Abstract
In mycobacteria, F420, a deazaflavin derivative, acts as a hydride transfer coenzyme for an F420-specific glucose-6-phosphate dehydrogenase (Fgd). Physiologically relevant reactions in the mycobacteria that use Fgd-generated reduced F420 (F420H2) are unknown. In this work, F420H2 was found to be oxidized by NO only in the presence of oxygen. Further analysis demonstrated that NO2, produced from NO and O2, was the oxidant. UV-visible spectroscopic and NO-sensor-based analyses proved that F420H2 reduced NO2 to NO. This reaction could serve as a defense system for Mycobacterium tuberculosis, which is more sensitive to NO2 than NO under aerobic conditions. Activated macrophages produce NO, which in acidified phagosomes is converted to NO2. Hence, by converting NO2 back to NO with F420H2, M. tuberculosis could decrease the effectiveness of antibacterial action of macrophages; such defense would correspond to active tuberculosis conditions where the bacterium grows aerobically. This hypothesis was consistent with the observation that a mutant strain of Mycobacterium smegmatis, a nonpathogenic relative of M. tuberculosis, which either did not produce or could not reduce F420, was ≈4-fold more sensitive to NO2 than the wild-type strain. The phenomenon is reminiscent of the anticancer activity of γ-tocopherol, which reduces NO2 to NO and protects human cells from NO2-induced carcinogenesis.
Keywords: macrophage, deazaflavin, nitrogen dioxide, nitric oxide, tuberculosis
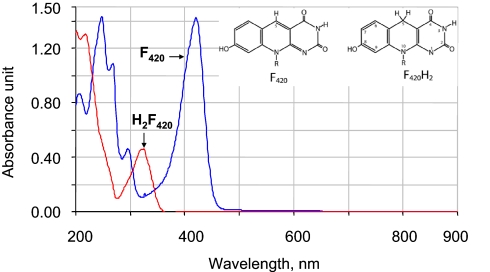
Coenzyme F420, a 7,8-didemethyl-8-hydroxy-5-deazaflavin derivative (Fig. 1), is a 2-electron or hydride transfer restricted redox catalyst (Eo′ = −360 mV) similar to the nicotinamides (Eo′ = −320 mV) (1, 2). F420 is found in all methanogenic and certain nonmethanogenic archaea, where it participates in energy metabolism, NADP reduction, oxygen detoxification, and sulfite reduction (3–6). In the bacterial domain, F420 is found in certain members of the Actinobacteria phylum, such as Mycobacterium species (7). These organisms express an F420-dependent glucose-6-phosphate dehydrogenase (Fgd, Reaction 1) (7, 8).
The physiological fate of F420H2 produced by Fgd is unknown. An insertional inactivation of fbiC, an essential gene for the synthesis of the deazaflavin chromophore or catalytic unit of F420 (9), renders Mycobacterium tuberculosis hypersusceptible to acidified nitrite (10). This in vitro treatment simulates an environment inside the phagosomes of an infected-activated macrophage, which produces nitric oxide (NO) by the action of inducible nitric oxide synthase (iNOS or NOS2) (11). Upon acidification of a phagosome, nitrite, a major product of NO oxidation, is converted to nitrous acid (HNO2; pKa = 3.16) (12), which in turn dismutates to NO and NO2 (10, 13); NO2 arises also from a reaction of NO with O2 (14). These observations suggest that the pathogenic mycobacteria could use F420H2 to combat an attack of reactive nitrogen intermediates generated by the activated macrophage. We have tested this hypothesis. The resulting data point toward a mechanism that M. tuberculosis may use to counter the bactericidal actions of macrophages.
Fig. 1.
UV-visible spectra of oxidized and reduced forms of coenzyme F420. F420 was reduced with sodium borohydride.
Results and Discussion
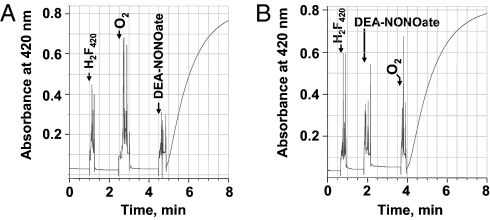
In a phosphate-buffered aqueous solution, F420H2 was rapidly oxidized by the NO-releasing compound diethylamine-NONOate (DEA-NONOate) and oxygen (Fig. 2).
