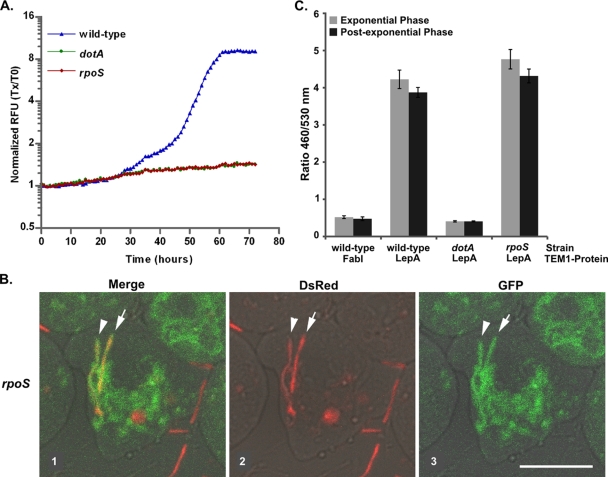
The Icm/Dot TFBSS is essential for the formation of the Legionella replicative vacuole and intracellular replication in all host cells (91). Mutations in the rpoS gene exhibit some, but not all, icm/dot mutant intracellular multiplication phenotypes (1, 38). Characterization of the phenotypes of the rpoS mutant required the use of multiple host cell types: A. castellanii was used to measure intracellular multiplication, D. discoideum was used for endocytic trafficking microscopy studies, and the J774 macrophage cell line was used to assay Icm/Dot-dependent translocation. To analyze the intracellular multiplication defect of an rpoS mutant, we measured intracellular growth by bacterial strains expressing GFP over 72 h using an automated fluorescence microplate spectrofluorometer. The data show that in A. castellanii, rpoS mutants have an intracellular multiplication defect that is indistinguishable from the Icm/Dot TFBSS mutant dotA (Fig. 1A; for supporting viable count data see Fig. S1A in the supplemental material). We tested whether rpoS mutants could block trafficking of the vacuolar ATPase to the LCV in the protozoan host D. discoideum. Using bacteria expressing cytoplasmic DsRed and D. discoideum expressing VatM-GFP, we observed for the icmT mutant and in contrast to the wild type (JR32) (18) that VatM-GFP association could be detected for some rpoS-containing phagosomes about 1 h after the cells and bacteria were mixed. By 4 h, most rpoS-containing phagosomes were surrounded by VatM-GFP, and many contained bacteria whose cytoplasmic DsRed marker had been largely destroyed, presumably because the bacteria were being digested by 4 h after infection (Fig. 1B), a behavior similar to the Icm/Dot TFBSS mutant icmT, a negative control (data not shown). If σS were required for the expression of a functional Icm/Dot system, it then would provide an explanation for the observed intracellular multiplication defects of rpoS mutants. Icm/Dot-dependent substrate translocation was tested using a previously described assay (24), in which Legionella strains harboring a TEM-1 (β-lactamase) fusion to an Icm/Dot substrate on a plasmid were used to infect host cells loaded with a fluorescent dye (CCF4-AM) containing a β-lactam ring, which, when cleaved by TEM-1, is converted from green (520 nm) to blue (460) fluorescence. We found that rpoS mutations did not result in reduced translocation of the Icm/Dot substrates LepA (Fig. 1C) and RalF (data not shown) into to J774 macrophages and that growth phase did not have a significant effect on Icm/Dot substrate translocation (Fig. 1C). This shows that the phenotypic similarities of rpoS and icm/dot mutants are not the result of an rpoS mutant Icm/Dot secretion defect. However, an alternative explanation is that an unknown rpoS-dependent factor may be required for Icm/Dot-dependent substrate translocation into amoebae but not macrophages.
FIG. 1.
Phenotypes of rpoS mutants. (A) Monolayers of A. castellanii were infected at an MOI of 1.0 with L. pneumophila harboring GFP on the plasmid pXDC31. Fluorescence was measured every 1 h for 72 h in a Tecan Infinite M200. Relative fluorescence units (RFU) were normalized to fluorescence at time zero for the wild type (JR32), the rpoS-null strain (blue line; LM1376), and the dotA-null strain (green line; LELA3118) Tx/T0, time x/time zero. (B) Dictyostelium amoeba expressing VatM-GFP were combined with the rpoS-null strain (LM1376) expressing cytoplasmic DsRed and analyzed after 4 h as described previously (18, 22). The left panel shows an overlay of GFP, monomeric red fluorescent protein, and a bright-field image; the middle panel shows monomeric red fluorescent protein and a bright-field overlay; and the right panel shows GFP and a bright-field overlay. Arrowheads indicate VatM-GFP-positive phagosomes containing bacteria. Scale marker, 10 μm. (C) Translocation of the Icm/Dot protein substrate LepA or the negative control FabI by KS79 (wild type), the dotA-null strain (GAH199), or ΔrpoS (GAH235) harboring TEM-1 fusion plasmids was measured by infecting J774 cells at an MOI of 50. Cells were loaded with CCF4-AM, and translocation was determined by measuring the ratio of cleaved (460 nm) to uncleaved (530 nm) CCF4-AM-labeled cells as described previously (17, 24). Error bars represent the standard deviation of triplicate samples from one experiment.
Mutation of the rpoS gene has widespread effects on L. pneumophila gene expression.
