Abstract
Our method concentrates on and constructs the distinguished single gene network. An integrated method was proposed based on linear programming and a decomposition procedure with integrated analysis of the significant function cluster using Kappa statistics and fuzzy heuristic clustering. We tested this method to identify ATF2 regulatory network module using data of 45 samples from the same GEO dataset. The results demonstrate the effectiveness of such integrated way in terms of developing novel prognostic markers and therapeutic targets.
1. Introduction
In the postgenomic era, with microarray technologies producing great deal of gene expression data, mining these data to get insight into biological processes at system-wide level has become a challenge for bioinformatics. On one hand, due to the complex and distribute nature of biological research, there is a great deal of methods for inferring gene regulatory networks. But all these methods focused on constructing the complicated entire network calculated from the given microarray data. The tremendous amounts of genes in those networks distribute analysts' attention, so it is hard to get any clear perception of valuable knowledge from such complicated networks, let alone further study of each single gene. On the other hand, the wide spread of knowledge over independent databases aggravates the hardness of integrating comprehensive annotation information for genes and lowers the study effectiveness. Thus, a novel method integrating both single molecular network construction and highly centralized gene-functional-annotation analysis is in demand for gene network and functional analysis.
This paper proposed an integrated method based on linear programming and a decomposition procedure with integrated analysis of the significant function cluster using Kappa statistics and fuzzy heuristic clustering. Our method concentrates on and constructs the distinguished single gene network integrated with function prediction analysis by DAVID. For the distinguished single molecular network, we did (1) control and experiment comparison, (2) identification of activation and inhibition networks, (3) construction of upstream and downstream feedback networks, and (4) functional module construction. We tested this method to identify ATF2 regulation network module using data of 45 samples from one and the same GEO dataset. The results demonstrate the effectiveness of such integrated way in terms of developing novel prognostic markers and therapeutic targets.
2. Methods
2.1. Distinguished Single Molecular Network Construction
The entire network was constructed using GRNInfer [1] and GVedit tools. GRNInfer is a novel mathematic method called gene network reconstruction (GNR) tool based on linear programming and a decomposition procedure that is used for inferring gene networks. The method theoretically ensures the derivation of the most consistent network structure with respect to all of the datasets, thereby not only significantly alleviating the problem of data scarcity but also remarkably improving the reconstruction reliability. The general solution for a single dataset is the following (1), which represents all of the possible networks:
| (1) |
where J = (Jij)n×n = ∂f(x)/∂x is an n × n Jacobian matrix or connectivity matrix, X = (x(t1),…, x(tm)), A = (a(t1),…, a(tm)), and X′ = (x′(t1),…, x′(tm)) are all n × m matrices with xi′(tj) = [xi(tj+1) − xi(tj)]/[tj+1 − tj] for i = 1,…, n; j = 1,…, m. X(t) = (x1(t),…, xn(t))T ∈ Rn, a = (a1,…, an)T ∈ Rn, xi(t) is the expression level (mRNA concentrations) of gene i at time instance t. y = (yij) is an n × n matrix, where yij is zero if ej ≠ 0 and is otherwise an arbitrary scalar coefficient. ⋀−1 = diag (1/ei) and 1/e is set to be zero if ei = 0. U is a unitary m × n matrix of left eigenvectors, ⋀ = diag (e1,…, en) is a diagonal n × n matrix containing the n eigenvalues, and VT is the transpose of a unitary n × n matrix of right eigenvectors.
But the entire network is too complex to get any clear perception of such complicated relationships among those genes, let alone further study of each single gene. We constructed the distinguished single molecular network by selecting the centered gene and its directly related genes based on the entire network for further study. We take into account the effectiveness of biology study in order to concentrate on single molecular network rather than the intricate entire network. It is helpful to get intensive and deep insight of the whole network. For the distinguished single molecular network, we did (1) control and experiment comparison, (2) identification of activation and inhibition networks, (3) construction of upstream and downstream feedback networks, and (4) functional module construction.
