Abstract
The Picornaviridae family comprises a diverse group of small RNA viruses that cause a variety of human and animal diseases. Some of these viruses are known to induce cleavage of components of the innate immune system and to inhibit steps in the interferon pathway that lead to the production of type I interferon. There has been no study of the effect of picornaviral infection on the events that occur after interferons have been produced. To determine whether members of the Enterovirus genus can antagonize the antiviral activity of interferon-stimulated genes (ISGs), we pretreated cells with alpha interferon (IFN-α) and then infected the cells with poliovirus type 1, 2, or 3; enterovirus type 70; or human rhinovirus type 16. We found that these viruses were able to replicate in IFN-α-pretreated cells but that replication of vesicular stomatitis virus, a Rhabdovirus, and encephalomyocarditis virus (EMCV), a picornavirus of the Cardiovirus genus, was completely inhibited. Although EMCV is sensitive to IFN-α, coinfection of cells with poliovirus and EMCV leads to EMCV replication in IFN-α-pretreated cells. The enteroviral 2A proteinase (2Apro) is essential for replication in cells pretreated with interferon, because amino acid changes in this protein render poliovirus sensitive to IFN-α. The addition of the poliovirus 2Apro gene to the EMCV genome allowed EMCV to replicate in IFN-α-pretreated cells. These results support an inhibitory role for 2Apro in the most downstream event in interferon signaling, the antiviral activities of ISGs.
Members of the Picornaviridae family are a diverse group of viruses that cause a variety of human and animal diseases. The family comprises eight genera: Aphthovirus, Cardiovirus, Enterovirus, Erbovirus, Hepatovirus, Kobuvirus, Parechovirus, and Teschovirus. The best-studied member of the family is poliovirus (PV), the etiologic agent of the paralytic disease poliomyelitis (39). The three PV serotypes are classified in the Enterovirus genus along with members such as enterovirus type 70 (EV70), which causes acute hemorrhagic conjunctivitis (52). The Enterovirus genus has now been expanded to include the previously separate Rhinovirus genus, which consists of more than 100 serotypes of rhinoviruses, causative agents of the common cold (52). A well-studied member of the Cardiovirus genus is encephalomyocarditis virus (EMCV), which causes disease in several mammalian species (52).
The genome of picornaviruses is a positive-strand RNA of 7,000 to 9,000 nucleotides that is translated into a polyprotein which yields structural and nonstructural proteins upon cleavage by viral proteinases. The genomes of enteroviruses encode a 2A proteinase (2Apro), which is structurally and functionally distinct from the 2A proteins encoded in the genomes of members of other genera. For example, the 2A protein of cardioviruses has no sequence homology to the enteroviral 2Apro and lacks protease activity (54). Enterovirus 2Apro processes the viral polyprotein (62), and it also cleaves a variety of host proteins, including the translation proteins eIF4GI (36), eIF4GII (43), and poly(A)-binding protein (27). Proteinase 2Apro has been linked to the inhibition of host translation that occurs during enterovirus infection (1, 63) and is also involved in processes as varied as viral RNA replication (42), viral translation (33), and viral RNA stability (30).
Interferons (IFNs) are pleiotropic cytokines that are well known for their role in the cellular antiviral response (4, 35, 57). Type I IFNs, which include alpha and beta IFNs (IFN-α and -β), are induced in response to viral infection (26). The replication of RNA viruses leads to the production of double-stranded RNA (dsRNA) intermediates that are recognized by Toll-like receptors or by the cytoplasmic retinoic acid-inducible gene I (RIG-I)-like helicases, RIG-I and MDA-5 (61). Upon activation, MDA-5 and RIG-I bind to mitochondrial antiviral signaling protein (MAVS), which then coordinates the activation of the transcription factors IFN regulatory factor 3 (IRF3) and NF-κB to induce the production of type I IFN (61). RIG-I-like helicase signaling has been shown to inhibit picornavirus replication (32). Type I IFNs bind to the type I IFN receptor and elicit a cascade of signaling events that lead to the formation of the IFN-stimulated gene factor 3 transcriptional complex, which binds consensus DNA sites to stimulate the transcription of IFN-stimulated genes (ISGs) (20). The importance of IFNs is underscored by the observation that mice that lack the type I IFN receptor or proteins of the Jak-Stat signaling pathway have an increased sensitivity to viral infections (16, 31, 45).
There are more than 300 ISGs, most of which have not been characterized (10). The antiviral properties of the better-known ISGs have been documented. Protein kinase R or dsRNA-dependent protein kinase (PKR) is activated upon binding to dsRNA and phosphorylates eukaryotic initiation factor 2α to inhibit both host and viral translation (46). The 2′-5′ oligoadenylate synthetase/RNase L pathway is also activated by dsRNA and leads to the destruction of viral RNA (14). ISG56 suppresses both host and viral translation by binding to eukaryotic initiation factor 3 (25).
The IFN system evolved to inhibit viral replication, but viruses have coevolved with the system to block this response. This antagonism can occur at three levels: (i) viral recognition and IFN production, (ii) IFN signaling and ISG production, and (iii) ISG activity. Examples of viral antagonism at the IFN production step include hepatitis C virus inhibition of IRF3 and NF-κB activation via the NS3/4A protease (19), vesicular stomatitis virus (VSV) inhibition of IFN-β transcription by the matrix protein (18), and the binding of human papillomavirus 16 E6 oncoprotein to IRF3 (58). IFN signaling is disrupted during many viral infections, including those with human cytomegalovirus (47), dengue virus (28), and human papillomavirus (2). Examples of viruses with direct antagonism of ISGs include herpes simplex virus, which targets RNaseL (6) and PKR (41), and hepatitis C virus, which blocks activation of PKR (21).
The IFN response is altered by a variety of mechanisms in cells infected with picornaviruses. The production of type I IFN is attenuated in cells infected with mengovirus (23), while the hepatitis A virus 3Cpro cleaves MAVS to inhibit type I IFN production (65). PV infection leads to inhibition of the cellular secretory pathway, leading to reduced secretion of cytokines, including IFN-β (13). MDA-5 and RIG-I are cleaved during PV infection (3; P. Barral, P. Fisher, and V. R. Racaniello, unpublished data), and the induction of IFN-β mRNA is inhibited by the L proteinase of foot-and-mouth disease virus (8). Type I IFN has been shown to regulate PV tropism (50, 66) and is known to regulate replication of EMCV replication (7, 32).
Here we investigate viral inhibition of the IFN response at the most downstream step of the pathway, when ISGs have already been produced. We show that picornaviruses that encode a 2Apro, i.e., members of the Enterovirus genus such as PV, human rhinovirus 16 (HRV16), and EV70, are able to replicate in cells that have been pretreated with IFN-α. Picornaviruses that do not encode this proteinase, such as EMCV, are exquisitely sensitive to IFN and are unable to replicate in IFN-pretreated cells. The synthesis of 2Apro rescues the replication of EMCV in IFN-pretreated cells, indicating that 2Apro functions as an ISG antagonist.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
HeLa S3 (human cervical carcinoma), Vero (African green monkey kidney), and Huh7 (human hepatoma) lines were maintained in Dulbecco's modified essential medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with a 1% penicillin-streptomycin solution (Invitrogen) and 10% bovine calf serum (HyClone, Logan, UT). The K562 (human myeloid leukemia) line was maintained in Iscove's modified medium (Invitrogen) supplemented with a 1% penicillin-streptomycin solution and 10% fetal bovine serum (Atlanta Biologicals, Atlanta, GA). Mrc5 cells were maintained in DMEM supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum.
