Abstract
Krüppel-like factor 4 is a zinc finger-type transcription factor expressed in a variety of tissues, including the epithelium of intestine and the skin, where it is important in differentiation and cell cycle arrest. KLF4 can both activate and repress transcription, depending on the gene targeted. Moreover, KLF4 can function as a tumor suppressor or an oncogene, depending on the cellular context. Finally, KLF4 is important in reprogramming differentiated fibroblasts into inducible pluripotent stem cells, which highly resemble embryonic stem cells. This review will summarize what is known about the diverse functions of KLF4, as well as their molecular mechanisms.
Keywords: Krüppel-like factor 4, colorectal cancer, stem cell
Krüppel-like factor 4 (KLF4) is a transcription factor expressed in a wide variety of tissues in humans, including the intestine and the skin, and is important in many different physiologic processes, including development, differentiation, and maintenance of normal tissue homeostasis. KLF4 is a bi-functional transcription factor that can either activate or repress transcription, depending on the target gene, and utilizing different mechanisms. In addition, KLF4 can function as an oncogene or a tumor suppressor depending on the type of cancer involved. In concert with three other transcription factors, KLF4 can reprogram differentiated fibroblasts into a state resembling embryonic stem cells in every possible manner tested so far. This review will provide a detailed summary of what is currently known about KLF4 and its role in the homeostasis of tissues, in cancer, and in stem cell reprogramming.
The Krüppel-like Factor Family
Krüppel-like factors are a family of transcription factors that play an important role in many fundamental biologic processes including development, proliferation, differentiation, and apoptosis (Fig. 1). Krüppel-like factor family members contain three C-terminal C2H2-type zinc fingers that bind DNA, and were named “Krüppel-like” due to strong homology in this region with the Drosophila gene product Krüppel. Krüppel is important in segmentation of the developing embryo, and genetic deletion of Krüppel results in complete absence of the thoracic and anterior abdominal segments [1]. KLF4 was cloned independently by two groups, and given two different names: gut-enriched Krüppel-like factor (GKLF) due to the fact that it was found to be highly expressed in the intestine [2], and epithelial zinc finger (EZF) due to its high expression in the skin epithelium [3]. GKLF/EZF was later renamed KLF4 to avoid confusion, as expression of KLF4 is also detectable in the lung, skin, testis [2–5], thymus [6], cornea [7], cardiac myocytes [8] and lymphocytes [9]. In addition, KLF4 is important in development, as it is detectable in the mouse embryo, with the highest expression occurring in the later stages [3,4].
Fig. 1.
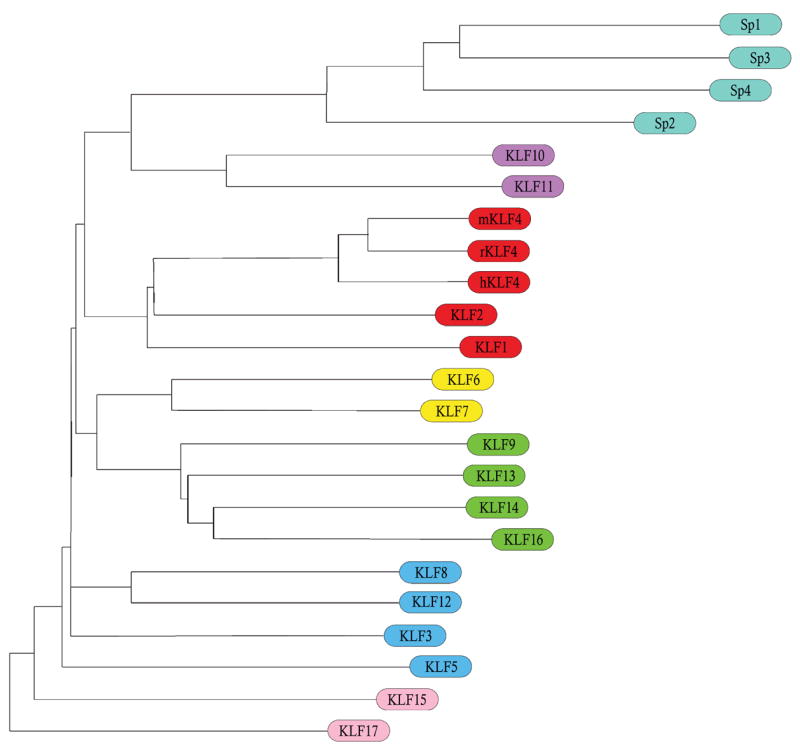
Phylogenetic tree of the Sp/KLF transcription factor family Amino acid sequence comparison between KLF/Sp family members. Note human, mouse, and rat KLF4 are included for comparison as well (hKLF4, mKLF4, and rKLF4, respectively). Horizontal distance on the tree is proportional to number of residue changes between adjacent members.
Roles of KLF4 in Homeostasis of the Colonic Epithelium
The colonic epithelium consists of three major types of differentiated cells, enterocytes, goblet cells, and enteroendocrine cells. Actively proliferating cells reside at the base of the crypts and migrate towards the luminal surface as they differentiate, eventually to be sloughed off. KLF4 inhibits proliferation and promotes differentiation, and consistent with this role, expression of KLF4 is greatest near the luminal surface and gradually decreases toward the base of the crypts [2,10]. Klf4−/− mice lack goblet cells, without affecting the total number of enterocytes, suggesting that KLF4 may be specifically required for goblet cell differentiation [11]. In addition, KLF4 can interact with β-catenin and antagonize Wnt signaling [10], a key pathway in driving proliferation of the intestinal epithelium [12–14]. Thus, KLF4 may also be important in mediating the switch from transit-amplifying cells to the various differentiated cell types in the colonic crypts.
Butyrate is constantly produced in the colon by bacterial fermentation of dietary fiber in the intestine [15], and can induce expression of KLF4 [5,16]. In cell culture, butyrate stimulates expression of the enterocyte-specific marker intestinal alkaline phosphatase (IAP) [17], and induces colon cancer cells to acquire a more differentiated, enterocyte-like phenotype [18]. KLF4 positively regulates expression of IAP [19,20], and overexpression of KLF4 in cell culture inhibits proliferation [2,5].
