Abstract
How the nucleolus is segregated during mitosis is poorly understood and occurs by very different mechanisms during closed and open mitosis. Here we report a new mechanism of nucleolar segregation involving removal of the nucleolar-organizing regions (NORs) from nucleoli during Aspergillus nidulans mitosis. This involves a double nuclear envelope (NE) restriction which generates three NE-associated structures, two daughter nuclei (containing the NORs), and the nucleolus. Therefore, a remnant nucleolar structure can exist in the cytoplasm without NORs. In G1, this parental cytoplasmic nucleolus undergoes sequential disassembly releasing nucleolar proteins to the cytoplasm as nucleoli concomitantly reform in daughter nuclei. By depolymerizing microtubules and mutating spindle assembly checkpoint function, we demonstrate that a cycle of nucleolar “segregation” can occur without a spindle in a process termed spindle-independent mitosis (SIM). During SIM physical separation of the NOR from the nucleolus occurs, and NE modifications promote expulsion of the nucleolus to the cytoplasm. Subsequently, the cytoplasmic nucleolus is disassembled and rebuilt at a new site around the nuclear NOR. The data demonstrate the existence of a mitotic machinery for nucleolar segregation that is normally integrated with mitotic spindle formation but that can function without it.
INTRODUCTION
The nucleolus is the most prominent subcompartment of the nucleus (Leung et al., 2006) and is the site of rRNA transcription, pre-rRNA processing and ribosome subunit assembly (Tschochner and Hurt, 2003; Boisvert et al., 2007). The nucleolus is generated around regions of the genome that encode the repeated rRNA genes, referred to as nucleolar-organizing regions (NORs; Shaw and Doonan, 2005). In lower eukaryotes, such as Saccharomyces cerevisiae, there is a single NOR per genome and hence a single nucleolus per nucleus (Fuchs and Loidl, 2004).
There are three subregions of the nucleolus starting in the middle with the fibrillar center (FC) and moving outward to the dense fibrillar component (DFC) and then finally to the granular component (GC; Boisvert et al., 2007). These subdomains of the nucleolus have been defined using electron microscopy (Jordan, 1991; Leger-Silvestre et al., 1999; Trumtel et al., 2000) and are thought to functionally reflect the different stages of ribosome biogenesis (Shaw and Doonan, 2005): rRNA transcription at the FC/DFC interface, pre-rRNA processing in the DFC, and ribosome subunit assembly in the GC.
During S. cerevisiae and Schizosaccharomyces pombe closed mitosis, the nucleolus and NOR remain associated and segregate later than bulk genomic DNA (Granot and Snyder, 1991; Fuchs and Loidl, 2004). In S. cerevisiae activation and regulated release of the Cdc14 phosphatase from the nucleolus plays a critical role in post-anaphase condensation of the NORs, which causes subsequent splitting and separation of the nucleolus to daughter nuclei (Strunnikov, 2005; Wang et al., 2006). Interestingly however, Robinow and Caten (1969) saw that the nucleolus of the model filamentous fungus Aspergillus nidulans remained intact after DNA segregation and suggested that the nucleolus is not divided but perhaps becomes “budded off” during the mitotic process. More recent data utilizing expression of a BIMG-green fluorescent protein (GFP) fusion also indicates the nucleolus of A. nidulans might remain intact into late mitosis (Fox et al., 2002). These data suggest that mitotic segregation of the nucleolus is significantly different between yeasts and filamentous fungi.
During open mitosis the nucleolus undergoes disassembly. This involves activation of cyclin-dependent kinase 1 (CDK1)-Cyclin B and phosphorylation of promoter selectivity factor SL1 components which prevents RNA polymerase I preinitiation complex formation and shuts down rDNA transcription (Heix et al., 1998; Sirri et al., 2002; Leung et al., 2004). Shutdown of rDNA transcription has been linked to nucleolar disassembly (Leung et al., 2004; Boisvert et al., 2007). Disassembled nucleolar proteins have several mitotic locations including the surface of condensed chromosomes and cytoplasmic bodies of variable size and number termed nucleolar-derived foci (NDF). These NDF release their constituents during mitotic progression. Released NDF components associate with rRNA processing proteins on the surface of chromatin, forming structures termed prenucleolar bodies (PNB), which eventually form the new nucleolus in G1. However, not all nucleolar proteins disperse during mammalian mitosis. For instance, some Topo I remains at the NORs throughout mitosis (Christensen et al., 2002).
In mammalian cells, postmitotic nucleolar reassembly requires association of RNA polymerase I transcription factors to the NOR (Dundr et al., 2000; Dousset et al., 2000; Leung et al., 2004; Angelier et al., 2005) and transcription of ribosomal DNA (Benavente et al., 1987). Live cell imaging of different nucleolar rRNA processing proteins has shown a stepwise reassociation of proteins to the developing nucleoli. For example, fibrillarin locates to NORs before Nop52. The stepwise reassembly is thought to reflect the order of function during the biogenesis pathway of ribosome subunits (Savino et al., 1999, 2001) and, as mentioned above, this order of function is also reflected in the substructure of nucleoli. Sequential release from the PNB is thought to be the mechanistic basis for stepwise nucleolar reassembly (Savino et al., 2001; Angelier et al., 2005). However, what regulates the stepwise release–reassembly remains to be determined.
Although studies of A. nidulans nuclear envelope (NE) structure using electron microscopy reveal that the NE remains intact during mitosis (Robinow and Caten, 1969; Oakley and Morris, 1983), we have previously shown that A. nidulans undergoes partially open mitosis (De Souza et al., 2004; Osmani et al., 2006a) during which core structural nuclear pore complex (NPC) proteins (nucleoporins or Nups) remain at the NE, whereas peripheral Nups disperse throughout the cell. This stops regulated nuclear transport and allows proteins to equilibrate across the NE by diffusion through a minimal core NPC structure during mitosis. As mitosis is completed the core NPC structure acts as a seed for dispersed Nups to return and reestablish regulated nuclear transport (Osmani et al., 2006a; De Souza and Osmani, 2007). However, how two nuclei are generated from one or how the nucleolus is segregated during mitosis has not been defined.
MATERIALS AND METHODS
Standard growth and genetic methodologies for A. nidulans were as described previously (Pontecorvo et al., 1953) with slight modifications. Strains used in this study are listed in Supplemental Table S1. A. nidulans orthologues of proteins were identified as described (Osmani et al., 2006a) and include An-Fib (AN0745.3, similar to S. cerevisiae Nop1 and vertebrate fibrillarin a rRNA 2′-O-methyltransferase), An-Bop1 (AN1367.3, similar to S. cerevisiae Erb1), An-Nrap (Utama et al., 2002; AN3455.3, similar to S. cerevisiae Utp22), An-CgrA (Bhabhra et al., 2004; similar to S. cerevisiae Cgr1), An-Erg24 (AN4094.3, similar to S. cerevisiae Erg24 a C-14 sterol reductase), An-Topo I (topoisomerase I, AN0253), and An-PolI (similar to S. cerevisiae Rpa190, the largest subunit of RNA polymerase I. The gene encoding An-PolI resides on linkage group VIII-R on contigs 1.12 and 1.180; see Table 5.1 in Clutterbuck, 2008). Gene deletions and endogenous chromosomal tagging was completed as previously described (Yang et al., 2004; Osmani et al., 2006b). All fluorescent tags were integrated at the endogenous gene locus. Fluorescence of GFP, monomeric red fluorescent protein (mRFP), mCherry Red Fluorescent Protein (chRFP), and DsRed was maintained after fixation in 1× PHEM buffer (45 mM PIPES, 45 mM HEPES, 10 mM EGTA, 5 mM MgCl2, pH 6.9) containing 6% paraformaldehyde (EM grade; Electron Microscopy Sciences, Hatfield, PA).
