Abstract
Bacillus subtilis contains two nitrogen transcription factors, GlnR and TnrA. The activities of GlnR and TnrA are regulated by direct protein-protein interactions with the feedback-inhibited form of glutamine synthetase (GS). To look for other factors involved in regulating GlnR activity, we isolated mutants with constitutive glnRA expression (GlnC). The twenty-seven GlnC mutants isolated in this mutant screen all contained mutations tightly linked to the glnRA operon which encodes GlnR (glnR) and GS (glnA). Four GlnC mutants contained mutations in the glnR gene that most likely impair the ability of GlnR to bind DNA. Three other GlnC mutants contained novel glnA mutations (S55F, V173I, and L174F). GlnR regulation was completely relieved in the three glnA mutants, while only modest defects in TnrA regulation were observed. In vitro enzymatic assays showed that the purified S55F mutant enzyme was catalytically defective while the V173I and L174F enzymes were highly resistant to feedback inhibition. The V173I and L174F GS proteins were found to require higher glutamine concentrations than the wild-type GS to regulate the DNA-binding activities of GlnR and TnrA in vitro. These results are consistent with a model where feedback-inhibited GS is the only cellular factor involved in regulating the activity of GlnR in B. subtilis.
Glutamine synthetase (GS) is a metalloenzyme that catalyzes the ATP-dependent synthesis of glutamine from glutamate and ammonium. While Mg2+ or Mn2+ can support GS activity in vitro, the Mg2+-dependent reaction is the physiologically significant catalytic activity (37, 50). Glutamine is a key nitrogen metabolite that serves as the nitrogen donor for the synthesis of 25% of the nitrogen-containing compounds in the cell (36). To ensure that cells contain sufficient glutamine for optimal growth under all conditions, both the activity and expression of GS are tightly regulated (36).
In the low-G+C, gram-positive bacterium Bacillus subtilis, the enzymatic activity of GS is controlled by feedback inhibition. Glutamine is the principal inhibitor of the physiologically relevant Mg2+-dependent reaction, although other nitrogen-containing compounds such as AMP inhibit GS activity in vitro (12). In B. subtilis, the GlnR and TnrA transcription factors control gene expression in response to nitrogen availability (15, 41, 49). The genes for GlnR (glnR) and GS (glnA) are located together within the glnRA operon, while the tnrA gene is monocistronic (45, 49). GlnR and TnrA are active under different growth conditions (41, 49). GlnR is active during growth with excess nitrogen, where it represses the expression of glnRA and several other genes (7, 22, 41, 48, 57). In contrast, TnrA is active during nitrogen-limited growth, where it activates and represses the expression of genes involved in the transport and metabolism of nitrogen compounds (4, 5, 14, 18, 29, 38, 47, 49, 56, 57).
Initial observations that GlnR- and TnrA-regulated genes are transcribed constitutively in glnA null mutants indicated that GS is required for the regulation of these transcription factors in response to cellular nitrogen availability (1, 10, 14, 17, 23, 29, 42, 44). Subsequently, the feedback-inhibited form of GS (FBI-GS) was shown to control the activities of TnrA and GlnR through direct protein-protein interactions. FBI-GS is only present in cells growing with excess nitrogen. TnrA is inactive under these growth conditions due to the formation of a stable complex between FBI-GS and TnrA that sequesters TnrA and inhibits its binding to DNA (55). In contrast, when nitrogen is in excess, FBI-GS activates GlnR DNA binding through a transient association where FBI-GS acts as a chaperone that stabilizes GlnR-DNA complexes (20). Thus, the feedback inhibition of GS plays a central role in nitrogen metabolism in B. subtilis because it not only controls glutamine synthesis but also serves as the nitrogen signal for regulating the activity of GlnR and TnrA.
Examination of gene expression in Escherichia coli revealed that the expression of nitrogen-regulated genes is activated sequentially during the transition from nitrogen-excess growth to nitrogen starvation. During this transition, the expression of GS is elevated prior to the increase in expression of gene products that generate ammonium due to the uptake and catabolism of nitrogen-containing compounds (3, 36). Nitrogen-regulated E. coli promoters are activated by the phosphorylated form of NRI (also called NtrC) which is encoded in the glnA-ntrBC operon (36). The differential response of NRI-regulated promoters during nitrogen limitation was found to result from alterations in the intracellular concentrations of NRI (36). NRI levels are low during growth with excess nitrogen. During the initial transition to nitrogen limitation, these low levels of NRI are sufficient to activate expression of the glnA-ntrBC operon, which contains two high-affinity NRI sites in the glnAp2 promoter (31, 40). The resulting elevated expression of the glnA-ntrBC operon increases the intracellular concentration of NRI and allows activation of promoters with low-affinity NRI-binding sites (3, 34).
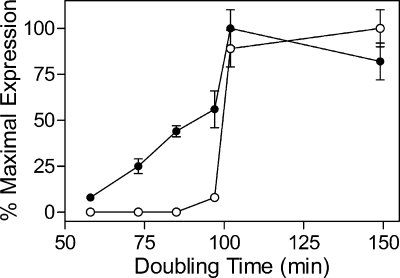
Differential activation of nitrogen-regulated gene expression during nitrogen limitation also occurs in B. subtilis. During growth in glucose minimal medium containing different nitrogen sources which support progressively slower growth rates, the expression of GS is activated before the expression of genes required for the production of ammonium from other nitrogen-containing compounds is increased (Fig. 1) (1). Although only FBI-GS is known to control the activity of GlnR and TnrA, the expression levels of GlnR- and TnrA-dependent regulated genes do not respond identically to growth on different nitrogen sources. For instance, in cells grown on glucose minimal medium with urea as the nitrogen source, the level of GlnR-regulated GS is 56% of its maximal value while the level of a TnrA-regulated amtB-lacZ gene fusion is only 8% of its maximal value (Fig. 1). TnrA is the only factor known to regulate amtB expression, while glnRA expression is repressed by GlnR during growth with excess nitrogen and weakly repressed by TnrA during nitrogen limitation (41, 49, 57).