Moreover, KLF4 appears to have inhibitory effect on a wide variety of cellular processes, including protein and cholesterol synthesis, transcription, cell growth, and DNA repair [21,22]. Consistent with its anti-proliferative role, KLF4 simultaneously induces the expression of cyclin-dependent kinase inhibitor proteins p21Cip1/WAF1 [23–25] and p57Kip2 [21], and represses the expression of cyclin D1 [5,26,27], cyclin D2 [28], cyclin E [29], and cyclin B1 [30] (Fig. 2). In addition, KLF4 represses expression of ornithine decarboxylase [7,31], an enzyme involved in the production of a class of molecules known as polyamines, which are also important in proliferation. KLF4 is required for both the G1/S-phase [32,33] and G2/M-phase [30] checkpoints. Finally, KLF4 represses expression of p53 and may be important in determining whether cells decided to undergo apoptosis or cell cycle arrest [34].
Fig. 2.
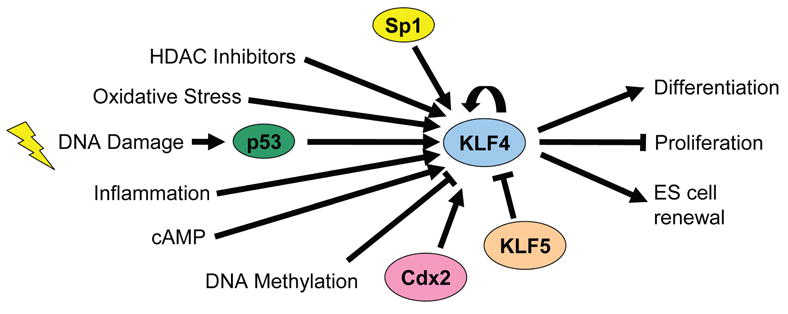
KLF4 signaling pathways Expression of KLF4 is upregulated by many stimuli, including DNA damage, inflammation, oxidative stress, and HDAC inhibitors. Sp1, Cdx2, and p53 positively regulate the KLF4 promoter, whereas KLF5 represses its expression. Overall, KLF4 functions to promote differentiation and inhibit proliferation. KLF4 is also important in ES cell renewal.
Roles in other homeostasis of other tissues
Although the importance of KLF4 in the intestine is well characterized, increasing evidence demonstrates its importance in other organs and tissues as well. For example, Klf4−/− mice die soon after birth of dehydration due to defects in the epidermal barrier of the skin [35], whereas targeted overexpression of KLF4 results in early formation of the epithelial permeability barrier [36]. These data clearly implicate KLF4 as an important molecule in differentiation of the skin epithelium.
Furthermore, overexpressed KLF4 can synergize with maternally-injected corticosteroids in accelerating the formation of the skin barrier. This is likely due to overlap between the genes targeted by KLF4 and the glucocorticoid receptor [37]. The utility of glucocorticoids in lung maturation of premature infants is well-established [38], thus it might be interesting to determine whether KLF4 or possibly other Krüppel-like factors could synergize with glucocorticoids in fetal lung maturation as well. Also in the developing fetus, KLF4 synergizes with Sp1 in up-regulating expression of PSG-5, a protein secreted into the maternal circulation by the placenta [39]. PSG-5 is thought be required for maintenance of a term pregnancy and may protect the fetus from attack by the maternal immune system. In addition, KLF4 and PSG-5 have closely overlapping patterns of expression in the placenta, suggesting an in vivo role for KLF4 in the regulation of PSG-5 expression [40].
Human KLF4 was isolated from a umbilical vein cDNA library and is expressed in the vascular endothelium [41]. Expression of KLF4 is induced by shear stress in endothelial cells [42], whereas KLF4 appears to block differentiation and is expressed at low levels in differentiated arterial smooth muscle cells [43]. However, expression of KLF4 is rapidly up-regulated in smooth muscle cells in response to vascular injury [44].
Overexpression of KLF4 in a pro-myelocytic cell line increases the expression of monocyte markers, whereas knockdown of KLF4 decreases TPA-induced overexpression of these same markers. In addition, Klf4−/− hematopoietic stem cells less frequently differentiate into monocytes [45]. When fetal liver cells from Klf4−/− mice were transplanted into lethally-irradiated wild-type mice, they had undetectable levels of circulating inflammatory monocytes [46]. Thus KLF4 appears to be important for both resident and inflammatory monocyte differentiation.
KLF4 is highly expressed in the corneal epithelium, where it is important in differentiation. Targeted deletion of KLF4 in the eye results in corneal fragility, edema, and a lack of goblet cells in the conjunctiva [47]. In a cell culture model of adipocyte differentiation using 3T3-L1 cells, siRNA-mediated knockdown of KLF4 completely blocked expression of several phenotypic markers of differentiated adipocytes [48]. Collectively, these data strongly implicate KLF4 as a factor involved in the differentiation of many tissues.
Roles of KLF4 in Cancers
As an anti-proliferative factor expressed in differentiated epithelia, it seems logical that KLF4 might act as a tumor suppressor, and indeed this appears to be the case in the gastrointestinal tract [49,50]. However, recent evidence suggests that KLF4 might also act as an oncogene in certain contexts [51]. This section will investigate these two contrasting roles.
KLF4 as a tumor suppressor
Increasing evidence implicates KLF4 as a tumor suppressor in the intestinal epithelium. In human colorectal carcinoma, expression of KLF4 is downregulated, with evidence of both hypermethylation and loss-of-heterozygosity [52–54]. However, no association has been found between downregulation of KLF4 and tumor staging or 5-year survival in patients with metastatic carcinoma, suggesting that loss of KLF4 in colorectal cancer may be an early event [53,54].