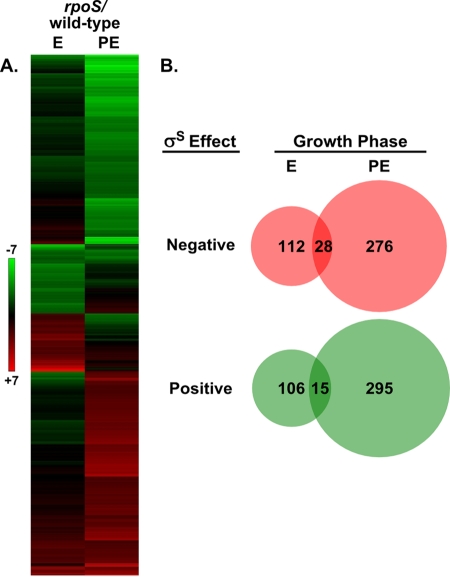
To investigate the regulatory networks associated with the intracellular multiplication defect of an rpoS mutant, we employed global gene expression analysis. RNA was isolated from six replicate cultures of both the wild type (JR32) and an isogenic rpoS mutant (LM1376) during exponential and postexponential growth in rich medium and used for microarray analysis (89). The log2 ratio of mutant/wild-type bacteria during exponential and postexponential growth for all the annotated ORFs in the L. pneumophila Philadelphia-1 genome is shown in Table S1 in the supplemental material. Genes whose steady-state transcript levels were significantly affected by the mutation of rpoS were defined as having a log2 ratio of mutant/wild-type bacteria greater than or equal to +2 or less than or equal to −2, i.e., a minimum of a fourfold change, with a P value of ≤0.005 (see Table S2 in the supplemental material). Positive values for the rpoS mutant/wild-type ratio indicate genes whose steady-state transcript level is negatively affected by σS, and negative values for the rpoS mutant/wild-type ratio indicate genes whose steady-state transcript level is positively affected by σS. This resulted in the identification of 739 rpoS-affected genes within the L. pneumophila genome (Fig. 2), which suggests that σS affects the transcription of approximately one-fourth of the 3,007 annotated genes (73). Sigma factors are generally associated with gene activation through transcription initiation (11); however, the data show that the global effects of σS are fairly balanced between positive and negative with 386 and 360 genes, respectively. This suggests that many of the observed effects might not occur through direct interaction of σS with the promoter of the affected gene but, rather, indirectly through additional regulators. As is true in other organisms, σS affects the expression of many more genes during the postexponential phase of growth (571) than during exponential phase (218) (42). This could be due to the postexponential phase accumulation of σS protein (38).
FIG. 2.
σS is a global regulator of L. pneumophila gene expression. The global gene expression profile of an rpoS-null mutant (LM1376) was compared to a wild-type strain (JR32) during exponential (E) and postexponential (PE) growth phases. Genes for which the ratio of the rpoS-null strain to the wild-type strain is −2 ≥ log2 ≥ +2 with a P value of ≤0.005 were clustered using Tibco Spotfire DecisionSite default settings (A). Green indicates reduced expression in the mutant, and red indicates increased expression in the mutant. The number of positively (green) and negatively (red) σS-affected genes in both growth phases (E and PE) are represented in a Venn diagram (B), which depicts all major types of σS regulation but does not illustrate the seven genes that are positively regulated in one phase of growth and negatively regulated in the other.
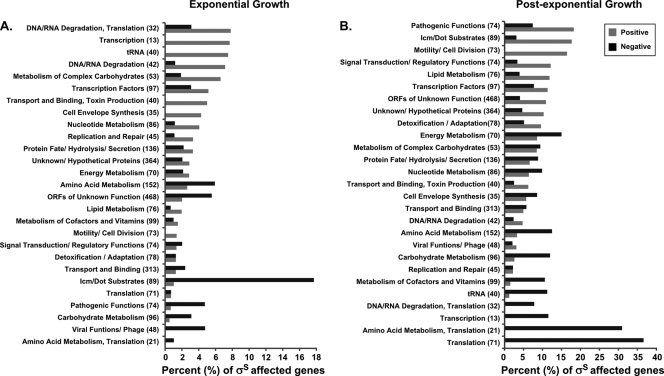
A global view of σS regulation was acquired by determining the percentage of σS-affected genes within each major category assigned to the annotated genome sequence of the L. pneumophila Philadelphila-1 (73) during exponential and postexponential growth (Fig. 3). An additional category was created to include the 89 proteins for which Icm/Dot-dependent translocation has been shown (3, 19, 23, 24, 52, 58, 61, 65, 75, 78, 94, 103, 108). In exponential phase, the steady-state transcript level of amino acid metabolism genes is negatively affected, whereas genes associated with DNA/RNA degradation, transcription, and tRNA synthesis are positively affected (Fig. 3A). In postexponential phase σS negatively affects the steady-state transcript level of genes in multiple categories associated with translation and metabolism while positively affecting genes categorized in pathogenic functions, motility, signal transduction, transcription factors, and substrates of the Icm/Dot system (Fig. 3B). Transcription of L. pneumophila secretion system genes (55, 66, 87), including icm/dot genes, was not significantly affected by mutation of rpoS in either growth phase (see Table S3 in the supplemental material). The absence of a defect in icm/dot gene expression is supported by the finding, shown above, that rpoS mutants are not defective in Icm/Dot-dependent substrate translocation (Fig. 1C).
FIG. 3.
σS affects the expression of genes in diverse functional categories. The percentage of σS-affected genes (x axis) within each functional category was identified from the rpoS mutant microarray based on the functional categories (y axis) assigned to the L. pneumophila Philadelphia-1 genome (20). The values are sorted from the highest percentage of positively affected genes (gray bars versus black bars for negatively affected) per category in both exponential and postexponential growth phases. The total number of genes present in each category is shown in parentheses; the σS-affected genes are shown as a percentage of this total. The category Icm/Dot substrates was created for this study. The difference in scale between panels A and B reflects σS regulatory effects.
Thus, during postexponential phase, σS does not affect L. pneumophila secretion system genes but participates in modulating the expression of genes associated with actively growing cells while enhancing the expression of motility genes, which are known to be induced during postexponential phase, and genes categorized with pathogenic functions. Many genes encoding Icm/Dot-translocated substrates were affected by σS during both growth phases.
σS affects the transcription of many genes encoding Icm/Dot-translocated substrates.