Wild-type and mutant PVs were isolated from HeLa cells transfected with in vitro-synthesized RNA using DEAE-dextran (51). RNA transcripts were produced from a PV type 1 Mahoney P1M infectious clone, pT7M (56); one of its derivatives, pT7M-2A-Y88S, pT7M-2A-Y88L, pT7M-3C-V54A; or a PV type 2 Lansing (P2L) infectious clone, pT7L (55), or the mutant R2-5NC-14 (P2L-5′UTR; a small-plaque mutant of P2L with a single nucleotide substitution in the 5′ untranslated region) (53). EMCV was isolated from HeLa cells transfected with in vitro-synthesized RNA from an EMCV infectious clone, pEC4 (15). All in vitro-synthesized RNAs were transcribed from linearized templates using T7 RNA polymerase (Promega, Madison, WI). The EMCV/P1M chimeric viruses and HRV16 were produced from the infectious clones pEC4-EP1, pEC4-EP3, pEC4-EP4, pEC4-EP5, and pRV16.11 (40). VSV was obtained from the American Type Culture Collection (Manassas, VA) and propagated in HeLa cells. EV70 was produced from an infectious clone (34) and propagated in HeLa cells. Virus titers were determined by plaque assay on HeLa cells.
Plaque assays.
To determine the titer of P1M and EV70, HeLa cells were covered with an overlay consisting of DMEM (Specialty Media, Philipsburg, NJ), 0.2% NaHCO3 (Sigma, St. Louis, MO), 5% bovine calf serum, 1% penicillin-streptomycin, and 0.8% Noble agar (Sigma). To determine the titer of HRV16, HeLa cells were covered with an overlay of DMEM, 0.2% NaHCO3, 1% bovine calf serum, 1% penicillin-streptomycin, 0.1 M MgCl2 (Sigma), and 0.8% Noble agar. After the overlay solidified, it was covered with liquid medium consisting of DMEM (Invitrogen), 0.04 M MgCl2 (Sigma), 0.002% glucose (Sigma), 0.1% bovine serum albumin (Sigma), 2 mM sodium pyruvate (Invitrogen), 4 mM glutamine (Invitrogen), 4 mM oxaloacetic acid (Sigma), 0.2% NaHCO3 (Sigma), and 1% penicillin-streptomycin. Plaque assays of EMCV were performed using 3T3 or HeLa cells in the same medium that was used for determining the titer of HRV16.
Construction of PVs with altered 2Apro or 3Cpro.
Mutations were introduced into cloned PV type 1 DNA by mutagenic PCR. All PCR amplifications were done with PfuUltra or PicoMaxx enzymes (Stratagene, La Jolla, CA). To change amino acid 88 of 2Apro from Y to either S or L, the BstEII fragment of PV DNA from nucleotide (nt) 3240 to 3930 was replaced with a PCR product containing the appropriate mutations. To change amino acid 54 of 3Cpro from V to A, a BseRI fragment (nt 4742 to 6148) was transferred from Se1-3C-02 DNA (11) to pT7M.
Construction of EMCV clones expressing PV 2Apro.
Sequences encoding 2Apro of P1M, with or without an amino-terminal FLAG epitope, were inserted between the coding region for P1 and 2A of EMCV in pEC4. The clone with the untagged version of 2Apro was named EP4, and the clone containing a FLAG epitope at the amino terminus of 2Apro was named EP1. A cleavage site for EMCV 3C proteinase, VLMLESPNAL, was placed at the N and C termini of PV 2Apro. The two versions of the 2Apro fragment were constructed by overlap or mutagenic PCR. The final PCR fragment was cloned into pCR2.1 using the TOPO TA cloning kit and cut using SpeI and SacI (New England BioLabs) enzymes. This fragment was used to replace the corresponding SpeI-SacI fragment in pEC4 between nt 2293 and 3609.
To construct EP5 and EP3, two versions of 2Apro of P1M, with or without an N-terminal FLAG epitope, were inserted before the L coding region in the genome of EMCV in pEC4. The clone with the untagged version of 2Apro was named EP5, and the clone containing a FLAG epitope at the amino terminus of 2Apro was named EP3. An AUG site was placed at the N terminus of 2Apro, and a cleavage site for EMCV 3Cpro, DEQEQGPYN, was placed at the C terminus of PV 2Apro. The two versions of the 2Apro fragment were constructed by overlap or mutagenic PCR. The final PCR fragment was cloned into pCR2.1 using the TOPO TA cloning kit and cut using AvrII and BssHII (New England BioLabs) enzymes. This fragment was used to replace the corresponding AvrII-BssHII fragment in pEC4 between nt 283 and 734.
Single-step growth analysis.
Confluent monolayers of cells in 35-mm plates that had been mock treated or pretreated with IFN-α for 24 h were infected at 37°C at the desired multiplicity of infection (MOI). After a 1-hour adsorption step, cells were rinsed once with phosphate-buffered saline (PBS; pH 7.5), followed by the addition of DMEM supplemented with 1% penicillin-streptomycin and 1% bovine calf serum and incubation at 37°C. Cells and supernatants were collected at each time point and frozen and then thawed to release the virus. Virus was clarified by centrifugation, and titrated on HeLa cells in the case of single-virus infections or on both HeLa cells and 3T3 cells in the case of coinfections. Because 3T3 cells do not allow entry of PV, these cells can be used to distinguish this virus from EMCV and its derivatives in the yield of mixed infections.
Protein labeling and Western blot analysis.
To visualize proteins in infected cells, confluent monolayers of HeLa cells in 35-mm plates were infected at an MOI of 10 with wild-type P1M or with the PV mutants P1M-2A-Y88L (a small-plaque mutant of P1M with a tyrosine-to-leucine substitution at amino acid 88 in 2Apro), P1M-2A-Y88S (a small-plaque mutant of P1M with a tyrosine-to-serine substitution at amino acid 88 in 2Apro), or P2L-5′UTR at 37°C. After a 1-hour adsorption step, cells were rinsed once with PBS, followed by the addition of serum-free DMEM. At each time point, the medium was removed from a batch of cells and the cells were rinsed with serum-free DMEM lacking methionine and cysteine. Serum-free DMEM containing 25 μCi 35S-labeled methionine and cysteine (Amersham, Piscataway, NJ) per ml was added to the cells for 15 min. The cells were then washed with PBS and lysed with radioimmunoprecipitation buffer (PBS containing 1% NP-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]). Cytoplasmic extracts were denatured in SDS buffer (250 mM Tris Cl [pH 6.8; Sigma], 50% glycerol [Sigma], 10% SDS [Sigma], 0.5% bromophenol blue [Sigma], β-mercaptoethanol [Bio-Rad, Hercules, CA]), and equal amounts of lysate were resolved on a 10% SDS-polyacrylamide gel. Gels were stained, fixed, and dried by standard protocols and then exposed to X-ray film overnight.
For Western blot analysis, confluent monolayers of HeLa cells in 35-mm plates were infected with virus at an MOI of 10 at 37°C. After a 1-hour adsorption step, cells were rinsed once with PBS (pH 7.5), followed by the addition of DMEM supplemented with 1% penicillin-streptomycin and 1% bovine calf serum and incubation at 37°C. At each time point, cells were washed with PBS and lysed in radioimmunoprecipitation buffer. Cytoplasmic extracts were denatured in SDS buffer, and equal amounts of lysate were resolved on a 6%, 10%, or 18% SDS-polyacrylamide gel, depending on the size of protein being visualized. For detection of eIF4GI, the proteins were transferred to a polyvinylidene fluoride membrane and subjected to Western blot analysis with rabbit anti-eIF4GI antibody (Abcam, Cambridge, MA) at a 1/1,000 dilution and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma) at a 1/10,000 dilution. For detection of the FLAG epitope, Western blot analysis was carried out with mouse M2 anti-FLAG antibody (Sigma) at a 1/4,000 dilution and horseradish peroxidase-conjugated anti-mouse immunoglobulin G (Promega) at a 1/3,000 dilution.
Efficiency of plaque formation.