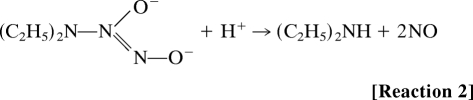 |
Neither DEA-NONOate nor O2 alone was able to perform this reaction (Fig. 2). To ascertain that NO was responsible for oxidizing F420H2, we examined whether the oxidation rate of F420H2 can be controlled by the NO release rate. For this purpose, two other derivatives of diazeniumdiolates, spermine-NONOate and DETA-NONOate, which release NO slowly, were used. The half-lives of DEA-, spermine-, and DETA-NONOate to release NO are 2 min, 39 min, and 20 h, respectively (15). Indeed, with the last two compounds, the F420H2 oxidation was much slower than that with DEA-NONOate. With 80 μM dissolved O2 and 25 μM F420H2, DEA-NONOate (0.5 mM), spermine-NONOate (0.5 mM), and DETA-NONOate (2.5 mM) oxidized F420H2 at the rates of 17.3, 0.94, and 0.22 μmol min−1 L−1, respectively. These data established that NO was one of the entities involved in the oxidation of F420H2.
Fig. 2.
Oxidation of F420H2 by nitric oxide in the presence of oxygen. Nitric oxide was generated in situ from NONOate derivatives. Into a 1-mL anaerobic solution of 50 mM potassium phosphate buffer (pH 7.0) in a stoppered cuvette with N2 in the headspace, the following were added to the final concentrations and in the sequence indicated: (A) Reduced F420 (20 μM), oxygen (80 μM), and DEA-NONOate (320 μM). (B) Reduced F420 or F420H2 (20 μM), DEA-NONOate (320 μM), and oxygen (80 μM). The time of each addition is shown by an arrow. Oxidation of F420H2 was monitored spectrophotometrically at 420 nm. DEA-NONOate, 2-(N,N-diethylamino)-diazenolate-2-oxide sodium salt; DETA-NONOate, 2,2′-(hydroxynitrosohydrazino)bisethanamine; spermine-NONOate, N-[4-[1-(3-aminopropyl)-2-hydroxy-2-nitrosohydrazino]butyl]-1,3-propanediamine.
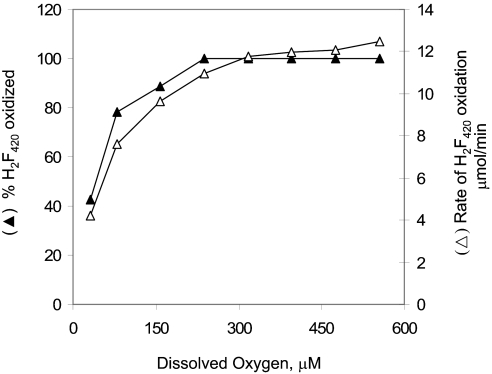
To understand the role of O2 in the NO-dependent F420H2 oxidation, the effect of O2 concentration on the reaction rate and amount of F420 produced at fixed concentrations of DEA-NONOate and F420H2 was studied. The results presented in Fig. 3 show that when DEA-NONOate was present in excess, the rate of F420H2 oxidation and the amount of F420 formed increased with the dissolved O2 concentration.
Fig. 3.
Reaction rate and amount of F420 produced at various dissolved oxygen concentrations and at fixed levels of DEA-NONOate and F420H2. The assay mixture contained 1 mL of anaerobic 100 mM potassium phosphate buffer (pH 7.0), with 20 μM F420H2 and varied volumes of O2 added to the nitrogen headspace. Before the assay, the mixture was held at room temperature for 15 min to ensure equilibration with the added oxygen. The reaction was initiated by the addition of DEA-NONOate (final concentration, 320 μM) and was followed spectrophotometrically at 420 nm. The dissolved oxygen concentration was calculated by using a value of 43,800 atm/mol fraction for the Henry's constant for oxygen (40).
The above results led to the question of how a combination of NO and O2 oxidized F420H2. In an aqueous solution, nitric oxide reacts rapidly and spontaneously with oxygen to produce mainly nitrite, via the intermediate formation of nitrogen dioxide (NO2) and dinitrogen trioxide (N2O3) (Reactions 3–5) (14).
