Abstract
Abiotic stresses are major limiting factors for growth, development, and productivity of crop plants. Here, we report on OsSKIPa, a rice homolog of human Ski-interacting protein (SKIP) that can complement the lethal defect of the knockout mutant of SKIP homolog in yeast and positively modulate cell viability and stress tolerance of rice. Suppression of OsSKIPa in rice resulted in growth arrest and reduced cell viability. The expression OsSKIPa is induced by various abiotic stresses and phytohormone treatments. Transgenic rice overexpressing OsSKIPa exhibited significantly improved growth performance in the medium containing stress agents (abscisic acid, salt, or mannitol) and drought resistance at both the seedling and reproductive stages. The OsSKIPa-overexpressing rice showed significantly increased reactive oxygen species-scavenging ability and transcript levels of many stress-related genes, including SNAC1 and rice homologs of CBF2, PP2C, and RD22, under drought stress conditions. More than 30 OsSKIPa-interacting proteins were identified, but most of these proteins have no matches with the reported SKIP-interacting proteins in animals and yeast. Together, these data suggest that OsSKIPa has evolved a specific function in positive modulation of stress resistance through transcriptional regulation of diverse stress-related genes in rice.
Keywords: abiotic stress, Oryza sativa, SKIP, transcriptional regulation
Abiotic stresses, such as drought and salinity, are major limiting factors for crops to reach their yield potential. Crop plants with enhanced resistance to abiotic stresses can broaden the spectrum of growth conditions, thereby increasing yield stability and productivity. Plants can develop numerous physiological and biochemical strategies to cope with adverse conditions. Cell viability is hypothetically important for plants to survive abiotic stresses. However, the regulatory mechanisms and the genes responsible for cell viability in plants are largely unknown. Here we report that a homolog of Ski-interacting protein (SKIP) in rice (Oryza sativa L.), OsSKIPa, can positively modulate cell viability and stress tolerance of rice.
SKIP protein, with its homolog Bx42 originally discovered in Drosophila (1), was identified in yeast two-hybrid screening using the oncogene v-Ski as bait (2). SKIP has been well characterized as a transcriptional coregulator as well as a spliceosome component in humans (3, 4). All the SKIP homologs identified so far contain a SNW/SKIP domain with an S-N-W-K-N peptide signature and may have conserved basic functions, such as acting as a cofactor in transcription and splicing (5). Nevertheless, the derived or additional functions of the SKIP homologs vary among species. The SKIP homolog Bx42 in Drosophila melanogaster is involved in ecdysone-stimulated transcription (6) and Notch signal transduction (7), and it is essential for the development of the nervous system (8) and many other tissues (7). In Saccharomyces cerevisiae, PRP45, a protein containing the SNW/SKIP domain, was found to be essential for cell viability (9). CeSKIP (Skp-1), the Caenorhabditis elegans homolog of SKIP, was characterized as an essential component of RNA polymerase II transcription complexes and is indispensable for viability and development of C. elegans (10). However, there has been no report on identification and functional analysis of SKIP homologs in plants.
Unlike animals, sessile plants have to respond and adapt to ever-changing environmental cues by arrays of internal molecular and physiological changes. Among these changes, transcriptional regulation of numerous stress-related genes by diverse families of transcription factors has been investigated extensively in plants (11, 12). Emerging evidence suggests that RNA processing is also involved in plant responses to abiotic stresses. So far, at least 8 genes involved in RNA processing have been implicated in plant responses to abiotic stresses or the phytohormone abscisic acid (ABA). STABILIZED1, an ortholog of PRP46 in Arabidopsis, is a pre-mRNA splicing factor for the splicing and turnover of unstable transcripts important for plant responses to abiotic stress (13). Although the functions of these genes in RNA processing have not been completely characterized, the available evidence strongly suggests that RNA metabolism plays important roles in stress response and tolerance in plants.
In the study reported in this article, we performed a functional analysis of OsSKIPa, a homolog of SKIP in rice. We show that OsSKIPa not only has the ability to complement the yeast mutant of SNW/SKIP homolog PRP45 but functions in regulating cell viability and stress tolerance. Among the 35 OsSKIPa-interacting proteins identified in this study, very few showed homology to the SKIP-interacting proteins reported in animals and yeast, indicating functional diversification of SKIP proteins in plants.
Results
Identification of SKIP Homologs in Rice.