Subconfluent 100-mm plates of HeLa cells were pretreated for 24 h with 2,000 or 100 units/ml IFN-α. Cells were then infected with a known quantity of virus and overlaid with plaquing medium for 3 days to allow plaque formation. Plaques were picked from each plate and expanded in HeLa cells, and RNA from these plaques was extracted using the RNeasy kit (Qiagen, Valencia, CA). The RNA was reverse transcribed to cDNA using Superscript III (Invitrogen) and amplified via PCR using PicoMaxx enzyme and primers flanking the 2Apro gene. The nucleotide sequence of the PCR products was determined to identify changes in the 2Apro sequence. Plaques on each plate were counted.
RESULTS
Replication of VSV, EMCV, and P1M in IFN-α-treated HeLa cells.
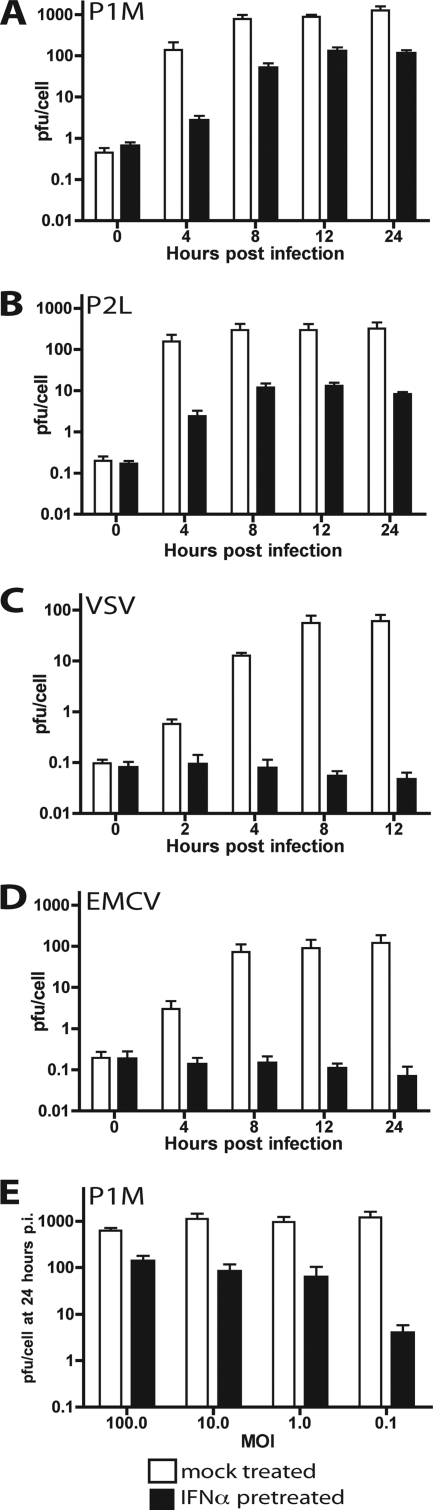
To determine the effect of IFN-α pretreatment on PV replication, HeLa cells were treated with 1,000 units/ml IFN-α for 24 h and then infected with PV. IFN-α was not added to the cells postinfection. Yields of PV were 1,000 PFU/cell in untreated cells and 100 PFU/cell in IFN-α-treated cells (Fig. 1A). P2L and PV type 3 Leon (P3L) also replicated to high titers in IFN-α-treated cells (see Fig. 1B for P2L data; data not shown for P3L). The replication of these serotypes was inhibited 5- to 10-fold by IFN-α pretreatment. In contrast, replication of VSV, which is known to be exquisitely sensitive to IFN (59), was blocked in cells that had been treated with IFN-α (Fig. 1C). Replication of EMCV was also blocked in IFN-α-treated cells (Fig. 1D).
FIG. 1.
Effect of IFN-α pretreatment on viral replication. Monolayers of HeLa cells were pretreated with 1,000 units/ml IFN-α for 24 h or were left untreated and then were infected with P1M (A), P2L (B), VSV (C), or EMCV (D) at an MOI of 10. Total virus was harvested at the indicated times postinfection, and viral titers were determined by plaque assay on HeLa cells. (E) Monolayers of HeLa cells were pretreated with 1,000 units/ml IFN-α for 24 h and then infected with P1M at an MOI of 0.1, 1, 10, or 100. Total virus was harvested at 24 h postinfection, and viral titers were determined by plaque assay on HeLa cells.
The experiments described above were done at an MOI of 10, which would result in infection of >99.99% of the cells. To determine the outcome of infection at a lower MOI, untreated or IFN-α-treated HeLa cells were infected with P1M at an MOI of 100, 10, 1, or 0.1, and viral titers were determined 24 h postinfection. P1M was relatively resistant to IFN pretreatment at an MOI of 10 or 1, but IFN resistance declined at an MOI of 0.1 (Fig. 1E). These results indicate that the IFN resistance of PV is dependent on the MOI.
Replication of EMCV and P1M in other cell lines treated with IFN-α.
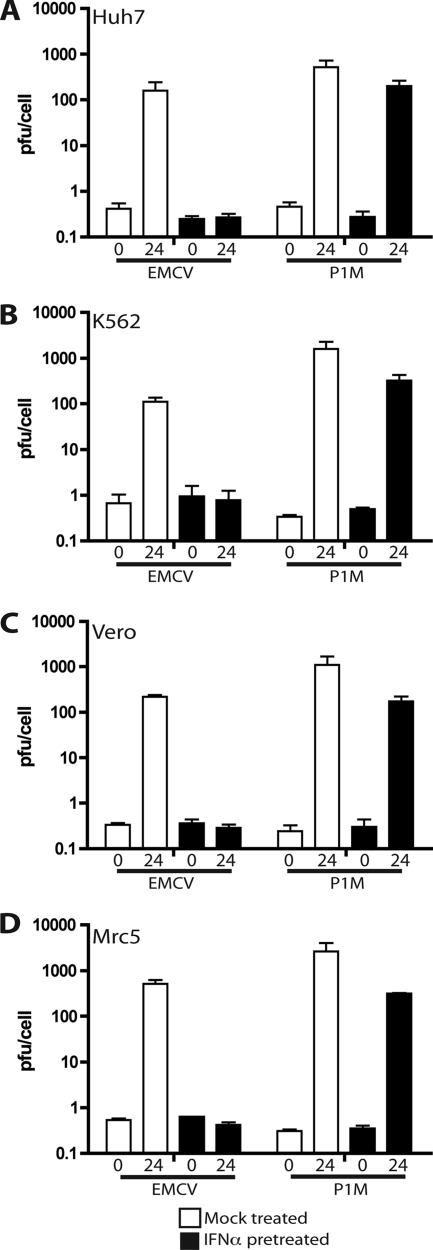
Since HeLa cells are transformed and aneuploid, the ability of PV to replicate in IFN-α-treated cells may not be representative of the results in other cell types. To determine if IFN resistance is cell type specific, Huh7, Vero, K562, and Mrc5 cells were treated with 1,000 units/ml IFN-α for 24 h and infected with P1M or EMCV. In all the cell lines tested, P1M titers were approximately 1,000 PFU/cell in untreated cells and 100 PFU/cell in cells treated with IFN (Fig. 2). EMCV titers were ∼100 PFU/cell in untreated Huh7, K562, and Vero cells, while replication was completely inhibited in cells that had been treated with IFN-α (Fig. 2). EMCV was completely inhibited in Mrc5 cells that had been treated with IFN-α, but yields were ∼500 PFU/cell in untreated Mrc5 cells. Because Mrc5 cells are not transformed, the results indicate that the transformed phenotypes of HeLa cells and other cell types do not contribute to PV IFN resistance and EMCV IFN sensitivity. The effects of IFN-α on EMCV and P1M replication are therefore independent of cell type.
FIG. 2.