To test whether nitrite was the F420H2-oxidizing agent, in an experiment similar to that represented in Fig. 2, DEA-NONOate was incubated in an aerobic potassium phosphate buffer for 60 min at 25 °C (allowing dissipation of NO, NO2, and N2O3 via Reactions 3–5), and then this solution was mixed with F420H2. This treatment did not result in F420H2 oxidation. Also, as determined from the UV-visible spectral characteristics, 1 mM sodium nitrite or 1 mM sodium nitrate did not oxidize F420H2. However, the subsequent addition of DEA-NONOate to these aerobic mixtures caused the oxidation of F420H2. Therefore, nitrite or nitrate was neither an F420H2-oxidizing agent nor an inhibitor of F420H2 oxidation. It is unlikely that N2O3 was responsible for F420H2 oxidation because this compound is not an oxidizing agent (16). Thus, it was hypothesized that NO2, a fairly potent oxidant (16), was the compound that oxidized F420H2. This hypothesis was tested as described below.
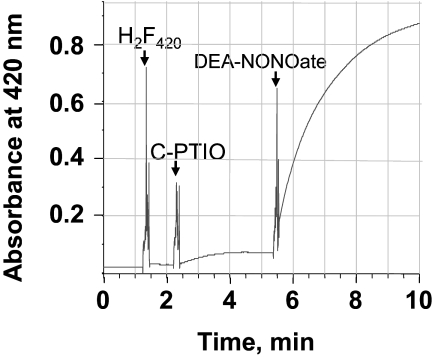
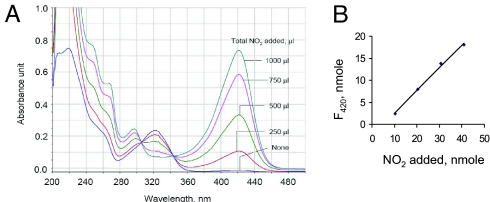
In the first assay, NO2 was generated in situ via a reaction between C-PTIO, a nitronyl nitroxide, and NO produced from DEA-NONOate in an anaerobic solution (17). The data in Fig. 4 show that the oxidation of F420H2 was observed only if both DEA-NONOate and C-PTIO were present in the reaction mixture, suggesting that F420H2 was oxidized by NO2. Because of an overlap between the UV-visible spectra of C-PTIO and F420H2 (17), this system was unsuitable for spectroscopic studies. Therefore, further experiments were performed using pure NO2 gas. The resulting data demonstrated that NO2 did indeed oxidize F420H2 directly (Fig. 5). The UV-visible spectra of the reaction mixture (Fig. 5A) that was recorded immediately after each addition of NO2 into an anaerobic solution of F420H2 in 100 mM potassium phosphate buffer (pH 7.0) showed the appearance of the oxidized form of F420. The amount of F420 produced was proportional to the amount of NO2 added into the reaction mixture (Fig. 5B); because NO2 would rapidly be converted to nitrite and nitrate in the phosphate-buffered solution we used (Reactions 6 and 7) (14), the ratio of NO2 added and F420 formed was not 1:1.
The UV-visible spectral changes shown in Fig. 5A clearly indicated that F420H2 was oxidized to F420. This oxidation is known to be a 2-electron or hydride transfer process (2, 18). The observation leads to the hypothesis that F420H2 oxidation was coupled to 2-electron reduction of NO2 that generated NO (Reaction 8).
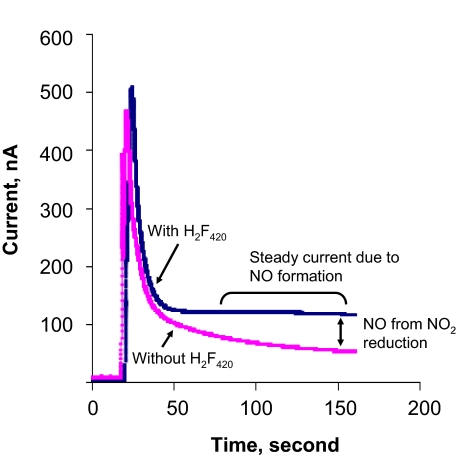
This possibility was tested via measurements with an ISO-NOPF100H NO probe from World Precision Instruments. The addition of NO2 in the nitrogen headspace of a sealed serum bottle containing anaerobic 100 mM potassium phosphate buffer (pH 7.0) created a high electrical current (Fig. 6) because the NO sensor detects NO2, albeit with much less sensitivity than that for NO (Michael McIntosh and Xueji Zhang, World Precision Instruments, personal communication). However, the current dissipated quickly because of the conversion of NO2 to nitrite and nitrate (Reactions 6 and 7). The overall result was a current spike (Fig. 6). When F420H2 was present in the reaction mixture, a similar current spike was also seen, but the residual current was higher than that recorded with NO2 alone (Fig. 6). This residual current represented some of the NO generated from NO2 by F420H2. NO is relatively stable in an anaerobic aqueous solution, but it can be dissipated via Reactions 4 and 5, if NO2 is present. Because added NO2 was lost quickly via Reactions 6 and 7, only a small amount of NO was generated. However, because of the lack of residual NO2, this NO was not sequestered and provided a higher residual current. The amount of F420 formed in this experiment was proportional to the amount of NO2 added. Therefore, it was clear that F420H2 reduced NO2 to NO. Based on the well-established ground-state chemistry of F420 (2, 18), we hypothesize that this reduction occurs via a hydride transfer mechanism; this possibility is currently under investigation.