For live cell imaging (De Souza et al., 2004) conidiospores were germinated in minimal medium containing glucose as the carbon source and urea as the nitrogen source in 35-mm glass-bottom microwell dishes (MatTek, Ashland, MA). Cells were imaged using Orca-ER cameras (Hamamatsu, Bridgewater, NJ) on TE300 inverted microscopes (Nikon, Melville, NJ) configured with Ultraview spinning disk confocal systems (Perkin Elmer-Cetus, Norwalk, CT) controlled by Ultraview software (Perkin Elmer-Cetus) utilizing Nikon Plan Apo 60× NA 1.40 oil objectives. Narrow bandpass filters were used that eliminated any bleed-through during dual color imaging even with bright signals such as fibrillarin-GFP/chRFP. To generate movies, lower-limit thresholds were either set just below the cytoplasmic levels or the background levels adjacent to cells were subtracted digitally using Ultraview software. RGB images and movies were generated by merging the appropriate grayscale images using Ultraview software. All imaging was carried out at room temperature at which strains containing temperature-sensitive mutations behave normally. Where indicated, benomyl was used at a concentration of 2.4 μg/ml (Ovechkina et al., 2003). Image analysis and kymograph generation was completed using ImageJ freeware (http://rsb.info.nih.gov/ij/). All images are represented as maximum intensity projections captured at 2 × 2 binning with the exception of data in Figure 3A in which images were collected at 0.2-μm intervals without binning. The z series stack was used to generate a 3D rendered volume using Slidebook (Intelligent Imaging Innovations, Denver, CO) and exported as a movie file. Views are as rotated around the y-axis.
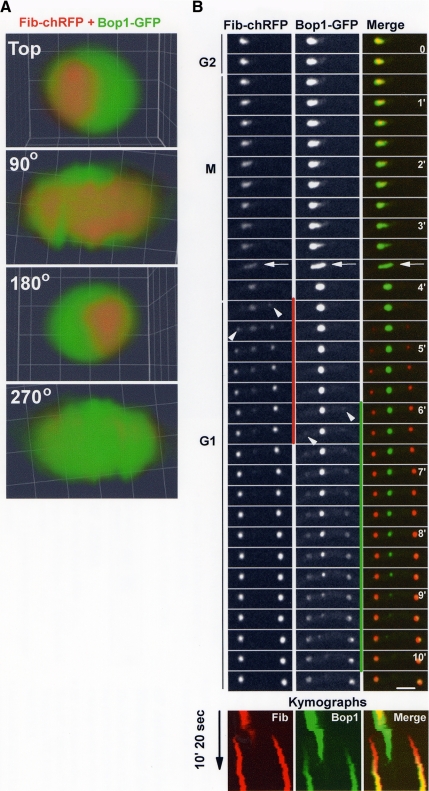
Figure 3.
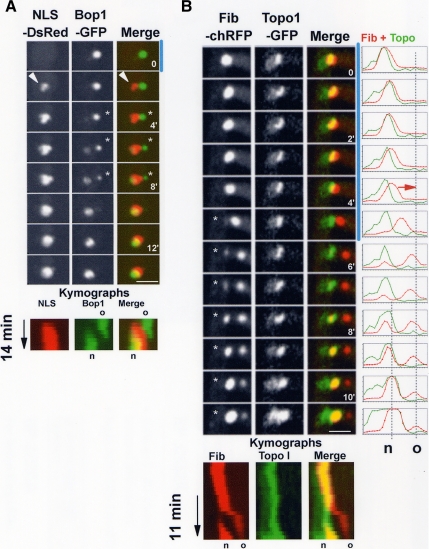
Sequential disassembly and reassembly of the nucleolus. (A) An interphase nucleolus showing the location of An-Fib-chRFP in relation to An-Bop1-GFP. Confocal images were collected at 0.2-μm intervals without binning, and the resulting z series stack was used to generate a 3D rendered volume. Views rotated around the y-axis are shown. (B) Live cell imaging of nucleolar segregation following An-Fib-chRFP and An-Bop1-GFP (Video3.mov). Although An-Fib-chRFP resides internal to An-Bop1, it undergoes its cycle of disassembly–reassembly (marked by a vertical red line) before that of An-Bop1-GFP (marked by a vertical green line). The mitotic nucleolar stretching (see also Figure 2A) is marked by an arrow, and the arrowheads indicate the initiation of reassembly of first An-Fib then An-Bop1 to daughter nucleoli. The kymographs show an extended time course and highlight the early segregation of red An-Fib compared with the more external green An-Bop1. Similarly, the pattern of yellow indicates the earlier release of An-Fib-chRFP (yellow turns to green in the parent nucleolus) and the later incorporation of the more peripheral An-Bop1-GFP (red turns to yellow in the daughter nucleoli) in the merged kymograph. The period shown in the montage is indicated on the kymographs. Bars, ∼5 μm.
RESULTS
Mitosis in A. nidulans Involves a Double Restriction of the Nuclear Envelope and Formation of a Transient Nuclear Remnant Devoid of DNA
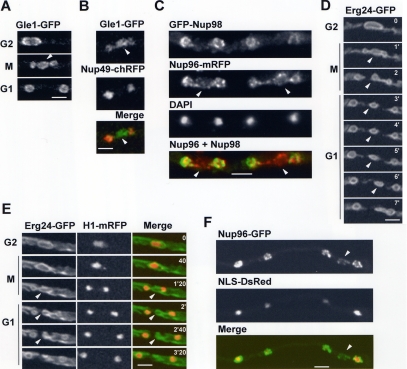
We were interested to define how two nuclei are generated from one during A. nidulans mitosis. Live cell imaging of the NPC protein An-Gle1-GFP indicated that the NE restricts during mitosis at two points to form daughter nuclei and a transient central nuclear remnant (Figure 1, A and B, arrowheads). The double restriction, as well as the generation of the nuclear remnant, was also detected in fixed cells observing An-Nup96-mRFP, another core NPC component (Figure 1C). Mitotically dispersed peripheral Nups were found to preferentially relocate back to nascent daughter nuclei, and not the nuclear remnant, during mitotic exit. This was seen for both An-Nup49-chRFP (Figure 1B) and GFP-An-Nup98 (Figure 1C).
Figure 1.
Nuclear division via double restriction and fission of the NE. (A) A nucleus undergoing mitosis observing An-Gle1-GFP. Arrowhead indicates the nuclear remnant generated by a double NE restriction. (B) A telophase nucleus displaying double NE restrictions (An-Gle1-GFP) around the nuclear remnant (arrowhead) and mitotically dispersed An-Nup49-chRFP located back to daughter nuclei but not the NE of the nuclear remnant. (C) Fixed cell image of two telophase nuclei displaying NE restrictions around the nuclear remnant (arrowhead) as revealed by the location of An-Nup96-mRFP. The mitotically dispersed GFP-Nup98 has returned to the NE of daughter nuclei but not the NE of the nuclear remnant. (D) The nuclear envelope (ER/NE marker An-Erg24-GFP) is shown through mitosis (Video1.mov). NE fission occurs first to the left and then to the right, generating the nuclear remnant (arrowheads). (E) A nucleus of a cell expressing An-Erg24-GFP and H1-mRFP progressing through mitosis. The nuclear remnant (arrowheads) does not contain DNA. (F) A fixed germling with nuclei in slightly different stages of mitotic exit. Although daughter nuclei import NLS-DsRed, the nuclear remnant (arrowhead) does not. Bars, ∼5 μm.