Examination of KLF4 expression in mouse models of colorectal cancer has yielded similar results. The APCmin/+ mouse develops hundred of intestinal adenomas early in life and is a widely-used model of intestinal tumorigenesis [55,56]. In adenomas from these mice, KLF4 is down-regulated, with expression inversely related to the size of the tumor [4,57]. As APC is a critical component of the Wnt/β-catenin pathway and APCmin/+ mice express a truncated form of the APC protein, these mice have deregulated Wnt signaling in their intestine [58,59]. Interestingly, KLF4 can interact with β-catenin in the nucleus and repress Wnt signaling in vivo, as well as inhibit tumor growth in tumor xenografts [10]. In addition, crossing APCmin/+ mice with KLF4+/− heterozygotes resulted in significantly more adenomas than in APCmin/+ mice alone [60]. Notably, this phenotype was similar to that found with another double mutant, APCMin/+/TCF1−/−. The most abundant isoform of TCF1 expressed in the intestine is also an antagonist of Wnt/β-catenin signaling, suggesting that an important effect of decreased KLF4 expression during colorectal tumorigenesis may be de-repression of Wnt signaling. In human colon cancer cell lines, several point mutations have been found in the KLF4 gene. One mutation had a significant effect on the ability to activate a p21Cip1/WAF1 reporter construct in NIH3T3 cells [52]. However, an investigation to identify mutations in tissue samples of human colorectal cancers has not yet been performed. In the HCT116 colorectal cancer cell line, KLF4 is required to prevent centrosome amplification after gamma-irradiation, and loss of KLF4 may promote chromosomal instability [29]. In addition, KLF4 represses expression of the enzyme ornithine decarboxylase [31], a proto-oncogene that alone is sufficient to transform NIH3T3 cells [61]. Collectively, these data strongly implicate KLF4 as a tumor suppressor in the colon. Strong evidence implicates KLF4 as tumor suppressor in the gastric epithelium as well. Similar to colorectal cancer, KLF4 is downregulated in gastric cancer, with evidence of loss-of-heterozygosity and hypermethylation [62–64]. Moreover, targeted loss of the Klf4 gene in the gastric mucosa of mice results in pre-cancerous changes in the stomach [65]. In examining both normal and cancerous gastric mucosal tissue from humans, one study found an inverse relationship between the expression of KLF4 and Sp1, a distantly related Krüppel-like factor family member (Fig. 1) [62]. In addition, the same study found that in gastric cancer cell lines, KLF4 can directly repress the expression of Sp1. Given that strong expression of Sp1 is correlated with poor survival in gastric cancer [66], loss of KLF4 may contribute to gastric cancer progression.
In addition to gastric and colorectal cancer, KLF4 is downregulated in esophageal cancer [67,68], bladder cancer [69], non-small-cell lung carcinoma [70], and leukemia [71,72].
KLF4 as an oncogene
Although these data clearly demonstrate that KLF4 can act as tumor suppressor in multiple tissues, the possibility that KLF4 might be an oncogene as well was first demonstrated almost one decade ago. Using E1A-immortalized rat kidney epithelial cells (RK3E) to screen for factors that could induce transformation, KLF4 was identified. Moreover, KLF4-transformed RK3E cells could produce tumors in xenografted mice [73]. KLF4 is overexpressed in laryngeal squamous cell carcinoma as an early event in its progression [73]. Expression of KLF4 is increased in ductal carcinoma of the breast [74] and increased nuclear staining is associated with a more aggressive phenotype and poorer prognosis [75]. In the skin, overexpression of KLF4 results in hyperplasia and dysplasia [76], eventually leading to squamous cell carcinoma [77].
Whether KLF4 acts as a tumor suppressor or an oncogene is likely due to differences in cell context, expression patterns of other genes, and the chromatin environment of individual cells, but a mechanism to fully explain these differences is lacking. Some insight was gained in a recent study where it was found that KLF4 could override RasV12-induced senescence in primary fibroblasts and induce transformation [34]. Furthermore, this study demonstrated that whether overexpression of KLF4 induced transformation or resulted in cell cycle arrest depended on the status of p21Cip1/WAF1, a transcriptional target of KLF4. Overexpression of KLF4 alone increases expression of p21Cip1/WAF1 and results in cell cycle arrest. However, the addition of RasV12 resulted in inhibition of p21Cip1/WAF1 expression, allowing the ability of KLF4 to repress p53 to predominate. Repression of p53 effectively blocked apoptosis and in concert with the decreased expression of p21Cip1/WAF1, eventually led to transformation. Thus, KLF4 can be added to a growing list of genes that have multiple, context-dependent roles in cancer, including CDKN1A (p21), TGF-β, Ras, and NOTCH1 [51].
Roles of KLF4 in Stem Cell Renewal and Reprogramming
Recently, it was found that overexpression of KLF4, in combination with three other transcription factors could transform mouse fibroblasts into a state resembling embryonic stem cells (ES cells). These cells have been termed “inducible pluripotent stem cells” (iPS cells) [78]. By replacing the open reading frame of Fbx15, a non-essential marker of embryonic stem cells, with a neomycin resistance gene, it was hypothesized that neomycin-resistant colonies might have somehow reprogrammed themselves into embryonic stem cells. After screening a short list of potential factors, it was found that the simultaneous infection of retroviruses expressing Oct3/4, Sox2, c-Myc, and KLF4 were able to produce resistant clones. These cells could form teratomas that contained differentiated tissues from all three germ layers, confirming their pluripotency. This approach was further refined by screening for neomycin resistance based on Nanog or Oct4 expression instead of Fbx15. Unlike Fbx15-iPS cells, Nanog and Oct4-iPS could produce chimeric mice and, could generate live late-term embryos when injected into tetraploid blastocysts [79–81]. Thus, Nanog and Oct4-iPS satisfy are even more stringent tests of pluripotency than Fbx15-iPS cells.
An area currently under intense investigation is understanding the molecular events that occur during stem cell reprogramming as well as the precise role of each of the four individual factors required. The importance of Oct3/4 and Sox2 in ES cell renewal is well established [82]. What is less clear is the function of the other two factors that make up the “magic brew”: c-Myc and KLF4. One possibility is that c-Myc and KLF4 confer increased proliferative capacity on potential iPS cells, since both can function as oncogenes [83]. Since c-Myc regulates a significant number of genes, its function may be to effect global changes in the chromatin environment by recruiting histone acetyl-transferase complexes. According this model, KLF4 may then function to inhibit apoptosis induced by overexpression of c-Myc. KLF4 represses expression of c-Myc expression in colon cancer cells through inhibiting Wnt signaling [10]. Thus, c-Myc may provide a balance for KLF4. The role of Wnt signaling in iPS cells is still an open question.
Overexpression of KLF4 in ES cells inhibited differentiation into erythroid progenitors, and increased their capacity to generate secondary embryoid bodies, suggesting a role for KLF4 in self-renewal [84]. In concert with Oct3/4 and Sox2, KLF4 activates expression of Lefty1, a gene expressed in ES cells, but lost during differentiation [85]. In addition, KLF4-null mice survive to term and have no detectable defects during embryogenesis in their pluripotent stem cell population [11,35], suggesting that in normal ES cells, KLF4 may be dispensable. More recently, human iPS have been produced using a slightly different mix of factors, substituting c-Myc and KLF4 with Nanog and LIN28 [86], further calling into question the overall importance of c-Myc and KLF4. It has even been suggested that c-Myc and KLF4 are merely molecular catalysts, in that they might accelerate or increase the efficiency of the reprogramming process, but are otherwise not absolutely required [87].