Fig. 4.
Oxidation of F420H2 under anaerobic condition by nitrogen dioxide generated from an in situ reaction between carboxy-PTIO and NO (from DEA-NONOate). Into 1 mL of anaerobic 50 mM potassium phosphate buffer (pH 7.0) in a stoppered cuvette with N2 in the headspace, the following were added: 25 μM F420H2, 100 μM C-PTIO, and 250 μM DEA-NONOate. The point of each addition is shown by an arrow and the name of the compound. Oxidation of F420H2 was monitored spectrophotometrically at 420 nm. Carboxy-PTIO or C-PTIO, 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide potassium salt. Other abbreviations are shown in the legend for Fig. 2.
Fig. 5.
Oxidation of F420H2 by nitrogen dioxide gas under anaerobic condition. (A) To 1 mL of an anaerobic 25 μM F420H2 solution in 50 mM potassium phosphate buffer (pH 7.0) in a 3-mL quartz cuvette, nitrogen dioxide gas was added by using a gas-tight syringe. The UV-visible spectrum of the solution was recorded before NO2 addition and then after each addition of 250 μL of (10.22 nmol) NO2. (B) Relationship between NO2 added and F420 generated. Note: Most of the NO2 added was likely converted into NO2− and NO3−.
Fig. 6.
Nitric oxide production from the reaction of nitrogen dioxide and F420H2. One milliliter of NO2 (1 atm) was added to the nitrogen headspace of a stoppered, 10-mL serum bottle containing 7.5 mL of anaerobic 100 mM potassium phosphate buffer (pH 7.0) with 20 μM F420H2 (blue) or without F420H2 (pink). NO production was measured by using an NO sensor (ISO-NOPF100H) (World Precision Instruments).
Because F420 catalyzes an NAD(P)H-type hydride transfer, the reactivity of the latter toward NO2 was examined. NADH reacts with NO2 in the presence of oxygen (19). In this process, NO2 is reduced to nitrite, and an NAD· radical is produced as an intermediate, and the latter reacts with oxygen to generate superoxide (
). We found that in an anaerobic assay system of the type shown in Fig. 5, NO2 does not oxidize NADH or NADPH; in each case the concentration of the reduced cofactor was 160 μM. Therefore, in its ability to reduce NO2 to NO, F420H2 was distinct from NAD(P)H.
To examine whether nitrogen dioxide caused nitrosation of the phenol moiety of F420, the products from a reaction similar to that shown in Fig. 2A, but with F420H2 and DEA-NONOate at 50 and 500 μM concentrations, respectively, were characterized. In a C18-silica column-based reversed-phase HPLC analysis employing a diode array detector (20), the deazaflavin product from the reaction and authentic F420 exhibited the same retention time and UV-visible spectrum [supporting information (SI) Fig. S1]. A nitrophenol generated from a reaction between a phenol and NO2 binds more tightly to C18-silica than the corresponding phenol (21, 22). These results ruled out the possibility of nitrosation of F420H2 with NO2 or NO.