Replication of EMCV and P1M in different cell lines treated with IFN-α. Monolayers of Huh7 (A), Vero (C), and Mrc5 (D) cells and suspensions of K562 (B) cells were pretreated with 1,000 units/ml IFN-α for 24 h or were left untreated before they were infected with P1M or EMCV at an MOI of 10. Total virus was harvested at 0 and 24 h postinfection, and viral titers were determined by plaque assay on HeLa cells.
Because Vero cells (17, 48) and K562 cells (12) do not produce IFN, these cells can be used to dissociate type I IFN induction from downstream events, such as ISG production and activity. Treatment of both cell lines with IFN-α results in partial inhibition of P1M and complete inhibition of EMCV. Since the extent of inhibition of viral replication in these cells is the same as in cell types that produce IFN, the phenomenon being studied here is not antagonism of the IFN induction that normally occurs upon viral infection and during the type I IFN feedback loop (44) but antagonism at the level of ISG production or activity.
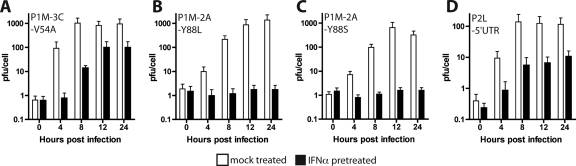
PV 2Apro is essential for replication in IFN-α-treated cells.
To identify viral proteins essential for PV replication in IFN-α-treated cells, viruses with amino acid changes in one of the two virus-encoded proteinases were studied. A single amino acid change in 3Cpro which leads to defects in trans but not cis cleavage has been described previously (11). A virus carrying this change, P1M-3C-V54A (a small-plaque mutant of P1M with a valine-to-alanine substitution at amino acid 54 in the 3Cpro proteinase), replicated to approximately 500 PFU/cell in untreated cells and to over 100 PFU/cell in IFN-treated cells (Fig. 3A). Amino acid changes at position 88 of 2Apro that affect cleavage of cellular but not viral substrates have been described previously (64, 67). The growth of PV mutants P1M-2A-Y88L and P1M-2A-Y88S was similar to that of wild-type virus in untreated cells (∼1,000 PFU/cell) but was completely inhibited in IFN-α-treated cells (Fig. 3B and C). These results suggest that 2Apro is essential for PV replication in IFN-α-treated cells.
FIG. 3.
Replication kinetics of small-plaque PV mutants in IFN-α-pretreated cells. Monolayers of HeLa cells were pretreated with 1,000 units/ml IFN-α for 24 h or were left untreated before they were infected with virus at an MOI of 10. Total virus was harvested at the indicated times postinfection, and viral titers were determined by plaque assay on HeLa cells. The mutant viruses were P1M-3C-V54A, a small-plaque mutant of P1M with a valine-to-alanine substitution at amino acid 54 in 3Cpro (A), P1M-2A-Y88L, a small-plaque mutant of P1M with a tyrosine-to- leucine substitution at amino acid 88 in 2Apro (B), P1M-2A-Y88S, a small-plaque mutant of P1M with a tyrosine-to-serine substitution at amino acid 88 in 2Apro (C), and P2L-5′UTR, a small-plaque mutant of P2L with a single nucleotide substitution in the 5′ untranslated region (D).
Other PV mutants were examined to rule out the possibility that any replication-defective virus would be sensitive to IFN-α. A small-plaque mutant of P2L with a nucleotide substitution in the 5′ UTR, P2L-5′UTR (53), replicated as well in the presence of IFN (Fig. 3D) as did the wild-type parental virus (Fig. 1B). The Sabin type 3 vaccine strain, whose genome has 10 mutations that contribute to a small-plaque phenotype (60), also replicated to high titers in mock-treated and IFN-α-treated cells (data not shown).
To determine whether changes in other PV proteins could confer the ability to replicate in IFN-treated cells, we attempted to isolate viruses that could form plaques on monolayers of IFN-α-treated HeLa cells. HeLa cell monolayers were treated with 2,000 units/ml of IFN-α for 24 h, infected with a known quantity of wild-type P1M, P1M-2A-Y88S, P1M-2A-Y88L, or EMCV, and covered with an agar overlay containing IFN-α. The efficiencies of plaque formation differed among the viruses (Table 1). The plaquing efficiency was highest for wild-type PV, at 50%. All viruses isolated from plaques arising on IFN-treated plates encoded wild-type 2Apro. The frequency of plaque formation was much lower for P1M-2A-Y88S and P1M-2A-Y88L than for wild-type PV and was zero for EMCV. The frequency of plaque formation for P1M-2A-Y88S was approximately 1/120,000 on IFN-treated cells, and all viruses within the plaques encoded the wild-type amino acid at residue 88. P1M-2A-Y88L had a higher efficiency of plaquing on IFN-treated cells (∼1/3,000), though it was still more than 1,000-fold lower than that observed for wild-type virus. The viruses within the P1M-2A-Y88L plaques all encoded a tyrosine or a phenylalanine at amino acid 88. Only one nucleotide change is required to convert leucine to phenylalanine, explaining why revertants arose with greater frequency with P1M-2A-Y88L than with P1M-2A-Y88S. A P1M-2A-Y88F mutant virus is able to replicate to wild-type titers in IFN-treated cells (data not shown).
TABLE 1.
Efficiency of plaque formation on IFNα-pretreated cells
| Virus | Efficiency of plaques recovereda | Change(s) at amino acid 88 of 2Apro | Nucleotide changes in codon 88 of 2Apro |
|---|---|---|---|
| P1M-WT | 1/2 | None | None |
| P1M-2A-Y88L | 1/3,000 | L to Y, L to F | CTC to TAC, TAT, or TTC |
| P1M-2A-Y88S | 1/120,000 | S to Y | TCG to TAC or TAT |
| EMCV | 0 | NAb | NA |
Number of plaques on cells treated with 2,000 units/ml IFNα relative to number of plaques on untreated cells.
NA, not applicable.
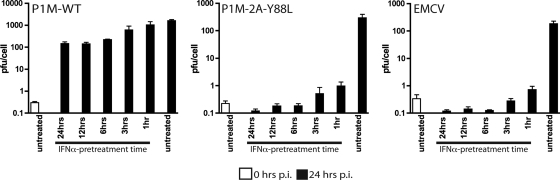
Effect of IFN treatment time on virus replication.
In all of the experiments described in this paper, cells were pretreated with IFN for 24 h. It has been shown that different sets of ISGs are induced at various times after treatment with IFN (24). To investigate the effect of IFN treatment times on viral replication, HeLa cells were treated for different lengths of time with IFN-α and then infected with virus at an MOI of 10. As in previous experiments, IFN was not added back to the cells after infection. Virus production was determined at 0 and 24 h postinfection. Complete inhibition of EMCV and P1M-2A-Y88L replication was observed after treatment of cells for 3 h or longer, indicating that some of the ISGs that are antagonized by enteroviruses are induced early after IFN-α treatment (Fig. 4).
FIG. 4.
Effect of IFN treatment time on the replication of EMCV, wild-type P1M (P1M-WT), and P1M-2A-Y88L. Monolayers of HeLa cells were pretreated with 1,000 units/ml IFN-α for 0, 1, 3, 6, 12, or 24 h and then infected with P1M-WT (A), P1M-2A-Y88L (B), or EMCV (C). Total virus was harvested at 24 h post infection (p.i.) for all conditions and also at 0 h postinfection for cells that had not been treated with IFN-α. Viral titers were determined by plaque assay on HeLa cells.
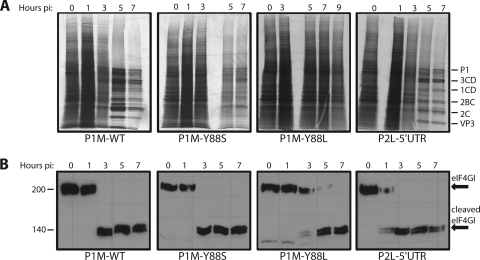
Protein synthesis and eIF4GI levels in cells infected with PV 2Apro mutants.