The double NE restriction, as well as generation of the nuclear remnant, was also revealed using An-Erg24-GFP, a marker of the ER/NE (Figure 1, D and E, Video1.mov). The restrictions on occasion occurred one after the other (Figure 1D). In some cases, NE surrounding the nuclear remnant appeared to be reabsorbed into one daughter nucleus, as in Figure 1D, but in others the remnant disappeared with no clear pattern, presumably by reincorporation into the ER (Figure 1E).
Nuclear transport, which can be followed in A. nidulans using an NLS-DsRed reporter construct (Suelmann et al., 1997), is turned off upon entry into mitosis (because NPCs undergo partial disassembly), and nuclear NLS-DsRed is released into the cytoplasm at prophase before being reimported to daughter G1 nuclei as functional NPCs are reassembled. By following NLS-DsRed reimport during G1, we determined that the nuclear remnant is transport incompetent (Figure 1F). We have never seen NLS-DsRed locate to a third structure during mitosis (100s of mitoses), confirming the nuclear remnant to be transport incompetent.
Mitotic Segregation of the A. nidulans Nucleolus Involves Its Disassembly and Reassembly
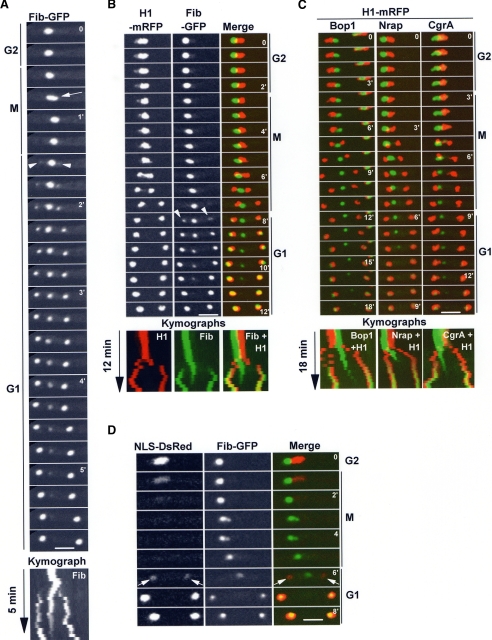
The nucleolus, as defined using functional endogenously tagged nucleolar proteins resides in a subregion of the nucleus, offset from the nuclear center, abutting a region of the NE. Chromatin is largely excluded from the nucleolar region (Figure 2, B and C, merged G2 images). One early event in the mitotic segregation of the nucleolus is its transient stretching (Figure 2A, arrow). This stretching was rapid and was observed following not only An-Fib-GFP but also An-Bop1-GFP during rapid time-point acquisitions; see Figure 3B, arrows). Starting in mitosis and continuing into G1 (Figure 2A), parental levels of nucleolar An-Fib-GFP diminish, and a slight increase in the cytoplasmic levels occurs. During telophase and G1, two new An-Fib-GFP foci appear within newly generated daughter nuclei, while the diminishing parental nucleolus is still present (Figure 2, A and B, arrowheads). At the end of the process two daughter nucleoli are generated, presumably from protein that originated from the parental nucleolus. To determine if the mitotic behavior of An-Fib-GFP was unique and to define the relationship between nucleolar segregation and DNA segregation, endogenously tagged nucleolar proteins An-Fib, An-Bop1, An-Nrap1, and An-CgrA were followed together with histone H1-mRFP. Representative data for each is presented (Figure 2, B and C; Video2.mov). During DNA segregation nucleolar Fib-GFP becomes positioned between the separating chromosomes (Figure 2B). After DNA is segregated, new nucleoli start to appear in a discrete region within daughter nuclei. The expansion of daughter nucleoli corresponds with a concomitant decrease in the parental An-Fib-GFP signal (Figure 2B; Video2.mov). During G1 the parental An-Fib-GFP signal is eventually lost and all An-Fib-GFP is located within the smaller daughter nuclei (Figure 2, A and B). The pattern of segregation of An-Bop1-GFP, An-Nrap1-GFP, and An-CgrA-GFP, in relationship to DNA, was similar to that of An-Fib-GFP (Figure 2C). Interestingly, although the nucleolus is typically located to one side of the nucleus in late G2, as mitosis proceeds the parental nucleolus always became positioned between the separating daughter nuclei.
Figure 2.
Nucleolar segregation occurs after DNA segregation. (A) Live cell imaging of An-Fib-GFP segregation. Mitotic nucleolar stretching is indicated by an arrow. The parental nucleolus undergoes disassembly, whilst daughter nucleoli reassemble (arrowheads). (B) Mitotic segregation of An-Fib-GFP in relationship with chromatin (H1-mRFP; Video2.mov). Note the nucleolus does not segregate with DNA. (C) Mitotic segregation of nucleolar proteins An-Bop1-GFP, An-Nrap1-GFP, and An-CgrA-GFP in relationship with H1-mRFP. For An-Bop1, in the earlier time points, the nucleus to the right can be ignored. (D) Mitotic nucleolar An-Fib-GFP segregation in relationship with nuclear transport. At mitosis nuclear transport stops, and NLS-DsRed is dispersed. On exit from mitosis, nuclear transport is reestablished, and daughter nuclei reimport NLS-DsRed, and nucleolar reassembly within daughter nuclei occurs (arrows). Kymographs (A–C) highlight that the parental nucleolus is present, while the daughter nuclei are reforming. Bars, ∼5 μm.
These data suggest that nucleolar proteins are imported to daughter nuclei after nuclear transport has been reestablished in G1. To investigate this, we followed nuclear transport using NLS-DsRed and formation of nucleoli using An-Fib-GFP or An-Bop1-GFP. We consistently detected nuclear accumulation of An-Fib-GFP after, or at the same time as, nuclear accumulation of NLS-DsRed (Figure 2D) but not before. However there was a marked lag between initiation of NLS-DsRed import and An-Bop1 import, suggesting that the disassembly–reassembly of An-Bop1-GFP occurs later than An-Fib-GFP (see below).
Sequential Mitotic Disassembly–Reassembly of the Nucleolus
To test if mitotic An-Bop1 disassembled after An-Fib, we followed their segregation in the same cells. High-resolution imaging through the z-axis and volume rendering revealed that An-Fib-chRFP occupied a more central position within the nucleolus and was surrounded unevenly by An-Bop1-GFP (Figure 3A). This indicates that, as in other organisms, fibrillarin is likely a protein of the more central FC and DFC nucleolar regions and An-Bop1 a protein of the more peripheral GC in A. nidulans. During mitotic exit and after an initial stretching of both signals (Figure 3B, arrows), An-Fib-chRFP disassembled before An-Bop1-GFP (Figure 3B). Even as An-Fib-chRFP initially reassembled in daughter nucleoli (Figure 3B, arrowheads for An-Fib-chRFP), all of An-Bop1-GFP remained at the parental nucleolus. Only after An-Fib-chRFP almost completes its reassembly in daughter nuclei does An-Bop1-GFP begin to disassemble. An-Bop1-GFP then undergoes its cycle of disassembly–reassembly. This demonstrates that the disassembly and reassembly of An-Fib precedes that of An-Bop1 (Figure 3B) and that the nucleolus of A. nidulans undergoes both stepwise disassembly and stepwise reassembly (Video3.mov).