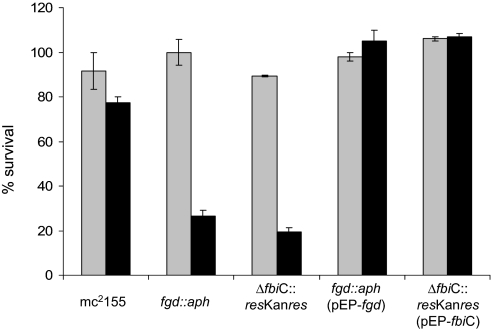
The ability to use F420H2 in reducing NO2 to NO would be useful to M. tuberculosis in combating the host defense system. Although both NO and NO2 exhibit antimycobacterial activities, NO2 is more potent in killing M. tuberculosis (23). Therefore, because an activated macrophage produces NO and converts this compound in its acidified phagosomes to NO2 either via the formation and dismutation of nitrous acid (13) or a reaction of NO with O2 (14) for bringing about a more aggressive attack, M. tuberculosis could use Fgd-generated F420H2 to reduce NO2 back to NO and lower the effectiveness of the antibacterial action of macrophages. This hypothesis is consistent with the observed hypersensitivity of M. tuberculosis fbiC strain to acidified nitrite (10). A mycobacterial cell contains a substrate level of glucose 6-phosphate (24). Therefore, F420H2 produced via the Fgd reaction (Reaction 1) is an effective vehicle for delivering substantial reducing power for NO2 detoxification. To provide an experimental validation for this deduction, the effect of NO2 on the viability of Mycobacterium smegmatis, a close relative of M. tuberculosis and a nonpathogen, was studied. In this experiment, actively growing cultures of wild-type, fgd::aph, and Δfbic::aph strains of M. smegmatis were exposed to NO2, and then the numbers of the living cells in these cultures were determined (Fig. 7). NO2 was generated in situ through a reaction between dissolved O2 and NO (derived from spermine-NONOate; half-life, 39 min); an exposure time of 6 h was selected because it provided an observable yet mild effect on the viability of the wild-type cells. The survivability of a strain was expressed in terms of the ratio of the colony-forming units from an NO2-exposed culture and that from a control culture (without NO2 exposure). The values for these ratios for the wild-type, fgd::aph, and Δfbic::aph strains were 0.8, 0.25, and 0.2, respectively (Fig. 7). Therefore, an inability to synthesize F420 or to produce F420H2 made the organism ≈4-fold more sensitive to NO2. When the fgd::aph and Δfbic::aph were complemented with Fgd and Fbic expression vectors, respectively, they were able to tolerate NO2 even better than the wild-type cells (Fig. 7); the increased tolerance was likely caused by above-normal level of F420H2 generated by the elevated levels of FbiC and Fgd proteins. These results established a requirement for F420H2 in the reduction of NO2. However, it remains possible that the observed phenotypes of the mutant strains were not solely the result of a loss of the chemical reaction described in this report but also in part caused by the lack of the activity of a yet to be identified NO2 detoxification enzyme that requires F420H2. The overall phenomenon is similar to the inhibition of neoplastic transformation of human cells by γ-tocopherol (25). It has been suggested that the nitrosation of primary amines of DNA bases by NO, which leads to cancer-causing mutations, requires oxidation of this oxide to NO2 (25). Because γ-tocopherol efficiently reduces NO2 back to NO, it protects cells from NO2-induced carcinogenesis (25).
Fig. 7.
Requirement for reduced F420 for NO2 detoxification in M. smegmatis. The sensitivities of wild-type, fgd::aph, Δfbic::aph, fgd::aph (pEP-fgd), and Δfbic::aph (pEP-fbiC) strains to NO2 were studied. In each case, the cells were exposed to NO2 by incubating a corresponding diluted suspension in growth medium with spermine-NONOate under air; a control culture received water in place of spermine-NONOate solution. In each case, including for the control, the number of surviving cells per milliliter in a culture is presented as a percentage of the value recorded for a sample drawn before incubation with spermine-NONOate (or water); as a result, the value even for the control culture was <100%. Each error bar was calculated from measurements in triplicate. Gray bar, control culture; black bar, NO2-exposed culture.
It has been known that PA-824, a bicyclic nitroimidazole and a promising candidate drug for the treatment of tuberculosis, acts as an antimycobacterial agent only when the organism produces F420 and reduces it to F420H2 (9, 20, 26). A recent report shows that the basis for this activity is the F420H2-dependent production of NO from PA-824 (27); NO kills M. tuberculosis anaerobically. The imidazole ring of PA-824 is reduced with F420H2 by a nitroreductase called Ddn (27). This reaction is followed by the release of the nitro group of the compound as nitrous acid that breaks down to NO (27). Therefore, F420-dependent reactions not only provide a protection to the mycobacteria from nitrosative stresses, but also can be exploited to impose such a stress on these organisms.