An important cellular substrate for 2Apro is eIF4G (5, 37, 38). Cleavage of this translation protein leads to inhibition of host protein synthesis during picornavirus infection (1, 5, 22, 63, 68). To determine whether translation inhibition plays a role in the ability of PV to replicate in cells treated with IFN-α, cellular protein synthesis was examined after labeling with [35S]methionine. Host protein synthesis was inhibited beginning at 3 h after infection with wild-type PV, and by 5 h postinfection, mainly viral proteins were detected (Fig. 5A). Inhibition of host protein synthesis coincides with the complete cleavage of eIF4GI by 3 h postinfection (Fig. 5B). Infection with the PV 2Apro mutant P1M-2A-Y88S, which is IFN sensitive, also leads to inhibition of cellular protein synthesis (Fig. 5A) and cleavage of eIF4GI (Fig. 5B) with kinetics similar to those observed during infection with wild-type virus. Cells infected with the other IFN-sensitive virus, P1M-2A-Y88L, display a different eIF4GI cleavage and host translation inhibition phenotype. During P1M-2A-Y88L infection, there is a lag in eIF4GI cleavage compared to that of the wild type, with some intact eIF4GI remaining until 7 h postinfection (Fig. 5B). No translation shutoff was observed even at 9 h postinfection (Fig. 5A). Protein was not harvested at points past 9 h postinfection due to the high level of cell death seen at those times. Infection with the IFN-resistant PV mutant P2L-5′UTR leads to cleavage of eIF4GI even earlier in infection than with wild-type virus (Fig. 5B) and inhibition of cellular translation, though not as effectively as with wild-type virus or P1M-2A-Y88S (Fig. 5A). These results show that neither PV-induced host translation inhibition or eIF4GI cleavage correlates with the ability to replicate in IFN-treated cells. This conclusion is highlighted by the different properties of the two IFN-sensitive mutants: infection with P1M-2A-Y88S, but not P1M-2A-Y88L, leads to inhibition of host translation and eIF4GI cleavage early in infection.
FIG. 5.
Protein synthesis and eIF4GI cleavage in HeLa cells infected with different PV mutants. (A) HeLa cell monolayers were infected at an MOI of 10 with wild-type P1M (P1M-WT) or with the PV mutant P1M-2A-Y88L, P1M-2A-Y88S, or P2L-5′UTR. At the indicated time postinfection (p.i.), cells were labeled with [35S]methionine for 15 min. Cytoplasmic extracts were analyzed by 10% SDS-polyacrylamide gel electrophoresis. (B) Monolayers of HeLa cells were infected with virus at an MOI of 10. Cytoplasmic extracts were prepared at each time point and fractionated on a 6% SDS-polyacrylamide gel. Western blot analysis was carried out with rabbit anti-eIF4GI antibody.
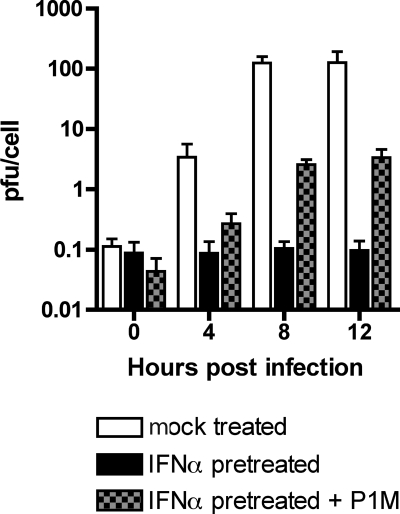
Effect of PV on EMCV replication in IFN-α-treated HeLa cells.
We hypothesize that replication of P1M in IFN-treated cells is a consequence of antagonism of the IFN response. To test this hypothesis, we attempted to rescue an IFN-sensitive virus by coinfection with P1M. This experiment was complicated by the inhibition of cap-dependent translation that takes place in cells infected with P1M. Consequently, VSV replication is completely inhibited in cells that are coinfected with P1M (data not shown). We used EMCV as the IFN-sensitive virus, because translation of the genome occurs via an internal ribosomal entry site and is not affected by cleavage of eIF4G. HeLa cells were coinfected with P1M at an MOI of 1 and EMCV at an MOI of 10 to minimize PV-mediated inhibition of EMCV replication at higher MOIs (data not shown). As expected, EMCV did not replicate in IFN-treated HeLa cells (Fig. 1 and 6). However, when IFN-α-treated HeLa cells were coinfected with P1M and EMCV, EMCV replicated to a titer of about 3 PFU/cell (Fig. 6). These results indicate that infection with P1M modifies the cells to allow EMCV replication in the presence of ISGs.
FIG. 6.
EMCV replication in HeLa cells coinfected with PV. HeLa cell monolayers were pretreated with 1,000 units/ml IFN-α for 24 h or were left untreated before they were infected with EMCV (MOI, 10) alone or coinfected with P1M (MOI, 1). Total virus was harvested at the indicated times postinfection, and the EMCV titers were determined by plaque assay on 3T3 cells. P1M does not form plaques on this cell line.
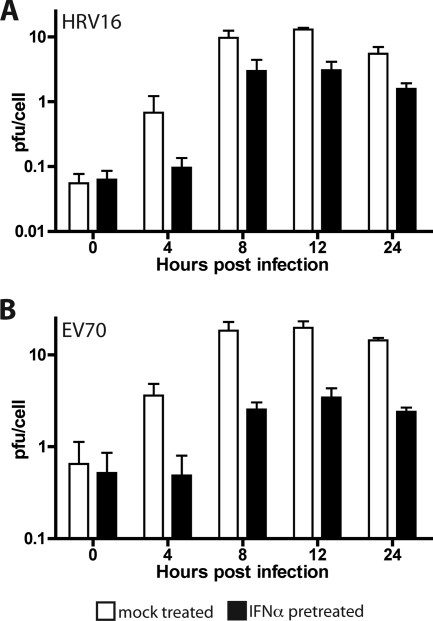
Replication of other enteroviruses in IFN-α-treated HeLa cells.
The picornavirus family includes diverse members in addition to PV and EMCV. It was therefore of interest to determine the effect of IFN-α pretreatment on the replication of other members of this virus family. When untreated cells were infected with EV70 or HRV16, both viruses replicated to about 15 PFU/cell (Fig. 7). Neither virus replicated as well in untreated cells as PV did but nevertheless showed IFN resistance, replicating to about 3 PFU/cell in IFN-treated HeLa cells.
FIG. 7.
Replication of HRV16 and EV70 in cells treated with IFN-α. HeLa cell monolayers were pretreated with 1,000 units/ml IFN-α for 24 h or were left untreated before they were infected with HRV16 (A) or EV70 (B) at an MOI of 10. Total virus was harvested at the indicated hours postinfection, and viral titers were determined by plaque assay on HeLa cells.
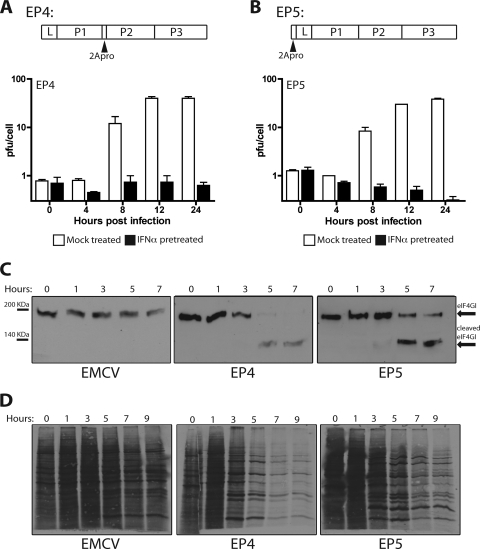
Replication of EMCV-P1M chimeras in IFN-α-treated cells.