FIG. 1.
Expression of GlnR- and TnrA-regulated genes in cultures grown with different nitrogen sources. The levels of GlnR-dependent GS (•) and TnrA-dependent amtB (○) expression determined in each culture is plotted with respect to the doubling time of the culture. The nitrogen sources and culture doubling times are as follows: glutamine, 58 min; glutamate plus ammonium, 73 min; ammonium, 85 min; urea, 97 min; proline, 100 min; and glutamate, 150 min. GS specific activity was determined in permeabilized cells, while amtB (formerly nrg-29) expression was determined using a amtB-lacZ fusion. The error bars correspond to the standard errors of the means. The data presented in this figure were taken from Atkinson and Fisher (1).
The mechanism(s) responsible for the differential response of GlnR- and TnrA-regulated genes during the transition to nitrogen-limited growth in B. subtilis is not understood. One possible explanation is that while there are additional factors which assist in the activation of GlnR DNA binding in vivo, these factors are not involved in the regulation of TnrA. To search for additional factors involved in GlnR-mediated regulation, we isolated B. subtilis mutants with derepressed levels of glnRA expression. Interestingly, the three novel glnA mutants isolated in this screen were found to relieve GlnR-dependent repression of gene expression but to have only a minor effect on TnrA-mediated regulation.
MATERIALS AND METHODS
Bacterial strains, cell growth, and media.
Table 1 lists the B. subtilis strains used in this study. The methods used for bacterial cultivation in the minimal medium of Neidhardt et al. (30) have been reported elsewhere (2). Balanced salt solution minimal medium agar plates were prepared as previously described (8). Glucose was added to all media at a final concentration of 0.5%. All nitrogen sources were added to a final concentration of 0.2%. 5-Bromo-4-chloro-3-indoyl-β-d-galactoside (X-Gal) was added to agar plates to give a final concentration of 40 μg/ml. The ability of mutant strains to crossfeed a Gln mutant was examined with a previously described plate assay (50).
TABLE 1.
Strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| 168 | trpC2 | Our laboratory |
| SF14A | ΔglnA14::spc trpC2 | 49 |
| SF21 | amyE::[(glnRA-lacZ)21 cam] trpC2 | 168 × pGLN21 DNA |
| SF21A | amyE::[(glnRA-lacZ)21 cam] ΔglnA14::spc trpC2 | SF21 × SF14A DNA |
| SF21AT | amyE::[(glnRA-lacZ)21 cam] ΔglnA14::spc tnrA62::Tn917 trpC2 | SF21A × SF62 DNA |
| SF62 | tnrA62::Tn917 trpC2 | 49 |
| SF402 | amyE::[(amtB-lacZ)402 cam] trpC2 | 168 × pNRG402 DNA |
| SF402A | amyE::[(amtB-lacZ)402 cam] ΔglnA14::spc trpC2 | SF402 × SF14A DNA |
| SF517 | lacA::[(glnRA-lacZ)517 neo] trpC2 | 51 |
| SF517T | lacA::[(glnRA-lacZ)517 neo] tnrA62::Tn917 trpC2 | 51 |
Plasmid constructions.
Plasmids pSFL1 and pSFL2 are chloramphenicol-resistant lacZ transcriptional fusion vectors that integrate into the amyE gene (52). Plasmid pGLN21 contains a glnRA-lacZ fusion that was constructed by inserting a glnRA promoter fragment from pGLN16 (57) into pSFL2. This glnRA promoter fragment extends from −77 to + 80 with respect to the transcriptional start site (+1). Plasmid pNRG402 contains an amtB-lacZ fusion that was constructed by inserting an amtB promoter fragment (−80 to + 47) from pNRG401 (53) into pSFL1.
Mutant isolation.
Strain SF517T was mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine as previously described (9) and plated onto glucose X-Gal minimal media that contained either ammonium or glutamate plus ammonium as the nitrogen source. After incubation at 37°C for 2 days, mutant colonies with increased levels of β-galactosidase expression were identified by their blue color and purified by two rounds of single-colony isolation. To determine if the mutations causing constitutive expression of the glnRA-lacZ fusion were genetically linked to the glnA gene, chromosomal DNA from the mutants was used to transform strain SF21AT with selection for glutamine prototrophy. Transformants were then screened for constitutive expression of the (glnRA-lacZ)21 fusion on glucose X-Gal minimal medium agar plates. Strains containing the glnA mutations and the (amtB-lacZ)402 fusion were constructed by transforming strain SF402A with chromosomal DNA from the mutants with selection for glutamine prototrophy.
Enzyme assays.
β-Galactosidase activity was assayed in crude extracts prepared from cells grown to mid-log growth phase (70 to 90 Klett units) as previously described (2). β-Galactosidase levels were corrected for the endogenous activity present in B. subtilis cells containing the promoterless lacZ fusion vectors integrated at the amyE site. One unit of β-galactosidase activity produced 1 nmol of o-nitrophenol per min.
The biosynthetic and reverse (transferase) enzymatic activities of GS were measured by the production of γ-glutamylhydroxamate as previously described (16). The kinetic constants for the Mg2+-dependent biosynthetic reaction were determined as previously described (50). The glutamine, AMP, and methionine sulfoximine concentrations necessary to reduce enzymatic activity by 50% (IC50) were determined with the Mg2+-dependent biosynthetic reaction, where the glutamate and ATP concentrations were 150 and 18 mM, respectively (50).
DNA and protein methods.