In summary, F420H2-dependent NO2 reduction reaction is a defense tool for the mycobacteria against NO2 stress. It should be noted that often NO has been cited as the agent that kills M. tuberculosis under both aerobic and low-oxygen or hypoxic conditions (28, 29), although, as mentioned above, NO2 has been shown to be a more potent antimycobacterial agent (23), and it is produced under aerobic conditions (10, 13, 14). Therefore, the F420H2-dependent defense against nitrosative stress would be useful to M. tuberculosis when it grows aerobically and causes active tuberculosis.
Materials and Methods
Chemicals.
Coenzyme F420 was purified from Methanothermobacter thermautotrophicus as described (30, 31). 2-(N,N-diethylamino)-diazenolate-2-oxide sodium salt (DEA-NONOate), N-[4-[1-(3-aminopropyl)-2-hydroxy-2-nitrosohydrazino]butyl]-1,3-propanediamine (spermine-NONOate), 2,2′-(hydroxynitrosohydrazino)bisethanamine (DETA-NONOate), 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide potassium salt (carboxy-PTIO or C-PTIO) were from Cayman Chemicals. Sodium borohydride was from Sigma–Aldrich. F420H2 was generated by reducing F420 with sodium borohydride in an anaerobic water solution under nitrogen atmosphere in a rubber stopper-sealed glass tube followed by neutralization with hydrochloric acid. Only freshly made F420H2 was used in the experiments. Nitrogen dioxide gas was prepared in microscale (32). The anaerobic techniques used in this work have been described in ref. 30.
F420H2 Oxidation Assays.
The assays were carried out as described in ref. 30. A 1-mL anaerobic aqueous solution containing 50 mM potassium phosphate buffer (pH 7.0) and 20 or 25 μM F420H2 under a N2 headspace in a 12-mm × 75-mm (8.5 mL) KIMAX borosilicate tube (Kimble/Kontes) sealed with a cutoff black butyl rubber stopper no. 00 was used for each assay. To this solution, DEA-NONOates, spermine-NONOate, and DETA-NONOate were added from respective 50 mM anaerobic stock solutions in 10 mM sodium hydroxide. For C-PTIO, a 50 mM stock was prepared in water. NaNO2 or NaNO3 was added from an anaerobic 100 mM stock solution in water. NO2 was added by using a gas-tight syringe. Oxidation of F420H2 was followed spectrophotometrically at 420 nm (at 25 °C), and the rates were calculated by using a value of 41.4 mM−1 cm−1 for the extinction coefficient of oxidized F420 at this wavelength (31).
NO Measurement Assay.
NO produced in an anaerobic F420H2 oxidation–NO2 reduction assay was measured with an Apollo 1000 single-channel, free radical detector employing an amperometric-type NO probe (ISO-NOPF100H; World Precision Instruments). The data were collected and analyzed by using a WPI DataTrax. The measurement was performed under anaerobic conditions. For this purpose, a 10-mL serum bottle containing 7.5 mL of 100 mM potassium phosphate buffer (pH 7.0), with 25 μM F420H2 was sealed with a 20-mm septum stopper (Bellco Glass), and the content was made anaerobic via 3 cycles of evacuation and pressurization with N2 (30); a control vial was prepared without F420H2. The final headspace gas was N2 at 1.3 atm. Then a 1-inch 18-gauge hypodermic needle with a Luer hub (Precision Glide needle; Becton Dickinson), which was shortened to 0.5 inches by cutting part of its tip end, was inserted into the bottle through the rubber stopper. Through this needle the probe was lowered in a manner where 2 mm of the probe tip was inside the buffer; before use, the probe was polarized according to the manufacturer's instructions. After the current output stabilized, NO2 gas was added to the headspace of the bottle through the rubber stopper by using a syringe, and the probe output was measured. To avoid contamination of air, the syringe was flushed with N2 before use.
Determining the Sensitivities of M. smegmatis to NO2.