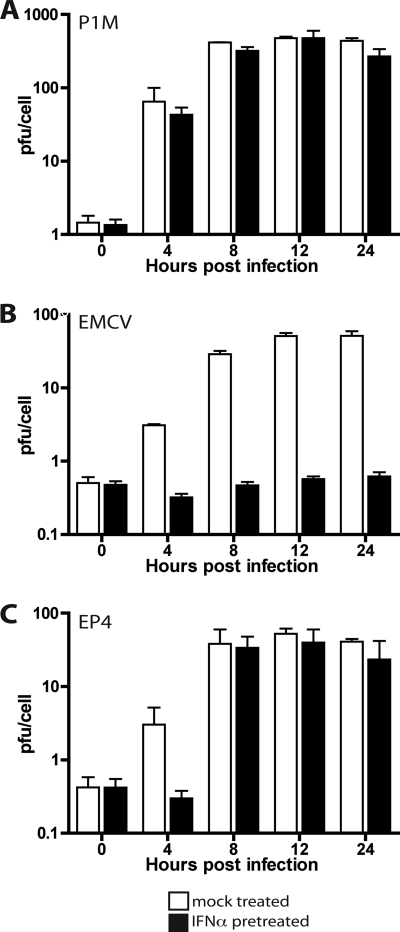
Although the genome of EMCV encodes a 2A protein, it is very different from enterovirus 2Apro and does not have proteolytic activity. We therefore investigated whether insertion of the coding sequence for PV 2Apro into the genome of EMCV would allow viral replication in IFN-α-treated cells. Chimeric viruses were constructed with 2Apro of P1M (with or without an N-terminal FLAG epitope) inserted between P1 and P2 of EMCV (EP1 and EP4) or upstream of L (EP3 and EP5). The results of Western blot analysis demonstrated that the 2Apro proteins were produced in HeLa cells infected with chimeric viruses EP1 and EP3 (data not shown). Cleavage of eIF4GI was observed in cells infected with all chimeric viruses (Fig. 8D and data not shown), and shutoff of cellular translation was observed in cells infected with EP4 and EP5 (Fig. 8C). These results demonstrate that 2Apro, when produced from the EMCV genome, is capable of cleaving at least one of its known host substrates. The chimeric viruses EP4 and EP5 replicated slightly less well than wild-type EMCV in untreated cells (Fig. 8A and B). However, replication of the chimeric viruses in HeLa cells was completely inhibited by IFN pretreatment. Interestingly, the levels of EP4 remained constant in IFN-treated HeLa cells while the levels of EP5 declined (Fig. 8A and B). This observation suggests that EP4, unlike EP5, might have a higher level of IFN resistance, which might allow it to replicate at low levels. We therefore examined replication of P1M, EMCV, and EP4 in cells treated with lower doses of IFN. A dose of 10 units/ml IFN-α was chosen for growth curve analysis, because EP4 replication was inhibited at higher doses of IFN-α pretreatment (data not shown). Yields of P1M were unaffected, while EMCV replication was completely blocked in HeLa cells treated with 10 units/ml IFN-α (Fig. 9). Under the same conditions, the yield of EP4 was approximately 40 PFU/cell. To determine the efficiency of plaquing, HeLa cell monolayers were treated with IFN-α for 24 h, infected with P1M, EMCV, or EP4, and covered with an agar overlay. EP4 failed to form plaques on cells that were pretreated with 1,000 or 2,000 units/ml IFN-α (data not shown) but had a plaquing efficiency of 1/450 on cells that had been pretreated with 100 units/ml IFN-α. The efficiency of plaquing of P1M at this concentration was 1, while that of EMCV was zero.
FIG. 8.
Replication, eIF4GI cleavage, and translation in HeLa cells. (A and B) Diagrams of EMCV polyprotein showing the locations of PV 2Apro in EMCV chimeras EP4 and EP5. Duplicate monolayers of HeLa cells were pretreated with 1,000 units/ml IFN-α for 24 h or were left untreated before they were infected at an MOI of 10 with EP4 or EP5. Total virus was harvested at different hours postinfection, and viral titers were determined by plaque assay on HeLa cells. (C) Cytoplasmic extracts were prepared from the second set of monolayers described for panels A and B and fractionated by electrophoresis on a 6% SDS-polyacrylamide gel, and Western analysis was carried out with rabbit anti-eIF4GI antibody. (D) HeLa cell monolayers were infected at an MOI of 10 with EMCV, EP4, or EP5. At the indicated times postinfection, cells were labeled with [35S]methionine for 15 min. Cytoplasmic extracts were fractionated by 12% SDS-polyacrylamide gel electrophoresis.
FIG. 9.
Effect of 10 units/ml IFN-α pretreatment on viral replication. Monolayers of HeLa cells were pretreated with 10 units/ml IFN-α for 24 h or were left untreated and then infected with P1M (A), EMCV (B), or EP4 (C) at a multiplicity of infection (MOI) of 10. Total virus was harvested at the indicated times postinfection, and viral titers were determined by plaque assay on HeLa cells.
DISCUSSION
We have shown that the enteroviruses EV70, HRV16, and PV types 1, 2, and 3 can replicate in type I IFN-pretreated cells. In contrast, replication of EMCV and VSV is completely inhibited by IFN pretreatment. In our experiments, IFN is not present postadsorption, allowing us to distinguish between antagonism of IFN signaling and antagonism of ISG activity. At least 3 h of IFN pretreatment is required for complete inhibition of EMCV replication. This observation indicates that the ISGs that inhibit picornaviral replication are produced soon after type I IFN treatment. Since IFN pretreatment leads to the production of ISGs, and enteroviruses can replicate in cells that have been pretreated with type I IFN, enteroviruses must have some means of neutralizing the activity of ISGs that target picornaviral replication. Infection of cells that do not produce type I IFNs provides further evidence for abrogation of ISG activity. Vero cells and K562 cells do not produce IFN but are able to produce ISGs in response to the cytokine. Replication of EMCV is completely inhibited in these cell lines when they are pretreated with IFN-α, and PV replication is inhibited to a level similar to that in cell types that produce IFN. If the ability of PV to replicate in IFN-α-pretreated cells were a consequence of blocking type I IFN production, we would expect that viral replication would be unencumbered or significantly less encumbered in IFN-α-pretreated K562 and Vero cells.
Enterovirus IFN resistance is dependent on MOI. At high or intermediate MOIs (100, 10, or 1), PV replication is relatively resistant to IFN, yielding ∼100 PFU/cell. At a lower MOI (0.1), resistance decreases and virus production is ∼4 PFU/cell. The number of virions that infect each cell may determine the efficiency of replication in cells treated with IFN. At an MOI of 100 or 10, more than 99.95% of cells receive more than 1 virus particle. In contrast, at an MOI of 0.1, only 9.5% of the cells are infected and only 0.47% of cells receive more than one particle. When more virions enter a cell, viral proteins more rapidly accumulate to higher levels. Consequently, the antiviral activities of ISGs are countered early in infection to allow more robust viral replication.
PV replicates only in specific cells and tissues in primates, even though the virus reaches many organs during the viremic phase (reviewed in reference 52). In mice, the IFN response is an important determinant of PV tissue tropism (50). PV infection of CD155 transgenic mice lacking the receptor for IFN-α/β leads to viral replication in the liver, spleen, and pancreas, in addition to the central nervous system. In contrast, PV replication is limited to the central nervous system of wild-type CD155 transgenic mice. Our finding that PV replicates in a variety of human and monkey cell lines treated with IFN-α suggests that the antiviral effect of the cytokine may vary depending on the species or cell type. Understanding the mechanism by which enteroviruses limit the antiviral effects of IFN will provide insight into the mechanism of viral pathogenesis.