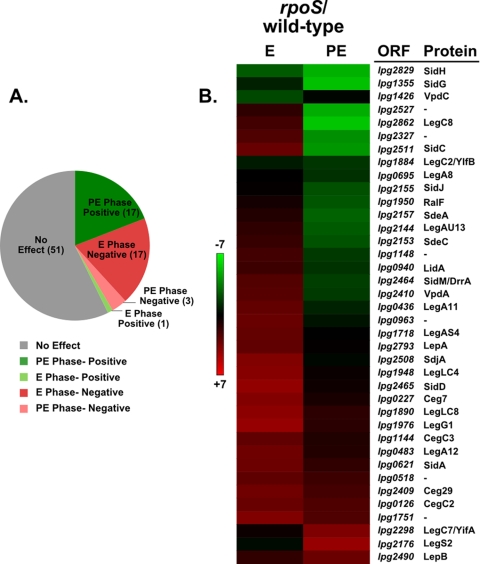
The Icm/Dot-dependent translocation of proteins from the bacterium to the host cell interior is proposed to determine the outcome of a Legionella infection (18, 24, 25). Some Icm/Dot-translocated proteins have established effects on host cell processes that are proposed to contribute to the establishment of the LCV (18, 48, 64, 75). Previous studies have implicated σS in the regulation of Icm/Dot substrate expression (25). In order to obtain a comprehensive understanding of the effects of σS on Icm/Dot substrate gene expression, a list of 89 proteins for which Icm/Dot-dependent translocation has been demonstrated was compiled (see Table S4 in the supplemental material). Analysis of this list showed that out of 89 Icm/Dot substrates, the transcription of 38 genes (43%) is significantly affected by mutation of rpoS (see Table S2 in the supplemental material). The major effects of σS are equally balanced between genes whose expression is negatively affected in exponential phase and those positively affected in postexponential phase, with 17 genes in each category (Fig. 4A). Transcription of many of the ceg genes is modulated by σS during exponential phase, whereas most sid and sde gene transcription is enhanced by σS in postexponential phase (Fig. 4B; see also Table S4 in the supplemental material). The transcription of only one Icm/Dot-translocated substrate gene, vpdC, was positively affected during exponential phase, and there were three negatively affected during postexponential phase, legC7, legS2, and lepB (Fig. 4; see also Table S4 in the supplemental material). Microarray results were validated by quantitative real-time qPCR for a subset of Icm/Dot-translocated substrate genes (see Table S5 in the supplemental material). The observed effect of σS on Icm/Dot substrate gene expression may contribute to the intracellular multiplication defects of rpoS mutants.
FIG. 4.
σS affects the expression of many Icm/Dot-translocated substrate genes. The expression of 89 genes whose protein products were previously shown to be translocated in an Icm/Dot-dependent manner (for references, see Table S4 in the supplemental material) were analyzed from the rpoS mutant microarray. The effects of σS on the 89 genes analyzed is depicted in a pie chart (A), and the 38 σS-affected Icm/Dot substrate genes are shown in a hierarchical clustering diagram along with ORF designations and protein names (if assigned) (B). E, exponential phase; PE, postexponential phase.
Transcription of the L. pneumophila csrB homologs, rsmY and rsmZ, requires σS.
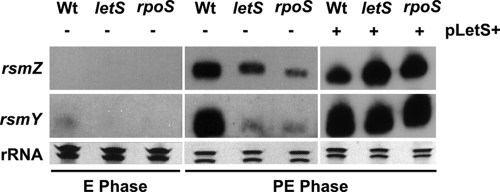
Efficient intracellular multiplication of L. pneumophila requires a functional LetAS two-component system (32, 62) as well as the regulator CsrA, which is responsible for the posttranscriptional repression of genes that promote postexponential phenotypes (5, 7, 71, 72). In other bacterial species, LetAS homologs (e.g., GacAS in P. aeruginosa [56, 104]) activate the postexponential transcription of homologs of the small RNA csrB, which sequesters CsrA proteins, thus relieving the repression of postexponential genes (4, 9, 41, 56, 71, 104). In L. pneumophila, a phenotypic link between LetAS and CsrA has been established and proposed to occur by the LetAS regulation of a csrB homolog (5, 7, 68, 71, 72). Recently, two L. pneumophila csrB homologs were predicted using a bioinformatics approach; these were named rsmY and rsmZ based on their structural similarity to the P. aeruginosa small RNAs (53). The microarray data showed a twofold reduction in letS transcripts in the rpoS mutant during postexponential phase (see Table S1 in the supplemental material). Although this suggests a possible role for σS in a pathway that controls CsrA, it is not considered significant based on the statistical criteria established in this study. To determine if σS affects the regulatory pathway that controls CsrA function in L. pneumophila, we investigated the expression of the crsB homologs.
To determine if the predicted small RNAs rsmY and rsmZ are transcribed in L. pneumophila, we performed Northern blot analysis on wild-type bacteria (JR32). These results demonstrated the postexponential production of rsmY and rsmZ transcripts near their predicted sizes of 106 nucleotides and 132 nucleotides, respectively (53) (Fig. 5). The L. pneumophila rsmY and rsmZ genes were further validated as homologs of csrB by demonstrating that their transcription is reduced in an letS mutant (GAH338) (Fig. 5). This is the expected result from studies of gacAS mutants in P. aeruginosa (56). We also found that overexpression of rsmY results in increased pigmentation (data not shown), consistent with the phenotype of an L. pneumophila csrA mutant (72). To determine if σS has a role in this pathway, we analyzed the amounts of rsmY and rsmZ transcripts in an rpoS mutant background (LM1376) and found that the levels of both transcripts were reduced in the mutant compared to the wild type during postexponential growth (Fig. 5). Interestingly, the levels of the rsmY and rsmZ transcripts were similarly reduced in both the letS and rpoS mutant backgrounds (Fig. 5). This suggests that LetS and σS might function in the same pathway. To analyze the relationship between σS and LetS in the csrB regulatory pathway, we overexpressed LetS in both the rpoS and letS mutant backgrounds. We found that this resulted in the recovery of wild-type rsmY and rsmZ transcript levels during postexponential phase in both mutants (Fig. 5). These data suggest that σS acts upstream of LetS in a regulatory circuit that results in increased postexponential transcription of L. pneumophila rsmY and rsmZ (see Fig. 7).
FIG. 5.
σS and LetS positively affect the transcription of the L. pneumophila csrB homologs rsmY and rsmZ. Transcripts of rsmY and rsmZ were detected by Northern blot analysis during exponential (E) and postexponential (PE) growth in wild-type (Wt; JR32), ΔletS (GAH338), and rpoS-null (LM1376) mutants. Wild-type (SPF38), ΔletS (SPF39), and rpoS-null (SPF40) strains harboring an inducible letS gene on the plasmid pSF21 (pLetS+) were induced with IPTG (1.0 mM), and transcripts were measured during the PE phase. Levels of RNA loaded are demonstrated by the amount of 23S and 16S rRNA observed from the ethidium bromide-stained gel.