2.2. Functional Annotation Clustering
For the function of genes that is neither determined by their sequence nor by the protein families they belong to [2], the function of those genes included in the same single molecular network should not be interpreted separately, but should be analyzed together according to the whole single molecular network. This method takes into account the network nature of biological annotation contents in order to concentrate on the larger biological picture rather than an individual gene. We used DAVID to do functional annotation clustering. It changes functional annotation analysis from term- or gene-centric to biological module-centric [2] in accordance with our network analysis aim.
The DAVID gene functional clustering tool provides typical batch annotation and gene-GO term enrichment analysis for highly throughput genes by classifying them into gene groups based on their annotation term co-occurrence [3]. DAVID uses a novel algorithm to measure relationships among the annotation terms based on the degrees of their coassociation genes to group similar annotation contents from the same or different resources into annotation groups. The grouping algorithm is based on the hypothesis that similar annotations should have similar gene members. The functional annotation clustering integrates the same techniques of Kappa statistics to measure the degree of the common genes between two annotations, and fuzzy heuristic clustering to classify the groups of similar annotations according kappa values [4, 5]. The tool also allows observation of the internal relationships of the clustered terms by comparing it to the typical linear, redundant term report, over which similar annotation terms may be distributed among many other terms.
3. Results and Discussion
We tested this method using microarrays containing 22215 genes in 40 MPM tumors and 5 normal pleural tissues from one and the same GEO datasets. We identified potential tumor molecular markers and chose the top 51 significant positive genes with normalization of log2, the minimum fold change = 3.5, delta = 1.59, and a false-discovery rate of 0% using SAM [6]. We selected activating transcription factor (ATF)-2 because it is one of the most distinguished genes in MPM. It is a member of the ATF/cyclic AMP-responsive element binding protein family of transcription factors.
3.1. Normal Tissues and Tumor Comparisons of Distinguished Single Molecular Network
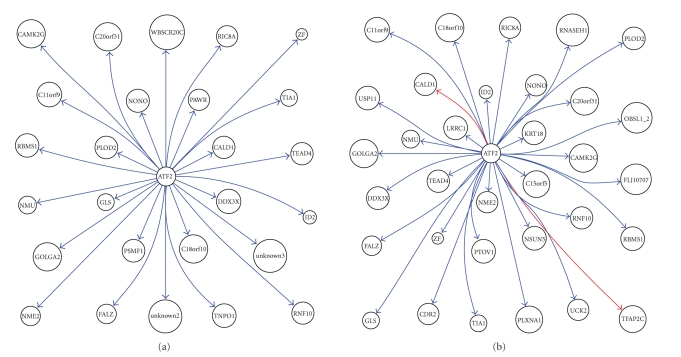
We, respectively, constructed the interaction network of the above 51 genes in healthy tissues and that in tumor using GRNInfer [1] and GVedit tools and selected the ATF2-centered downstream subnetworks. With comparison of these ATF2-centered subnetworks, we can get a more clear perception of the notable differences between normal tissues and tumor, as shown in Figure 1. It appeared that ATF2 inhibits C11orf9, C18orf10, C20orf31, CALD1, CAMK2G, DDX3X, FALZ, GLS, GOLGA2, ID2, NME2, NMU, NONO, PAWR, PLOD2, PSMF1, RBMS1, RIC8A, RNF10, TEAD4, TIA1, TNPO1, unknown2, unknown3, WBSCR20C, and ZF in normal tissues, as shown in Figure 1(a). It appeared that ATF2 inhibits C11orf9, C15orf5, C18orf10, C20orf31, CAMK2G, CDR2, DDX3X, FALZ, FLJ10707, GLS, GOLGA2, ID2, KRT18, LRRC1, NME2, NMU, NONO, NSUN5, OBSL1_2, PLOD2, PLXNA1, PTOV1, RBMS1, RIC8A, RNASEH1, RNF10, TEAD4, TIA1, UCK2, USP11, and ZF, while it activates CALD1 and TFAP2C in tumor, as shown in Figure 1(b).
