Abstract
Brucellae are worldwide bacterial pathogens of livestock and wildlife, but phylogenetic reconstructions have been challenging due to limited genetic diversity. We assessed the taxonomic and evolutionary relationships of five Brucella species—Brucella abortus, B. melitensis, B. suis, B. canis, and B. ovis—using whole-genome comparisons. We developed a phylogeny using single nucleotide polymorphisms (SNPs) from 13 genomes and rooted the tree using the closely related soil bacterium and opportunistic human pathogen, Ochrobactrum anthropi. Whole-genome sequencing and a SNP-based approach provided the requisite level of genetic detail to resolve species in the highly conserved brucellae. Comparisons among the Brucella genomes revealed 20,154 orthologous SNPs that were shared in all genomes. Rooting with Ochrobactrum anthropi reveals that the B. ovis lineage is basal to the rest of the Brucella lineage. We found that B. suis is a highly divergent clade with extensive intraspecific genetic diversity. Furthermore, B. suis was determined to be paraphyletic in our analyses, only forming a monophyletic clade when the B. canis genome was included. Using a molecular clock with these data suggests that most Brucella species diverged from their common B. ovis ancestor in the past 86,000 to 296,000 years, which precedes the domestication of their livestock hosts. Detailed knowledge of the Brucella phylogeny will lead to an improved understanding of the ecology, evolutionary history, and host relationships for this genus and can be used for determining appropriate genotyping approaches for rapid detection and diagnostic assays for molecular epidemiological and clinical studies.
Highly contagious infections from bacteria in the genus Brucella are among the most ubiquitous and prevalent zoonotic diseases worldwide. Brucellae are a group of facultative intracellular alphaproteobacteria (32) that infect a range of mammalian livestock and wildlife, from cattle and pigs to seals and rodents, with most Brucella species occurring primarily in one or a few hosts. Establishing relationships within the genus has been challenging because of the relatively few genetic polymorphisms that distinguish each species (31). In fact, the genus was for a time classified as containing only one species, with a series of biovars (46). The genus can be distinguished by its 16S rRNA sequence (16), and the species and biovars can be differentiated with a range of traditional microbiological tests, serology, and phenotypic traits (8). Early DNA fragment analysis and sequencing demonstrated that Brucella typically contains distinct species-specific lineages (3, 11, 30). The weight of DNA evidence from an array of different loci upheld the traditional division of Brucella species (7), which led to a readoption of the classical species with a series of biovars (35). Nonetheless, the phylogenetic relationships among the Brucella species have remained poorly examined.
Determining the relationships among Brucella species is essential to understanding its ecology, evolutionary history, and host relationships and for developing accurate genotyping methods. Multilocus sequence typing, which assesses single nucleotide polymorphisms (SNPs) and other mutations in housekeeping genes, has revealed considerable variation among Brucella isolates that is taxonomically informative (48). Single SNPs can then be used to identify Brucella species because they are evolutionarily stable and can be incorporated into genotyping methods (13, 14, 41). Multilocus sequencing, however, does not capture enough variation in many species because conserved genomes often have too few polymorphic loci. Highly resolved phylogenies therefore depend on many loci, particularly in highly conserved genomes such as the brucellae.
Fortunately, the ability to create highly accurate, high-resolution phylogenies is rapidly increasing with ongoing developments in new sequencing technologies (17). Because of the relatively small size of their genomes, whole-genome phylogenies for bacteria show the greatest immediate potential for deciphering evolutionary histories at the species or genus level (2, 12, 19, 37). Rather than drawing phylogenetic inferences from a small portion of the genome, entire genomes can now be compared. Moreover, such in-depth work on a single genus differs from studies attempting to draw the tree of life using many but phylogenetically diverse genomes because of the far greater extent of genome coverage from SNP comparisons in similar genomes. Traditional whole-genome phylogenies involve comparisons of homologous genes (10, 27, 42). Among closely related species, SNPs appear to be a better choice for phylogenetics because of their coverage of the entire genome, relative stability over evolutionary time, ease of comparison, and inclusion of intergenic regions (4, 33). The sheer number of SNPs present between the genomes of closely related species can provide hundred to thousands of characters for phylogenetic reconstructions to resolve problems associated with character state conflict and create topologies with fine resolution. Furthermore, selecting only orthologous SNPs rather than including paralogous SNPs for analysis improves phylogenetic inference. Currently, whole-genome comparisons have only been done on a limited scale in Brucella, involving comparisons of two to three genomes (5, 9, 18, 36).
We compared the whole genomes of 13 Brucella isolates of five species: five genomes of Brucella suis from four of the five recognized biovars, three B. melitensis genomes from each of the three recognized biovars, three B. abortus genomes from the most widespread biovar, and one each from B. canis and B. ovis. We utilized only orthologous SNPs that were shared among all genomes. The phylogeny of these genomes was rooted with the closely related soil bacterium, Ochrobactrum anthropi, to polarize each SNP into ancestral or derived states. Finally, in pairwise comparisons of the genomes we utilized a molecular clock based on the accumulation of synonymous mutations to assess the relative age of the genus and divergence times of each species. The present study provides a solid and comprehensive phylogenetic framework that will serve as the basis for a detailed understanding of the evolution and ecology of Brucella, which is crucial for research in nearly all aspects Brucella biology.