DNA sequencing of the mutations and construction of mutant GS overexpression plasmids were performed as previously reported (16). Purifications of GlnR, TnrA, and GS were done by published procedures (51, 54, 55). The concentrations of TnrA and GS were determined by measuring their absorbance at 280 nm. The molar absorption coefficients of the proteins were calculated from their amino acid sequences (32). The concentration of GlnR was determined by the Advanced protein assay (Cytoskeleton, Inc.). Gel mobility shift experiments to examine the abilities of wild-type and mutant GS proteins to alter the DNA binding activities of GlnR and TnrA were performed as previously described (20, 55).
RESULTS
Isolation and characterization of mutants that constitutively express glnRA.
To determine whether any gene products other than GlnR and GS are involved in the regulation of glnRA expression, mutants with constitutive glnRA expression (GlnC) were isolated. Chemically mutagenized cells containing a glnRA-lacZ fusion were spread onto glucose minimal medium plates containing excess nitrogen. These plates did not contain glutamine, so that all of the mutants would retain at least some GS biosynthetic enzymatic activity, thus preventing the isolation of glnA null mutants. To avoid the isolation of glnA mutants with significant defects in GS enzymatic activity, small colonies with obvious growth defects were not selected for further characterization. Expression of the glnRA-lacZ fusion was visualized by including X-Gal, a chromogenic substrate for β-galactosidase, in the plates. Mutants with high-level constitutive glnRA expression were identified as blue colonies. Genetic mapping experiments revealed that all 27 of the isolated GlnC mutants contained mutations that were tightly linked to the glnA gene. Sequencing of the glnRA operon in 13 of the GlnC mutants revealed four different glnR (T21I, P39S, R47Q, and G72K) and three novel glnA (S55F, V173I, and L174F) amino acid substitutions. The other six mutants contained mutations in glnA that had been isolated previously as mutants which constitutively expressed the TnrA-dependent amtB promoter (16, 19, 50).
The growth properties of the three novel glnA mutants (S55F, V173I, and L174F) were examined on glucose minimal medium plates containing four different nitrogen sources: glutamine, ammonium, glutamate, and glutamate plus ammonium. All three glnA mutants were found to exhibit growth phenotypes that were identical to that of the wild-type strain on these nitrogen sources (data not shown). It has been shown previously that unlike the wild-type strain, B. subtilis glnA mutants that encode feedback-resistant GS enzymes can crossfeed Gln mutants (that lack GS activity) on solid medium (19, 50). While the glnA(V173I) and glnA(L174F) mutants were able to crossfeed a ΔglnA mutant in a plate assay, the glnA(S55F) mutant did not have this phenotype (data not shown). These observations suggest that the glnA(V173I) and glnA(L174F) mutants encode feedback-resistant GS enzymes.
GlnR- and TnrA-dependent regulation in vivo.
To examine the effect of the mutant glnR alleles on the regulation of glnRA expression, the β-galactosidase levels produced by a glnRA-lacZ fusion were determined in wild-type and mutant strains. In cells containing the wild-type glnR gene, glnRA expression was 140-fold lower in cells grown with the excess nitrogen source glutamine than in cells grown with the limiting nitrogen source glutamate (Table 2). The repression of glnRA expression was relieved in strains containing the mutant glnR alleles, where β-galactosidase levels in glutamine-grown cultures were 26- to 220-fold higher in the glnR mutants than in the wild-type strain (Table 2). All four glnR mutations generate amino acid substitutions that are located within the N-terminal DNA-binding domain of GlnR and most likely relieve glnRA repression due to impaired DNA-binding activity of the mutant GlnR proteins.
TABLE 2.
GlnR- and TnrA-dependent regulation in wild-type and mutant strainsa
| Relevant genotype | Amino acid change | Codon change | GlnR-dependent regulationb
|
TnrA-dependent regulationc
|
||
|---|---|---|---|---|---|---|
| Glutamine | Glutamate | Glutamine | Glutamate | |||
| Wild type | 7.7 | 1,100 | 0.05 | 70 | ||
| glnR(T21I) | Thr21 → Ile | ACT → ATT | 200 | 1,400 | NDd | ND |
| glnR(P39S) | Pro39 → Ser | CCA → TCA | 400 | 970 | ND | ND |
| glnR(R47Q) | Arg47 → Gln | CGA → CAA | 1,700 | 1,600 | ND | ND |
| glnR(G72K) | Gly72 → Lys | GGA → AAA | 240 | 740 | ND | ND |
| glnA(S55F) | Ser55 → Phe | TCT → TTT | 1,300 | 1,500 | 1.1 | 90 |
| glnA(V173I) | Val173 → Ile | GTA → ATA | 640 | 850 | 3.3 | 68 |
| glnA(L174F) | Leu174 → Phe | CTT → TTT | 730 | 1,100 | 1.4 | 88 |
Cells were grown in glucose minimal medium containing the indicated nitrogen sources. Values are the averages of two or more determinations and did not vary by more than 20%. Strains contained either the (glnRA-lacZ)21 fusion or the (amtB-lacZ)402 fusion. Strains with the glnRA-lacZ fusion also contained a tnrA null mutation.
β-Galactosidase specific activity (U/mg protein) in a glnRA-lacZ fusion strain grown on glutamine or glutamate.
β-Galactosidase specific activity (U/mg protein) in an amtB-lacZ fusion strain grown on glutamine or glutamate.
ND, not determined.
The effect of the three glnA alleles on glnRA regulation was also examined. Constitutive expression of the glnRA-lacZ fusion was observed in the strains containing the glnA mutations (Table 1). Because the activity of TnrA is also controlled by GS, the effect of the mutant glnA alleles on the TnrA-dependent gene expression of an amtB-lacZ fusion was also examined. In wild-type cells, amtB expression was regulated 1,400-fold in response to nitrogen availability (Table 2). In cells grown with the excess nitrogen source glutamine, amtB expression was 16- to 47-fold higher in strains containing the mutant glnA genes that in wild-type cells (Table 2).