Small-plaque PV mutants with changes at amino acid 88 in 2Apro cannot replicate in IFN-pretreated cells. Other small-plaque PV mutants with changes elsewhere in the genome can replicate under the same conditions, indicating that IFN sensitivity is not conferred by any mutation that leads to a small-plaque phenotype. These data suggest that 2Apro functions as an IFN antagonist. When we attempted to select for IFN-resistant viruses by plaque assay of the two 2Apro mutants on IFN-α-treated cells, the only viruses that were recovered had changes at residue 88 of 2Apro to the tyrosine of the wild type or to phenylalanine. Tyrosine and phenylalanine are structural similar, and as expected, a Y88F 2Apro mutant replicated to wild-type levels in IFN-pretreated cells. These observations strongly implicate 2Apro in IFN antagonism.
Our results indicate that 2Apro is necessary for replication in cells pretreated with IFN but that it is not apparently sufficient for full expression of this phenotype. An EMCV recombinant that synthesizes 2Apro of PV (EP4) has a higher level of IFN resistance than wild-type EMCV, as measured by plaquing efficiency on HeLa cells pretreated with 100 units/ml IFN-α and by analysis of growth in cells pretreated with 10 units/ml IFN-α. However, EP4 is unable to replicate in cells that have been treated with high levels (1,000 units/ml) of IFN, in contrast to enteroviruses such as PV, EV70, and HRV16. This observation suggests that other enteroviral proteins, in addition to 2Apro, are required for replication in cells treated with high levels of IFN. Other viral proteins might be required together with 2Apro to overcome the higher levels of ISGs produced after treatment with 1,000 units/ml IFN-α. Alternatively, efficient antagonism by 2Apro may require the proteinase to be part of a membrane-bound precursor polyprotein. There are precedents for such a requirement. Initiation of PV negative-strand synthesis in HeLa extracts does not occur when a mixture of processed forms of 2A, 2B, 2C, 2AB, and 2BC are synthesized (29). Instead initiation occurs only when the precursor, P23, or a mix of 2A and 2BCP3, or of P2 and P3, is added to the extracts. The 3Cpro of hepatitis A virus cleaves the mitochondrial outer membrane protein MAVS only when it is part of the precursor, 3ABC (65). The precursor is required for cleavage because the hydrophobic protein 3A targets the proteinase to the mitochondrial membrane.
Viral proteases have been implicated as antagonists of several steps in the IFN response. The NS3/4A protease of hepatitis C virus cleaves MAVS to abrogate IFN production (19). 3Cpro and 2Apro of PV induce apoptosis, which leads to MDA-5 cleavage in a proteasome- and caspase-dependent manner (3). 3Cpro of hepatitis A virus, as part of the 3ABC precursor, cleaves MAVS (65), and the p65-RelA component of the NF-κB complex is cleaved by PV 3Cpro (49). Foot-and-mouth disease virus leader proteinase also subverts type I IFN production (8) by cleaving p65-RelA (9). These examples illustrate antagonism of upstream events in the IFN response. Until now, no viral proteases have been implicated in blocking the events that occur after IFN binds to cells. We have shown here that there is an inhibitory role for 2Apro in the most downstream event in IFN signaling, ISG activity. Which ISG proteins inhibit PV replication and how their activities are blocked by 2Apro remain to be elucidated. It has been reported that PKR, an ISG induced during infection with many viruses, is degraded in cells infected with PV (5). However, we found no change in the amount of PKR during PV infection of HeLa cells when the protein was examined by Western blot analysis (J. M. Morrison and V. R. Racaniello, unpublished data). Because there are more than 300 ISGs, identification of specific proteins whose activities are modulated during enterovirus infection will require the use of large-scale proteomic or genomic screening.
Acknowledgments
This work was supported in part by Public Health Service grants AI50754, AI068017, and T32AI07161 from the National Institute of Allergy and Infectious Diseases.
We thank Bert Semler for the PV 3Cpro mutant Se1-3C-02 and Ann Palmenberg for EMCV EC4.
Footnotes
Published ahead of print on 11 February 2009.
REFERENCES
- 1.Barco, A., E. Feduchi, and L. Carrasco. 2000. A stable HeLa cell line that inducibly expresses poliovirus 2Apro: effects on cellular and viral gene expression. J. Virol. 742383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, P., and N. A. McMillan. 1999. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology 259305-313. [DOI] [PubMed] [Google Scholar]
- 3.Barral, P. M., J. M. Morrison, J. Drahos, P. Gupta, D. Sarkar, P. B. Fisher, and V. R. Racaniello. 2007. MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 813677-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biron, C. A., and G. C. Sen. 2007. Innate responses to viral infection, p. 249-278. In P. M. Howley, D. M. Knipe, P. M. Howley, D. E. Griffin, and R. A. Lamb (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 5.Black, T. L., B. Safer, A. Hovanessian, and M. G. Katze. 1989. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J. Virol. 632244-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cayley, P. J., J. A. Davies, K. G. McCullagh, and I. M. Kerr. 1984. Activation of the ppp(A2′p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2′p)nA-dependent RNase. Eur. J. Biochem. 143165-174. [DOI] [PubMed] [Google Scholar]
- 7.Chebath, J., P. Benech, M. Revel, and M. Vigneron. 1987. Constitutive expression of (2′-5′) oligo A synthetase confers resistance to picornavirus infection. Nature 330587-588. [DOI] [PubMed] [Google Scholar]
- 8.de Los Santos, T., S. de Avila Botton, R. Weiblen, and M. J. Grubman. 2006. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J. Virol. 801906-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Los Santos, T., F. Diaz-San Segundo, and M. J. Grubman. 2007. Degradation of nuclear factor kappa B during foot-and-mouth disease virus infection. J. Virol. 8112803-12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69912-920. [PubMed] [Google Scholar]
- 11.Dewalt, P. G., and B. L. Semler. 1987. Site-directed mutagenesis of proteinase 3C results in a poliovirus deficient in synthesis of viral RNA polymerase. J. Virol. 612162-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA 855259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 758158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, B., L. Xu, A. Zhou, B. A. Hassel, X. Lee, P. F. Torrence, and R. H. Silverman. 1994. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J. Biol. Chem. 26914153-14158. [PubMed] [Google Scholar]
- 15.Duke, G. M., and A. C. Palmenberg. 1989. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J. Virol. 631822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durbin, J. E., R. Hackenmiller, M. C. Simon, and D. E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84443-450. [DOI] [PubMed] [Google Scholar]
- 17.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43247-252. [DOI] [PubMed] [Google Scholar]
- 18.Ferran, M. C., and J. M. Lucas-Lenard. 1997. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J. Virol. 71371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 1022986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, X. Y., D. S. Kessler, S. A. Veals, D. E. Levy, and J. E. Darnell, Jr. 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA 878555-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 185208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glaser, W., A. Triendl, and T. Skern. 2003. The processing of eIF4GI by human rhinovirus type 2 2Apro: relationship to self-cleavage and role of zinc. J. Virol. 775021-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hato, S. V., C. Ricour, B. M. Schulte, K. H. Lanke, M. de Bruijni, J. Zoll, W. J. Melchers, T. Michiels, and F. J. van Kuppeveld. 2007. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell Microbiol. 92921-2930. [DOI] [PubMed] [Google Scholar]
- 24.Holko, M., and B. R. Williams. 2006. Functional annotation of IFN-alpha-stimulated gene expression profiles from sensitive and resistant renal cell carcinoma cell lines. J. Interferon Cytokine Res. 26534-547. [DOI] [PubMed] [Google Scholar]