However, a recent study has found that the function in ES cell self-renewal of KLF4 is partially redundant with KLF2 and KLF5, as knockdown of all three Krüppel-like factors, but not any one individually, resulted in spontaneous ES cell differentiation [88]. In addition, significant overlap was found between genes regulated by Nanog and the three Krüppel-like factors. Clearly, a complete understanding of the role of KLF4 in ES cell self-renewal and iPS cell reprogramming awaits further study
Molecular Mechanisms of KLF4
Human and mouse KLF4 are 470 and 483 amino acids in length, respectively, and produce a 55 kDa protein. KLF4 can be roughly divided into three separate domains: an N-terminal activation domain [3,41,89], a central repressive domain [41], and a C-terminal DNA binding domain (Fig. 3). The DNA binding domain consists of three successive zinc fingers. Each zinc finger contains an anti-parallel β-sheet, followed by a short loop and an α-helix. Two cysteines within the β-sheet and two histidines within the α-helix work together to coordinate a single zinc ion, which acts to stabilize the fold. Each zinc finger interacts with three consecutive nucleotides on a target DNA sequence, and the sequence specificity of a zinc finger protein can be increased simply by adding additional zinc fingers [90].
Fig. 3.
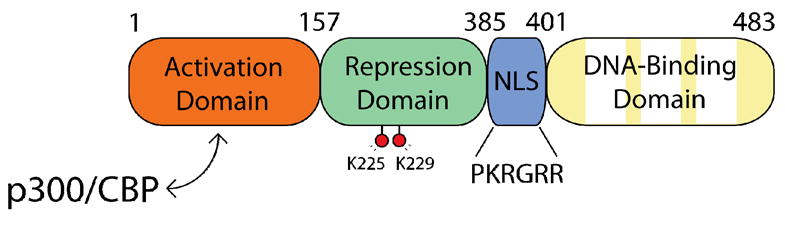
Functional domains of the KLF4 protein N-terminus of KLF4 contains a transactivation domain known to interact with the co-activators p300/CBP. The central region contains a repression domain, as well as two lysines that are acetylated by p300/CBP, followed by a hexapeptide nuclear localization sequence (NLS). Finally, the C-terminus contains the DNA binding domain, consisting of three sequential zinc fingers (each zinc finger is colored in white).
In general, KLF4 interacts with GT-rich or CACCC elements on target genes [41,91]. Although one report suggests that KLF4 prefers to bind a RRGGYGY sequence (where R=A/G and Y=C/T) [92], it is still not clear whether this is a true consensus in vivo. KLF4 is exclusively nuclear, like many other transcription factors, and appears to contain two discrete nuclear localization sequences (NLS). The first is an basic hexapeptide sequence just N-terminal to the three C-terminal zinc fingers, whereas the second is contained within the first two zinc fingers themselves [93].
Given the large number of genes regulated by KLF4, it is not unexpected that expression of KLF4 itself should be highly regulated (Table 1). In the colon cancer cell line HCT116, KLF4 has a half-life of only 2 hours and is quickly degraded by the proteasome [94]. However, a variety of stimuli can induce KLF4 expression including serum starvation, contact inhibition [3], interferon- [31,95], sodium butyrate [5,16], cAMP [48], gastrin [96], DNA damage [24,33], and oxidative stress [8,25]. The precise mechanism of how the majority these stimuli increase the expression of KLF4 is unclear, although possibilities include increased transcription of the KLF4 gene, increased mRNA stability, and/or increased protein stability.
Table 1.
Factors and conditions that modulate expression of KLF4
| Factor/condition | References |
|---|---|
| Increase expression | |
| Butyrate | [5,16] |
| Cdx2 | [23,97] |
| Contact inhibition | [2,3] |
| Endothelin-1 | [8] |
| γ-irradiation | [33] |
| H2O2 | [8,25] |
| Interferon-γ | [31,95,104] |
| IBMX | [48] |
| KLF4 | [98] |
| LPS | [104] |
| Methyl methanesulfonate | [24] |
| p53 | [24] |
| Serum starvation | [2,3] |
| Shear stress | [110] |
| Sp1 | [23] |
| Sp3 | [23] |
| TNF-α | [104] |
| Trichostatin A | [5,16] |
| TGF-β | [111] |
| Decrease expression | |
| KLF5 | [98] |
| Promoter methylation | [52,57] |
Although much remains to be known about how expression KLF4 is regulated, several transcription factors have been found to regulate its promoter. For example, p53 transactivates the KLF4 gene, and p53 is required for the induction of KLF4 after DNA damage [24,33]. CDX2, another protein important in differentiation of the intestinal epithelium, can activate a KLF4 reporter construct [97]. This suggests that KLF4 may act downstream of CDX2, although more work is necessary to demonstrate this in vivo. KLF4 up-regulates its own expression by binding to its promoter, whereas KLF5 inhibits KLF4 expression and blocks the binding of KLF4 to its promoter [98]. Although KLF4 and KLF5 are closely related transcription factors, expression of KLF5 is found in a completely opposite pattern in the colonic intestine, with strongest expression found in the actively proliferating cells at the base of the crypts and absent expression in differentiated cells at the luminal surface [99,100]. In fact, KLF4 and KLF5 have several antagonizing roles in the intestinal epithelium, as reviewed elsewhere [49].
Mechanism of activation
A major function of KLF4 is to activate transcription of target genes (Table 2). Consistent with this function, the N-terminus of KLF4 contains a strong transactivation domain [3,41,89]. This domain alone, when directly fused to its three C-terminal zinc fingers, is sufficient to activate a synthetic reporter construct [89]. In addition, the N-terminal domain interacts with the transcriptional co-activators p300 and CBP, and this interaction is required for its function, as point mutations that block interactions with CBP also completely abrogate its ability to activate transcription [20,89]. p300/CBP are histone acetyltransferase (HAT) proteins, and recruitment of p300/CBP results in an increase in localized histone acetylation at the promoter. Acetylation of histones facilitates the recruitment of other transcription factors as well as the basal transcriptional machinery. In addition, KLF4 itself is acetylated by p300/CBP at lysine residues 225 and 229. Mutation of these two lysines to arginine significantly decreases the ability of KLF4 to transactivate target genes, as well as its ability to inhibit proliferation [20], suggesting that acetylation of KLF4 is important for its function.