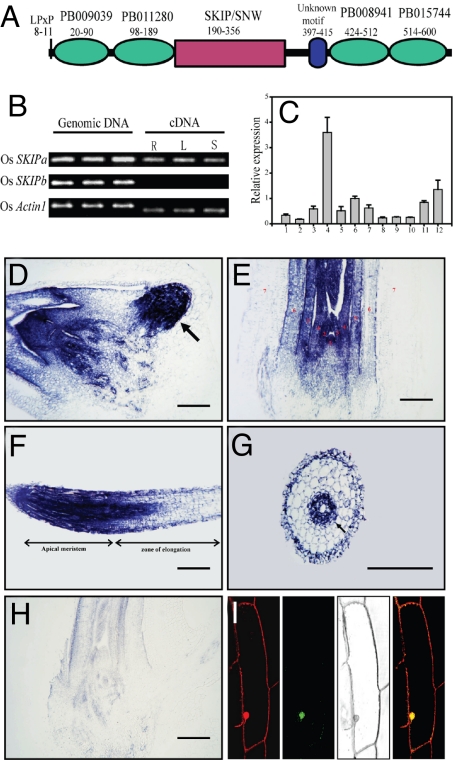
A sequence similarity search of SKIP against The Institute for Genomic Research (TIGR) genomic annotation database (http://rice.plantbiology.msu.edu/) resulted in two putative SKIP homologs in rice, named OsSKIPa (LOC_Os02g52250) and OsSKIPb (LOC_Os06g11420). The predicted protein sequences of OsSKIPa and OsSKIPb have 49% and 46% identity to SKIP, respectively. The longest ORF of OsSKIPa encodes a protein of 607 aa. Sequence alignment indicated that SKIP proteins are highly conserved in eukaryotes [supporting information (SI) Fig. S1A]. The SKIP/SNW protein domain of OsSKIPa is located between amino acids 190 and 356 (Fig. 1A). By searching the OsSKIPa sequence against the Pfam database (14), 4 additional putative domains were identified: PB009039, PB011280, PB008941, and PB015744. Two conserved motifs were identified: one is LPxP located between amino acids 8 and 11, and the other is located between amino acids 397 and 415 (Fig. 1A and Fig. S1A). We could not detect the transcript of OsSKIPb (Fig. 1B), and no cDNA sequence of this gene was available in the current databases. Moreover, an SSW sequence was found in OsSKIPb that replaced the conserved SNW signature in OsSKIPa and other SKIP homologous proteins. Thus, OsSKIPb is most likely nonfunctional or a pseudogene.
Fig. 1.
Identification of SKIP homologs in rice. (A) Predicted domains and motifs of rice SKIP homolog, OsSKIPa. (B) RT-PCR analysis of OsSKIPb. The genomic DNA and cDNA templates from root (R), young leaf (L), and shoot (S) were used for amplification. Rice Actin1 gene was used as a control. (C) Expression of OsSKIPa in different tissues and organs: 1–4: root, culm, leaf sheath, and leaf lamina, respectively, at heading stage; 5: young panicles (length <1 cm); 6: panicles (length <5 cm); 7: panicles at heading stage; 8: stamen; 9: pistil; 10: etiolated shoot at 1 week after germination; 11: shoot at 1 week after germination; 12: shoot at tillering stage. OsSKIPa transcript in different tissues at 10 days after germination detected by in situ hybridization: radicle (arrow indicates the primordia of lateral root) (D); shoot (shoot apicel meristem labeled with no. 1 and the leaf primordia or mature leaves labeled with nos. 2 to 7 according to the initiation order) (E); root tips (F); cross section of mature zone of root (G); and in situ hybridization control using a sense of OsSKIPa as a probe (H). Bars = 200 μm. (I) Confocal microscopic images of an onion epidermis cell transiently expressing OsSKIPa-GFP fusion protein (Left to Right: propidium iodide stained, GFP, light, merged image).
A BLAST search was performed against the assembled genome databases (http://www.ncbi.nlm.nih.gov/BLAST/) to investigate the SKIP sequences in various organisms. The intron/exon structures of SKIP genes are quite diverse among the eukaryotic species (Fig. S1B). The phylogenetic tree based on the SKIP sequences agrees well with the evolutionary relation among these species (Fig. S1C). Although there are 3 and 5 SKIP sequences in the human and mouse genomes, respectively, only 1 sequence in each genome has cDNA support in the current databases, suggesting that the additional sequences might be pseudogenes like OSKIPb in rice.
The results of a quantitative PCR analysis indicated that OsSKIPa is expressed in all the tissues or organs investigated, although the expression level is slightly higher in young leaves than in other tissues or organs (Fig. 1C). To characterize the expression pattern of OsSKIPa further, RNA in situ hybridization was performed in the shoot and root of 10-day-old seedlings. High expression of this gene was detected in all the tissues except senescent tissues, where its expression was low (Fig. 1 D–H). These results suggest that OsSKIPa is constitutively expressed in living cells.
Confocal results showed that the OsSKIPa-GFP fusion protein was localized in the nucleus (Fig. 1I), suggesting that OsSKIPa is a nuclear protein.
OsSKIPa Can Complement the Yeast Mutant of SKIP Homolog.