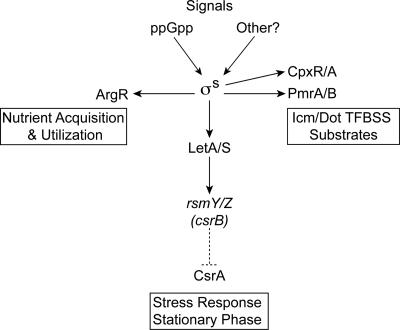
FIG. 7.
Model of σS effects on multiple pathways associated with intracellular multiplication. In the model, solid arrows extending from σS illustrate transcriptional effects, which may occur directly, indirectly, or through feed-forward control, demonstrated in this study by microarray, qPCR, or Northern blot analysis. The dashed line to CsrA indicates the predicted posttranscriptional effect of the csrB homologs, rsmY and rsmZ, identified in this study. The boxes suggest the predicted outcomes of the σS-affected pathway.
Identification of σS-affected transcription factors required for intracellular multiplication.
The widespread effects of σS can occur through its direct interaction with the promoter of an affected gene or indirectly by way of its effect on additional regulators. We mined the rpoS mutant microarray to identify σS-affected regulatory genes that may be required for intracellular multiplication. Genes encoding transcription factors whose expression is affected by σS were identified from the rpoS mutant global gene expression profile (see Table S2 in the supplemental material). Initially, 25 genes annotated as encoding transcription factors from the L. pneumophila genome (20) were identified. Of these, the 13 encoding putative DNA-binding domains were considered further (Table 2). Of the 13 putative transcription factors, 9 had not been studied previously and were deleted by allelic exchange (see Table S6 in the supplemental material). The four previously described transcription factors whose expression is affected by σS are fleQ, fliA, cpxR, and pmrA. It has been previously demonstrated that the postexponential transcription of the flagellar regulator genes fleQ and fliA is σS dependent and is not critical for L. pneumophila intracellular multiplication (50, 71). A new finding from this study is that the steady-state transcript levels of the response regulator genes of the two-component systems cpxRA and pmrAB are decreased by more than fourfold in an rpoS mutant compared to the wild type (Table 2; see also Table S5 in the supplemental material). CpxRA and PmrAB directly regulate the transcription of specific Icm/Dot-translocated substrate genes (3, 108), some of which were also shown to be affected by σS in this study. Although L. pneumophila cpxR mutants are not defective for intracellular replication, pmrA mutants are unable to replicate in A. castellanii (3, 108). Thus, reduced pmrA expression may contribute to the inability of the rpoS mutant to replicate in A. castellanii.
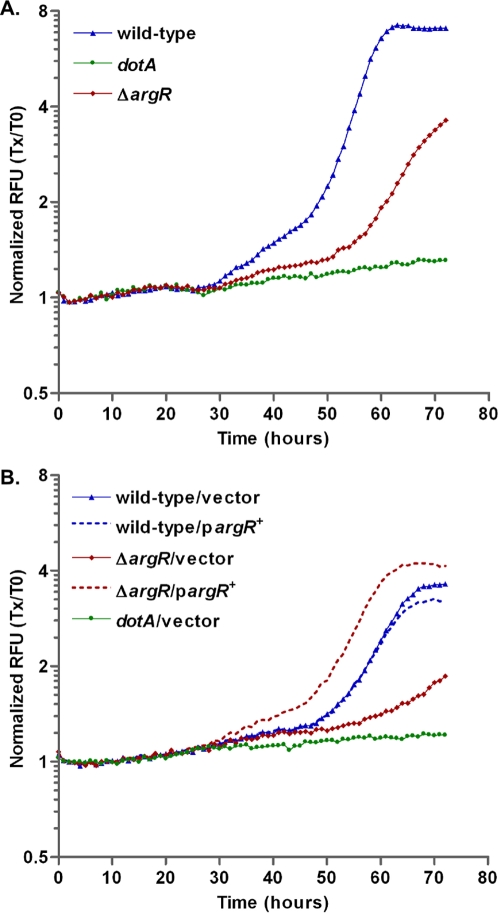
During the course of this investigation we developed a fluorescence-based method for the measurement of intracellular multiplication in which bacteria constitutively expressing a fluorescent marker (e.g., GFP) are used to infect host cells in a multiwell microplate. The level of fluorescence in each well can be monitored automatically at programmed intervals in a microplate fluorimeter (Fig. 1A). We found that the data obtained by this method agreed with data obtained by the traditional method of determining the number of CFU over time (see Fig. S1 in the supplemental material). The primary advantage of the fluorescence-based method, in contrast to viable-count assays, is that it permits the detailed side-by-side comparison of a large number of strains or conditions. We used this method to analyze the ability of the nine putative transcription factor mutants to replicate within the protozoan host A. castellanii. We found that mutation of the L. pneumophila arginine repressor homolog, argR (lpg0490), resulted in a reduction in intracellular multiplication in A. castellanii (Fig. 6A). None of the other mutants constructed in this study exhibited defects in intracellular multiplication (data not shown). The rpoS microarray data show that the steady-state transcript level of argR is up more than 12-fold during exponential phase and down by more than twofold during postexponential phase in the mutant (Table 2; see also Table S5 in the supplemental material). The intracellular multiplication defect of the argR mutant in A. castellanii was complemented in strains harboring an inducible argR gene (Fig. 6B). It is interesting that when the argR gene is expressed under ptac promoter control in the ΔargR mutant, there is an increase in intracellular multiplication compared to wild-type L. pneumophila (Fig. 6B). In contrast, when the same construct is present in the wild-type background, it results in a small but consistent reduction in intracellular multiplication (Fig. 6B). Although the mechanism for these phenotypes requires further investigation, it is possible that native transcriptional regulation of argR is required to accurately recapitulate the wild-type phenotypes. We conclude that derepression of the arginine regulon may contribute to the intracellular multiplication defect of the rpoS mutant. The role of the L. pneumophila arginine repressor and the argR regulon in intracellular multiplication is unclear at the present time and will require further investigation.
FIG. 6.