MATERIALS AND METHODS
SNP discovery.
Orthologous SNPs were discovered by sequence comparisons of the 13 Brucella genomes available at the time of analysis (Table 1). Eight genomes from GenBank were generated by Sanger shotgun sequencing. Whole-genome sequences from an additional five unpublished genomes were determined by 454 pyrosequencing (40). Because these five genomes are currently unpublished and occur as contigs, we present the positions for SNPs from all genomes relative to the sequence of B. melitensis 16M (see Table S1 in the supplemental material).
TABLE 1.
Brucella genomes and O. anthropi outgroup used in phylogenetic comparisonsa
| Species | NCBI genome(s)b | Sequencing center |
|---|---|---|
| Brucella abortus 2308 | NC_007618, NC_007624 | Oak Ridge National Laboratory |
| Brucella abortus 9-941 | NC_006932, NC_006933 | University of Minnesota |
| Brucella abortus S19 | NC_010742, NC_010740 | Virginia Bioinformatics Institute |
| Brucella canis ATCC 23365 | NC_010103, NC_010104 | Joint Genome Institute |
| Brucella melitensis 16M | NC_003317, NC_003318 | Integrated Genomics |
| Brucella melitensis 63-9 | Unpublished data* | U.S. Department of Homeland Security |
| Brucella melitensis Ether | Unpublished data* | U.S. Department of Homeland Security |
| Brucella ovis ATCC 25840 | NC_009505, NC_009504 | The Institute for Genomic Research |
| Brucella suis 1330 | NC_004310.3, NC_004311.2 | The Institute for Genomic Research |
| Brucella suis 40 | Unpublished data* | U.S. Department of Homeland Security |
| Brucella suis 686 | Unpublished data* | U.S. Department of Homeland Security |
| Brucella suis ATCC 23445* | NC_010169, NC_010167 | Joint Genome Institute & Los Alamos National Lab |
| Brucella suis Thomsen* | Unpublished data* | U.S. Department of Homeland Security |
| Ochrobactrum anthropi ATCC 49188 | NC_009667, NC_009668 | Joint Genome Institute |
All of these genomes, except for B. suis 686, have two chromosomes.
*, SNPs are listed in Table S1 in the supplemental material.
We utilized an in-house pipeline for SNP discovery that used both PERL and Java Scripts for sequence comparisons and data parsing. Briefly, the pipeline compares genome sequences pairwise for sequence alignment using MUMmer (24) and then groups the SNPs by shared location for comparisons across all taxa. A sliding window for comparison of regions was used with the potential SNP flanked by 100 bases on each side. Repeated regions and paralogous genes were excluded from analysis. We defined homologous SNPs as those found in all genomes and paralogous if any SNP came from a region that had been duplicated. Orthologous SNPs were those homologous SNPs that remained after the paralogous SNPs were removed. We only included SNPs in the phylogeny that were present (both orthologous and shared) in all 13 genomes. For molecular clock estimations, the number of SNPs varied because pairwise comparisons were made between all genomes (see below).
To root the phylogeny, we performed the same comparison procedures but also included the O. anthropi genome. This allow us to polarize the Brucella SNP characters and precisely identify the most basal taxon (i.e., the root) of the phylogeny. We recognize that the choice of O. anthropi as an outgroup may affect which taxon is most basal due to potential issues of long-branch attraction (21), but it is the most closely related species that is currently known. The Brucella phylogeny itself was constructed using only SNPs shared among all Brucella genomes, increasing the number of shared loci and therefore allowing more detailed and accurate depiction of topology and branch lengths within the genus. Two genomes of B. suis are from the same isolate but were sequenced by two different labs with different sequencing strategies, providing a direct comparison of the sequencing/assembly approaches and a validation of our SNP discovery technique.
Phylogenetic reconstructions.
We generated a matrix of the SNP state for each genome that included the SNP position in the B. melitensis 16M genome as a reference and a mismatch cutoff value that indicated the proximity of the closest SNP. For the phylogeny, we excluded all SNPs with a mismatch cutoff of eight bases, meaning that if there were two SNPs within eight bases of each other, neither SNP was included in the analyses. This cutoff level allowed for the exclusion of potential sequencing errors typical in pyrosequencing such as issues with homopolymeric repeats and also excluded potential alignment errors, but it allowed retention of the majority of the data. As discussed more fully in Results, this mismatch cutoff did not affect the topology of the tree. We generated a nexus text file of concatenated SNP sequence for each sample. We analyzed the aligned sequence by using the neighbor-joining (NJ) and maximum-parsimony (MP) algorithms in PAUP* (43). The best substitution model was selected by using ModelTest (38). We used the following conditions for the analyses with the substitution model and parameters selected by ModelTest: NJ, general time reversible model, MP, full heuristic search with a random seed, and 1,000 bootstrap repetitions.