The Nuclear Remnant Formed by Double NE Restrictions Contains Nucleolar Proteins But Not the Nucleolar-organizing Region
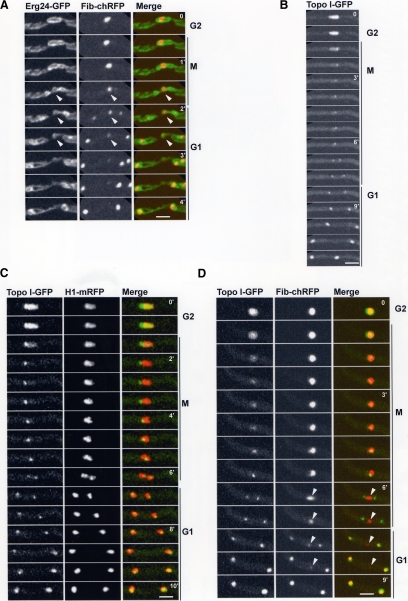
We considered the possibility that the transient cytoplasmic nucleolar structure observed during mitotic exit (Figures 2 and 3B) might reside inside the nuclear remnant formed by the double restriction of the NE (Figure 1). We visualized An-Erg24-GFP and An-Fib-chRFP in the same cells to investigate this possibility, revealing that indeed the nuclear remnant contained the parental nucleolus (Figure 4A).
Figure 4.
The nuclear remnant contains the nucleolus but not the NOR. (A) Live cell imaging of the NE (An-Erg24-GFP) and nucleolus (An-Fib-chRFP) during mitosis. The nuclear remnant (arrowheads) contains the parental nucleolus during late mitosis and early G1. (B) Time course showing that during mitosis most Topo I disperses, although a single dot remains at the nucleolus. The single Topo I dot divides and segregates to daughter nuclei where Topo I then reaccumulates to its interphase levels. (C) Segregation of the Topo I foci together with DNA (H1-mRFP) during mitosis. (D) The NOR (Topo I-GFP) is removed from the nucleolus (An-Fib-chRFP) during mitosis (Video4.mov). Note that at anaphase the NORs segregate and are removed from the nucleolus (arrowheads) and that their removal corresponds with the mitotic nucleolar stretching (6′, arrowhead). As nuclei exit mitosis the daughter nucleoli reassemble exclusively around Topo I foci, confirming that they represent NORs. Bars, ∼5 μm.
Our data indicate that the NORs are removed from the nucleolus at some stage during A. nidulans mitosis. In mammalian cells some topoisomerase I (Topo I) remains associated with NORs during mitosis (Christensen et al., 2002). We therefore followed An-Topo I-GFP as a potential marker for mitotic NORs. During interphase An-Topo I-GFP is predominantly a nuclear protein, and higher levels are present at the nucleolus (Figure 4B, G2). At mitosis most An-Topo I-GFP is released from nuclei when NPCs disassemble, but a fraction remains as a single discrete dot where the nucleolus resides. As mitosis proceeds, the An-Topo I-GFP dot divides into two and segregates to daughter nuclei (Figure 4B, M). During exit from mitosis dispersed An-Topo I-GFP returns to daughter nuclei to reestablish its interphase nuclear configuration (Figure 4B, G1). This pattern of segregation is distinct from the other nucleolar proteins we have examined that do not segregate to daughter nuclei until G1. Examination of An-Topo I-GFP together with H1-mRFP indicated that the An-Topo I-GFP foci segregated with chromatin in anaphase, consistent with the An-Topo I-GFP foci representing NORs (Figure 4C, M-G1). To further demonstrate the An-Topo I foci represent the NORs, we endogenously tagged RNA polymerase I (An-PolI) and followed its distribution during mitosis (Supplemental Figure S1A). A pool of An-PolI remained associated with the An-Topo I foci during anaphase, and these two proteins continued to colocalize as foci and further accumulated as cells progressed into G1 (Supplemental Figure S1B). Importantly, the segregated Topo-I foci define the point at which An-Fib returns to nuclei during G1 (see below, Figure 4D; Supplemental Figure S1C). Together, these data indicate that a pool of Topo I remains associated with the NORs during mitosis in A. nidulans as observed in higher eukaryotes.
To test when the NORs are removed from the nucleolus during mitosis, we followed the distribution of Topo I-GFP in relationship to An-Fib-chRFP. During anaphase, when the single dot of Topo I-GFP separates into two, the nucleolar An-Fib-chRFP becomes transiently extended in the direction of the NOR movements but remains between the two separated Topo I-GFP foci (Figure 4D, 6′, arrowhead; Video4.mov). Thus, when the NORs segregate during mitosis, they are removed from the nucleolus, which is eventually excluded to the cytoplasm. As the cell cycle proceeds, the nucleolus then undergoes disassembly and subsequent reassembly occurs around the NORs within the daughter nuclei. An-Fib-chRFP integrates specifically at the Topo I-GFP foci to reform nucleoli (Figure 4D, G1). This phenomena was observed even when using rapid 7-s time-point delays to monitor Topo I foci and the earliest stages of nucleolar formation following fibrillarin (Supplemental Figure S1C). These data confirm the Topo I foci as the sites of mitotic NORs.
Nucleolar Segregation and the Spindle Assembly Checkpoint
To begin to understand how nucleolar segregation is regulated we wanted to determine if the process was influenced by the spindle assembly checkpoint (SAC) system. Treatment of cells with microtubule poisons, such as benomyl, does not prevent entry into mitosis but hinders anaphase due to lack of spindle function, which causes activation of the SAC and mitotic arrest (Musacchio and Salmon, 2007). To follow mitosis and activation of the SAC, we monitored NLS-DsRed, which, as mentioned above, is released from nuclei at prophase and reimported during early G1. We have found that nuclear reimport of NLS-DsRed during exit from mitosis is under control of the SAC. Thus continued dispersal of NLS-DsRed from nuclei in the presence of benomyl indicates cells are in mitosis with the SAC engaged. On the other hand, nuclear reimport of NLS-DsRed, in the continued presence of benomyl, indicates the SAC is no longer engaged.
When cells enter mitosis in the presence of benomyl, NLS-DsRed is dispersed and remains dispersed for an extended period of time compared with the control (Figure 5A, WT) due to SAC activation (Figure 5A, + benomyl). During this mitotic arrest we consistently observed slight dispersal of An-Fib-GFP from the nucleolus into the cytoplasm. Notably however, all An-Bop1-GFP remained associated with the nucleolus at the SAC arrest point (cf. Figure 5B, WT, with Figure 5B, + benomyl). This indicates that disassembly of the nucleolus is influenced by the SAC with early stages of disassembly (shown by An-Fib) occurring during SAC activation, but later stages remaining arrested (shown by An-Bop1).
Figure 5.