The phenotype of these three glnA mutations is unique in that GlnR regulation is relieved, but there is only a modest defect in TnrA regulation. In contrast, all of the glnA mutants previously isolated by screening for constitutive expression of TnrA-regulated genes or for feedback-resistant GS enzymes were found to be significantly defective in both TnrA- and GlnR-dependent regulation (16, 19, 50). The one exception to the generalization that glnA mutations have similar TnrA and GlnR regulatory phenotypes is a mutant that was isolated by screening for resistance to the GS inhibitor l-methionine-S-sulfoximine (MetSox) (43). This mutant GS was found to contain an alanine substitution for residue Val190 (43). GlnR-dependent repression is relieved in the glnA(V190A) mutant, but no significant defect in TnrA-dependent regulation is observed (16, 43). Thus, the glnA(S55F), glnA(V173I), glnA(L174F), and glnA(V190A) alleles belong to a novel class of glnA mutations that relieve GlnR regulation but have few or no defects in TnrA regulation.
Enzymatic properties of the mutant enzymes.
To examine the catalytic and feedback properties of the mutant enzymes, the S55F, V173I, L174F, and V190A GS proteins were overexpressed and purified to homogeneity. The specific activities of the Mg2+-dependent biosynthetic and transferase (reverse) reactions were determined for each enzyme. The S55F GS enzyme had specific activities that were 20- to 30-fold lower than that of wild-type GS (Table 3). This lack of enzymatic activity is surprising in that the glnA(S55F) mutant exhibited no observable growth defect in vivo. Due to the low in vitro enzymatic activity of S55F GS, this mutant protein was not characterized further. The specific activities of the mutant V173I, L174F, and V190A GS enzymes were all similar to that of the wild-type enzyme (Table 3). The kinetic properties of the biologically significant Mg2+-dependent biosynthetic reaction were also determined. Compared to wild-type GS, the V173I, L174F, and V190A enzymes had only modest (less than threefold) differences in their kinetic constants (Table 3).
TABLE 3.
Kinetic parameters of wild-type and mutant GS
| Enzyme | Mg2+-dependent biosynthetic reactiona
|
Transferase sp act (μmol/min/mg) | |||
|---|---|---|---|---|---|
| Km glutamate (mM) | Km ATP (mM) | Vmax (μmol/min/mg) | Sp actb (μmol/min/mg) | ||
| Wild type | 27 ± 2.2 | 2.4 ± 0.1 | 3.7 ± 0.2 | 3.6 ± 0.4 | 90 ± 3 |
| S55F | NDc | ND | ND | 0.3 ± 0.1 | 3 ± 0.4 |
| V173I | 33 ± 5 | 2.6 ± 0.3 | 6.6 ± 0.2 | 3.1 ± 0.1 | 81 ± 3 |
| L174F | 16 ± 2 | 3.0 ± 0.8 | 4.4 ± 0.1 | 4.0 ± 0.2 | 130 ± 10 |
| V190A | 17 ± 2 | 3.9 ± 0.2 | 8.4 ± 0.2 | 5.9 ± 1.2 | 120 ± 20 |
Values are the averages of at least two determinations ± standard errors of the means.
Specific activities were determined in the presence of 100 mM glutamate, 7.5 mM ATP, and 40 mM hydroxylamine.
ND, not determined.
The V173I and L174F mutant enzymes were highly resistant to feedback inhibition by glutamine (Table 4). These data agree with the in vivo observation that the glnA(V173I) and glnA(L174F) mutants were able to crossfeed Gln mutant cells. In contrast, the mutant V190A GS was only slightly more resistant to glutamine inhibition (fourfold) than the wild-type enzyme (Table 4). This result is consistent with the observation that the glnA(V190A) mutant was unable to crossfeed Gln mutant cells in a plate assay (data not shown). The activities of the three mutant enzymes were three- to ninefold more resistant to AMP inhibition than the wild-type enzyme (Table 4). As would be expected for a mutant that was isolated by screening for increased MetSox resistance, the V190A GS is 10-fold more resistant to MetSox inhibition than is wild-type GS (Table 4). In contrast, the V173I and L174F mutant enzymes had sensitivities to MetSox inhibition that were similar to that of the wild-type enzyme (Table 4).
TABLE 4.
Sensitivities of wild-type and mutant GS for inhibitiors
| Enzyme | IC50 (mM) for Mg2+-dependent biosynthetic reactiona
|
||
|---|---|---|---|
| Glutamine | AMP | MetSox | |
| Wild type | 2.4 ± 0.1 | 0.52 ± 0.02 | 0.13 ± 0.01 |
| V173I | >140 | 4.8 ± 0.2 | 0.19 ± 0.01 |
| L174F | >140 | 2.6 ± 0.1 | 0.12 ± 0.01 |
| V190A | 9.6 ± 0.4 | 1.4 ± 0.1 | 1.3 ± 0.1 |
Values are the averages of at least two determinations ± standard errors of the means.
Regulation of GlnR and TnrA in vitro.
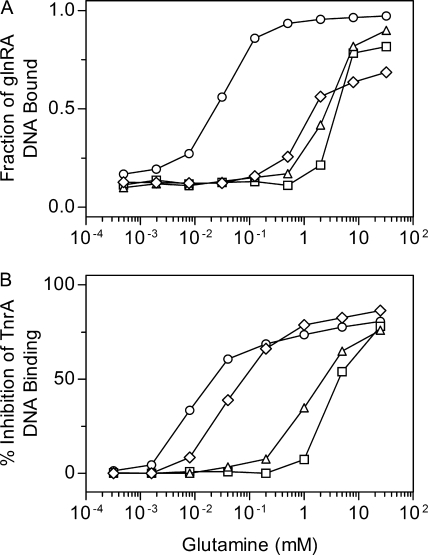
The ability of the wild-type and mutant enzymes to activate GlnR DNA binding in vitro was determined with a DNA gel mobility shift assay. In these experiments, the levels of GlnR and GS were held constant while the concentration of the inhibitor glutamine was varied. As would be expected for mutant enzymes that are resistant to glutamine inhibition, the V173I, L174F, and V190A GS proteins required significantly higher levels of glutamine than did the wild-type GS in order to activate GlnR DNA binding (Fig. 2A).