Molecular clock.
Estimation of the rate of evolution for a molecular clock requires knowledge of the number of synonymous SNPs (sSNPs), the number of potential sSNP sites, the mutation rate, and the number of generations per year. We first pared down the sequences of the genomes to include only coding regions, using genes from the B. melitensis 16M genome as the reference. The potential sSNP sites were calculated by first finding all three-base codons in the genes and determining which SNPs did not result in an amino acid change. All SNPs from potential SNP sites within these regions were summed to give a total number of sSNPs. The potential sSNP sites for each codon were calculated from a lookup table of codon possibilities and added together to give the number of potential synonymous SNP sites for all codons in the sequence. We chose sSNPs because presumably they are selectively neutral or nearly neutral and therefore allow for a relatively unbiased estimation of SNP accumulation.
We then made pairwise comparisons between all genomes, with the absolute base count being the total number of bases in all of the genes included in the pairwise comparisons and a filtered base count that included only bases of the genes that are shared, excluding indels. Thus, the SNPs used in these comparisons were slightly different than those used in the phylogeny because of the different requirements for SNP inclusion. The following equation was used to roughly determine the age of divergence for each pairwise comparison: the number of sSNPs/(the number of sSNP sites × the mutation rate × the number of generations per year × 2).
We used a synonymous mutation rate of 1.4 × 10−10 mutations per base pair per generation based on mutation rates from Escherichia coli (26). Age estimates are sensitive to the mutation rate because mutation rate estimates can exhibit considerable variation. The number of generations per year of Brucella species in natural hosts is not known, so we have given a range of possible generation times from 50 to 150, with the actual value yet to be determined in the wild. We recognize that this also introduces variation in the age estimates but that these values are between 22 and 43 generations per year in Bacillus anthracis (44) and between 100 and 300 generations per year in E. coli (34), which is consistent with Brucella biology. The “2” in the denominator of the equation is introduced to account for the time to divergence of the two genomes (1).
RESULTS
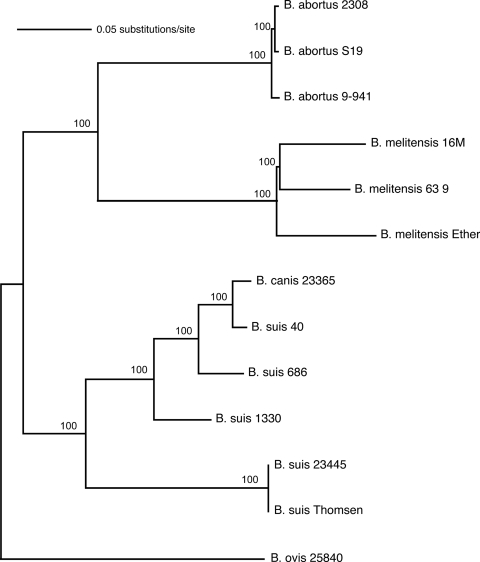
Comparisons of the 13 Brucella genomes with the outgroup O. anthropi yielded 181,685 polymorphic nucleotides that were shared among all genomes. Of this total, 172,598 SNPs separated O. anthropi from the Brucella genus, leaving only ∼9,000 SNPs among the Brucella species. Phylogenetic analysis including O. anthropi indicated that the B. ovis lineage was the first to split from the rest of the Brucella. Therefore, B. ovis was used to root subsequent trees constructed using only Brucella genomes. The exclusion of Ochrobactrum for SNP discovery within brucellae reduced homoplasy and yielded more SNPs for resolution of the genus. Alignments of the 13 Brucella genomes yielded 20,154 SNPs that were present in all genomes (Table 2). Of this total, 16,803 SNPs were in coding regions, and 3,351 were in noncoding regions. At least 1,398 SNPs were found on a different chromosome in at least one of the genomes (excluding B. suis 686, which has one genome). The reduced data set with a mismatch cutoff of eight bases (i.e., ignoring SNPs that are within 8 bp of one another) gave 17,032 SNPs, 9,021 of which were parsimony informative. In this data set, the incidence of homoplasy or possible sequencing error was extremely low (homoplasy index = 0.0104), excluding SNPs found only on terminal branches. The resulting Brucella phylogenetic tree shows strong differentiation by species (Fig. 1). Phylogenetic trees drawn with data having SNP mismatch cutoffs of 0 to 30 gave an identical topology but with slightly different branch lengths (data not shown), indicating that selecting a mismatch cutoff of 8 bp did not affect relationships. Only one possible tree emerged in NJ and MP analyses. Bootstrap support for MP was 100% for all nodes within the Brucella data set. With the O. anthropi outgroup in the analysis, support for the basal B. ovis clade was 99%. Maximum-likelihood analyses gave similar results (data not shown).
TABLE 2.