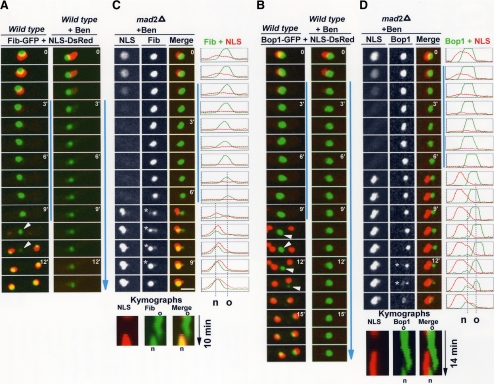
Nucleolar segregation and the spindle assembly checkpoint. (A) Time-lapse images of An-Fib-GFP and NLS-DsRed during mitosis without or with addition of the microtubule poison benomyl. Mitotic entry with depolymerized microtubules activates the SAC, which maintains the mitotic dispersal of NLS-DsRed. (C) Location of An-Fib-GFP and NLS-DsRed during a spindle-independent mitosis (SIM) in a SAC-deficient strain (mad2 deletion) after addition of benomyl (Video5.mov). Line profiles of pixel intensities along a cross section and kymographs are also shown. Nuclei enter and exit mitosis as indicated by the dispersal and return of NLS-DsRed, but nuclear segregation cannot occur because of lack of spindle formation (Figure 6B). An-Fib-GFP disassembles from the old nucleolus and reassembles at a different site forming a new nucleolus (o and n in the kymographs and pixel profiles). (B and D) As for A and C, but following An-Bop1-GFP. Spindle-independent formation of a new nucleolus occurs as in C, but An-Bop1 accumulation at this site occurs significantly after NLS-DsRed reimport (see text; Video6.mov). Time points in which both the old and new nucleolus are resolved are indicated by an asterisk. Vertical blue lines indicate the period of time that cells are in mitosis. Bars, ∼5 μm.
To further analyze the role of the SAC in controlling nucleolar segregation, we followed the behavior of An-Fib and An-Bop1 when SAC-deficient cells entered mitosis with depolymerized microtubules. This was done using strains expressing NLS-DsRed and either An-Fib-GFP or An-Bop1-GFP in a mad2Δ (A. nidulans gene designation for the Mad2 orthologue is md2A, but for ease of understanding we will use mad2; Prigozhina et al., 2004) SAC-deficient background. In such cells, entry into mitosis correlates with release of NLS-DsRed from nuclei as normal (Figure 5, C and D). However, rather than remaining dispersed, as occurs when the SAC is engaged, mad2Δ cells fail to arrest in mitosis, and NLS-DsRed is reimported. This indicates that the mad2Δ cells had transited mitosis even though, because of lack of spindle function, nuclear division cannot occur. This demonstrates the SAC is not functional when mad2 is deleted, as previously shown (Prigozhina et al., 2004), and provides us with an experimental system to study mitotic events that might occur independently of the mitotic spindle.
We next investigated what aspects of nucleolar segregation can occur in the absence of the mitotic spindle. As indicated by both An-Fib-GFP (Figure 5C; Video5.mov) and An-Bop1-GFP (Figure 5D; Video6.mov), mad2Δ cells treated with benomyl transit mitosis and complete a cycle of nucleolar disassembly and reassembly without nuclear division. In these experiments the temporal order of An-Fib reassembling to nucleoli before An-Bop1 was maintained (Figure 5, C and D). These experiments also reveal that a dynamic mitotic redistribution of the nucleolus occurs in the absence of the mitotic spindle. This phenomenon is clear from the kymographs and pixel profiles showing that the position at which the “new” nucleolus (designated n in Figure 5) is formed is distinct from the location of the “old” nucleolus (designated o). Notably, for both An-Fib and An-Bop1, the old and the new nucleolus can be seen in the same cell at several time points (Figure 5, C and D, asterisk). We will refer to the mitotic events that occur without spindle formation as Spindle Independent Mitosis or SIM.
To confirm that the above mitotic redistribution of nucleolar proteins occurs without spindle function, we followed mitosis with and without benomyl addition in mad2Δ cells containing GFP-tagged tubulin and An-Fib-chRFP (Figure 6). Without benomyl addition, nucleolar segregation occurred after anaphase spindle elongation (Figure 6A). This is similar to the mad2+ wild-type situation (data not shown). After addition of benomyl and microtubule disassembly, soluble GFP-tubulin was excluded from interphase nuclei as previously demonstrated (Ovechkina et al., 2003; Figure 6B). On mitotic entry, tubulin entered nuclei. After a delay tubulin was reexcluded as cells exit mitosis (because of lack of the SAC), and nuclei resume regulated nuclear transport. At no time during SIM was spindle formation apparent. However An-Fib clearly underwent a round of disassembly from one location and reassembly at a different location (Figure 6B; asterisk indicates a time point at which the old and new nucleolus is present), confirming that mitotic redistribution of the nucleolus can occur without spindle formation.
Figure 6.
Nucleolar segregation and the mitotic spindle. (A) Time-lapse images of mitosis in a mad2Δ strain containing GFP-tagged tubulin and An-Fib-chRFP. Arrowheads indicate the onset of daughter nucleoli formation, which occurs while the parental nucleolus is still present. (B) The same strain as in A undergoing a SIM in the presence of benomyl. In the Fib and Merge panels, the conditions and contrast are the same as in A. In the Tub* and Merge* panels, the contrast for tubulin has been adjusted to show the nuclear exclusion of depolymerized tubulin in interphase but not mitosis. A cycle of An-Fib disassembly from the parental nucleolus and reassembly at a distinct new nucleolus occurs (*) in the absence of a spindle. Kymographs are shown to highlight these changes. The period shown in the above montage is indicated on the kymographs. Bar, ∼5 μm.
Expulsion of the Nucleolus to the Cytoplasm without a Mitotic Spindle
In the SIMs described above, the location of the new nucleolus is distinct from the location of the old nucleolus, suggesting the existence of a “segregation” mechanism for the nucleolus that is independent of spindle formation. After monitoring many cells passing through SIM, we found all displayed separation between the position of the old and new nucleolus (Figure 7A, An-Bop1-GFP shows a particularly clear example).
Figure 7.
Mitotic-driven partitioning of the nucleolus without mitotic spindle formation. Time-lapse images, kymographs, and line profiles following the indicated tagged proteins in the presence of benomyl during SIM. (A) NLS reimport occurs in an area distinct (arrowhead) from the old nucleolus (An-Bop1-GFP). As G1 continues, An-Bop1-GFP disassembles from the old nucleolus (kymograph o) and reassembles at the opposite side of the nucleus forming the new nucleolus (n). (B) The nucleolus (An-Fib-chRFP) initially contains the unsegregated NORs (An-Topo I-GFP) but begins to separate from the NORs at 4 min (line profile, red arrow). Subsequently, An-Fib disassembles from the old nucleolus and reassembles back at the unsegregated NORs (Video7.mov). Bars, ∼5 μm.
Postmitotic G1 nuclei are defined by the volume newly occupied by imported NLS-DsRed (Figure 2D, for example). Similarly, during SIM the postmitotic nucleus is also defined by the volume newly occupied by NLS-DsRed (Figure 7A, NLS-DsRed). At this point in SIM exit, the old nucleolus is still present but resides outside the NLS-DsRed defined nucleus (Figure 7A, asterisk), in the cytoplasm, similar to what occurs at the early G1 stage during normal mitosis. As cell cycle progression continues, the old cytoplasmic nucleolus undergoes disassembly and reassembles at a new location residing within the nucleus defined by NLS-DsRed (Figure 7A).