Figure 1.
ATF2 downstream network in (a) normal tissue and (b) MPM tissue.
With comparison between the two results, notable differences can be shown clearly in order to get further perception of pathological changes in MPM. For example, ATF2 target genes appeared in ATF2 activation to CALD1, TFAP2C in MPM, as only shown in Figure 2(b). Caldesmon (CALD1) is a potential actomyosin regulatory protein found in smooth muscle and nonmuscle cells [7]. Transcription factor AP2-gamma (TFAP2C) is alternatively titled AP2. Families of related transcription factors are often expressed in the same cell lineages but at different times or sites in the developing embryo. The AP2 family appears to regulate the expression of genes required for development of tissues of ectodermal origin such as neural crest and skin [8]. AP2 may also be involved in the overexpression of c-erbB-2 in human breast cancer cells [9].
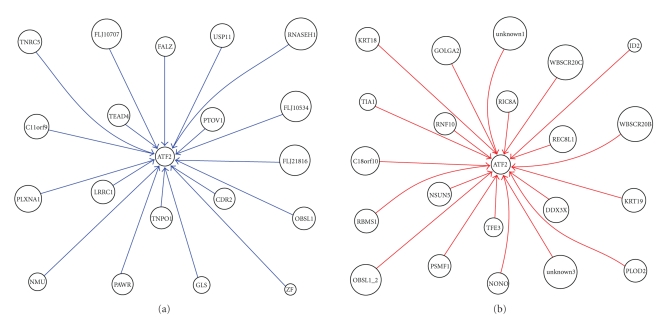
Figure 2.
(a) ATF2 upstream inhibition network of MPM; (b) ATF2 upstream activation network of MPM.
3.2. Identification of Activation and Inhibition Networks for the Distinguished Single Molecule
We also identified the activation and inhibition networks, respectively, in order to simplify and intensify the analysis process. For example, in ATF2 upstream network of MPM, as shown in Figure 2, it appeared that C11orf9, CDR2, FALZ, FLJ10534, FLJ10707, FLJ21816, GLS, LRRC1, NMU, OBSL1, PAWR, PLXNA1, PTOV1, RNASEH1, TEAD4, TNPO1, TNRC5, USP11, and ZF inhibit ATF2, as shown in Figure 2(a), whereas C18orf10, DDX3X, GOLGA2, ID2, KRT18, KRT19, NONO, NSUN5, OBSL1_2, PLOD2, PSMF1, RBMS1, REC8L1, RIC8A, RNF10, TFE3, TIA1, unknown1, unknown3, WBSCR20B, and WBSCR20C activate ATF2, as shown in Figure 2(b).
ATF2 upstream genes TFE3, REC8L1 showed activation to ATF2. TFE3 is a member of the helix-loop-helix family of transcription factors and binds to the mu-E3 motif of the immunoglobulin heavy-chain enhancer and is expressed in many cell types [10]. Nakagawa et al. [11] identified TFE3 as a transactivator of metabolic genes that are regulated through an E box in their promoters which led to metabolic consequences such as activation of glycogen and protein synthesis, but not lipogenesis, in liver [11]. REC8L1 is the human homolog of yeast Rec8, a meiosis-specific phosphoprotein involved in recombination events [12]. Brar et al. (2006) showed that phosphorylation of the cohesin subunit REC8 contributes to stepwise cohesin removal [13].