Number of SNPs defining the branches of Brucella phylogenya
| Genome group | No. of SNPs | SNP group |
|---|---|---|
| B. suis Thomsen | 3 | 1 |
| B. suis 686 | 577 | 2 |
| B. suis 40 | 184 | 3 |
| B. suis 23445 | 3 | 6 |
| B. suis Thomsen + B. suis 23445 | 2,370 | 7 |
| B. suis 1330 | 739 | 10 |
| B. ovis 25840 | 3,945 | 15 |
| B. melitensis Ether | 1,303 | 19 |
| B. melitensis 63-9 | 908 | 27 |
| B. melitensis 16M | 1,363 | 32 |
| B. melitensis 16M + B. melitensis 63-9 | 43 | 41 |
| B. melitensis 16M + B. melitensis 63-9 + B. melitensis Ether | 1,666 | 42 |
| B. canis 23365 | 253 | 54 |
| B. canis 23365 + B. suis 40 | 429 | 55 |
| B. canis 23365 + B. suis 40 + B. suis 686 | 584 | 56 |
| B. canis 23365 + B. suis 40 + B. suis 686 + B. suis 1330 | 893 | 61 |
| All B. suis + B. canis | 865 | 63 |
| All B. suis + B. canis + B. ovis | 968 | 69 |
| B. abortus 2308 + B. abortus 9-941 + B. abortus S19 | 2,295 | 98 |
| B. abortus S19 | 46 | 107 |
| B. abortus 9-941 | 91 | 130 |
| B. abortus S19 + B. abortus 2308 | 43 | 139 |
| B. abortus 2308 | 41 | 143 |
| Total | 19,612 |
The entire data set of 20,154 SNPs was used without any mismatch cutoff, indicating that 542 SNPs did not fall into these groupings. The SNP group is an arbitrary assignment number that designates each genome grouping and is based on information presented in Table S1 in the supplemental material.
FIG. 1.
Rooted phylogeny of the genus Brucella, including 13 genomes of five species. Tree was constructed by using neighbor joining. MP analysis gave the same topology, and percent bootstrap support based on 1,000 repetitions is shown at each node. The outgroup O. anthropi was used to root this tree but is not shown because of the long branch length.
The B. suis clade exhibited considerable genetic diversity, with the Thomsen strain (biovar 2, ATCC 23445) as the most basal and distantly related to the other strains in the clade. B. canis arose as a clone from within the B. suis clade and is relatively closely related to B. suis strain 40 (biovar 4). Only 253 SNPs (1.3% of total) from the full data set separate B. canis from the last common ancestor it shares with B. suis strain 40. This split was recent, occurring only ca. 7,500 to 22,500 years ago (Table 3). B. abortus and B. melitensis are sister species, with B. ovis more distantly related to all other sampled species. Within B. abortus, there is minimal genetic diversity within the three genomes from biovar 1, the most common and widespread biovar. The vaccine strain S19 is most closely related to strain 2308, although only 43 SNPs from the full data set separate this group from strain 9-941; S19 and 2308 strains diverged ca. 2,200 to 6,500 years ago based on pairwise comparisons. Genomes from the three biovars of B. melitensis have experienced considerable diversification since splitting from their common ancestor. B. melitensis 16M (biovar 1) is a sister taxon to B. melitensis 63-9 (biovar 2), but these two taxa diverged soon after their split from B. melitensis Ether (biovar 3).
TABLE 3.
Mean divergence times in years since the last common ancestor for five Brucella speciesa
| Common ancestor | Mean divergence time (yr) at:
|
||
|---|---|---|---|
| 50 G/yr | 100 G/yr | 150 G/yr | |
| B. abortus | 10,215 | 5,107 | 3,405 |
| B. canis/B. suis | 22,555 | 11,277 | 7,518 |
| B. melitensis | 92,474 | 46,237 | 30,825 |
| B. suis | 161,317 | 80,658 | 53,772 |
| B. abortus/B. melitensis | 200,404 | 100,202 | 66,801 |
| B. abortus/B. suis | 229,569 | 114,785 | 76,523 |
| B. melitensis/B. suis | 248,859 | 124,430 | 82,953 |
| B. ovis/B. suis | 259,214 | 129,607 | 86,405 |
| B. abortus/B. ovis | 278,347 | 139,174 | 92,782 |
| B. melitensis/B. ovis | 295,977 | 147,989 | 98,659 |
| B. suis/O. anthropi | 17,238,545 | 8,619,273 | 5,746,182 |
| B. melitensis/O. anthropi | 17,239,489 | 8,619,745 | 5,746,496 |
| B. abortus/O. anthropi | 17,246,753 | 8,623,376 | 5,748,918 |
| B. ovis/O. anthropi | 17,405,066 | 8,702,533 | 5,801,689 |
Times were estimated based on the molecular clock equation in the text, which includes a range of 50 to 150 generations (G) per year and a mutation rate per base pair per generation of 1.4 × 10−10.