Our data suggest that the NOR can be removed from the nucleolus in a spindle-independent manner. To test this, we followed An-Topo I location in relationship with An-Fib-chRFP during SIM (Figure 7B; Video7.mov). Before the transition out of mitosis, the NOR (An-Topo I-GFP) partially colocalizes within the old nucleolus (An-Fib-chRFP), resulting in a yellow color (Figure 7B, 0–4 min). As cells exit mitosis separation between the old nucleolus (An-Fib-chRFP, now clearly red) and the NOR (An-Topo I-GFP, now clearly green) occurs beginning at 5 min (Figure 7B). After separation of the NOR from the old nucleolus has occurred, An-Fib-chRFP disassembles from the old nucleolus and reassembles around the separated NOR to form the new nucleolus, their resulting colocalization once again giving a yellow color. This demonstrates that the nucleolus and NOR can be separated in the absence of spindle function or disassembly of nucleolar proteins.
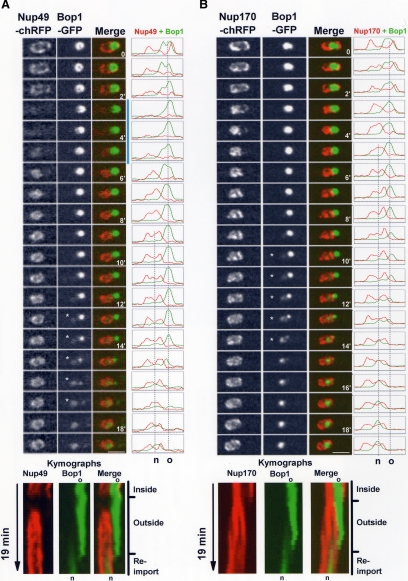
Peripheral Nups such as An-Nup49 disperse from NPCs at mitosis and return to the NE surrounding the daughter nuclei to reestablish nuclear transport in G1 nuclei (e.g., Figure 1, B, C, and F; De Souza et al., 2004). If the nucleolus is indeed expelled from the nucleus during SIM, the return of An-Nup49 to the postmitotic NE should be informative. For instance, if mitotically dispersed An-Nup49 does not return around the old nucleolus, but does return to the rest of the nuclear periphery, this would demonstrate the old nucleolus had been partitioned into the cytoplasm. In G2, An-Nup49-chRFP surrounds the nucleus including the nucleolar An-Bop1-GFP (Figure 8A, time 0). As cells enter SIM, An-Nup49-chRFP disperses from the NE as expected. Importantly, upon SIM exit An-Nup49-chRFP returns to the NE but fails to locate around the nucleolus (Figure 8A, 6′; Video8.mov). As exit from mitosis continues, An-Bop1-GFP disperses from the old cytoplasmic nucleolus and is imported into the new nucleus defined by An-Nup49-chRFP. The exclusion of the old nucleolus from the new nucleus can clearly be seen in the kymograph and pixel profile representations of the time course (Figure 8A).
Figure 8.
Time-lapse images, kymographs, and line profiles following the indicated proteins in the presence of benomyl during SIM (mad2Δ + benomyl). (A) An-Nup49-chRFP and An-Bop1-GFP distribution during SIM (Video8.mov). (B) An-Nup170-chRFP and An-Bop1-GFP distribution during SIM (Video9.mov). In both series, An-Bop1-GFP is excluded to the cytoplasm before disassembling from the old parental nucleolus (o) and reassembling within the “new” (n) nucleolus (kymograph, reimport). Time points in which both the old and new nucleolus are spatially resolved are indicated by asterisk. Bars, ∼5 μm.
To further define that the nucleolus is expelled to the cytoplasm during SIM, we followed the location of An-Bop1-GFP together with the core nucleoporin An-Nup170-chRFP, which remains at the nuclear envelope during mitosis (Osmani et al., 2006a). As cells progress through SIM the distribution of An-Nup170-chRFP changes from encompassing the entire nucleus, including the nucleolus, to excluding the nucleolus (Figure 8B, Video9.mov). This is clearly seen in the merged micrograph images, the line profiles of cross sections and also in the kymograph representation of the data. From 6 to 12 min in this time course, changes in the nuclear envelope effectively expels the old nucleolus from the nucleus to the cytoplasm (Kymograph, outside). Subsequent to this, the old cytoplasmic nucleolus disassembles and the new nucleolus reassembles in the nucleus from which it was just removed (Figure 8B). These data demonstrate that in the absence of spindle function, the nucleolus is removed from the nucleus, undergoes disassembly in the cytoplasm, and is then reassembled back within the nucleus.
DISCUSSION
This work defines a new mitotic mechanism by which the nucleolus is segregated via a cytoplasmic intermediate. The processes, which involve removal of the NOR from the nucleolus and NE restructuring, are normally integrated with the mitotic spindle but can function independently of it.
Double Restriction and Fission of the NE during Mitosis
Chromatin condensation and segregation of DNA to daughter nuclei on the mitotic spindle are universal aspects of mitosis. However, there is considerable variation in the mitotic behavior of the NE and the mechanisms by which two nuclei are generated from one. During closed mitoses, the NE remains intact, and two nuclei are generated via a restriction and fission of the NE between segregated DNA. How this is achieved or regulated is currently unknown, although in S. cerevisiae cytokinesis plays a nonessential role (Lippincott and Li, 2000). During open mitosis, the NE is disassembled in a process in part involving mechanical tearing of the NE followed by its disassembly (Rabut et al., 2004). New nuclei are generated by reassembly of the NE around segregated DNA. Although mechanistically very distinct, in both open and closed mitoses the location of DNA provides the spatial information dictating generation of daughter nuclei.
In A. nidulans the mitotic NE behaves in a manner not typical of closed or open mitosis but shares aspects of both (Figure 9). Generation of daughter nuclei involves NE restriction and fission, as in closed mitosis, but two restrictions are involved instead of one, resulting in three NE defined structures, not two. The third central postmitotic structure contains the nucleolus associated with a modified NE. Marked differences exist between the NE of the daughter nuclei and the nucleolus. For instance, nuclear transport is reestablished only in daughter nuclei. This is achieved by the reestablishment of functional NPCs in the NE of the daughter nuclei but not the nuclear remnant. The basis for this and its regulation is unknown but is of great interest and presumably involves spatial information provided by segregated DNA.
Figure 9.
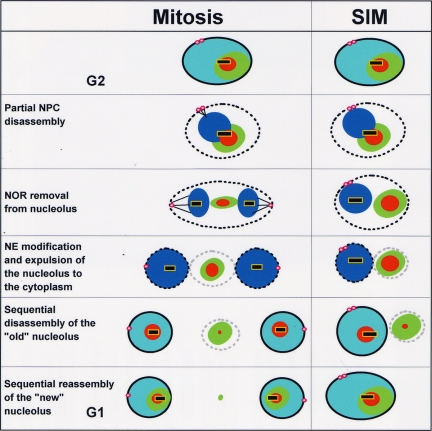
Nucleolar segregation with and without spindle formation. Changes in nuclear structure during normal mitotic progression and SIM are depicted. In G2, DNA (blue) is surrounded by the nuclear envelope (solid black circle) in which is embedded the duplicated spindle pole bodies (pink spheres). At entry into mitosis the NPCs undergo partial disassembly, permeabilizing the NE (dotted black line), and DNA condenses (darker blue). At anaphase, the NORs (yellow around black) are removed from the nucleolus (red, An-Fib; green, An-Bop1), and a similar event occurs during SIM. Modifications of the NE (gray dotted line) result in exclusion of the nucleolus to the cytoplasm and occur during both a normal telophase and a SIM. As nuclei progress into G1 the NPCs reassemble, nuclear transport is reestablished, and DNA decondenses. The “old” cytoplasmic nucleolus undergoes stepwise disassembly and the “new” nucleolus is generated in a stepwise manner. All of these events occur in both normal mitosis and SIM, indicating that they are independent of microtubules. However, as DNA segregation is dependent on microtubules, NORs do not segregate during a SIM resulting in generation of a single new nucleolus. Thus mitosis involves integrated spindle dependent and independent processes for nucleolar segregation that are able to function independent of each other.