The L. pneumophila arginine repressor homolog, argR, is required for efficient intracellular replication in A. castellanii. Intracellular multiplication of L. pneumophila strains harboring GFP on plasmids was measured by monitoring fluorescence in a Tecan Infinite M200 plate reader every 2 h for 72 h. Monolayers of A. castellanii were infected at an MOI of 1.0 with wild-type (KS79), dotA-null strain, or ΔargR bearing pXDC31 (GFP+) (A). For complementation studies (B), GFP was produced from pGA123 (GFP+), and strains harbored either empty vector (vector; pMMB207c) or an argR overexpression vector (pargR+; pGAH125). All strains were induced with 0.5 mM IPTG. RFU, relative fluorescence units; Tx/T0, time x/time zero.
DISCUSSION
The σS factor has been widely studied in multiple species of gram-negative bacteria and has been shown to be important for the expression of virulence-related genes in several pathogens (49, 69, 76, 86). Although loss of the L. pneumophila rpoS gene has no noticeable effect on growth in complex medium, it results in severe defects in intracellular replication (5, 6, 38, 109) and endocytic trafficking that are not the result of impaired Icm/Dot TFBSS function (Fig. 1) or icm/dot gene expression (see Table S3 in the supplemental material). This study demonstrates that, in addition to genome-wide transcriptional effects on basic cellular processes (Fig. 2 and 3), σS affects the expression of genes associated with multiple pathways (Fig. 7) required for L. pneumophila intracellular multiplication including the following: (i) Icm/Dot-translocated substrates (Fig. 3), (ii) the two-component systems CpxRA and PmrAB (Table 2; see also Table S5 in the supplemental material), (iii) the small RNA csrB homologs rsmY and rsmZ (Fig. 5), and (iv) the L. pneumophila arginine repressor gene, argR, whose importance in intracellular multiplication was discovered in this study (Fig. 6).
The fact that the expression of more than one-fourth of all of the L. pneumophila predicted ORFs are affected in the rpoS mutant indicates that σS is a major regulator of L. pneumophila gene expression (Fig. 2). In E. coli, σS can partially replace the vegetative sigma factor σ70 under many stress conditions, resulting in the transcriptional activation of numerous σS-dependent genes (42). Expression of σS in E. coli is regulated by a complex array of signals including cell density, temperature, osmolarity, pH, and limiting concentrations of multiple nutrients (36, 42). However, unlike E. coli's survival, L. pneumophila's ability to survive stress in postexponential phase is largely σS independent (38), a phenotype similar to that observed in P. aeruginosa (98, 104), which suggests that the function of σS varies among bacterial genera. Despite this difference, the global gene expression patterns of E. coli and L. pneumophila rpoS mutants are comparable with respect to the large number of genes that are both positively and negatively regulated in both phases of growth (27, 80, 107). The extent of σS negative regulation may be due, in part, to the activation of transcriptional repressors such as the argR homolog identified in this study. Although the mechanism of σS action is likely conserved among bacterial species, the signals that control σS activity and the outcomes of that response vary depending on the lifestyle of the bacterium (2, 12, 38, 86, 104).
Although the focus of this study was the rpoS mutant intracellular replication defect, we found that σS also has notable effects on the transcription of genes associated with translation and metabolism. The transcription of more than 50 ribosomal protein, tRNA synthesis, and tRNA genes is negatively affected by σS during postexponential phase (Fig. 3B; see also Table S2 in the supplemental material). Reduced transcription of ribosomal protein genes and tRNA genes has been linked to the postexponential accumulation of the signaling molecule ppGpp and the stringent response to limiting amino acid availability (26, 79). In both L. pneumophila and E. coli, ppGpp results in σS accumulation; however, the stringent response and σS regulatory networks appear to be independent (1, 39, 81). It is interesting that with respect to amino acid biosynthetic gene transcription, the roles of ppGpp, which functions as a direct activator (81), and σS, whose effect is predominantly negative (Fig. 3B; see also Table S2 in the supplemental material), are opposite. The importance of amino acid metabolism regulation may be especially critical in Legionella because amino acids are its sole source of carbon and nitrogen and meet most of the bacterium's energy needs (34, 95). The transcription of more than 70 genes required for cellular metabolism is negatively affected during postexponential phase by σS (see Table S2 in the supplemental material), and approximately 40 of these genes are associated with amino acid metabolism (see Table S2 in the supplemental material). Understanding the biological consequences of σS effects on metabolism and translation will require further investigation.
The positive effects of σS during postexponential phase may determine the fate of L. pneumophila infection. During postexponential phase, σS positively affects the transcription of many genes categorized as “pathogenic function,” such as the enhanced entry system, as well as many genes encoding Icm/Dot-translocated substrates of L. pneumophila (Fig. 3B). Expression of the operon containing enhC, a Sel1-like tricopeptide repeat-containing protein that promotes the entry of L. pneumophila into host cells (21, 57), is reduced on the rpoS mutant microarray eightfold compared to the wild type during postexponential phase (see Table S2 in the supplemental material). Mutants of enhC also demonstrate increased stress sensitivity (57). Although rpoS mutants have a functional Icm/Dot TFBSS (Fig. 1B), they exhibit extensive alterations in the transcription of genes encoding Icm/Dot-translocated substrates (Fig. 4). This study shows that there are two major divisions of σS transcriptional effects on Icm/Dot-translocated substrates: genes negatively affected in exponential phase and those positively affected in postexponential phase (Fig. 5). It has been proposed that the large number of Icm/Dot substrates encoded in the L. pneumophila genome correlates with the broad host range of Legionella species and that subsets of the translocated proteins may influence vesicular trafficking in different hosts (77). In contrast to icm/dot mutants, rpoS mutants are able to replicate in stabilized macrophage cell lines (38). One possible explanation for this observation is that the rpoS mutant is defective for expression of genes encoding specific Icm/Dot substrates required for replication in amoebae and primary macrophages that are not required for replication in stabilized macrophage cell lines.