3.3. Constructing Feedback Network of the Distinguished Single Upstream and Downstream Gene
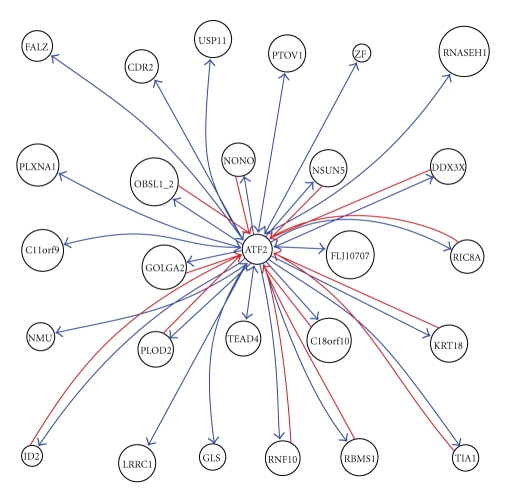
We took into account the feedback relationship and setup ATF2 feedback network, as shown in Figure 3. ATF2 target genes appeared in ATF2 inhibition to CDR2, GLS, and USP11, consistently, its upstream genes also appeared in CDR2, GLS, and USP11 inhibition to ATF2. CDR2 is also called CDR62, where CDR means cerebellar degeneration-related. On Western blot analysis of Purkinje cells and tumor tissue, the anti-Yo sera react with at least 2 antigens, a major species of 62 kD called CDR62 and a minor species of 34 kD called CDR34 [14]. Sahai (1983) demonstrated phosphate-activated glutaminase (GLS) in human platelets [15]. It is the major enzyme yielding glutamate from glutamine. Significance of the enzyme derives from its possible implication in behavior disturbances in which glutamate acts as a neurotransmitter [16]. USP11 is also called UHX1. Swanson et al. (1996) cited evidence indicating that ubiquitin hydrolases play a role in oncogenesis (oncogenes and tumor suppressor gene products are degraded in ubiquitin-dependent pathways) [17]. The relationship of ATF2 with CDR2, GLS, and USP11 represents a negative feedback loop.
Figure 3.
ATF2 feedback subnetwork of MPM.
3.4. Functional Module Construction of the Distinguished Single Gene
According to ATF2 upstream network, we did DAVID analysis of function cluster, respectively. The DAVID functional annotation clustering results appeared that one ATF2 regulation network was identified as consisting of the ATF2 upstream genes including RBMS1, RNASEH1, PTOV1, NONO, C11orf9, PSMF1, TIA1, TEAD4, GLS, ID2, USP11, TNPO1, PAWR, PLOD2, and TFE3, as shown in Figure 4.
Figure 4.
One ATF2 upstream gene metabolic network including RBMS1, RNASEH1, PTOV1, NONO, C11orf9, PSMF1, TIA1, TEAD4, GLS, ID2, USP11, TNPO1, PAWR, PLOD2, and TFE3.
According to Figure 2, it appeared that RBMS1, NONO, PSMF1, TIA1, ID2, PLOD2, TFE3 activate ATF2; whereas RNASEH1, PTOV1, C11orf9, TEAD4, GLS, USP11, TNPO1, and PAWR inhibit ATF2.
RBMS1, NONO, TIA1, ID2, and TFE3 enhance nucleoside, nucleotide, and nucleic acid metabolism because RBMS1, NONO, TIA1, ID2, and TFE3 are involved in these metabolism; PSMF1 activation to ATF2 means the increase of Acyl-CoA metabolism and porphyrin metabolism; PLOD2 activation to ATF2 indicates the progress of cholesterol metabolism and other protein metabolism, as shown in Figure 5.
Figure 5.
Molecular function and biological process from DAVID.
RNASEH1, PTOV1, and TEAD4 inhibition to ATF2 decreases nucleoside, nucleotide, and nucleic acid metabolism mediated by the three genes; C11orf9 inhibition to ATF2 means the decline of polysaccharide metabolism, whereas GLS represents the weakness of amino acid and cyclic nucleotides metabolism; USP11 inhibition to ATF2 indicates the fall-off in protein metabolism and modification, whereas PAWR in glycogen metabolism, as shown in Figure 5.