Mitotic Segregation of the Nucleolus
The nucleolus is segregated differently during open and closed mitosis. During closed mitosis in S. cerevisiae, the nucleolus remains intact and associated with the NORs. Thus segregation of DNA on the mitotic spindle is sufficient to segregate the nucleolus. During open mitosis the nucleolus undergoes disassembly, and the majority of its constituents are removed from the NORs. The NORs are segregated via the mitotic spindle and act as focal points for reassembly of daughter nucleoli. During A. nidulans mitosis the nucleolus acts, in many ways, like the nucleolus during open mitosis. The NORs are removed from the mitotic nucleolus, which then undergoes disassembly, and this process is sequential. Internal, early functioning, nucleolar proteins disassemble first before those located more peripherally, which function later in ribosome formation (Figure 9, Mitosis). This sequential disassembly is coordinated with the sequential reassembly of G1 nucleoli on the NORs of daughter nuclei, similar to the sequential nucleolar reassembly after open mitosis (Figure 9, Mitosis).
During A. nidulans mitosis partial disassembly of NPCs opens the central NPC channels, allowing rapid nuclear location of tubulin and other proteins required for the quick ∼5-min mitoses (Ovechkina et al., 2003; De Souza and Osmani, 2007). As multiple nuclei undergo synchronous mitoses in a common cytoplasm, the remaining NE protects kinetochores of individual nuclei from interference from spindles of other nuclei. This provides an explanation for why A. nidulans opens its NPCs but does not undergo completely open mitoses. However, because the mitotic NE is permeable, the nucleoplasm and cytoplasm mingle, contaminating the nucleolus with cytoplasmic components. This is similar to higher eukaryotes, which then excludes cytoplasmic components from daughter nuclei by rebuilding the NE from the surface of chromatin. To fulfill a similar requirement, we suggest the nucleolus in A. nidulans is expelled to the cytoplasm and forced to undergo a round of disassembly before nuclear import and reassembly. In this manner the nucleolus will be cleansed of cytoplasmic contaminates. Earlier cytological studies of fixed cells (Heath, 1980) indicate that many other fungi and protists might segregate their nucleoli via a cytoplasmic intermediate, indicating this pattern of nucleolar segregation will not be unique to A. nidulans.
Uncoupling Spindle-dependent and Spindle-independent Mitotic Mechanisms
The key events in the normal cycle of nucleolar segregation in A. nidulans are removal of the NORs from the parental nucleolus and functional changes in the NE, promoting expulsion of the nucleolus to the cytoplasm (Figure 9, Mitosis). Quite remarkably both of these events can occur in the absence of a mitotic spindle (Figure 9, SIM). We suggest the prime function of the spindle during nucleolar segregation is to segregate DNA, and thus the NORs, to two different locations to provide spatial cues for the generation of separated daughter nuclei and nucleoli. However, without spindle function, the process that separates the nucleolus from DNA, and drives it to the cytoplasm, still occurs. The only difference regarding the nucleolus between normal mitosis and SIM is that daughter NOR separation cannot occur during SIM. Thus A. nidulans mitosis consists of two parallel, but independent, processes that are normally coupled to orchestrate successful segregation of DNA and the nucleolus. The nature of the nucleolar segregation mechanisms remains to be determined, but our data suggest that the NE, along with associated proteins, is likely to play a key role and could perhaps provide the mechanical forces required to separate the NOR from the nucleolus and expel the nucleolus into the cytoplasm. The molecular nature of these mitotic forces, and how they are regulated, could be related to ancient membrane based cell divisions that occur in organisms lacking mitotic spindles (Heath, 1980). However, it is possible that the nucleolar cycle may be driven by movement mediated through other non–tubulin-based mitotic structures, such as the spindle matrix for instance (Johansen and Johansen, 2007).
Regulation of Nucleolar Disassembly–Reassembly
Our current understanding of the regulation of mitotic nucleolar segregation in higher eukaryotes is at an early stage (Boisvert et al., 2007). Mitotic phosphorylation of the rDNA transcription machinery results in shutdown of rDNA transcription and may initiate nucleolar disassembly (Leung et al., 2004; Boisvert et al., 2007). We find that nucleolar structure does not require ongoing NOR transcription and that surprisingly a nucleolus can exist in the cytoplasm in the absence of NORs. The disassembly of the nucleolus is likely a regulated process in A. nidulans, controlled by regulatory systems unique to organisms that disassemble their nucleoli during mitosis. Our data suggest the SAC might play a role, yet other regulatory mechanisms are also likely to be involved. It will be interesting to define the nature of the forces driving separation of the nucleolus from the NOR and how this is normally regulated and integrated with spindle driven aspects of mitosis and with NE restructuring.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Osmani lab for help and input to this work, particularly Shahr Hashmi and Aysha Osmani. We thank Berl Oakley for help with image processing and John Doonan for insightful discussions and sharing of unpublished data. This work was supported by National Institutes of Health Grant GM042564 to S.A.O. and a National Research Service Award T32 fellowship to C.P.C.D.
Abbreviations used:
- DFC
dense fibrillar component
- FC
fibrillar center
- GC
granular component
- NE
nuclear envelope
- NORs
nucleolar organizing regions
- NPC
nuclear pore complex
- rRNA
ribosomal RNA
- SAC
spindle assembly checkpoint
- SIM
spindle independent mitosis.
Note added in proof.
The late mitotic behavior of the NE and nucleolus described in this work must be highly regulated and integrated with other aspects of mitosis. In an accompanying paper on p. 2146 of this issue of Mol. Biol. Cell (De Souza et al., 2009), we demonstrate that several mitotic regulators transiently localize in between daughter nuclei in the vicinity of the cytoplasmic nucleolus. We speculate that such localization of mitotic regulators could be important to coordinate the post mitotic disassembly-reassembly of the nucleolus.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-10-1046) on February 11, 2009.