Table 2.
Targets regulated by KLF4
| Factor/condition | References | Factor/condition | References |
|---|---|---|---|
| Activation targets | Repression targets | ||
| 1200015N20Rik | [85] | Bax | [60] |
| A33 antigen | [112] | CD11d | [108] |
| B2R | [113] | Cyclin B1 | [30] |
| CYP1A1 | [105] | Cyclin D1 | [5,26,27] |
| Cytokeratin 4 | [67] | Cyclin E | [29] |
| EBV ED-L2 | [114] | Fgf5 | [88] |
| hSMVT | [115] | Histidine decarboxylase | [106] |
| IAP | [19,21,116] | KLF2 | [85,88] |
| iNOS | [104] | Laminin α1 | [117] |
| Keratin 4 | [114] | Nes | [88] |
| Keratin 19 | [118] | Ornithine decarboxylase | [31] |
| KLF4 | [23,98] | p53 | [34] |
| Laminin-α 3A | [119] | PAI-1 | [104] |
| Laminin-γ 1 | [120] | SM22α | [43] |
| Lefty1 | [85] | SM α-actin | [121] |
| Nanog | [85,88] | Sp1 | [62] |
| Oct4 | [88] | ||
| p21Cip1 | [23–25] | ||
| p27Kip1 | [25] | ||
| p57Kip2 | [21] | ||
| PKG-Iα | [122] | ||
| Rb | [25] | ||
| Sox2 | [88] | ||
| SPRR1A | [67] | ||
| SPRR2A | [67] | ||
| Tbx3 | [88] | ||
| u-PAR | [123] |
One report found that KLF4 can interact with Tip60, a bi-functional cofactor that contains intrinsic HAT activity, but can also recruit HDAC7 [96]. Tip60 is a co-activator for several nuclear hormone receptors [101] as well as APP [102], but appears to function as a co-repressor for STAT3 by recruiting HDAC7 [103]. Another zinc finger protein Krox20, can directly interact with KLF4 and synergistically activate the C/EBPβgene in 3T3-L1 cells [48]. KLF4 interacts with the NF-κB subunit p65/RelA and synergistically activates expression of iNOS [104]. Thus, the mechanisms of transactivation mediated by KLF4 may be gene-dependent.
Mechanism of repression
One mechanism for repression by a transcription factor is to simple competition with an activator for binding to a target DNA sequence. This mechanism is known as a form of passive repression. On the CYP1A1, HDC, and SP1 genes, KLF4 binds to a sequence overlapping that recognized by the activator Sp1, displacing Sp1 from the promoter and resulting in repression of the target gene [62,105,106]. Since Sp1 is ubiquitously expressed and positively regulates many genes [107], it is likely this is mechanism is used by KLF4 to repress many of its target genes. GAL4 fusion assays demonstrate that KLF4 contains central repressive domain in addition to its more fully characterized transactivation domain [41]. This suggests that KLF might actively repress expression of some genes, in addition to, or instead of passive repression via competition with a transcriptional activator. In KLF4-mediated repression of the CD11d gene, KLF4 interacts with and recruits HDAC1 and HDAC2 [108], whereas KLF4 represses cyclin B1 via specifically recruiting HDAC3 [20]. On the TP53 gene, MUC1-C recruits KLF4, as well as HDAC1 and HDAC3, to mediate repression [109]. KLF4 inhibits Smad3-mediated activation of PAI-1 by directly competing with Smad3 for p300 binding [104]. Finally, KLF4 represses transcriptional targets of Wnt signaling by directly interacting with β-catenin/TCF4 [10]. These data strongly suggest that KLF4-mediated activation and repression is complex and gene-dependent.
Final Thoughts
KLF4 is complex transcription factor that can act as a transcriptional activator, a transcriptional repressor, an oncogene, and a tumor suppressor, depending on the context. A question that commonly arises when learning about such a transcription factor is how it can switch between these modes. Another important question is what molecular mechanisms govern the function of KLF4 in normal cells, in cancer, and in stem cell reprogramming. Although this review discusses much of what is already known in regards to these questions, more work is needed to fully answer them. Attaining a greater understanding of the molecular function of KLF4 will ultimately give deeper insight into these many different fundamental processes.