Previously, the response regulators CpxR and PmrA have been shown to directly regulate the transcription of genes encoding subsets of Icm/Dot substrates in L. pneumophila (3, 31, 108). This study links σS to both cpxR and pmrA (Table 2 and Fig. 7; see also Table S5 in the supplemental material) by showing that their transcription and the transcription of some of their Icm/Dot substrate target genes are affected by the mutation of rpoS. In L. pneumophila PmrA enhances the transcription of a number of genes encoding Icm/Dot-translocated substrates including sdeA, sdeC, sidG, and ceg23 (108), which we found are also positively affected by σS (Fig. 5B; see also Table S4 in the supplemental material). Transcripts of pmrA are reduced significantly in an rpoS mutant during postexponential growth (Table 2; see also Table S5 in the supplemental material), and both rpoS and pmrA mutants are defective for intracellular multiplication in unicellular protozoa (38, 108). The cpxR gene, whose transcription is also positively affected by σS (Table 2; see also Table S5 in the supplemental material) but is reported to not play a role in intracellular multiplication (3, 31), has the ability to activate or repress the transcription of its targets (3). Transcripts of the Icm/Dot-translocated substrate genes cegC2, ceg7, and legA11 are increased in both cpxR (3) and rpoS mutant backgrounds (Fig. 4B; see also Table S4 in the supplemental material), whereas transcription of the gene that encodes sidM-drrA, an Icm/Dot substrate that recruits Rab1 to Legionella-containing vacuoles (48, 64), is decreased in both cpxR and rpoS null mutants (3) (Fig. 4B; see also Table S4 in the supplemental material). Further mapping of the regulatory networks identified here will require comparative microarray analysis of cpxR, pmrA, and rpoS mutants.
It has been proposed that the postexponential accumulation of ppGpp observed in L. pneumophila activates two parallel pathways (1): one controlled by σS (1, 38, 68, 109) and another in which activation of the two-component system LetAS results in a reduction in functional CsrA protein (5, 6, 10, 39, 40, 62, 71, 72). However, previous studies have shown an overlap in the LetAS and σS regulatory outcomes that suggest that the pathways might be interconnected (5, 10, 68, 70, 71). In other bacteria the global regulator CsrA inhibits the translation of its targets by binding their mRNAs (82). CsrA repression is released following the transcription of the small RNA csrB, which can bind and inactivate multiple copies of CsrA protein (4, 82). Studies of CsrA in L. pneumophila suggest that it functions by a similar mechanism (5, 7, 29, 71, 72); however, a csrB homolog of the system had not been characterized. In this study we showed that the postexponential transcription of the two predicted L. pneumophila csrB (53) homologs, rsmY and rsmZ, is dependent upon both of the letS and rpoS genes and that overexpression of the letS gene can restore the transcription of rsmYZ in letS and rpoS mutant backgrounds (Fig. 5). These data are consistent with a unified pathway in which σS acts upstream of LetS in the postexponential induction of csrB homolog transcription, which is predicted to result in CsrA repression (5, 29, 41, 71, 72, 82) (Fig. 7).
To identify previously undiscovered pathways required for intracellular multiplication, putative transcription factor genes affected by σS (Table 2) were deleted and analyzed for their intracellular growth phenotypes. We discovered that the L. pneumophila arginine repressor homolog, encoded by the argR gene, is required for efficient intracellular replication in A. castellanii (Fig. 6). In other bacterial species the arginine repressor argR oligomerizes into a hexamer that binds DNA in the presence of arginine, resulting primarily in the repression of genes required for arginine biosynthesis (35, 63, 101, 102). Additional reported functions of argR include the positive regulation of arginine catabolism (59) and the resolution of ColE1 plasmid multimers (97). Legionella species are arginine auxotrophs that do not possess the N-acetlyglutamate synthetase enzyme required to complete the first step of arginine biosynthesis from glutamic acid (34, 100). Subsequent steps of arginine biosynthesis can be accomplished (100), and the genes required for the conversion of ornithine to arginine, argFGH, are divergent from argR (lpg0490) in a region that also includes a putative ABC transporter (lpg0491-lpg0496). In E. coli and P. aeruginosa, ArgR regulates the expression of a number of transporters required for the uptake of amino acids and other compounds (13, 60), and ArgR has been predicted to regulate arginine uptake from the host cell in some members of the Chlamydiaceae (88). It is intriguing to speculate that ArgR regulates one or more L. pneumophila transporters required for the acquisition of critical nutrients from the host cell. Determining the role of ArgR in Legionella biology and intracellular multiplication is the subject of further investigation.
This investigation utilized rpoS mutant phenotypes and global gene expression data to identify and connect regulatory networks required for L. pneumophila intracellular multiplication. The data presented here allow us to construct a basic model (Fig. 7) in which σS responds to signals, which likely include ppGpp (5, 71, 104, 109), to control the expression of downstream regulators such as the two-component systems CpxRA and PmrAB, which affect Icm/Dot substrate gene expression (3, 108), the arginine repressor ArgR, which we hypothesize may be involved in nutrient acquisition in the LCV, and the small RNAs rsmY and rsmZ, which are predicted to inhibit the function of the regulator CsrA and result in postexponential phase gene induction (4, 71, 82).
Supplementary Material
Acknowledgments
We thank Xavier Charpentier for supplying plasmids used in this work and for his constant encouragement and intellectual input. We thank Gloria Recio for technical assistance.