DISCUSSION
Whole-genome analyses provide unprecedented phylogenetic resolution and the power to distinguish even extremely closely related isolates. For species or genera that have emerged relatively recently, whole genomes are necessary to provide fine-scale differentiation. With next-generation sequencing technologies making sequencing cheaper and faster, whole-genome phylogenies will soon become a reality for a growing number of bacterial species, as well as for Archaea, and eukaryotes with smaller genome sizes. In clonal or nearly clonal organisms, simple phylogenetic methods such as NJ or MP are optimal for tree reconstructions due to low levels of homoplasy. However, our approach of using shared orthologous SNPs from whole genomes will allow for phylogenetic reconstructions even in species that frequently recombine, such as Burkholderia spp. (T. Pearson, unpublished data).
SNP genotyping and analysis.
Our SNP discovery in these genomes allows for thousands of potential assays to differentiate between the various species. For example, we identified as many as 253 SNPs that distinguish B. canis 23365 from its closest sequenced relative B. suis 40, which includes the previously identified distinguishing mutation in outer membrane proteins (47), that could be used as targets for assays. For most branches, the SNPs defining them will be redundant and interchangeable for genotyping. Due to the extremely low rate of mutational change in SNPs, only a single SNP is necessary to define a particular clade and can then be designated as a canonical SNP (23).
The SNP data set identified in the present study contains a decided lack of evidence for recombination among Brucella species. Using the full data set with no SNP mismatch cutoff, we had one major pattern of shared SNPs (n = 248) that was inconsistent with the phylogeny. The following isolates grouped together: B. abortus 2308 and 9-941, B. canis 23365, B. ovis 25840, and B. suis 1330 and Thomsen. Notably, B. suis 23445 did not fall into this same group even though it is the same strain as Thomsen. None of these SNPs were retained with a mismatch cutoff of eight bases, and we know of no biological mechanism that would cause this pattern to occur. Lateral gene transfer from other organisms cannot be ruled out with this approach, although it is challenging to conceive a scenario where such anomalous results would be limited to a few taxa.
The three SNP differences between B. suis 23445 and B. suis Thomsen, the same type strain, are the result of either sequencing/alignment errors or mutations that have arisen during laboratory passage. Mutational differences from whole genome comparisons of the same strain are known to occur (45). In our case, the exact same archival isolate sample was not used. The true test is to sequence the exact same strain on different platforms (19). Nonetheless, such a small number of differences lends support to the accuracy of the 454 sequencing platform and to our SNP discovery methods.
Brucella phylogeny.
Ever since early microbiological work by Wilson (50), researchers have been developing increasingly sophisticated methods of classifying Brucella species. However, despite technical advances in genotyping, most methods have been able to roughly generate the same evolutionary relationships seen in whole genome phylogenies. For instance, the close relationship of B. abortus and B. melitensis and the more distant grouping of B. suis was suggested by restriction mapping (30). The basal position of B. ovis in the Brucella phylogeny was suggested based on the likely inheritance of certain genes (11). Multilocus sequence typing trees of Brucella roughly approximate the whole-genome phylogeny but use only seven housekeeping genes (48). Variable number tandem repeat analyses correctly group and depict the taxonomic relationships of all of the major Brucella clades, such as the close relationship of B. suis biovars 3 and 4 to B. canis and the close but more distant relationship of B. suis biovar 1 (20, 25, 49).
Although each of these approaches has its value, particularly when low-cost genotyping is the goal, only whole-genome sequencing can capture the full extent of genetic variation. Furthermore, only whole-genome phylogenies allow us to gauge the accuracy of previous genetic methods. Understanding the evolutionary framework of the genus Brucella is essential for designing assays that differentiate the various strains or biovars, and only by rooting our phylogeny can we understand the directionality of the evolutionary process. Incorrect conclusions about the relationships among Brucella isolates have inevitably been made because all prior attempts at phylogenetic constructions using data with reduced sets of markers are less accurate approximations of the “true” phylogeny than can be deduced from whole-genome analysis.
B. suis is the most diverse species within the Brucella thus far examined. Exceptional diversity in this clade was expected because our data set contained B. suis from four of the five recognized biovars. Furthermore, a range of genetic analyses have indicated considerable diversification within the B. suis clade and have even suggested likely relationships among the biovars (14, 20, 25). Most studies looking at variation within B. suis have had difficulty differentiating isolates from those of B. canis (3, 6, 14, 15), suggesting the close relationship of these two species. Using whole-genome comparisons, it is clear that B. canis, B. suis 686 (biovar 3), and B. suis 40 (biovar 4) are all highly similar at the nucleotide level. The species B. canis appears to have arisen directly from a B. suis ancestor, making currently defined B. suis isolates paraphyletic. Therefore, no single DNA-based assay will be able to distinguish all of the isolates in B. suis from the other Brucella species because the paraphyly of B. suis will cause such assays to also identify B. canis. Early fragment analysis by Allardet-Servent et al. (3) using restriction endonucleases also suggested that B. canis likely evolved from a strain of B. suis. Interestingly, SNPs were able to readily resolve the relationship of B. suis biovar 3 to the other Brucella species and B. suis biovars even though it contains only one large chromosome rather than the two chromosomes seen in all other Brucella (22). Regardless of the genome arrangement, B. suis biovar 3 is a subclade of B. suis. The genome of the only B. suis biovar not included in our analyses, biovar 5 (strain 513), which was isolated from rodents in the former Union of Soviet Socialist Republics, is likely quite different genetically than the other four biovars (25, 49). Previous research has suggested that B. suis biovar 5 is most closely related to the brucellae of marine mammals (14, 28, 48), but whole-genome-based phylogenetic analyses are needed to confirm this hypothesis.