Acknowledgments
PME is supported by a Multidisciplinary Training in Cancer Research pre-doctoral training grant from the Sealy Center for Cancer Cell Biology and the National Institutes of Health Grant T32CA117834; CL is supported by grants from the National Institutes of Health and Charlotte Geyer Foundation
References
- 1.Rreiss A, Rosenberg UB, Kienlin A, Seifert E, Jackle H. Molecular genetics of Kruppel, a gene required for segmentation of the Drosophila embryo. Nature. 1985;313:27–32. doi: 10.1038/313027a0. [DOI] [PubMed] [Google Scholar]
- 2.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 4.Ton-That H, Kaestner KH, Shields JM, Mahatanankoon CS, Yang VW. Expression of the gut-enriched Kruppel-like factor gene during development and intestinal tumorigenesis. FEBS Lett. 1997;419:239–243. doi: 10.1016/s0014-5793(97)01465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shie JL, Chen ZY, O’Brien MJ, Pestell RG, Lee ME, Tseng CC. Role of gut-enriched Kruppel-like factor in colonic cell growth and differentiation. Am J Physiol Gastrointest Liver Physiol. 2000;279:G806–G814. doi: 10.1152/ajpgi.2000.279.4.G806. [DOI] [PubMed] [Google Scholar]
- 6.Panigada M, Porcellini S, Sutti F, Doneda L, Pozzoli O, Consalez GG, Guttinger M, et al. GKLF in thymus epithelium as a developmentally regulated element of thymocyte-stroma crosstalk. Mech Dev. 1999;81:103–113. doi: 10.1016/s0925-4773(98)00237-8. [DOI] [PubMed] [Google Scholar]
- 7.Chiambaretta F, De Graeve F, Turet G, Marceau G, Gain P, Dastugue B, Rigal D, et al. Cell and tissue specific expression of human Kruppel-like transcription factors in human ocular surface. Mol Vis. 2004;10:901–909. [PubMed] [Google Scholar]
- 8.Cullingford TE, Butler MJ, Marshall AK, Tham EL, Sugden PH, Clerk A. Differential regulation of Kruppel-like factor family transcription factor expression in neonatal rat cardiac myocytes: effects of endothelin-1, oxidative stress and cytokines. Biochim Biophys Acta. 2008;1783:1229–1236. doi: 10.1016/j.bbamcr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fruman DA, Ferl GZ, An SS, Donahue AC, Satterthwaite AB, Witte ON. Phosphoinositide 3-kinase and Bruton’s tyrosine kinase regulate overlapping sets of genes in B lymphocytes. Proc Natl Acad Sci USA. 2002;99:359–364. doi: 10.1073/pnas.012605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Chen X, Kato Y, Evans PM, Yuan S, Yang J, Rychahou PG, et al. Novel cross talk of Kruppel-like factor 4 and β-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26:2055–2064. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 13.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 14.Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, et al. β-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 15.Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen ZY, Rex S, Tseng CC. Kruppel-like factor 4 is transactivated by butyrate in colon cancer cells. J Nutr. 2004;134:792–798. doi: 10.1093/jn/134.4.792. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Wang X, Hernandez A, Kim S, Evers BM. Inhibition of the phosphatidylinositol 3-kinase pathway contributes to HT29 and Caco-2 intestinal cell differentiation. Gastroenterology. 2001;120:1381–1392. doi: 10.1053/gast.2001.24044. [DOI] [PubMed] [Google Scholar]
- 18.Heerdt BG, Houston MA, Augenlicht LH. Potentiation by specific short-chain fatty acids of differentiation and apoptosis in human colonic carcinoma cell lines. Cancer Res. 1994;54:3288–3293. [PubMed] [Google Scholar]
- 19.Hinnebusch BF, Siddique A, Henderson JW, Malo MS, Zhang W, Athaide CP, Abedrapo MA, et al. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Kruppel-like factor. Am J Physiol Gastrointest Liver Physiol. 2004;286:G23–G30. doi: 10.1152/ajpgi.00203.2003. [DOI] [PubMed] [Google Scholar]
- 20.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–4002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitney EM, Ghaleb AM, Chen X, Yang VW. Transcriptional profiling of the cell cycle checkpoint gene Kruppel-like factor 4 reveals a global inhibitory function in macromolecular biosynthesis. Gene Expr. 2006;13:85–96. doi: 10.3727/000000006783991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahatan CS, Kaestner KH, Geiman DE, Yang VW. Characterization of the structure and regulation of the murine gene encoding gut-enriched Kruppel-like factor (Kruppel-like factor 4) Nucleic Acids Res. 1999;27:4562–4569. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, et al. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickenig G, Baudler S, Muller C, Werner C, Werner N, Welzel H, Strehlow K, et al. Redox-sensitive vascular smooth muscle cell proliferation is mediated by GKLF and Id3 in vitro and in vivo. FABSEB J. 2002;16:1077–1086. doi: 10.1096/fj.01-0570com. [DOI] [PubMed] [Google Scholar]
- 26.Shie JL, Pestell RG, TC C. Repression of the Cyclin D1 promoter by Gut-Enriched Kruppel-like Factor. Gastroenterology. 1999;11:A520. [Google Scholar]
- 27.Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klaewsongkram J, Yang Y, Golech S, Katz J, Kaestner KH, Weng NP. Kruppel-like factor 4 regulates B cell number and activation-induced B cell proliferation. J Immunol. 2007;179:4679–4684. doi: 10.4049/jimmunol.179.7.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon HS, Ghaleb AM, Nandan MO, Hisamuddin IM, Dalton WB, Yang VW. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene. 2005;24:4017–4025. doi: 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon HS, Yang VW. Requirement of Kruppel-like factor 4 in preventing entry into mitosis following DNA damage. J Biol Chem. 2004;279:5035–5041. doi: 10.1074/jbc.M307631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen ZY, Shie JL, Tseng CC. Gut-enriched Kruppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J Biol Chem. 2002;277:46831–46839. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, et al. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowland BD, Bernards R, Peeper DS. The KLF4 tumor suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 35.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–260. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 36.Jaubert J, Cheng J, Segre JA. Ectopic expression of Kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–2777. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- 37.Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc Natl Acad Sci USA. 2006;103:18668–18673. doi: 10.1073/pnas.0608658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Effect of corticosteroids for fetal maturation on perinatal outcomes. NIH consensus development panel on the effect of corticosteroids for fetal maturation on perinatal outcomes. JAMA. 1995;273:413–418. doi: 10.1001/jama.1995.03520290065031. [DOI] [PubMed] [Google Scholar]
- 39.Blanchon L, Nores R, Gallot D, Marceau G, Borel V, Yang VW, Bocco JL, et al. Activation of the human pregnancy-specific glycoprotein PSG-5 promoter by KLF4 and Sp1. Biochem Biophys Res Commun. 2006;343:745–753. doi: 10.1016/j.bbrc.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Blanchon L, Bocco JL, Gallot D, Gachon AM, Lemery D, Dechelotte P, Dastugue B, et al. Co-localization of KLF6 and KLF4 with pregnancy-specific glycoproteins during human placenta development. Mech Dev. 2001;105:185–189. doi: 10.1016/s0925-4773(01)00391-4. [DOI] [PubMed] [Google Scholar]