FIG. 2.
Effect of glutamine on the in vitro ability of wild-type and mutant GS proteins to alter DNA binding by GlnR and TnrA. (A) The activation of GlnR binding to glnRA promoter DNA was determined with a gel mobility shift assay. The GlnR and GS promoter concentrations were 25 nM and 20 μM, respectively. (B) The inhibition of TnrA binding to tnrA promoter DNA was determined with a gel mobility shift assay. The TnrA dimer and GS subunit concentrations were 100 nM and 1 μM, respectively. The symbols for the different GS proteins are as follows: wild type, ○; V173I, □; L174F, ▵; and V190A, ⋄. Each data point is the mean of at least two independent experiments and is reproducible to ±10%.
The inhibition of TnrA DNA binding by the wild-type and mutant GS proteins was also examined in vitro with a DNA gel mobility shift assay where the concentrations of TnrA and GS were fixed while the glutamine concentration was varied. Compared to wild-type GS, all of the mutant proteins required higher levels of glutamine to inhibit DNA binding by TnrA (Fig. 2B). The proteins with the highest levels of resistance to glutamine inhibition, V173I and L174F GS, required higher glutamine concentrations than the low-level resistant V190A GS. These results correlate well with the in vivo regulation phenotypes of these mutants, where the V190A GS is able to fully regulate TnrA-dependent expression while the V173I and L174F proteins are partially defective in this regulation (Table 2) (16).
DISCUSSION
All of the mutations isolated in the screen for GlnC mutants were found to be located in the glnR and glnA genes. This result argues that only GlnR and GS are required for regulation of GlnR-dependent gene expression, and it is consistent with the results of in vitro experiments which demonstrated that FBI-GS alone is sufficient to activate GlnR DNA binding (20). However, these observations do not unambiguously rule out the possibility that additional factors participate in the in vivo activation of GlnR. For instance, mutants that lack a factor with an auxiliary role in GlnR regulation could only have a relatively minor defect in glnRA expression. This class of mutants would not have been identified in these experiments, because only GlnC mutants with high-level constitutive glnRA expression were characterized. In addition, mutations in factors which are essential for growth or redundantly encoded in the genome would not have been identified in this mutant hunt. Mutations in metabolic enzymes that caused a reduction in the levels of intracellular glutamine, and thus the levels of FBI-GS were also not isolated. Any GlnC mutant that results from reduced intracellular glutamine pools would be expected to have a significant growth defect. This class of GlnC mutants would not have been characterized because small colonies with obvious growth defects were not selected for further analysis in our mutant screen. Nonetheless, the results of this mutant hunt provide no evidence for trans-acting factors, other than FBI-GS being involved in the control of GlnR-dependent repression of gene expression.
The glnA(S55F), glnA(V173I), glnA(L174F), and glnA(V190A) alleles belong to a novel class of glnA mutations that relieve GlnR regulation but have little or no defects in TnrA regulation. To understand the altered enzymatic and regulatory properties of the amino acid substitutions, a previously described homology model of the B. subtilis GS structure (19) was used to analyze these four mutant GS proteins. This model is based on the crystal structure of the Salmonella enterica serovar Typhimurium GS protein (21). GS from these two bacteria contains twelve identical subunits arranged as two hexameric rings (11, 13). The active sites are located at the subunit-subunit interfaces within each hexameric ring (13).
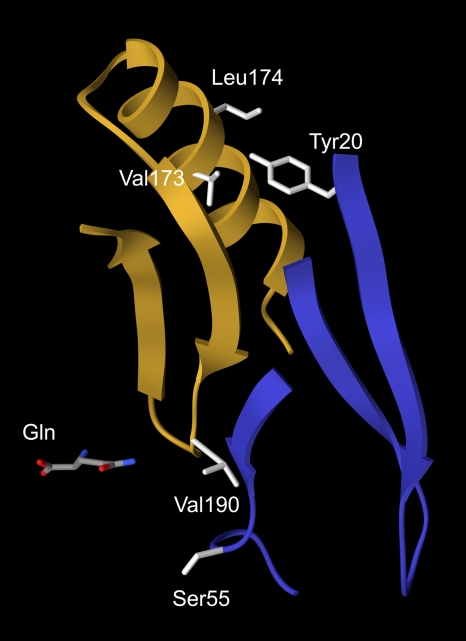
The mutant S55F GS had a significant defect in enzymatic activity (Table 3). Residue Ser55 is located in the active site and positioned so that its side chain extends into the active site cavity (Fig. 3). The S55F substitution replaces the small serine side chain with a much larger phenylalanine side chain. The intrusion of this large side chain into the active site would be expected to interfere with substrate binding and result in reduced enzymatic activity. Although the structural model provides an explanation for the in vitro enzymatic defects of the S55F enzyme, it does not provide any insight into why the glnA(S55F) mutant has a wild-type growth phenotype on different nitrogen sources. One possibility is that because glnRA expression is derepressed in the glnA(S55F) mutant, the partially active mutant S55F enzyme may be synthesized at high enough levels to support wild-type growth rates. Alternatively, the S55F mutant enzyme may be stabilized in vivo by high concentrations of GS substrates but then become inactive during the in vitro purification where the substrates are not present.
FIG. 3.