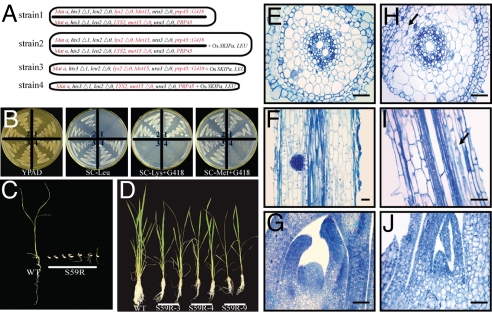
The SKIP homolog of yeast, PRP45, was identified as a component of spliceosome complexes and shown to be essential for pre-mRNA splicing (9). The prp45 mutant is lethal in haploid yeast. To determine if OsSKIPa is functionally conserved as a splicing factor, OsSKIPa was used to rescue the lethal phenotype of yeast that is null for PRP45. The diploid yeast strain BY4743, heterozygous for the PRP45 locus (Fig. 2A, strain 1), was transformed with pDEST32-OsSKIPa. The transformants (Fig. 2A, strain 2) were sporulated to produce haploid progeny. If OsSKIPa could functionally complement prp45 mutant in which PRP45 has been replaced by a genetin (G418) resistance gene, diploid strain 2 could produce both G418-resistant prp45 mutant haploid (Fig. 2A, strain 3) and G418-sensitive PRP45 haploid (Fig. 2A, strain 4). Both strain 3 and strain 4 genotypes of haploid colonies were produced in synthetic complete Leu (SC-Leu) medium, whereas in the controls (nontransformed or pDEST32 vector-transformed diploid strain heterozygous for the PRP45 locus), no haploid was produced. The genotypes of the 4 strains were further confirmed in other media, including YPAD, SC-Lys with G418, and SC-Met with G418 (Fig. 2B). These results indicate that OsSKIPa can functionally complement the prp45 mutation and rescue the lethal phenotype.
Fig. 2.
Complementation of yeast prp45 mutant by OsSKIPa and growth arrest of OsSKIPa-suppressed transgenic plants (S59R). (A) Genotypes of all strains used. Genes that can be used to identify each strain are shown in red. (B) The growth of strains 1–4 in different media. (C) Lethal phenotype by some of the OsSKIPa-suppressed lines at the seedling stage. (D) Growth retardation of S59R plants at the tillering stage. Sections of WT (E–G) and S59R (H–J): cross sections of mature zone of root (E and H), longitudinal sections of root mature zones (H and I; arrow in I indicates the dead zone of the cortex), and longitudinal sections of shoot apex (G and J). Bars = 100 μm.
Suppression of OsSKIPa Resulted in Growth Arrest of Rice.
The capability of OsSKIPa to complement the mutant of yeast SKIP homolog suggests that OsSKIPa may have important functions in rice. Because a homozygous mutant of OsSKIPa (probably lethal) is not available, we used RNAi to suppress the expression of this gene. Among the 37 OsSKIPa-RNAi transgenic plants generated, 22 (hereafter referred to as S59R plants) showed significantly decreased expression of OsSKIPa (Fig. S2A). The seeds from OsSKIPa-suppressed plants had a significantly (t test, P < 0.01; 6 lines tested) lower germination rate (<25%) than the WT or negative transgenic plants (> 99%). About 60% of the T1 S59R plantlets died within 1 week after germination in the Murashige and Skoog (MS) medium (Fig. 2C), and 18.8–86.7% of the surviving S59R plants in MS medium died at the seedling or tillering stage after transplanting in soil, whereas all WT plants grew normally under the same conditions. The growth of the S59R plants was severely arrested even if they survived (Fig. 2D). At the seedling stage, the growth rate was significantly lower than that of the WT or negative transgenic control (Table S1). At maturity, the tiller number and biomass of S59R plants were significantly lower than those of the WT or negative transgenic control (Table S1).
To determine whether the growth arrest was attributable to defects of cell viability in S59R plants, different zones of shoot and root from S59R plants were observed under a microscope. Compared with WT plants (Fig. 2 E–G), S59R plants showed abnormal cell shape in the mature zone (Fig. 2H), decreased number of cortex cell layers and some shrunken cortex cells in the primary roots (Fig. 2I), and abnormal cell shape and organization in the primordial regions in the shoot apical meristem (Fig. 2J). A TUNEL assay revealed that the hypocotyl and leaf primordium of S59R plants had more dead cells than WT plants (Fig. S2 B–E). These results suggest that the growth arrest of S59R plants mainly resulted from the reduced cell viability in the active growth regions.
Overexpression of OsSKIPa in Rice-Enhanced Stress Resistance.
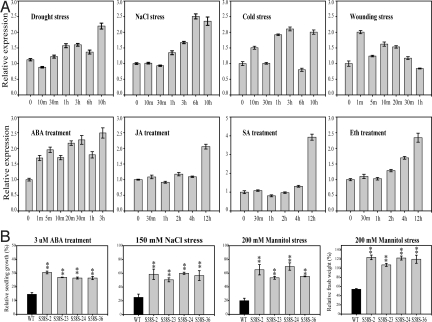
Growth arrest of the OsSKIPa-suppressing plants prompted us to investigate the expression of OsSKIPa under phytohormone and stress treatments. The results indicated that the transcript level of OsSKIPa increased after drought, high-salinity, cold, and wounding treatments (Fig. 3A). OsSKIPa was also induced (fold of induction) by ABA (100 μM), jasmonic acid (100 μM), salicylic acid (1 mM), and ethylene (1 mM; Fig. 3A) but not much (<2-fold) by gibberellic acid (100 μM), indole-3-acetic acid (50 μM), kinetin (450 μM), or brassinosteroid (10 μM) (data not shown). These data suggest that the expression of OsSKIPa is responsive mainly to stresses and stress-related phytohormones.
Fig. 3.