- 41.Yet SF, McA’Nulty MM, Folta SC, Yen HW, Yoshizumi M, Hsieh CM, Layne MD, et al. Human EZF, a Kruppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. 1998;273:1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 42.McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, Chittur KK. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci USA. 2001;98(16):8955–8960. doi: 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22α in vivo. J Biol Chem. 2000;275:37798–37806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 45.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alder JK, Georgantas RW, 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, et al. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. 2008;180:5645–5652. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swamynathan SK, Katz JP, Kaestner KH, Ashery-Padan R, Crawford MA, Piatigorsky J. Conditional deletion of the mouse Klf4 gene results in corneal epithelial fragility, stromal edema, and loss of conjunctival goblet cells. Mol Cell Biol. 2007;27:182–194. doi: 10.1128/MCB.00846-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 51.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 52.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi BJ, Cho YG, Song JW, Kim CJ, Kim SY, Nam SW, Yoo NJ, et al. Altered expression of the KLF4 in colorectal cancers. Pathol Res Pract. 2006;202(8):585–589. doi: 10.1016/j.prp.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Xu J, Lu B, Xu F, Gu H, Fang Y, Huang Q, Lai M. Dynamic down-regulation of Kruppel-like factor 4 in colorectal adenoma-carcinoma sequence. J Cancer Res Clin Oncol. 2008 Feb 9; doi: 10.1007/s00432-008-0353-y. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 56.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 57.Dang DT, Bachman KE, Mahatan CS, Dang LH, Giardiello FM, Yang VW. Decreased expression of the gut-enriched Kruppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS Lett. 2000;476:203–207. doi: 10.1016/s0014-5793(00)01727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of βcatenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 59.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 60.Ghaleb AM, McConnell BB, Nandan MO, Katz JP, Kaestner KH, Yang VW. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis. Cancer Res. 2007;67:7147–7154. doi: 10.1158/0008-5472.CAN-07-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Auvinen M, Paasinen A, Andersson LC, Holtta E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360:355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 62.Kanai M, Wei D, Li Q, Jia Z, Ajani J, Le X, Yao J, et al. Loss of Kruppel-like factor 4 expression contributes to Sp1 overexpression and human gastric cancer development and progression. Clin Cancer Res. 2006;12:6395–6402. doi: 10.1158/1078-0432.CCR-06-1034. [DOI] [PubMed] [Google Scholar]
- 63.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, Wu TT, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 64.Cho YG, Song JH, Kim CJ, Nam SW, Yoo NJ, Lee JY, Park WS. Genetic and epigenetic analysis of the KLF4 gene in gastric cancer. Apmis. 2007;115:802–808. doi: 10.1111/j.1600-0463.2007.apm_643.x. [DOI] [PubMed] [Google Scholar]
- 65.Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Wei D, Huang S, Peng Z, Le X, Wu TT, Yao J, et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- 67.Luo A, Kong J, Hu G, Liew CC, Xiong M, Wang X, Ji J, et al. Discovery of Ca2+-relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene. 2004;23:1291–1299. doi: 10.1038/sj.onc.1207218. [DOI] [PubMed] [Google Scholar]
- 68.Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN, Wu M. Down-regulation of gut-enriched Kruppel-like factor expression in esophageal cancer. World J Gastroenterol. 2002;8:966–970. doi: 10.3748/wjg.v8.i6.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, Kanai Y, Haga K, et al. Downregulation and growth inhibitory effect of epithelial-type Kruppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251–256. doi: 10.1016/s0006-291x(03)01356-1. [DOI] [PubMed] [Google Scholar]
- 70.Bianchi F, Hu J, Pelosi G, Cirincione R, Ferguson M, Ratcliffe C, Di Fiore PP, et al. Lung cancers detected by screening with spiral computed tomography have a malignant phenotype when analyzed by cDNA microarray. Clin Cancer Res. 2004;10:6023–6028. doi: 10.1158/1078-0432.CCR-04-0619. [DOI] [PubMed] [Google Scholar]
- 71.Yasunaga J, Taniguchi Y, Nosaka K, Yoshida M, Satou Y, Sakai T, Mitsuya H, et al. Identification of aberrantly methylated genes in association with adult T-cell leukemia. Cancer Res. 2004;64:6002–6009. doi: 10.1158/0008-5472.CAN-04-1422. [DOI] [PubMed] [Google Scholar]
- 72.Kharas MG, Yusuf I, Scarfone VM, Yang VW, Segre JA, Huettner CS, Fruman DA. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood. 2007;109:747–755. doi: 10.1182/blood-2006-03-011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, McKie-Bell P, Grizzle W, Hayes MR, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-Myc and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 74.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 75.Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.ccr-03-0484. [DOI] [PubMed] [Google Scholar]
- 76.Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK, Frost AR, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang CC, Liu Z, Li X, Bailey SK, Nail CD, Foster KW, Frost AR, et al. KLF4 and PCNA identify stages of tumor initiation in a conditional model of cutaneous squamous epithelial neoplasia. Cancer Biol Ther. 2005;4:1401–1408. doi: 10.4161/cbt.4.12.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 79.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 80.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 81.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 82.Lewitzky M, Yamanaka S. Reprogramming somatic cells towards pluripotency by defined factors. Curr Opin Biotechnol. 2007;18:467–473. doi: 10.1016/j.copbio.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 83.Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008;41:51–56. doi: 10.1111/j.1365-2184.2008.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Y, McClintick J, Zhong L, Edenberg HJ, Yoder MC, Chan RJ. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood. 2005;105:635–637. doi: 10.1182/blood-2004-07-2681. [DOI] [PubMed] [Google Scholar]
- 85.Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, et al. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006;26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 87.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 89.Geiman DE, Ton-That H, Johnson JM, Yang VW. Transactivation and growth suppression by the gut-enriched Kruppel-like factor (Kruppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 2000;28:1106–1113. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang VW. Eukaryotic transcription factors: identification, characterization and functions. J Nutr. 1998;128:2045–2051. doi: 10.1093/jn/128.11.2045. [DOI] [PubMed] [Google Scholar]