REFERENCES
- Angelier N., Tramier M., Louvet E., Coppey-Moisan M., Savino T. M., De Mey J. R., Hernandez-Verdun D. Tracking the interactions of rRNA processing proteins during nucleolar assembly in living cells. Mol. Biol. Cell. 2005;16:2862–2871. doi: 10.1091/mbc.E05-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavente R., Rose K. M., Reimer G., Hugle-Dorr B., Scheer U. Inhibition of nucleolar reformation after microinjection of antibodies to RNA polymerase I into mitotic cells. J. Cell Biol. 1987;105:1483–1491. doi: 10.1083/jcb.105.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabhra R., Miley M. D., Mylonakis E., Boettner D., Fortwendel J., Panepinto J. C., Postow M., Rhodes J. C., Askew D. S. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect. Immun. 2004;72:4731–4740. doi: 10.1128/IAI.72.8.4731-4740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Christensen M. O., Barthelmes H. U., Boege F., Mielke C. The N-terminal domain anchors human topoisomerase I at fibrillar centers of nucleoli and nucleolar organizer regions of mitotic chromosomes. J. Biol. Chem. 2002;277:35932–35938. doi: 10.1074/jbc.M204738200. [DOI] [PubMed] [Google Scholar]
- Clutterbuck J. Aspergillus nidulans linkage map and genome sequence: closing gaps and adding telomeres. In: Goldman Gustavo H., Osmani Stephen A., editors. The Aspergilli, Genomics, Medical Aspects, Biotechnology, and Research Methods. Boca Raton, FL: CRC Press; 2008. pp. 57–73. [Google Scholar]
- De Souza C. P., Osmani A. H., Hashmi S. B., Osmani S. A. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- De Souza C. P., Osmani S. A. Mitosis, not just open or closed. Eukaryot. Cell. 2007;6:1521–1527. doi: 10.1128/EC.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset T., Wang C., Verheggen C., Chen D., Hernandez-Verdun D., Huang S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell. 2000;11:2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Misteli T., Olson M. O. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox H., Hickey P. C., Fernandez-Abalos J. M., Lunness P., Read N. D., Doonan J. H. Dynamic distribution of BIMG(PP1) in living hyphae of Aspergillus indicates a novel role in septum formation. Mol. Microbiol. 2002;45:1219–1230. doi: 10.1046/j.1365-2958.2002.03092.x. [DOI] [PubMed] [Google Scholar]
- Fuchs J., Loidl J. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 2004;12:427–438. doi: 10.1023/B:CHRO.0000034726.05374.db. [DOI] [PubMed] [Google Scholar]
- Granot D., Snyder M. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil. Cytoskelet. 1991;20:47–54. doi: 10.1002/cm.970200106. [DOI] [PubMed] [Google Scholar]
- Heath I. B. Variant mitoses in lower eukaryotes: indicators of the evolution of mitosis? Int. Rev. Cytol. 1980;64:1–80. doi: 10.1016/s0074-7696(08)60235-1. [DOI] [PubMed] [Google Scholar]
- Heix J., Vente A., Voit R., Budde A., Michaelidis T. M., Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen K. M., Johansen J. Cell and molecular biology of the spindle matrix. Int. Rev. Cytol. 2007;263:155–206. doi: 10.1016/S0074-7696(07)63004-6. [DOI] [PubMed] [Google Scholar]
- Jordan E. G. Interpreting nucleolar structure: where are the transcribing genes? J. Cell Sci. 1991;98(Pt 4):437–442. doi: 10.1242/jcs.98.4.437. [DOI] [PubMed] [Google Scholar]
- Leger-Silvestre I., Trumtel S., Noaillac-Depeyre J., Gas N. Functional compartmentalization of the nucleus in the budding yeast Saccharomyces cerevisiae. Chromosoma. 1999;108:103–113. doi: 10.1007/s004120050357. [DOI] [PubMed] [Google Scholar]
- Leung A. K., Gerlich D., Miller G., Lyon C., Lam Y. W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A. I. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K., Trinkle-Mulcahy L., Lam Y. W., Andersen J. S., Mann M., Lamond A. I. NOPdb: nucleolar proteome database. Nucleic Acids Res. 2006;34:D218–D220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. Nuclear envelope fission is linked to cytokinesis in budding yeast. Exp. Cell Res. 2000;260:277–283. doi: 10.1006/excr.2000.5021. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Morris N. R. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J. Cell Biol. 1983;96:1155–1158. doi: 10.1083/jcb.96.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A. H., Davies J., Liu H. L., Osmani S. A. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell. 2006a;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A. H., Oakley B. R., Osmani S. A. Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat. Protoc. 2006b;1:2517–2526. doi: 10.1038/nprot.2006.406. [DOI] [PubMed] [Google Scholar]
- Ovechkina Y., Maddox P., Oakley C. E., Xiang X., Osmani S. A., Salmon E. D., Oakley B. R. Spindle formation in Aspergillus is coupled to tubulin movement into the nucleus. Mol. Biol. Cell. 2003;14:2192–2200. doi: 10.1091/mbc.E02-10-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J. A., Hemmons L. M., MacDonald K. D., Bufton A. W. The genetics of Aspergillus nidulans. In: Demerec M., editor. Advances in Genetics. New York: Academic Press; 1953. pp. 141–238. [DOI] [PubMed] [Google Scholar]
- Prigozhina N. L., Oakley C. E., Lewis A. M., Nayak T., Osmani S. A., Oakley B. R. γ-tubulin plays an essential role in the coordination of mitotic events. Mol. Biol. Cell. 2004;15:1374–1386. doi: 10.1091/mbc.E03-06-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Lenart P., Ellenberg J. Dynamics of nuclear pore complex organization through the cell cycle. Curr. Opin. Cell Biol. 2004;16:314–321. doi: 10.1016/j.ceb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Robinow C. F., Caten C. E. Mitosis in Aspergillus nidulans. J. Cell Sci. 1969;5:403–431. doi: 10.1242/jcs.5.2.403. [DOI] [PubMed] [Google Scholar]
- Savino T. M., Bastos R., Jansen E., Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J. Cell Sci. 1999;112(Pt 12):1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Savino T. M., Gebrane-Younes J., De Mey J., Sibarita J. B., Hernandez-Verdun D. Nucleolar assembly of the rRNA processing machinery in living cells. J. Cell Biol. 2001;153:1097–1110. doi: 10.1083/jcb.153.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P., Doonan J. The nucleolus. Playing by different rules? Cell Cycle. 2005;4:102–105. doi: 10.4161/cc.4.1.1467. [DOI] [PubMed] [Google Scholar]
- Sirri V., Hernandez-Verdun D., Roussel P. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J. Cell Biol. 2002;156:969–981. doi: 10.1083/jcb.200201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov A. V. A case of selfish nucleolar segregation. Cell Cycle. 2005;4:113–117. doi: 10.4161/cc.4.1.1488. [DOI] [PubMed] [Google Scholar]
- Suelmann R., Sievers N., Fischer R. Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol. Microbiol. 1997;25:757–769. doi: 10.1046/j.1365-2958.1997.5131873.x. [DOI] [PubMed] [Google Scholar]
- Trumtel S., Leger-Silvestre I., Gleizes P. E., Teulieres F., Gas N. Assembly and functional organization of the nucleolus: ultrastructural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell. 2000;11:2175–2189. doi: 10.1091/mbc.11.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Utama B., Kennedy D., Ru K., Mattick J. S. Isolation and characterization of a new nucleolar protein, Nrap, that is conserved from yeast to humans. Genes Cells. 2002;7:115–132. doi: 10.1046/j.1356-9597.2001.00507.x. [DOI] [PubMed] [Google Scholar]
- Wang B. D., Butylin P., Strunnikov A. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle. 2006;5:2260–2267. doi: 10.4161/cc.5.19.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ukil L., Osmani A., Nahm F., Davies J., De Souza C. P., Dou X., Perez-Balaguer A., Osmani S. A. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.