Response of OsSKIPa to various stresses and phytohormones and performance of seedlings overexpressing OsSKIPa under stress conditions. (A) Expression level of OsSKIPa in leaf of rice under various stress and phytohormone treatments. (B) Quantification of the growth rates of the WT and 4 S58S lines with the treatments as in Fig. S2I. Error bar indicates SE based on 4 replicates. **t test, with P < 0.01. Eth, ethylene; JA, jasmonic acid; SA, salicylic acid.
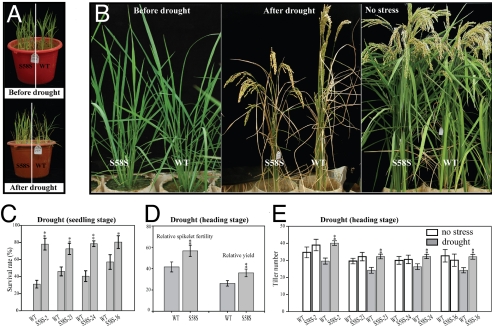
Up-regulation of OsSKIPa by abiotic stress and the role of this gene in maintaining cell viability prompted us to investigate whether overexpression of this gene could enhance stress resistance. Of the 43 OsSKIPa-overexpressing transgenic plants generated, 22 (named S58S plants) showed overexpression of the transgene (Fig. S2F), with normal morphology under normal growth conditions. Four randomly selected independent S58S lines were chosen to test their tolerance to various stresses. Western blot analysis indicated significant increases of OsSKIPa protein in all the selected S58S lines (Fig. S2G). In the MS medium containing 3 μM ABA, the relative shoot growth (ratio of growth under stress conditions to growth under normal conditions) of S58S plants (28–32%) was significantly higher than that of WT plants (16%; P < 0.01; Fig. 3B and Fig. S2I), suggesting that overexpression of OsSKIPa can reduce the growth-suppression effect of ABA. In the MS medium containing 150 mM NaCl, the relative shoot growth of all the S58S lines tested (>50%) was also significantly higher than the WT (about 25%; P < 0.01; Fig. 3B and Fig. S2I). However, the concentrations of Na+ (and K+) in the plants showed no significant difference between S58S and WT under both saline and normal conditions (data not shown), suggesting that the improved salt tolerance may not be attributable to the regulation of Na+ or K+ concentration. In the medium containing 200 mM mannitol, which can cause physiological dehydration stress, the relative growth and relative fresh weight of S58S lines were also significantly higher than that of WT (Fig. 3B and Fig. S2I). When 4-leaf–stage S58S and WT plants growing in soil were stressed by drought (no watering for 7 days), 70–80% of the transgenic plants recovered at 5 days after rewatering, whereas only 30–50% of WT plants recovered (Fig. 4 A and C), significantly (P < 0.05) lower than the transgenic plants. Drought tolerance was also tested at the panicle development stage using the method described previously (15), in which plants were individually stressed to the same degree in terms of relative water content in leaves. All the assayed transgenic plants were confirmed by PCR (Fig. S2H). The S58S lines had significantly higher relative spikelet fertility and grain yield (t tests, P < 0.01), 2 commonly used parameters for evaluation of drought resistance of rice at the reproductive stage, than the WT in the drought treatment (Fig. 4 B and D). After drought stress, S58S lines generated significantly more tillers than the WT if irrigation was resumed (t test, P < 0.01; Fig. 4E). Nevertheless, no significant differences in other agronomic traits were detected between S58S transgenic lines and WT under normal growth conditions (data not shown). These results clearly suggest that overexpression of OsSKIPa in rice can improve drought resistance.
Fig. 4.
Improved drought resistance of rice plants overexpressing OsSKIPa. (A and C) Improved drought resistance of OsSKIPa-overexpressing plants at the seedling stage. (C) The drought tolerance was evaluated by survival rate. Error bar indicates SE based on 3 replicates. (B and D) Increased relative spikelet fertility and relative yield of OsSKIPa-overexpressing plants at the heading stage. (E) Significantly increased tiller number of OsSKIPa-overexpressing plants after drought stress. Data represent mean ± SD (n = 12). **t test, with P < 0.01; *t test, with P < 0.05.
Enhanced Cell Viability, Reactive Oxygen Species-Scavenging Ability, and Expression of Stress Resistance-Related Genes in S58S Plants Under Stress Conditions.
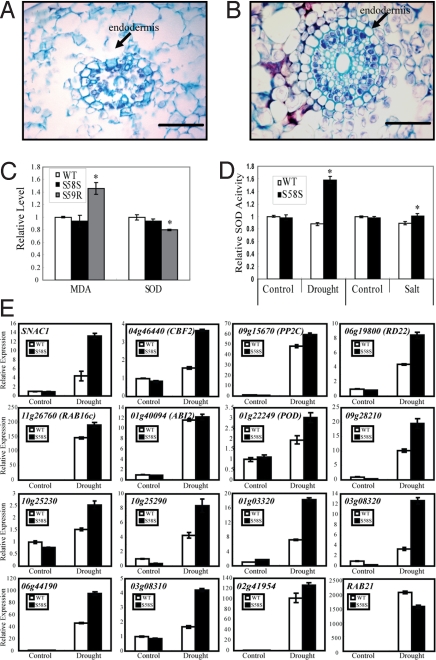
Because the S59R plants showed significantly reduced cell viability, we checked the cell viability of S58S plants as well. Under normal growth conditions, no obvious difference in cell viability was observed between S58S and WT plants. Under salt stress (medium with 150 mM NaCl for 7 days), almost all the cells of the primary root of WT plants lost viability (Fig. 5A), whereas most of the cells in the corresponding regions of S58S plants remained alive and only some of the cortex cells lost viability (Fig. 5B). These results suggest that S58S plants can retain cell viability better than WT plants under stress conditions.