Locations of mutated residues in the structural model of GS. The backbone residues 163 to 200 from one subunit are shown as a gold ribbon, while the backbone residues 20 to 36 and 51 to 58 from the adjacent subunit are shown as a blue ribbon. Residue side chains are colored light gray. Glutamine bound to the active site is colored by atomic elements: carbon, gray; nitrogen, blue; and oxygen, red. This figure was prepared with UCSF Chimera (33).
The mutant V190A GS was slightly more resistant to inhibition by glutamine and MetSox than was the wild-type enzyme (Table 4). MetSox is a glutamate analogue that inhibits GS enzymatic activity by a different mechanism than glutamine. While glutamine is a simple competitive inhibitor that binds to the glutamate substrate site (19, 50), MetSox is a substrate for GS that is phosphorylated in the presence of ATP (46). Phosphorylated MetSox is a GS transition state analogue that binds tightly to the active site and irreversibly inhibits GS activity (26). Structural models of GS from Mycobacterium tuberculosis and S. enterica serovar Typhimurium indicate that residue Val190 is located within the active site and is positioned 4 to 6 Å from ligands bound to the glutamate substrate site (Fig. 3) (21, 26, 27). The Val190 side chain may weakly interact with and stabilize the binding of glutamine and MetSox. The V190A substitution would remove the valine side chain and abolish these interactions. This change in the V190A enzyme presumably reduces the affinity of glutamine and MetSox for the active site and thus results in a low level of resistance to these inhibitors.
The V173I and L174F enzymes were highly resistant to inhibition by glutamine (Table 4). These residues are not located in the active site and are positioned 17 to 24 Å from the glutamate binding site (Fig. 3). The Val173 and Leu174 residues are located at the subunit-subunit interface and have proximity to residue Tyr20 from the adjacent subunit (Fig. 3.). The V173I and L174F substitutions most likely alter the subunit-subunit interaction and generate subtle long-range structural perturbations that reduce the affinity of the active site for glutamine. It has been shown for several other enzymes that amino acid substitutions positioned remotely from the active site can alter catalytic activity and inhibitor binding (24, 25, 28, 35, 39). Interestingly, although the V173I and L174F substitutions confer high-level resistance to glutamine, these replacements do not significantly alter the sensitivity to inhibition by MetSox (Table 4). This difference is most likely a reflection of the fact that glutamine and MetSox inhibit GS by different mechanisms.
All our previously isolated glnA mutants encoding feedback-resistant GS enzymes expressed both TnrA- and GlnR-regulated genes constitutively (19, 50). Surprisingly, even though the glnA(V173I) and glnA(L174F) mutants encode mutant GS enzymes that are highly resistant to feedback inhibition, these mutants have a different phenotype in that GlnR-dependent gene regulation is relieved but there is only a modest defect in TnrA-dependent gene regulation (Table 2). One possible explanation for this difference is that the previously isolated feedback-resistant GS proteins may contain amino acid substitutions that indirectly disrupt both the binding of glutamine and TnrA while the amino acid residue changes in the V173I and L174F enzymes inhibit glutamine binding but do not alter the TnrA binding interface on FBI-GS.
TnrA has previously been shown to be able to stabilize the binding of glutamine to GS (50). The glutamine IC50 of GS is sixfold lower in the presence of TnrA than when TnrA is absent (50). Since TnrA is thought to bind to GS at the glutamate entrance to the active site (16), TnrA most likely stabilizes glutamine bound at the active site by blocking the glutamate entrance to the active site and preventing the release of glutamine. Unfortunately, the effect of GlnR on the glutamine inhibition of GS cannot be determined due to the limited solubility of GlnR. Nonetheless, because GlnR only interacts weakly with FBI-GS, it is unlikely that GlnR significantly stabilizes the binding of glutamine to GS. We hypothesize that the V173I and L174F mutant enzymes must be able to adopt the GS conformation required for optimal interaction between GS and TnrA and that the TnrA-dependent stabilization of glutamine binding allows TnrA to interact with the V173I and L174F mutant enzymes, albeit with reduced affinity. In contrast, the TnrA-binding interface would be disrupted in the previously described feedback-resistant mutant proteins, and thus the interaction with TnrA would not have sufficient affinity to even partially stabilize glutamine binding. Since GlnR presumably does not stabilize glutamine binding to GS, GlnR-dependent regulation would be expected to be defective in all feedback-resistant glnA mutants.
Surprisingly, even though the glnA(V190A), glnA(V173I), and glnA(L174F) mutants cannot significantly activate GlnR DNA binding in vivo, all three mutant GS proteins were able to activate GlnR DNA binding in vitro in the presence of high levels of glutamine (Fig. 2A). In these in vitro assays, GS and GlnR were always present at high levels while the concentration of glutamine was varied. One explanation for the ability of the mutant GS proteins to activate GlnR DNA binding in vitro is that the levels of GS and/or GlnR present in the in vitro assays are higher than their levels in growing cells. As a result, high concentrations of glutamine are able to convert the mutant GS proteins to the FBI-GS form in vitro and thus activate GlnR DNA binding. Nonetheless, the observation that high levels of glutamine are required for activation of GlnR DNA binding in vitro by these three mutant GS proteins argues that the defective GlnR regulation seen in these glnA mutants in vivo results from the mutant GS proteins having a reduced affinity for glutamine rather than a defect in GlnR binding.
One possible explanation for the in vivo difference in the responses of GlnR- and TnrA-mediated regulation to the growth on different nitrogen sources is that GlnR-dependent regulation is more sensitive to the level of GS feedback inhibition than is the TnrA-dependent regulation. One of the determinants for this differential sensitivity would be that GlnR and TnrA have different affinities for FBI-GS. While TnrA forms a tight stable complex with FBI-GS, GlnR only transiently interacts with FBI-GS (20, 55). As a result of this difference, GlnR-dependent regulation would be more sensitive to fluctuations in the level of FBI-GS than TnrA-dependent regulation. Moreover the ability of TnrA to stabilize the binding of glutamine to GS would augment this differential sensitivity to feedback inhibition by moderating changes in the amount of glutamine bound to GS as the concentration of glutamine fluctuates (50). In contrast, GlnR would not be capable of mediating this effect. As a consequence, GlnR-mediated regulation would be more sensitive to changes in glutamine levels than TnrA-mediated regulation.