4. Conclusions
Our method concentrates on and constructs the distinguished single gene network integrated with function prediction analysis by DAVID. For the distinguished single molecular network, we did (1) control and experiment comparison, (2) identification of activation and inhibition networks, (3) construction of upstream and downstream feedback networks, and (4) functional module construction. We tested this method to identify ATF2 regulation network module using data of 45 samples from one and the same GEO dataset. The results demonstrate the effectiveness of such integrated way in terms of developing novel prognostic markers and therapeutic targets.
Acknowledgments
This work was supported by the National Natural Science Foundation in China (no. 60673109 and no. 60871100) and the Teaching and Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, State Key Lab of Pattern Recognition Open Foundation.
References
- 1.Wang Y, Joshi T, Zhang X-S, Xu D, Chen L. Inferring gene regulatory networks from multiple microarray datasets. Bioinformatics. 2006;22(19):2413–2420. doi: 10.1093/bioinformatics/btl396. [DOI] [PubMed] [Google Scholar]
- 2.Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biology. 2007;8(9, article R183):1–16. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Research. 2007;35, web server issue:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20(1):37–46. [Google Scholar]
- 5.Byrt T, Bishop J, Carlin JB. Bias, prevalence and kappa. Journal of Clinical Epidemiology. 1993;46(5):423–429. doi: 10.1016/0895-4356(93)90018-v. [DOI] [PubMed] [Google Scholar]
- 6.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphrey MB, Herrera-Sosa H, Gonzalez G, Lee R, Bryan J. Cloning of cDNAs encoding human caldesmons. Gene. 1992;112(2):197–204. doi: 10.1016/0378-1119(92)90376-z. [DOI] [PubMed] [Google Scholar]
- 8.Williamson JA, Bosher JM, Skinner A, Sheer D, Williams T, Hurst HC. Chromosomal mapping of the human and mouse homologues of two new members of the AP-2 family of transcription factors. Genomics. 1996;35(1):262–264. doi: 10.1006/geno.1996.0351. [DOI] [PubMed] [Google Scholar]
- 9.Bosher JM, Williams T, Hurst HC. The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(3):744–747. doi: 10.1073/pnas.92.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henthron PS, Stewart CC, Kadesch T, Puck JM. The gene encoding human TFE3, a transcription factor that binds the immunoglobulin heavy-chain enhancer, maps to Xp11.22. Genomics. 1991;11(2):374–378. doi: 10.1016/0888-7543(91)90145-5. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa Y, Shimano H, Yoshikawa T, et al. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nature Medicine. 2006;12(1):107–113. doi: 10.1038/nm1334. [DOI] [PubMed] [Google Scholar]
- 12.Parisi S, McKay MJ, Molnar M, et al. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Molecular and Cellular Biology. 1999;19(5):3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brar GA, Kiburz BM, Zhang Y, Kim J-E, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441(7092):532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- 14.Fathallah-Shaykh H, Wolf S, Wong E, Posner JB, Furneaux HM. Cloning of a leucine-zipper protein recognized by the sera of patients with antibody-associated paraneoplastic cerebellar degeneration. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(8):3451–3454. doi: 10.1073/pnas.88.8.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahai S. Glutaminase in human platelets. Clinica Chimica Acta. 1983;127(2):197–203. doi: 10.1016/s0009-8981(83)80004-7. [DOI] [PubMed] [Google Scholar]
- 16.Prusiner SB. Disorders of glutamate metabolism and neurological dysfunction. Annual Review of Medicine. 1981;32:521–542. doi: 10.1146/annurev.me.32.020181.002513. [DOI] [PubMed] [Google Scholar]
- 17.Swanson DA, Freund CL, Ploder L, McInnes RR, Valle D. A ubiquitin C-terminal hydrolase gene on the proximal short arm of the X chromosome: implications for X-linked retinal disorders. Human Molecular Genetics. 1996;5(4):533–538. doi: 10.1093/hmg/5.4.533. [DOI] [PubMed] [Google Scholar]