Fig. 5.
Enhanced cell viability, ROS-scavenging ability, and expression of stress-related genes in S58S plants under stress conditions. The cell viability of S58S compared with WT. After salt stress, the root (cross section) cells of WT plants were collapsed (A), whereas most of the cells of S58S plants remain viable (B). Bars = 100 μm. (C) Relative MDA level and SOD activity in S58S and S59R (compared with WT) under normal growth. *t test, with P < 0.05 (n = 4). (D) Relative SOD activity in S58S (compared with WT under control) under drought and salt stress conditions. Control, normal growth condition.*t test, with P < 0.05 (n = 4). (E) Expression of stress resistance-related genes in S58S and WT under drought stress. Leaf samples with similar relative water content (>95% for control and 75% for drought) from S58S and WT plants were used for expression analysis of stress-related genes by real-time PCR. 09g28210, putative DNA-binding protein; 01g03320, trypsin inhibitor; 01g22249, peroxidase; 02g41954, gibberellin 2-beta-dioxyenase; 06g44190, unknown stress protein; 10g25230, 10g25290, 03g08310, and 03g08320 are TIFY/ZIM transcription factors.
ABA and abiotic stresses may increase the level of reactive oxygen species (ROS), and accumulation of ROS can decrease cell viability (16). Superoxide dismutases (SODs) are important ROS-scavenging enzymes. Monodehydroascorbate (MDA) is an important intermediate in ROS scavenging (16, 17), and a high level of MDA is toxic to plant cells. Under normal growth conditions, the MDA and SOD levels in the S59R plants were 150% and 80% of the WT plants (Fig. 5C), respectively, suggesting reduced ROS-scavenging activity in the S59R plants. In the S58S plants, the SOD and MDA levels were not significantly different from those of WT plants under the normal growth conditions (Fig. 5C). However, the S58S plants had a significantly higher level of SOD than WT plants after drought or salt stress treatment (Fig. 5D). These results suggested that the increased stress tolerance of the OsSKIPa-overexpressing plants may be partially attributable to the enhanced ROS-scavenging activity.
To gain further insight into the mechanism of the enhanced stress resistance of S58S plants, transcript levels of 16 stress-responsive genes were assayed in the S58S and WT plants under normal and drought stress conditions. These genes, including the drought resistance gene SNAC1 (18) and homologs of a few well-documented stress-related genes (e.g., CBF2, PP2C, RD22), were selected based on the literature and our unpublished microarray data. Most of these genes showed no significant difference in transcript level between S58S and WT plants under normal conditions. However, 14 genes, excluding ABI2 homolog and RAB21, showed significantly higher expression level in S58S than in WT under drought stress conditions (P < 0.05; Fig. 5E). These results suggested that overexpression of OsSKIPa can increase the transcript levels of some stress-resistance genes under drought stress conditions, thus improving the drought resistance of the transgenic rice. Among the genes with significantly higher induction in S58S, quite a few (SNAC1, 10g25230, 10g25290, 03g08310, and 03g08320) encode stress-responsive transcription factors, indicating that OsSKIPa may indirectly regulate the expression of a large number of stress-responsive genes.
Global Expression Changes in S58S and S59R Plants.
Considering the nature of SKIP as a transcription regulator, we compared the expression profiles of S58S, S59R, and WT plants under normal growth conditions using the Affymetrix gene chip of rice. According to linear model analysis, 635 genes were detected with more than a 2-fold change (P < 0.01) of transcript level in S59R plants (336 and 299 were up- and down-regulated, respectively) and 115 genes exhibited more than a 2-fold change (P < 0.01) in S58S plants (57 and 58 were up- and down-regulated, respectively). About 95% (19 of 20) of the genes showing expression level change could be confirmed by real-time PCR analysis (data not shown). These results suggest that OsSKIPa can affect the transcript levels of a large number of genes.
Gene ontology analysis of the genes up- or down-regulated in the S58S and S59R plants revealed that genes in 3 categories of biological processes were significantly overrepresented (hypergeometric test, P < 0.01): response to stimulus (biotic, abiotic, and endogenous stimuli), metabolism, and cell communication (Fig. S3). Among the 661 genes with expression levels changed by more than 2-fold in the transgenic plants, 216, 199, and 120 genes are responsive to drought, salt, and cold stresses, respectively (data not shown), based on the published microarray analysis (19). Of note, several genes encoding enzymes for ROS reactions were changed in S58S or S59R plants. Bioinformatic analysis of the cis-elements in the promoters of these genes suggested that 29 cis-elements deposited in the promoter database (PLACE) were enriched (Table S2), especially stress- and ABA-specific cis-elements, including several ABA responsive element-related elements and CANNTG box. These results further suggested that OsSKIPa may participate in the transcriptional regulation of numerous stress-related genes.