The phenotype of the glnA(V190A) mutant supports the idea that GlnR and TnrA have different sensitivities to regulation by FBI-GS. While the expression of GlnR-regulated genes is derepressed in the glnA(V190A) mutant, no defect in TnrA-dependent gene expression is observed (16, 43). Compared to wild-type GS, the V190A enzyme has a fourfold increase in its resistance to glutamine inhibition (Table 4). The observation that a mutant GS with a small increase in the resistance to feedback inhibition has a much larger effect on GlnR-dependent regulation than on TnrA-dependent regulation is consistent with the hypothesis that GlnR-dependent regulation is more sensitive to reduced levels of GS feedback inhibition than is the TnrA-dependent regulation.
Acknowledgments
This research was supported by Public Health Service research grant GM051127 from the National Institutes of Health.
Footnotes
Published ahead of print on 20 February 2009.
REFERENCES
- 1.Atkinson, M. R., and S. H. Fisher. 1991. Identification of genes and gene products whose expression is activated during nitrogen-limited growth in Bacillus subtilis. J. Bacteriol. 17323-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 1724758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson, M. R., T. A. Blauwkamp, V. Bondarenko, V. Studitsky, and A. J. Ninfa. 2002. Activation of the glnA, glnK, and nac promoters as Escherichia coli undergoes the transition from nitrogen excess growth to nitrogen starvation. J. Bacteriol. 1845358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier, L., P. Nygaard, H. Jamer, and H. H. Saxild. 2002. Transcriptional analysis of the Bacillus subtilis PucR regulon and identification of a cis-acting site required for PucR-regulated expression of genes involved in purine catabolism. J. Bacteriol. 1843232-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belitsky, B. R., L. V. Wray, Jr., S. H. Fisher, D. E. Bohannon, and A. L. Sonenshein. 2000. Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J. Bacteriol. 1825939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biaudet, V., F. Samson, C. Anagnostopoulos, S. D. Erlich, and P. Bessières. 1996. Computerized map of Bacillus subtilis. Microbiology 1422669-2729. [DOI] [PubMed] [Google Scholar]
- 7.Brown, S. W., and A. L. Sonenshein. 1996. Autogenous regulation of the Bacillus subtilis glnRA operon. J. Bacteriol. 1782450-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chasin, L. A., and B. Magasanik. 1968. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J. Biol. Chem. 2435165-5178. [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., New York, NY.
- 10.Dean, D. R., J. A. Hoch, and A. I. Aronson. 1977. Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J. Bacteriol. 131981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deuel, T. F., A. Ginsburg, J. Yeh, E. Shelton, and E. R. Stadtman. 1970. Bacillus subtilis glutamine synthetase. Purification and physical characterization. J. Biol. Chem. 2455195-5205. [PubMed] [Google Scholar]
- 12.Deuel, T. F., and S. Prusiner. 1974. Regulation of glutamine synthetase from Bacillus subtilis by divalent cations, feedback inhibitors, and L-glutamine. J. Biol. Chem. 249257-264. [PubMed] [Google Scholar]
- 13.Eisenberg, D., H. S. Gill, G. M. U. Pfluegel, and S. H. Rotstein. 2000. Structure-function relationships of glutamine synthetases. Biochim. Biophys. Acta 1477122-145. [DOI] [PubMed] [Google Scholar]
- 14.Ferson, A. E., L. V. Wray, Jr., and S. H. Fisher. 1996. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol. Microbiol. 22693-701. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol. Microbiol. 32223-232. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, S. H., J. L. Brandenburg, and L. V. Wray, Jr. 2002. Mutations in Bacillus subtilis glutamine synthetase that block its interaction with transcription factor TnrA. Mol. Microbiol. 45627-635. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, S. H., and A. L. Sonenshein. 1984. Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression. J. Bacteriol. 157612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher, S. H., and L. V. Wray, Jr. 2002. Bacillus subtilis 168 contains two differentially regulated genes encoding l-asparaginase. J. Bacteriol. 1842148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher, S. H., and L. V. Wray, Jr. 2006. Feedback-resistant mutations in Bacillus subtilis glutamine synthetase are clustered in the active site. J. Bacteriol. 1885966-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher, S. H., and L. V. Wray, Jr. 2008. Bacillus subtilis glutamine synthetase regulates its own synthesis by acting as a chaperone to stabilize GlnR-DNA complexes. Proc. Natl. Acad. Sci. USA 1051014-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gill, H. S., and D. Eisenberg. 2001. The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry 401903-1912. [DOI] [PubMed] [Google Scholar]
- 22.Gutowski, J. C., and H. J. Schreier. 1992. Interaction of the Bacillus subtilis glnRA repressor with operator and promoter regions in vivo. J. Bacteriol. 174671-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, P., T. Leighton, G. Iskhanova, and S. Kustu. 1999. Sensing of nitrogen limitation by Bacillus subtilis: comparison to enteric bacteria. J. Bacteriol. 1815042-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, L. K., J. Baldwin, R. Akella, E. J. Goldsmith, and M. A. Phillips. 2004. Multiple active site conformations revealed by distant site mutation in ornithine decarboxylase. Biochemistry 4312990-12999. [DOI] [PubMed] [Google Scholar]
- 25.Kawate, H., D. M. Landis, and L. A. Loeb. 2002. Distribution of mutations in human thymidylate synthase yielding resistance to 5-fluorodeoxyuridine. J. Biol. Chem. 27736304-36311. [DOI] [PubMed] [Google Scholar]