The radiation of the three recognized biovars of B. melitensis occurred rapidly. These three strains are now clearly differentiated, but diverged at roughly the same time, and have undergone considerable evolution since divergence. The B. abortus clade was minimally differentiated, but the three strains from biovar 1 represent only a small portion of the diversity within this species.
Extremely low levels of character conflict within the tree suggest that all alternate phylogenetic methods give the same topology and similar branch lengths, indicating that the results are not an artifact of the analysis algorithm. Low amounts of genetic variation in Brucella are likely due to the relative youth of the lineage, as well as the lack of evidence of lateral gene transfer among Brucella species, although a few genomic islands consistent with horizontal transfer from other bacteria have been observed (36, 39). The genetic isolation of Brucella species is a result of their limited ecological niche, with fastidious growth only in hosts, few known mechanisms of genetic exchange, and virulence restricted to one or a few hosts (30). Whether the degree of differentiation in Brucella warrants species status for each traditional group has been debated over the years. The data from whole-genome phylogenies presented here resolve this issue; Brucella species are reproductively isolated and, with the exception of B. suis and B. canis, constitute reciprocally monophyletic lineages, separated by relatively long branches within the genus, and thus all species, including B. canis, are deserving of species status. In fact, several biovars within B. suis may be categorized as additional species, although all would be identical based on 16S rRNA, the standard method of bacterial identification.
Age and origin of Brucella species.
Previous whole-genome comparisons have indicated the close relationship of B. abortus and B. melitensis (5, 18), but the full phylogeny of the genus with B. ovis as the most basal species has not been previously described. Our rooted phylogeny suggests that brucellosis in animals such as pigs, goats, and cattle emerged from contact with infected sheep. Furthermore, this contact was recent, occurring roughly in the past 86,000 to 296,000 years. Our estimates, however, predate livestock domestication in the Middle East within the past 10,000 years (51), indicating that this disease was endemic within wildlife populations rather than emerging due to domestication. The coevolution of brucellae with their respective hosts (5, 31) is not consistent with the whole-genome phylogeny based on both topology and the likely rate of mutational change. For instance, it has been hypothesized that B. abortus and B. melitensis diverged roughly 20 million years ago with the divergence of their bovine and caprine (goats only) hosts, respectively (31). Similarly, the early differentiation of the genus has been speculated to have occurred 20 to 25 million years ago (29). However, the basal position of B. ovis in our phylogeny is distant from B. melitensis, even though their goat and sheep hosts are very closely related. Our data indicate a much more recent association, meaning independent acquisition of B. abortus, B. melitensis, and B. suis infections in their respective hosts after host speciation. Furthermore, Brucella as a genus is exceptionally monomorphic with relatively few SNPs, which strongly suggests that the entire lineage is considerably younger than previous estimates. Transmittal of brucellae from pigs to canids likely stemmed from infection of wolves or other canids feeding on the ancestor of B. suis 40 within the past 22,500 years. Why other Brucella species have not evolved within canids despite likely infections is unknown.
How the genomes of other Brucella species fit into the phylogeny described here will be extremely revealing for the evolutionary history of the genus. Our phylogeny and analyses provide the paradigm for phylogenetic differentiation among the Brucella. Genomes of other Brucella species such as B. neotomae, B. ceti, B. pinnipedialis, B. microti, and additional biovars of B. abortus will provide a more complete understanding of diversity and relationships in the genus; sequencing of these genomes is in progress (David O'Callaghan, unpublished data). In addition, sequencing of bacteria more closely related than O. anthropi will be able to resolve potential issues of long-branch attraction that can arise when distantly related taxa are used as outgroups (21). Among the many interesting avenues for future research in Brucella are the mechanisms of speciation. How did the various species adapt and become isolated in their respective hosts? Of particular interest is the relationship of marine and terrestrial brucellae, the timing of the emergence of the disease in marine organisms, and the evolutionary history of Brucella species that are currently limited to wildlife populations such as B. neotomae in wood rats, B. suis in caribou, and B. microti in voles. The genus also poses a challenge as to why the various brucellae have exhibited such distinct host preferences in some species but not in others.
Supplementary Material
Acknowledgments
This study was supported by the U.S. Department of Homeland Security. Sequencing of the B. canis genome was funded by the Intelligence Technology Innovation Center.
We thank Jim Burans and the staff at the National Bioforensics Analysis Center for the 454 pyrosequencing data.
Use of product or trade names does not constitute endorsement by the U.S. Government.