- 91.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 1998;26:796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shields JM, Yang VW. Two potent nuclear localization signals in the gut-enriched Kruppel-like factor define a subfamily of closely related Kruppel proteins. J Biol Chem. 1997;272:18504–18507. doi: 10.1074/jbc.272.29.18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen ZY, Wang X, Zhou Y, Offner G, Tseng CC. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65:10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- 95.Chen ZY, Shie J, Tseng C. Up-regulation of gut-enriched Kruppel-like factor by interferon-gamma in human colon carcinoma cells. FEBS Lett. 2000;477:67–72. doi: 10.1016/s0014-5793(00)01764-6. [DOI] [PubMed] [Google Scholar]
- 96.Ai W, Zheng H, Yang X, Liu Y, Wang TC. Tip60 functions as a potential corepressor of KLF4 in regulation of HDC promoter activity. Nucleic Acids Res. 2007;35:6137–6149. doi: 10.1093/nar/gkm656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 2002;30:2736–2741. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Watanabe N, Kurabayashi M, Shimomura Y, Kawai-Kowase K, Hoshino Y, Manabe I, Watanabe M, et al. BTEB2, a Kruppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B (SMemb/NMHC-B) gene. Circ Res. 1999;85:182–191. doi: 10.1161/01.res.85.2.182. [DOI] [PubMed] [Google Scholar]
- 100.Conkright MD, Wani MA, Anderson KP, Lingrel JB. A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999;27:1263–1270. doi: 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gaughan L, Brady ME, Cook S, Neal DE, Robson CN. Tip60 is a co-activator specific for class I nuclear hormone receptors. J Biol Chem. 2001;276:46841–46848. doi: 10.1074/jbc.M103710200. [DOI] [PubMed] [Google Scholar]
- 102.Cao X, Sudhof TC. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 103.Xiao H, Chung J, Kao HY, Yang YC. Tip60 is a co-repressor for STAT3. J Biol Chem. 2003;278:11197–11204. doi: 10.1074/jbc.M210816200. [DOI] [PubMed] [Google Scholar]
- 104.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 105.Zhang W, Shields JM, Sogawa K, Fujii-Kuriyama Y, Yang VW. The gut-enriched Kruppel-like factor suppresses the activity of the CYP1A1 promoter in a Sp1-dependent fashion. J Biol Chem. 1998;273:17917–17925. doi: 10.1074/jbc.273.28.17917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ai W, Liu Y, Langlois M, Wang TC. Kruppel-like factor 4 (KLF4) represses histidine decarboxylase gene expression through an upstream Sp1 site and downstream gastrin responsive elements. J Biol Chem. 2004;279:8684–8693. doi: 10.1074/jbc.M308278200. [DOI] [PubMed] [Google Scholar]
- 107.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and Kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 108.Noti JD, Johnson AK, Dillon JD. The leukocyte integrin gene CD11d is repressed by gut-enriched Kruppel-like factor 4 in myeloid cells. J Biol Chem. 2005;280:3449–3457. doi: 10.1074/jbc.M412627200. [DOI] [PubMed] [Google Scholar]
- 109.Wei X, Xu H, Kufe D. Human mucin 1 oncoprotein represses transcription of the p53 tumor suppressor gene. Cancer Res. 2007;67:1853–1858. doi: 10.1158/0008-5472.CAN-06-3063. [DOI] [PubMed] [Google Scholar]
- 110.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, et al. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 111.King KE, Iyemere VP, Weissberg PL, Shanahan CM. Kruppel-like factor 4 (KLF4/GKLF) is a target of bone morphogenetic proteins and transforming growth factor β1 in the regulation of vascular smooth muscle cell phenotype. J Biol Chem. 2003;278:11661–11669. doi: 10.1074/jbc.M211337200. [DOI] [PubMed] [Google Scholar]
- 112.Mao Z, Song S, Zhu Y, Yi X, Zhang H, Shang Y, Tong T. Transcriptional regulation of A33 antigen expression by gut-enriched Kruppel-like factor. Oncogene. 2003;22:4434–4443. doi: 10.1038/sj.onc.1206508. [DOI] [PubMed] [Google Scholar]
- 113.Saifudeen Z, Dipp S, Fan H, El-Dahr SS. Combinatorial control of the bradykinin B2 receptor promoter by p53, CREB, KLF-4, and CBP: implications for terminal nephron differentiation. Am J Physiol Renal Physiol. 2005;288:F899–F909. doi: 10.1152/ajprenal.00370.2004. [DOI] [PubMed] [Google Scholar]
- 114.Jenkins TD, Opitz OG, Okano J, Rustgi AK. Transactivation of the human keratin 4 and Epstein-Barr virus ED-L2 promoters by gut-enriched Kruppel-like factor. J Biol Chem. 1998;273:10747–10754. doi: 10.1074/jbc.273.17.10747. [DOI] [PubMed] [Google Scholar]
- 115.Reidling JC, Said HM. Regulation of the human biotin transporter hSMVT promoter by KLF-4 and AP-2: confirmation of promoter activity in vivo. Am J Physiol Cell Physiol. 2007;292:C1305–C1312. doi: 10.1152/ajpcell.00360.2006. [DOI] [PubMed] [Google Scholar]
- 116.Siddique A, Malo MS, Ocuin LM, Hinnebusch BF, Abedrapo MA, Henderson JW, Zhang W, et al. Convergence of the thyroid hormone and gut-enriched Kruppel-like factor pathways in the context of enterocyte differentiation. J Gastrointest Surg. 2003;7:1053–1061. doi: 10.1016/j.gassur.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 117.Piccinni SA, Bolcato-Bellemin AL, Klein A, Yang VW, Kedinger M, Simon-Assmann P, Lefebvre O. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J Biol Chem. 2004;279:9103–9114. doi: 10.1074/jbc.M305804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brembeck FH, Rustgi AK. The tissue-dependent keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J Biol Chem. 2000;275:28230–28239. doi: 10.1074/jbc.M004013200. [DOI] [PubMed] [Google Scholar]
- 119.Miller KA, Eklund EA, Peddinghaus ML, Cao Z, Fernandes N, Turk PW, Thimmapaya B, et al. Kruppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. J Biol Chem. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- 120.Higaki Y, Schullery D, Kawata Y, Shnyreva M, Abrass C, Bomsztyk K. Synergistic activation of the rat laminin gamma1 chain promoter by the gut-enriched Kruppel-like factor (GKLF/KLF4) and Sp1. Nucleic Acids Res. 2002;30:2270–2279. doi: 10.1093/nar/30.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu Y, Sinha S, Owens G. A transforming growth factor-β control element required for SM α-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J Biol Chem. 2003;278:48004–48011. doi: 10.1074/jbc.M301902200. [DOI] [PubMed] [Google Scholar]
- 122.Zeng Y, Zhuang S, Gloddek J, Tseng CC, Boss GR, Pilz RB. Regulation of cGMP-dependent protein kinase expression by Rho and Kruppel-like transcription factor-4. J Biol Chem. 2006;281:16951–16961. doi: 10.1074/jbc.M602099200. [DOI] [PubMed] [Google Scholar]
- 123.Wang H, Yang L, Jamaluddin MS, Boyd DD. The Kruppel-like KLF4 transcription factor, a novel regulator of urokinase receptor expression, drives synthesis of this binding site in colonic crypt luminal surface epithelial cells. J Biol Chem. 2004;279:22674–22683. doi: 10.1074/jbc.M401257200. [DOI] [PubMed] [Google Scholar]