This work was supported by Public Health Service Grants AI 064481 and AI 23549 (H.A.S.) from the National Institutes of Health and NSF grant MCB 344541 (M.C.). G.A.H. was supported by Training Grant T32 AI007161. S.P.F. was supported by a postdoctoral fellowship from National Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abu-Zant, A., R. Asare, J. E. Graham, and Y. Abu Kwaik. 2006. Role for RpoS but not RelA of Legionella pneumophila in modulation of phagosome biogenesis and adaptation to the phagosomal microenvironment. Infect. Immun. 743021-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 691739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman, E., and G. Segal. 2008. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J. Bacteriol. 1901985-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babitzke, P., and T. Romeo. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10156-163. [DOI] [PubMed] [Google Scholar]
- 5.Bachman, M. A., and M. S. Swanson. 2004. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 722468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 401201-1214. [DOI] [PubMed] [Google Scholar]
- 7.Bachman, M. A., and M. S. Swanson. 2004. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect. Immun. 723284-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banga, S., P. Gao, X. Shen, V. Fiscus, W. X. Zong, L. Chen, and Z. Q. Luo. 2007. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. USA 1045121-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertani, I., M. Sevo, M. Kojic, and V. Venturi. 2003. Role of GacA, LasI, RhlI, Ppk, PsrA, Vfr and ClpXP in the regulation of the stationary-phase sigma factor rpoS/RpoS in Pseudomonas. Arch. Microbiol. 180264-271. [DOI] [PubMed] [Google Scholar]
- 10.Broich, M., K. Rydzewski, T. L. McNealy, R. Marre, and A. Flieger. 2006. The global regulatory proteins LetA and RpoS control phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of Legionella pneumophila JR32. J. Bacteriol. 1881218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79743-746. [DOI] [PubMed] [Google Scholar]
- 12.Caimano, M. J., C. H. Eggers, K. R. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 726433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldara, M., D. Charlier, and R. Cunin. 2006. The arginine regulon of Escherichia coli: whole-system transcriptome analysis discovers new genes and provides an integrated view of arginine regulation. Microbiology 1523343-3354. [DOI] [PubMed] [Google Scholar]
- 14.Campodonico, E. M., L. Chesnel, and C. R. Roy. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 56918-933. [DOI] [PubMed] [Google Scholar]
- 15.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 361165-1173. [DOI] [PubMed] [Google Scholar]
- 16.Charpentier, X., S. P. Faucher, S. Kalachikov, and H. A. Shuman. 2008. Loss of RNase R induces competence development in Legionella pneumophila. J. Bacteriol. 1908126-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 β-lactamase as a new fluorescence-based reporter. J. Bacteriol. 1865486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 3031358-1361. [DOI] [PubMed] [Google Scholar]
- 19.Chen, J., M. Reyes, M. Clarke, and H. A. Shuman. 2007. Host cell-dependent secretion and translocation of the LepA and LepB effectors of Legionella pneumophila. Cell Microbiol. 91660-1671. [DOI] [PubMed] [Google Scholar]
- 20.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 3051966-1968. [DOI] [PubMed] [Google Scholar]
- 21.Cirillo, S. L., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 1461345-1359. [DOI] [PubMed] [Google Scholar]
- 22.Clarke, M., J. Kohler, Q. Arana, T. Liu, J. Heuser, and G. Gerisch. 2002. Dynamics of the vacuolar H+-ATPase in the contractile vacuole complex and the endosomal pathway of Dictyostelium cells. J. Cell Sci. 1152893-2905. [DOI] [PubMed] [Google Scholar]
- 23.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48305-321. [DOI] [PubMed] [Google Scholar]
- 24.de Felipe, K. S., R. T. Glover, X. Charpentier, O. R. Anderson, M. Reyes, C. D. Pericone, and H. A. Shuman. 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 1877716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis, P. P., and M. Nomura. 1974. Stringent control of ribosomal protein gene expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 713819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong, T., M. G. Kirchhof, and H. E. Schellhorn. 2008. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol. Genet. Genomics 279267-277. [DOI] [PubMed] [Google Scholar]
- 28.Fields, B. S. 1996. The molecular ecology of Legionellae. Trends Microbiol. 4286-290. [DOI] [PubMed] [Google Scholar]
- 29.Forsbach-Birk, V., T. McNealy, C. Shi, D. Lynch, and R. Marre. 2004. Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int. J. Med. Microbiol. 29415-25. [DOI] [PubMed] [Google Scholar]
- 30.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, et al. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 2971189-1197. [DOI] [PubMed] [Google Scholar]
- 31.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 1854908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gal-Mor, O., and G. Segal. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34187-194. [DOI] [PubMed] [Google Scholar]
- 33.Gautier, L., L. Cope, B. M. Bolstad, and R. A. Irizarry. 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20307-315. [DOI] [PubMed] [Google Scholar]
- 34.George, J. R., L. Pine, M. W. Reeves, and W. K. Harrell. 1980. Amino acid requirements of Legionella pneumophila. J. Clin. Microbiol. 11286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grandori, R., T. A. Lavoie, M. Pflumm, G. Tian, H. Niersbach, W. K. Maas, R. Fairman, and J. Carey. 1995. The DNA-binding domain of the hexameric arginine repressor. J. Mol. Biol. 254150-162. [DOI] [PubMed] [Google Scholar]
- 36.Gyaneshwar, P., O. Paliy, J. McAuliffe, A. Jones, M. I. Jordan, and S. Kustu. 2005. Lessons from Escherichia coli genes similarly regulated in response to nitrogen and sulfur limitation. Proc. Natl. Acad. Sci. USA 1023453-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habyarimana, F., S. Al-Khodor, A. Kalia, J. E. Graham, C. T. Price, M. T. Garcia, and Y. A. Kwaik. 2008. Role for the ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ. Microbiol. 101460-1474. [DOI] [PubMed] [Google Scholar]
- 38.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 1814879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33721-731. [DOI] [PubMed] [Google Scholar]
- 40.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44107-118. [DOI] [PubMed] [Google Scholar]
- 41.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 141351-1363. [DOI] [PubMed] [Google Scholar]
- 42.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72165-168. [DOI] [PubMed] [Google Scholar]
- 44.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 1581319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 1582108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 991936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Investig. 66441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingmundson, A., A. Delprato, D. G. Lambright, and C. R. Roy. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450365-369. [DOI] [PubMed] [Google Scholar]
- 49.Iriarte, M., I. Stainier, and G. R. Cornelis. 1995. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun. 631840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobi, S., R. Schade, and K. Heuner. 2004. Characterization of the alternative sigma factor σ54 and the transcriptional regulator FleQ of Legionella pneumophila, which are both involved in the regulation cascade of flagellar gene expression. J. Bacteriol. 1862540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector PBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 52.Kubori, T., A. Hyakutake, and H. Nagai. 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 671307-1319. [DOI] [PubMed] [Google Scholar]