Footnotes
Published ahead of print on 6 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 10117837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 1853392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allardet-Servent, A., G. Bourg, M. Ramuz, M. Pages, M. Bellis, and G. Roizes. 1988. DNA polymorphism in strains of the genus Brucella. J. Bacteriol. 1704603-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumfield, R. T., P. Beerli, D. A. Nickerson, and S. V. Edwards. 2003. The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol. Evol. 18249-256. [Google Scholar]
- 5.Chain, P. S. G., D. J. Comerci, M. E. Tolmasky, F. W. Larimer, S. A. Malfatti, L. M. Vergez, F. Aguero, M. L. Land, R. A. Ugalde, and E. Garcia. 2005. Whole-genome analyses of speciation events in pathogenic brucellae. Infect. Immun. 738353-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloeckaert, A., J. M. Verger, M. Grayon, and O. Grepinet. 1995. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology 1412111-2121. [DOI] [PubMed] [Google Scholar]
- 7.Cloeckaert, A., and N. Vizcaino. 2004. DNA polymorphism and taxonomy of Brucella species, p. 1-24. In I. Lopez-Goni and I. Moriyon (ed.), Brucella: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 8.Corbel, M. J., and W. J. Brinley-Morgan. 1984. Genus Brucella Meyer and Shaw 1920. The Williams & Wilkins Co., Baltimore, MD.
- 9.Crasta, O. R., O. Folkerts, Z. Fei, S. P. Mane, C. Evans, S. Martino-Catt, B. Bricker, G. Yu, L. Du, and B. W. Sobral. 2008. Genome sequence of Brucella abortus vaccine strain S19 compared to virulent strains yields candidate virulence genes. PLoS ONE 3e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen, J. A. 1998. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 8163-167. [DOI] [PubMed] [Google Scholar]
- 11.Ficht, T. A., H. S. Husseinen, J. Derr, and S. W. Bearden. 1996. Species-specific sequences at the omp2 locus of Brucella type strains. Int. J. Syst. Evol. Microbiol. 46329-331. [DOI] [PubMed] [Google Scholar]
- 12.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis. J. Bacteriol. 1883162-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, J., R. Okinaka, R. Svensson, K. Shaw, B. De, R. Robison, W. Probert, L. Kenefic, W. Brown, and P. Keim. 2008. Real-time PCR assays of single-nucleotide polymorphisms defining the major Brucella clades. J. Clin. Microbiol. 46296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fretin, D., A. M. Whatmore, S. Al Dahouk, H. Neubauer, B. Garin-Bastuji, D. Albert, M. Van Hessche, M. Menart, J. Godfroid, K. Walravens, and P. Wattiau. 2008. Brucella suis identification and biovar typing by real-time PCR. Vet. Microbiol. 131376-385. [DOI] [PubMed] [Google Scholar]
- 15.Gandara, B., A. L. Merino, M. A. Rogel, and E. Martinez-Romero. 2001. Limited genetic diversity of Brucella spp. J. Clin. Microbiol. 39235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gee, J. E., B. K. De, P. N. Levett, A. M. Whitney, R. T. Novak, and T. Popovic. 2004. Use of 16S rRNA gene sequencing for rapid confirmatory identification of Brucella isolates. J. Clin. Microbiol. 423649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, N. 2007. Advanced sequencing technologies and their wider impact in microbiology. J. Exp. Biol. 2101518-1525. [DOI] [PubMed] [Google Scholar]
- 18.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 1872715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holt, K. E., J. Parkhill, C. J. Mazzoni, P. Roumagnac, F. X. Weill, I. Goodhead, R. Rance, S. Baker, D. J. Maskell, J. Wain, C. Dolecek, M. Achtman, and G. Dougan. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 40987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huynh, L. Y., M. N. Van Ert, T. Hadfield, W. S. Probert, B. H. Bellaire, M. Dobson, R. J. Burgess, R. S. Weyant, T. Popovic, S. Zanecki, D. M. Wagner, and P. Keim. 2008. Multiple locus variable number tandem repeat (VNTR) analysis (MLVA) of Brucella spp. identifies species-specific markers and provides insights into phylogenetic relationships, p. 47-54. In V. St. Georgiev (ed.), National Institute of Allergy and Infectious Disease, NIH: frontiers in research. Humana Press, Totowa, NJ.