Diverse Functions of OsSKIPa-Interacting Proteins.
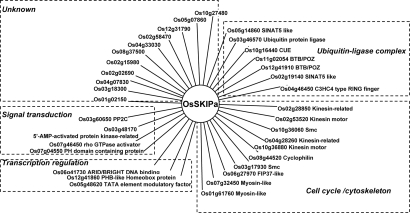
SKIP has been considered as a cofactor for transcription regulation in humans and yeast (5, 9). To address how OsSKIPa regulates transcription in rice, OsSKIPa was used as a bait to screen a prey cDNA library of rice. The yeast two-hybrid screening resulted in 35 unique genes encoding putative OsSKIPa-interacting proteins (SIPs; Fig. 6, Table S3, and Fig. S4A). Among 19 putative SIPs checked by GST pull-down assay, 18 were confirmed to interact with OsSKIPa in vitro (Fig. S4 B and C). All the SIP genes can be mapped to the japonica rice genome, and 21 of them were supported by full-length cDNAs in the database (Table S3).
Fig. 6.
OsSKIPa-interacting proteins identified by yeast two-hybrid screening. The OsSKIPa-interacting proteins were classified into 5 categories (as indicated in dashed frame boxes) based on their putative functions (more detail information in Table S3). Locus identifications of the proteins are from the TIGR rice annotation release 5 (http://rice.tigr.org).
The predicted functions of SIPs are quite diverse and can be classified into 5 categories: cell cycle/cytoskeleton proteins, ubiquitin ligase complex proteins, transcription regulation proteins, signal transduction proteins, and unknown proteins (Fig. 6). The transcription regulator category includes SIP5 (containing ARIT/BRIGHT DNA binding domain), SIP22 (HD-zip transcription factor), and SIP25 (homologous to TMF1, TATA element modulation factor 1, which plays an important role in the initiation of transcription) (20). The proteins SIP11 and SIP21 contain a pleckstrin homology domain that has been reported to have a role in lipid signaling (21). Of note, there are 11 rice SIPs without homology to known proteins, and 2 of them even have no homolog in the protein database of Arabidopsis. The functional diversity of OsSKIPa-interacting proteins suggested that OsSKIPa may be involved in diverse cellular processes in addition to being a transcription cofactor.
Discussion
Essential Roles of OsSKIPa for Cell Viability and Plant Growth.
Studies of SKIP homologs in yeast (PRP45) and C. elegans (CeSKIP) have indicated that SNW/SKIP protein, as a spliceosome component, plays a vital role in maintaining cell viability in these species (10, 22). Our data show that suppression of OsSKIPa resulted in severe growth arrest and even death of the plant, which is similar to the phenotype of embryonic arrest of the CeSKIP loss-of-function mutant in C. elegans (10), suggesting that OsSKIPa is also required for maintaining cell viability and normal growth in rice. Such an indispensable role of SKIP homolog in keeping normal cell viability and growth may be conserved in plants, which is supported by the facts that only 1 transcript of SKIP homolog was detected in rice and other plant species (not shown) and the plant SKIP homologs possess the conserved SNW domain for cell viability identified in PRP45 (22).
The vital role of OsSKIPa for plant growth and development is also supported by the important functions of a few SIP homologs in Arabidopsis. Knockout mutant of FIP37 (SIP2 homolog) showed an embryolethal phenotype caused by a strong delay of endosperm development and embryo arrest (23). Knockdown mutants of HUB1 (homolog of SIP20) have defects in the cell cycle during leaf and root growth (24). SINAT5 (homolog of SIP14 and SIP 33) promotes the ubiquitin-related degradation of NAC1 and down-regulation of auxin signal (25). SIP35 is similar to PLL4 (At2G28890), which regulates the meristem and organ development (26).
OsSKIPa Positively Modulates Stress Tolerance in Rice.
Sessile plants and motile animals have evolved arrays of distinct mechanisms to respond and adapt to abiotic stresses. We found that overexpression of OsSKIPa in rice can enhance tolerance to drought and high salinity. The specific function of OsSKIPa in conferring drought resistance may be especially useful in producing green super rice as proposed by Zhang (27). Although the sequences of SKIP homologs are highly conserved between animals and plants, such an improved stress tolerance resulting from overexpression of a SKIP homolog has not been reported in other species. This function may have evolved specifically in sessile plants.
Drought and high-salinity stresses can result in retarded growth or even cell damage in plants (28, 29) and can produce ROS in plant cells. Overaccumulation of ROS can lead to cell damage and even death. In S58S plants, the activity of SOD, a key ROS eliminator, was increased under stress conditions. These results suggested that the increased stress tolerance of the OsSKIPa-overexpressing plants may be partially attributable to the enhanced ROS-scavenging activity. Quite a few genes with putative functions in ROS scavenging were up-regulated in the S58S plants, which also supports this hypothesis.