- 53.Kulkarni, P. R., X. Cui, J. W. Williams, A. M. Stevens, and R. V. Kulkarni. 2006. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 343361-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laguna, R. K., E. A. Creasey, Z. Li, N. Valtz, and R. R. Isberg. 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. USA 10318745-18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lammertyn, E., and J. Anne. 2004. Protein secretion in Legionella pneumophila and its relation to virulence. FEMS Microbiol. Lett. 238273-279. [DOI] [PubMed] [Google Scholar]
- 56.Lapouge, K., M. Schubert, F. H. Allain, and D. Haas. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behavior. Mol. Microbiol. 67241-253. [DOI] [PubMed] [Google Scholar]
- 57.Liu, M., G. M. Conover, and R. R. Isberg. 2008. Legionella pneumophila EnhC is required for efficient replication in tumor necrosis factor alpha-stimulated macrophages. Cell Microbiol. 101906-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu, Y., and Z. Q. Luo. 2007. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect. Immun. 75592-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu, C. D., and A. T. Abdelal. 1999. Role of ArgR in activation of the ast operon, encoding enzymes of the arginine succinyltransferase pathway in Salmonella typhimurium. J. Bacteriol. 1811934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu, C. D., Z. Yang, and W. Li. 2004. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J. Bacteriol. 1863855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lynch, D., N. Fieser, K. Gloggler, V. Forsbach-Birk, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219241-248. [DOI] [PubMed] [Google Scholar]
- 63.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Machner, M. P., and R. R. Isberg. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318974-977. [DOI] [PubMed] [Google Scholar]
- 65.Machner, M. P., and R. R. Isberg. 2006. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 1147-56. [DOI] [PubMed] [Google Scholar]
- 66.Marra, A., S. J. Blander, M. A. Horwitz, and H. A. Shuman. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. USA 899607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDade, J. E., C. C. Shepard, D. W. Fraser, T. R. Tsai, M. A. Redus, and W. R. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 2971197-1203. [DOI] [PubMed] [Google Scholar]
- 68.McNealy, T. L., V. Forsbach-Birk, C. Shi, and R. Marre. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J. Bacteriol. 1871527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Merrell, D. S., A. D. Tischler, S. H. Lee, and A. Camilli. 2000. Vibrio cholerae requires rpoS for efficient intestinal colonization. Infect. Immun. 686691-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Molofsky, A. B., L. M. Shetron-Rama, and M. S. Swanson. 2005. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect. Immun. 735720-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molofsky, A. B., and M. S. Swanson. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 5329-40. [DOI] [PubMed] [Google Scholar]
- 72.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50445-461. [DOI] [PubMed] [Google Scholar]
- 73.Morozova, I., X. Qu, S. Shi, G. Asamani, J. E. Greenberg, H. A. Shuman, and J. J. Russo. 2004. Comparative sequence analysis of the icm/dot genes in Legionella. Plasmid 51127-147. [DOI] [PubMed] [Google Scholar]
- 74.Muder, R. R., V. L. Yu, and G. D. Fang. 1989. Community-acquired Legionnaires' disease. Semin. Respir. Infect. 432-39. [PubMed] [Google Scholar]
- 75.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295679-682. [DOI] [PubMed] [Google Scholar]
- 76.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ninio, S., and C. R. Roy. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15372-380. [DOI] [PubMed] [Google Scholar]
- 78.Ninio, S., D. M. Zuckman-Cholon, E. D. Cambronne, and C. R. Roy. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55912-926. [DOI] [PubMed] [Google Scholar]
- 79.Nomura, M., R. Gourse, and G. Baughman. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 5375-117. [DOI] [PubMed] [Google Scholar]
- 80.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272580-591. [DOI] [PubMed] [Google Scholar]
- 81.Potrykus, K., and M. Cashel. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 6235-51. [DOI] [PubMed] [Google Scholar]
- 82.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 291321-1330. [DOI] [PubMed] [Google Scholar]
- 83.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 331179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28663-674. [DOI] [PubMed] [Google Scholar]
- 85.Roy, C. R., and L. G. Tilney. 2002. The road less traveled: transport of Legionella to the endoplasmic reticulum. J. Cell Biol. 158415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rychlik, I., and P. A. Barrow. 2005. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 291021-1040. [DOI] [PubMed] [Google Scholar]
- 87.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 615361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schaumburg, C. S., and M. Tan. 2006. Arginine-dependent gene regulation via the ArgR repressor is species specific in chlamydia. J. Bacteriol. 188919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270467-470. [DOI] [PubMed] [Google Scholar]
- 90.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 951669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Segal, G., and H. A. Shuman. 1998. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol. 6253-255. [DOI] [PubMed] [Google Scholar]
- 92.Sexton, J. A., and J. P. Vogel. 2004. Regulation of hypercompetence in Legionella pneumophila. J. Bacteriol. 1863814-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shin, S., and C. R. Roy. 2008. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 101209-1220. [DOI] [PubMed] [Google Scholar]
- 94.Shohdy, N., J. A. Efe, S. D. Emr, and H. A. Shuman. 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. USA 1024866-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sinai, A. P., and K. A. Joiner. 1997. Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu. Rev. Microbiol. 51415-462. [DOI] [PubMed] [Google Scholar]
- 96.Steinert, M., U. Hentschel, and J. Hacker. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26149-162. [DOI] [PubMed] [Google Scholar]
- 97.Stirling, C. J., G. Szatmari, G. Stewart, M. C. Smith, and D. J. Sherratt. 1988. The arginine repressor is essential for plasmid-stabilizing site-specific recombination at the ColE1 cer locus. EMBO J. 74389-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 1813890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54567-613. [DOI] [PubMed] [Google Scholar]
- 100.Tesh, M. J., and R. D. Miller. 1983. Arginine biosynthesis in Legionella pneumophila: absence of N-acetylglutamate synthetase. Can. J. Microbiol. 291230-1233. [DOI] [PubMed] [Google Scholar]
- 101.Tian, G., and W. K. Maas. 1994. Mutational analysis of the arginine repressor of Escherichia coli. Mol. Microbiol. 13599-608. [DOI] [PubMed] [Google Scholar]
- 102.Van Duyne, G. D., G. Ghosh, W. K. Maas, and P. B. Sigler. 1996. Structure of the oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol. 256377-391. [DOI] [PubMed] [Google Scholar]
- 103.VanRheenen, S. M., Z. Q. Luo, T. O'Connor, and R. R. Isberg. 2006. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect. Immun. 743597-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Venturi, V. 2003. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different? Mol. Microbiol. 491-9. [DOI] [PubMed] [Google Scholar]
- 105.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279873-876. [DOI] [PubMed] [Google Scholar]
- 106.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12259-265. [DOI] [PubMed] [Google Scholar]
- 107.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 1871591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zusman, T., G. Aloni, E. Halperin, H. Kotzer, E. Degtyar, M. Feldman, and G. Segal. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 631508-1523. [DOI] [PubMed] [Google Scholar]
- 109.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 18467-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.