- 21.Johannes, B. 2005. A review of long-branch attraction. Cladistics 21163-193. [DOI] [PubMed] [Google Scholar]
- 22.Jumas-Bilak, E., S. Michaux-Charachon, G. Bourg, M. Ramuz, and A. Allardet-Servent. 1998. Unconventional genomic organization in the alpha subgroup of the proteobacteria. J. Bacteriol. 1802749-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keim, P., M. N. Van Ert, T. Pearson, A. J. Vogler, L. Y. Huynh, and D. M. Wagner. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4205-213. [DOI] [PubMed] [Google Scholar]
- 24.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Fleche, P., I. Jacques, M. Grayon, S. Al Dahouk, P. Bouchon, F. Denoeud, K. Nockler, H. Neubauer, L. A. Guilloteau, and G. Vergnaud. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenski, R. E., C. L. Winkworth, and M. A. Riley. 2003. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J. Mol. Evol. 56498-508. [DOI] [PubMed] [Google Scholar]
- 27.Lerat, E., V. Daubin, and N. A. Moran. 2003. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-proteobacteria. PLoS Biol. 1E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marianelli, C., F. Ciuchini, M. Tarantino, P. Pasquali, and R. Adone. 2006. Molecular characterization of the rpoB gene in Brucella species: new potential molecular markers for genotyping. Microbes Infect. 8860-865. [DOI] [PubMed] [Google Scholar]
- 29.Meyer, M. E. 1976. Evolution and taxonomy in the genus Brucella: concepts on the origins of the contemporary species. Am. J. Vet. Res. 37199-202. [PubMed] [Google Scholar]
- 30.Michaux-Charachon, S., G. Bourg, E. Jumas-Bilak, P. Guigue-Talet, A. Allardet-Servent, D. O'Callaghan, and M. Ramuz. 1997. Genome structure and phylogeny in the genus Brucella. J. Bacteriol. 1793244-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno, E., A. Cloeckaert, and I. Moriyon. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90209-227. [DOI] [PubMed] [Google Scholar]
- 32.Moreno, E., E. Stackebrandt, M. Dorsch, J. Wolters, M. Busch, and H. Mayer. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 1723569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morin, P. A., G. Luikart, R. K. Wayne, and the SNP workshop group. 2004. SNPs in ecology, evolution, and conservation. Trends Ecol. Evol. 19208-216. [Google Scholar]
- 34.Ochman, H., S. Elwyn, and N. A. Moran. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 9612638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osterman, B., and I. Moriyon. 2006. International committee on systematics of prokaryotes: subcommittee on the taxonomy of Brucella. Int. J. Syst. Evol. Microbiol. 561173-1175. [Google Scholar]
- 36.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 9913148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson, T., J. D. Busch, J. Ravel, T. D. Read, S. D. Rhoton, J. M. U'Ren, T. S. Simonson, S. M. Kachur, R. R. Leadem, M. L. Cardon, M. N. Van Ert, L. Y. Huynh, C. M. Fraser, and P. Keim. 2004. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl. Acad. Sci. USA 10113536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 39.Ratushna, V. G., D. M. Sturgill, S. Ramamoorthy, S. A. Reichow, Y. Q. He, R. Lathigra, N. Sriranganathan, S. M. Halling, S. M. Boyle, and C. J. Gibas. 2006. Molecular targets for rapid identification of Brucella spp. BMC Microbiol. 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronaghi, M., M. Uhlen, and P. Nyren. 1998. A sequencing method based on real-time pyrophosphate. Science 281363. [DOI] [PubMed] [Google Scholar]
- 41.Scott, J. C., M. S. Koylass, M. R. Stubberfield, and A. M. Whatmore. 2007. Multiplex assay based on single-nucleotide polymorphisms for rapid identification of Brucella isolates at the species level. Appl. Environ. Microbiol. 737331-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snel, B., P. Bork, and M. A. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21108-110. [DOI] [PubMed] [Google Scholar]
- 43.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0. Sinauer Associates, Sunderland, MA.
- 44.Van Ert, M. N., W. R. Easterday, L. Y. Huynh, R. T. Okinaka, M. E. Hugh-Jones, J. Ravel, S. R. Zanecki, T. Pearson, T. S. Simonson, J. M. U'Ren, S. M. Kachur, R. R. Leadem-Dougherty, S. D. Rhoton, G. Zinser, J. Farlow, P. R. Coker, K. L. Smith, B. Wang, L. J. Kenefic, C. M. Fraser-Liggett, D. M. Wagner, and P. Keim. 2007. Global genetic population structure of Bacillus anthracis. PLoS ONE 2e461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velicer, G. J., G. Raddatz, H. Keller, S. Deiss, C. Lanz, I. Dinkelacker, and S. C. Schuster. 2006. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc. Natl. Acad. Sci. USA 1038107-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verger, J. M., F. Grimont, P. A. D. Grimont, and M. Grayon. 1985. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 35292-295. [Google Scholar]
- 47.Vizcaino, N., P. Caro-Hernandez, A. Cloeckaert, and L. Fernandez-Lago. 2004. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6821-834. [DOI] [PubMed] [Google Scholar]
- 48.Whatmore, A. M., L. L. Perrett, and A. P. MacMillan. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whatmore, A. M., S. J. Shankster, L. L. Perrett, T. J. Murphy, S. D. Brew, R. E. Thirlwall, S. J. Cutler, and A. P. MacMillan. 2006. Identification and characterization of variable-number tandem-repeat markers for typing of Brucella spp. J. Clin. Microbiol. 441982-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, G. S. 1933. The classification of the Brucella group: a systematic study. J. Hyg. 33516-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeder, M. A. 2008. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc. Natl. Acad. Sci. USA 10511597-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.