- 25.Hui, D. J., C. R. Bhasker, W. C. Merrick, and G. C. Sen. 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP. Met-tRNAi. J. Biol. Chem. 27839477-39482. [DOI] [PubMed] [Google Scholar]
- 26.Isaacs, A., and J. Lindenmann. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147258-267. [PubMed] [Google Scholar]
- 27.Joachims, M., P. C. Van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, M., A. Davidson, L. Hibbert, P. Gruenwald, J. Schlaak, S. Ball, G. R. Foster, and M. Jacobs. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 795414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurgens, C., and J. B. Flanegan. 2003. Initiation of poliovirus negative-strand RNA synthesis requires precursor forms of p2 proteins. J. Virol. 771075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurgens, C. K., D. J. Barton, N. Sharma, B. J. Morasco, S. A. Ogram, and J. B. Flanegan. 2006. 2Apro is a multifunctional protein that regulates the stability, translation and replication of poliovirus RNA. Virology 345346-357. [DOI] [PubMed] [Google Scholar]
- 31.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. Virgin IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 2991575-1578. [DOI] [PubMed] [Google Scholar]
- 32.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 33.Kempf, B. J., and D. J. Barton. 2008. Poliovirus 2A(Pro) increases viral mRNA and polysome stability coordinately in time with cleavage of eIF4G. J. Virol. 825847-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, M. S., and V. R. Racaniello. 2007. Enterovirus 70 receptor utilization is controlled by capsid residues that also regulate host range and cytopathogenicity. J. Virol. 818648-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovarik, P., I. Sauer, and B. Schaljo. 2007. Molecular mechanisms of the anti-inflammatory functions of interferons. Immunobiology 212895-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krausslich, H. G., M. J. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 612711-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 27021975-21983. [DOI] [PubMed] [Google Scholar]
- 38.Lamphear, B. J., R. Yan, F. Yang, D. Waters, H. D. Liebig, H. Klump, E. Kuechler, T. Skern, and R. E. Rhoads. 1993. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 26819200-19203. [PubMed] [Google Scholar]
- 39.Landsteiner, K., and E. Popper. 1908. Mikroscopische präparate von einem menschlichen und zwei affentückermarker. Wien Klin. Wochenschr. 211830. [Google Scholar]
- 40.Lee, W. M., W. Wang, and R. R. Rueckert. 1995. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes 9177-181. [DOI] [PubMed] [Google Scholar]
- 41.Leib, D. A., M. A. Machalek, B. R. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 976097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, X., H. H. Lu, S. Mueller, and E. Wimmer. 2001. The C-terminal residues of poliovirus proteinase 2Apro are critical for viral RNA replication but not for cis- or trans-proteolytic cleavage. J. Gen. Virol. 82397-408. [DOI] [PubMed] [Google Scholar]
- 43.Liebig, H. D., J. Seipelt, E. Vassilieva, A. Gradi, and E. Kuechler. 2002. A thermosensitive mutant of HRV2 2A proteinase: evidence for direct cleavage of eIF4GI and eIF4GII. FEBS Lett. 52353-57. [DOI] [PubMed] [Google Scholar]
- 44.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 176660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meraz, M. A., J. M. White, K. C. Sheehan, E. A. Bach, S. J. Rodig, A. S. Dighe, D. H. Kaplan, J. K. Riley, A. C. Greenlund, D. Campbell, K. Carver-Moore, R. N. DuBois, R. Clark, M. Aguet, and R. D. Schreiber. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84431-442. [DOI] [PubMed] [Google Scholar]
- 46.Meurs, E. F., Y. Watanabe, S. Kadereit, G. N. Barber, M. G. Katze, K. Chong, B. R. Williams, and A. G. Hovanessian. 1992. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J. Virol. 665805-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, D. M., B. M. Rahill, J. M. Boss, M. D. Lairmore, J. E. Durbin, J. W. Waldman, and D. D. Sedmak. 1998. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J. Exp. Med. 187675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 62279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neznanov, N., K. M. Chumakov, L. Neznanova, A. Almasan, A. K. Banerjee, and A. V. Gudkov. 2005. Proteolytic cleavage of the p65-RelA subunit of NF-kappaB during poliovirus infection. J. Biol. Chem. 28024153-24158. [DOI] [PubMed] [Google Scholar]
- 50.Ohka, S., H. Igarashi, N. Nagata, M. Sakai, S. Koike, T. Nochi, H. Kiyono, and A. Nomoto. 2007. Establishment of a poliovirus oral infection system in human poliovirus receptor-expressing transgenic mice that are deficient in alpha/beta interferon receptor. J. Virol. 817902-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pagano, J. S., and A. Vaheri. 1965. Enhancement of infectivity of poliovirus RNA with diethylaminoethyl-dextran (DEAE-D). Arch. Gesamte Virusforsch. 17456-464. [DOI] [PubMed] [Google Scholar]
- 52.Pallansch, M., and R. Roos. 2007. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 839-894. In P. M. Howley, D. M. Knipe, P. M. Howley, D. E. Griffin, and R. A. Lamb (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 53.Pelletier, J., M. E. Flynn, G. Kaplan, V. Racaniello, and N. Sonenberg. 1988. Mutational analysis of upstream AUG codons of poliovirus RNA. J. Virol. 624486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Racaniello, V. 2007. Picornaviridae: the viruses and their replication, p. 795-838. In P. M. Howley, D. M. Knipe, P. M. Howley, D. E. Griffin, and R. A. Lamb (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 55.Racaniello, V. 1984. Poliovirus type II produced from cloned cDNA is infectious in mice. Virus Res. 1669-675. [Google Scholar]
- 56.Racaniello, V. R., and D. Baltimore. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214916-919. [DOI] [PubMed] [Google Scholar]
- 57.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 891-47. [DOI] [PubMed] [Google Scholar]
- 58.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 122061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubinstein, S., P. C. Familletti, and S. Pestka. 1981. Convenient assay for interferons. J. Virol. 37755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanway, G., P. J. Hughes, R. C. Mountford, P. Reeve, P. D. Minor, G. C. Schild, and J. W. Almond. 1984. Comparison of the complete nucleotide sequences of the genomes of the neurovirulent poliovirus P3/Leon/37 and its attenuated Sabin vaccine derivative P3/Leon 12a1b. Proc. Natl. Acad. Sci. USA 811539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeuchi, O., and S. Akira. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 2017-22. [DOI] [PubMed] [Google Scholar]
- 62.Toyoda, H., M. J. Nicklin, M. G. Murray, C. W. Anderson, J. J. Dunn, F. W. Studier, and E. Wimmer. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45761-770. [DOI] [PubMed] [Google Scholar]
- 63.Ventoso, I., A. Barco, and L. Carrasco. 1998. Mutational analysis of poliovirus 2Apro. Distinct inhibitory functions of 2apro on translation and transcription. J. Biol. Chem. 27327960-27967. [DOI] [PubMed] [Google Scholar]
- 64.Ventoso, I., and L. Carrasco. 1995. A poliovirus 2Apro mutant unable to cleave 3CD shows inefficient viral protein synthesis and transactivation defects. J. Virol. 696280-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang, Y., Y. Liang, L. Qu, Z. Chen, M. Yi, K. Li, and S. M. Lemon. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. USA 1047253-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshikawa, T., T. Iwasaki, M. Ida-Hosonuma, M. Yoneyama, T. Fujita, H. Horie, M. Miyazawa, S. Abe, B. Simizu, and S. Koike. 2006. Role of the alpha/beta interferon response in the acquisition of susceptibility to poliovirus by kidney cells in culture. J. Virol. 804313-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, S. F., and R. E. Lloyd. 1991. Identification of essential amino acid residues in the functional activity of poliovirus 2A protease. Virology 182615-625. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, X., B. J. Lamphear, D. Xiong, K. Knowlton, and R. E. Rhoads. 2003. Protection of cap-dependent protein synthesis in vivo and in vitro with an eIF4G-1 variant highly resistant to cleavage by coxsackievirus 2A protease. J. Biol. Chem. 2784449-4457. [DOI] [PubMed] [Google Scholar]