At the level of gene expression, numerous stress-related genes can be induced by drought and high-salinity stresses. The expression level of OsSKIPa is also responsive to abiotic stresses (Fig. 3A). In the OsSKIPa-overexpressing plants, the expression levels of more than 200 stress-associated genes were affected. In particular, the mRNA levels for several stress-tolerance genes are significantly higher under drought stress conditions, including SNAC1 as a key transcription factor recently reported as conferring drought resistance in rice (18). Because OsSKIPa itself is unlikely a transcription factor, the change in expression level of so many genes might result from the interaction of OsSKIPa with other transcription regulators. This is evidenced by the fact that quite a few putative transcription regulators were identified as OsSKIPa-interacting proteins (Fig. 6), including SIP5 (ARIT/BRIGHT DNA binding protein), SIP22 (HD-zip transcription factor), and SIP25 (homolog of TMF1). We have preliminary results showing that overexpression of SIP25 can also enhance abiotic stress tolerance in rice (not shown). These results suggest that OsSKIPa is involved in the regulation of stress response and adaptation along with other stress-related transcription factors, although the details of the regulation processes require further investigation.
Recent reports indicated that mRNA processing (13), mRNA decay rates (30), and protein ubiquitination (31) play important roles in stress response and adaptation. Here, we show that OsSKIPa may be a putative splicing component in rice because it can complement the splicing factor PRP45 in yeast. Our data indicate that OsSKIPa can interact with the components of the ubiquitin ligase complex (Table S3), suggesting that the stability of OsSKIPa may be regulated by an ubiquitin-proteasome system. Further investigation of how OsSKIPa recruits different proteins to the transcriptional machine is required to elucidate the molecular mechanism of OsSKIPa in conferring stress tolerance in plants.
Diversification of SKIP-Interacting Proteins.
We investigated whether the SNW/SKIP-interacting proteins are conserved in different species. According to the Human Protein Reference Database (32), SKIP interacts with 21 proteins. According to the IntAct database (33), the SNW/SKIP homologs PRP45 (yeast), Bx42 (Drosophila), and CeSKIP (C. elegans) interact with 34, 13 (including 2 human proteins), and 5 (including 2 human proteins) proteins, respectively (Table S4). Strangely, very few of the SIPs in rice have matches with the SKIP-interacting proteins in humans, Drosophila, C. elegans, or yeast. Only CUE and cyclophilin type peptidyl-prolyl cis-trans isomerase domains were found in the SKIP-interacting proteins both in rice and humans but not in the PRP45-interacting proteins. This is not surprising, because PRP45 is distinctly different in the N-terminal domain (missing in PRP45) and the SNW domain compared with human SKIP or OsSKIPa (Fig. S1A), and a full-length SKIP homolog (SpPRP45) from yeast, Schizosaccharomyces pombe, can actually interact with cyclophilin proteins (34). On the other hand, most of the SKIP-interacting proteins in animals and yeast had no or very low homologous matches in rice (Table S4). We isolated a few rice homologs of SKIP-interacting protein genes (Q92769, P43355, RbA, and RbB) and tested the interactions of the proteins of these genes with OsSKIPa in yeast, but the result suggested that none of them interacted with OsSKIPa (data not shown). In fact, there are few common SKIP-interacting proteins even among humans, other animals, and yeast (Table S4). These results suggest that the SKIP lineage may have evolved with diverse functions through interacting with different proteins in different species. Nevertheless, some conserved SKIP-interacting proteins among eukaryotes may still remain to be identified, especially considering the incomplete coverage of the yeast two-hybrid screening libraries used for identification of SKIP-interacting proteins in this study.
Methods
General Methods.
See SI Text for details.
Stresses Treatment.
Positive transgenic plants of S58S T1 or T2 families were selected by germinating seeds on MS medium containing 50 mg/L hygromycin. After germination, the positive seedlings were transplanted in MS medium containing 3 μM ABA, 150 mM NaCl, or 200 mM mannitol and grown for 10 days. S58S transgenic plants (at the 4-leaf stage) growing in sandy soil were drought-stressed (no watering for 7 days), followed by recovery. The drought stress at the panicle development stage was applied to plants growing in PVC tubes (15); genotypes of the plants were checked by PCR using primers specific for hygromycin resistance gene (Hpt) (Table S5). Each stress test was repeated 3 or 4 times. Details are provided in SI Text.
Complementation Assay of Yeast prp45Δ Mutant.
The heterozygous diploid yeast strain BY4743 (MATa/α, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, lys2Δ0/LYS2, MET15/met15Δ0, ura3Δ0/ura3Δ0 prp45::G418/PRP45) was obtained from Invitrogen (cat. no. 95400). The strain was transformed with vector pDEST32-OsSKIPa and induced for sporulation to produce haploid progeny by plating the yeast colonies onto MacConkey medium for 5 days. After sporulation, yeast cells were checked for their viability on SC medium lacking Leu, SC medium containing G418 but without Met, and SC medium containing G418 but lacking Lys, respectively.
Supplementary Material
Acknowledgments.
This work was supported by grants from the National Special Key Project of China on Functional Genomics of Major Plants and Animals; the National Program on the Development of Basic Research; the National Natural Science Foundation of China, Ministry of Education of China; and the European Commission Sixth Framework Programme (EC FP6) Project (Grant 015468).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) database [accession nos. EU368691–EU368725 (cDNAs) and AK067153 (genomic)]. The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE10054).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901940106/DCSupplemental.