- 26.Krajewski, W. W., T. A. Jones, and S. L. Mowbray. 2005. Structure of Mycobacterium tuberculosis glutamine synthetase in complex with a transition-state mimic provides functional insights. Proc. Natl. Acad. Sci. USA 10210499-10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liaw, S.-H., and D. Eisenberg. 1994. Structural model for the reaction mechanism of glutamine synthetase, based on five crystal structures of enzyme-substrate complexes. Biochemistry 33675-681. [DOI] [PubMed] [Google Scholar]
- 28.Muzammil, S., P. Ross, and E. Freire. 2003. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry 42631-638. [DOI] [PubMed] [Google Scholar]
- 29.Nakano, M. M., F. Yang, P. Hardin, and P. Zuber. 1995. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J. Bacteriol. 177573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ninfa, A. J., L. J. Reitzer, and B. Magasanik. 1987. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell 501039-1046. [DOI] [PubMed] [Google Scholar]
- 32.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 42411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 34.Pahel, G., D. M. Rothstein, and B. Magasanik. 1982. Complex glnA-glnL-glnG operon of Escherichia coli. J. Bacteriol. 150202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reetz, M. T. 2004. Controlling the enantioselectivity of enzymes by directed evolution: practical and theoretical ramifications. Proc. Natl. Acad. Sci. USA 1015716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57155-176. [DOI] [PubMed] [Google Scholar]
- 37.Rhee, S. G., B. Chock, and E. R. Stadtman. 1989. Regulation of Escherichia coli glutamine synthetase. Adv. Enzymol. Relat. Areas Mol. Biol. 6237-92. [DOI] [PubMed] [Google Scholar]
- 38.Robichon, D., M. Arnaud, R. Gardan, Z. Pragai, M. O'Reilly, G. Rapoport, and M. Débarbouille. 2000. Expression of a new operon from Bacillus subtilis ykzB-ykoL under the control of the TnrA and PhoO-PhoR global regulators. J. Bacteriol. 1821226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rod, T. H., J. L. Radkiewicz, and C. L. Brooks. 2003. Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc. Natl. Acad. Sci. USA 1006980-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothstein, D. M., G. Pahel, B. Tyler, and B. Maganasik. 1980. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc. Natl. Acad. Sci. USA 777372-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreier, H. J., S. W. Brown, K. D. Hirschi, J. F. Nomellini, and A. L. Sonenshein. 1989. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J. Mol. Biol. 21051-63. [DOI] [PubMed] [Google Scholar]
- 42.Schreier, H. J., S. H. Fisher, and A. L. Sonenshein. 1985. Regulation of expression from the glnA promoter of Bacillus subtilis requires the glnA gene product. Proc. Natl. Acad. Sci. USA 823375-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreier, H. J., C. A. Rostkowski, and E. M. Kellner. 1993. Altered regulation of the glnRA operon in a Bacillus subtilis mutant that produces methionine sulfoximine-tolerant glutamine synthetase. J. Bacteriol. 175892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreier, H. J., and A. L. Sonenshein. 1986. Altered regulation of the glnA gene in glutamine synthetase mutants of Bacillus subtilis. J. Bacteriol. 16735-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauch, M. A., A. I. Aronson, S. W. Brown, H. J. Schreier, and A. L. Sonenshein. 1988. Sequence of the Bacillus subtilis glutamine synthetase gene region. Gene 71257-265. [DOI] [PubMed] [Google Scholar]
- 46.Weisbrod, R. E., and A. Meister. 1973. Studies on glutamine synthetase from Escherichia coli. Formation of pyrrolidone carboxylate and inhibition by methionine sulfoximine. J. Biol. Chem. 2483997-4002. [PubMed] [Google Scholar]
- 47.Wray, L. V., Jr., M. R. Atkinson, and S. H. Fisher. 1994. The nitrogen-regulated Bacillus subtilis nrgAB operon encodes a membrane protein and a protein highly similar to the Escherichia coli glnB-encoded PII protein. J. Bacteriol. 176108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 1795494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wray, L. V., Jr., A. E. Ferson, K. Rohrer, and S. H. Fisher. 1996. TnrA, a transcriptional factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 938841-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wray, L. V., Jr., and S. H. Fisher. 2005. A feedback-resistant mutant of Bacillus subtilis glutamine synthetase with pleiotropic defects in nitrogen-regulated gene expression. J. Biol. Chem. 28033298-33304. [DOI] [PubMed] [Google Scholar]
- 51.Wray, L. V., Jr., and S. H. Fisher. 2008. Bacillus subtilis GlnR contains an autoinhibitory C-terminal domain required for the interaction with glutamine synthetase. Mol. Microbiol. 68277-285. [DOI] [PubMed] [Google Scholar]
- 52.Wray, L. V., Jr., F. K. Pettengill, and S. H. Fisher. 1994. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription start site. J. Bacteriol. 1761894-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wray, L. V., Jr., J. M. Zalieckas, A. E. Ferson, and S. H. Fisher. 1998. Mutational analysis of the TnrA-binding sites in the Bacillus subtilis nrgAB and gabP promoter regions. J. Bacteriol. 1802943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2000. Purification and in vitro activities of the Bacillus subtilis TnrA transcription factor. J. Mol. Biol. 30029-40. [DOI] [PubMed] [Google Scholar]
- 55.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2001. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 107427-435. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida, K., H. Yamaguchi, M. Kinehara, Y. Ohki, Y. Nakaura, and Y. Fujita. 2003. Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol. Microbiol. 49157-165. [DOI] [PubMed] [Google Scholar]
- 57.Zalieckas, J. M., L. V. Wray, Jr., and S. H. Fisher. 2006. Cross-regulation of the Bacillus subtilis glnRA and tnrA genes provides evidence for DNA binding site discrimination by GlnR and TnrA. J. Bacteriol. 1882578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]