A strain to be tested was grown in 7H9-glycerol medium with 0.05% Tween 80 at 37 °C until the early-logarithmic stage of growth. Then a sample of this culture was diluted with fresh medium to a 600-nm optical density of 0.5 [as measured with a Lambda 25 UV-visible spectrophotometer (PerkinElmer)]. One hundred microliters of this diluted culture was added to 900 μL of fresh medium, and this mixture was supplemented with 10 μL of 100 mM spermine-NONOate right after the latter was prepared in water; a control culture received water in place of the spermine-NONOate solution. The tubes with dilute cell suspensions (±spermine-NONOate) were incubated aerobically in 13-mm × 100-mm borosilicate culture tubes (catalog no. 73750-13100; Kimble/Kontes) for 6 h at 37 °C in a C25 incubator shaker (New Brunswick Scientific) operating at 240 rpm. Then the number of surviving cells in each such suspension was estimated via plating on 7H9-glycerol-agar medium and determining the number of colony-forming units.
Construction of M. smegmatis ΔfbiC::aph and fgd::aph Strains.
A ΔfbiC strain was generated by replacing the corresponding coding sequence with a kanamycin resistance cassette (aph) via homologous recombination (33). For this purpose, a 926-bp upstream region (UP) and 920-bp downstream region (DN) of M. smegmatis fbiC coding sequence (Msmeg_5126) were PCR-amplified with the following primer pairs: for the UP element, ccgaggatcctcgagaaggcaagggtctcggtcggcgtggtgag (forward) and gctcgaattccaaagccactcccgaataactccacgag (reverse) (underlined sequences, BamHI and EcoRI sites, respectively); for the DN element, cggccgaattcaccacatccagaccagctggg (forward) and gcccggctcgaggccgtcgaacacgagattca (reverse) (underlined sequences, EcoRI and XhoI sites, respectively). The UP and DN amplicons were digested with the above listed restriction enzymes. These restricted DNA fragments and a res-Km-res element that was excised from pCG122 (34) as an EcoRI fragment were cloned into BamHI and XhoI sites of pBluescript to obtain pEPfbiCKO1. Then the fbiC knockout construct was excised from pEPfbiCKO1 with XhoI and was cloned into the XhoI site of pJQ200KS (35) to obtain the suicide plasmid pEPfbiCKO2; pJQ200KS does not replicate in mycobacteria, but in an integrated form it allows a sacB-mediated sucrose counterselection and confers resistance to gentamicin in mycobacteria. In pEPfbiCKO2, the cloned fragments formed the following arrangement: 5′UP3′…5′res-Km-res3′…5′DN3′. Transformation of M. smegmatis mc2155 (wild-type strain) with pEPfbiCKO2 via electroporation (36) and selection of the transformants on gentamicin and kanamycin provided a merodiploid strain. By counterselecting segregants derived from the merodiploid strain on sucrose in the presence of kanamycin, a M. smegmatis ΔfbiC::res-Km-res strain was obtained; we call this strain ΔfbiC::aph. A Southern blot with EcoRI-digested chromosomal DNA and PshAI-digested pEPfbiCKO2 as the probe confirmed this genotype and showed that the mutant did not carry a remnant of pJQ200KS. An fgd::aph strain was constructed by inserting a kanamycin resistance cassette (Amersham Pharmacia Biotech) as a BamHI fragment at the BglII site of the gene; the details of this construction will be presented elsewhere. A Southern blot analysis confirmed the fgd::aph genotype of this strain. Assays for F420 and Fgd in cell lysates as described (24, 37) showed that the fgd::aph and ΔfbiC::aph strains lacked Fgd activity and F420, respectively.
Construction of Complementing of Plasmids for M. smegmatis fbiC and fgd.
The constructs were based on pSMT3, a mycobacteria–Escherichia coli shuttle vector that allows selection for hygromycin resistance and expression of cloned genes constitutively from the hsp60 promoter in mycobacteria (38). To generate the fbiC complementing plasmid (pEP-fbiC), the corresponding coding sequence along with a 222-bp upstream region (bearing the fbiC promoter and ribosome-binding site) was PCR-amplified from M. smegmatis chromosomal DNA with primers GATATCCGCGGACAGAGACAG (forward) and AAGCTTGACCTCGCGGTGAAG (reverse) (underlined sequence: EcoRV and HindIII sites, respectively) and cloned into EcoRV and HindIII sites of pSMT3; the selection of the upstream element was based on an earlier report (39). Similarly, the Fgd expression plasmid (pEP-fgd) was constructed by amplifying the corresponding coding sequence along with the 264-bp upstream region with the primers GGTTTTCGGGATCCAGATGGCAGTGCCGAAG (forward, with a BamHI site) and GAGCACTGCAGCTCGCGCGAGCACCCGCACG (reverse, with a PstI site) and cloning the amplicon into BamHI- and PstI-digested pSMT3.