References
- 1.Saumweber H, Frasch M, Korge G. Two puff-specific proteins bind within the 2.5 kb upstream region of the Drosophila melanogaster Sgs-4 gene. Chromosoma. 1990;99:52–60. doi: 10.1007/BF01737289. [DOI] [PubMed] [Google Scholar]
- 2.Dahl R, Wani B, Hayman MJ. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene. 1998;16:1579–1586. doi: 10.1038/sj.onc.1201687. [DOI] [PubMed] [Google Scholar]
- 3.Figueroa JD, Hayman MJ. Differential effects of the Ski-interacting protein (SKIP) on differentiation induced by transforming growth factor-beta1 and bone morphogenetic protein-2 in C2C12 cells. Exp Cell Res. 2004;296:163–172. doi: 10.1016/j.yexcr.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 4.Leong GM, et al. Ski-interacting protein, a bifunctional nuclear receptor coregulator that interacts with N-CoR/SMRT and p300. Biochem Biophys Res Commun. 2004;315:1070–1076. doi: 10.1016/j.bbrc.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Folk P, Puta F, Skruzny M. Transcriptional coregulator SNW/SKIP: The concealed tie of dissimilar pathways. Cell Mol Life Sci. 2004;61:629–640. doi: 10.1007/s00018-003-3215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wieland C, Mann S, von Besser H, Saumweber H. The Drosophila nuclear protein Bx42, which is found in many puffs on polytene chromosomes, is highly charged. Chromosoma. 1992;101:517–525. doi: 10.1007/BF00352475. [DOI] [PubMed] [Google Scholar]
- 7.Negeri D, Eggert H, Gienapp R, Saumweber H. Inducible RNA interference uncovers the Drosophila protein Bx42 as an essential nuclear cofactor involved in Notch signal transduction. Mech Dev. 2002;117:151–162. doi: 10.1016/s0925-4773(02)00193-4. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov AI, et al. Genes required for Drosophila nervous system development identified by RNA interference. Proc Natl Acad Sci USA. 2004;101:16216–16221. doi: 10.1073/pnas.0407188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albers M, Diment A, Muraru M, Russell CS, Beggs JD. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA. 2003;9:138–150. doi: 10.1261/rna.2119903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostrouchova M, Housa D, Kostrouch Z, Saudek V, Rall JE. SKIP is an indispensable factor for Caenorhabditis elegans development. Proc Natl Acad Sci USA. 2002;99:9254–9259. doi: 10.1073/pnas.112213799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 12.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee BH, Kapoor A, Zhu J, Zhu JK. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell. 2006;18:1736–1749. doi: 10.1105/tpc.106.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue B, et al. Genetic basis of drought resistance at reproductive stage in rice: Separation of drought tolerance from drought avoidance. Genetics. 2006;172:1213–1228. doi: 10.1534/genetics.105.045062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 17.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plants Sci. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 18.Hu H, et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain M, et al. F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007;143:1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia JA, et al. Cloning and chromosomal mapping of a human immunodeficiency virus 1 “TATA” element modulatory factor. Proc Natl Acad Sci USA. 1992;89:9372–9376. doi: 10.1073/pnas.89.20.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Leeuwen W, Okresz L, Bogre L, Munnik T. Learning the lipid language of plant signalling. Trends Plants Sci. 2004;9:378–384. doi: 10.1016/j.tplants.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Martinkova K, Lebduska P, Skruzny M, Folk P, Puta F. Functional mapping of Saccharomyces cerevisiae Prp45 identifies the SNW domain as essential for viability. J Biochem (Tokyo) 2002;132:557–563. doi: 10.1093/oxfordjournals.jbchem.a003257. [DOI] [PubMed] [Google Scholar]
- 23.Vespa L, et al. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2004;134:1283–1292. doi: 10.1104/pp.103.028050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleury D, et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell. 2007;19:417–432. doi: 10.1105/tpc.106.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Q, et al. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature. 2002;419:167–170. doi: 10.1038/nature00998. [DOI] [PubMed] [Google Scholar]
- 26.Song SK, Clark SE. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev Biol. 2005;285:272–284. doi: 10.1016/j.ydbio.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q. Strategies for developing Green Super Rice. Proc Natl Acad Sci USA. 2007;104:16402–16409. doi: 10.1073/pnas.0708013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zidan I, Azaizeh H, Neumann PM. Does salinity reduce growth in maize root epidermal cells by inhibiting their capacity for cell wall acidification? Plant Physiol. 1990;93:7–11. doi: 10.1104/pp.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neves-Piestun BG, Bernstein N. Salinity-induced inhibition of leaf elongation in maize is not mediated by changes in cell wall acidification capacity. Plant Physiol. 2001;125:1419–1428. doi: 10.1104/pp.125.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narsai R, et al. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. SDIR1 is a RING Finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell. 2007;19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra GR, et al. Human protein reference database: 2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerrien S, et al. IntAct: Open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skruzny M, et al. Cyclophilins of a novel subfamily interact with SNW/SKIP coregulator in Dictyostelium discoideum and Schizosaccharomyces pombe. Biochim Biophys Acta. 2001;1521:146–151. doi: 10.1016/s0167-4781(01)00301-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.