Supplementary Material
Acknowledgments.
We thank Robert H. White and Pablo Sobrado for discussions and Darleen Baker for editing. This work was supported by start-up funds (to E.P. and B.M.) from the Virginia Bioinformatics Institute. The F420 biochemistry research in B.M.'s laboratory is supported by NASA Astrobiology: Exobiology and Evolutionary Biology Grant NNG05GP24G.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812883106/DCSupplemental.
References
- 1.Eirich LD, Vogels GD, Wolfe RS. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry. 1978;17:4583–4593. doi: 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT. Naturally occurring 5-deazaflavin coenzymes: Biological redox roles. Acc Chem Res. 1986;19:216–221. [Google Scholar]
- 3.Johnson EF, Mukhopadhyay B. A new type of sulfite reductase, a novel coenzyme F420-dependent enzyme, from the methanarchaeon Methanocaldococcus jannaschii. J Biol Chem. 2005;280:38776–38786. doi: 10.1074/jbc.M503492200. [DOI] [PubMed] [Google Scholar]
- 4.Seedorf H, Dreisbach A, Hedderich R, Shima S, Thauer RK. F420H2 oxidase (FprA) from Methanobrevibacter arboriphilus, a coenzyme F420-dependent enzyme involved in O2 detoxification. Arch Microbiol. 2004;182:126–137. doi: 10.1007/s00203-004-0675-3. [DOI] [PubMed] [Google Scholar]
- 5.Yamazaki S, Tsai L, Stadtman TC, Jacobson FS, Walsh C. Stereochemical studies of 8-hydroxy-5-deazaflavin-dependent NADP+ reductase from Methanococcus vannielii. J Biol Chem. 1980;255:9025–9027. [PubMed] [Google Scholar]
- 6.Schauer NL, Ferry JG, Honek JF, Orme-Johnson WH, Walsh C. Mechanistic studies of the coenzyme F420 reducing formate dehydrogenase from Methanobacterium formicicum. Biochemistry. 1986;25:7163–7168. doi: 10.1021/bi00370a059. [DOI] [PubMed] [Google Scholar]
- 7.Purwantini E, Gillis TP, Daniels L. Presence of F420-dependent glucose-6-phosphate dehydrogenase in Mycobacterium and Nocardia species, but absence from Streptomyces and Corynebacterium species and methanogenic Archaea. FEMS Microbiol Lett. 1997;146:129–134. doi: 10.1111/j.1574-6968.1997.tb10182.x. [DOI] [PubMed] [Google Scholar]
- 8.Purwantini E, Daniels L. Molecular analysis of the gene encoding F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J Bacteriol. 1998;180:2212–2219. doi: 10.1128/jb.180.8.2212-2219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi KP, Kendrick N, Daniels L. Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F420 and FO biosynthesis. J Bacteriol. 2002;184:2420–2428. doi: 10.1128/JB.184.9.2420-2428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 11.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 12.da Silva G, Kennedy EM, Dlugogorski BZ. Ab initio procedure for aqueous-phase pKa calculation: The acidity of nitrous acid. J Phys Chem A. 2006;110:11371–11376. doi: 10.1021/jp0639243. [DOI] [PubMed] [Google Scholar]
- 13.Stuehr DJ, Nathan CF. Nitric oxide: A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ignarro LJ, Fukuto JM, Griscavage JM, Rogers NE, Byrns RE. Oxidation of nitric oxide in aqueous solution to nitrite but not nitrate: Comparison with enzymatically formed nitric oxide from l-arginine. Proc Natl Acad Sci USA. 1993;90:8103–8107. doi: 10.1073/pnas.90.17.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzhugh AL, Keefer LK. Diazeniumdiolates: Pro- and antioxidant applications of the “NONOates.”. Free Radic Biol Med. 2000;28:1463–1469. doi: 10.1016/s0891-5849(00)00251-3. [DOI] [PubMed] [Google Scholar]
- 16.Pryor WA, et al. Free radical biology and medicine: It's a gas, man! Am J Physiol. 2006;291:R491–R511. doi: 10.1152/ajpregu.00614.2005. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein S, Russo A, Samuni A. Reactions of PTIO and carboxy-PTIO with *NO, *NO2, and O2−*. J Biol Chem. 2003;278:50949–50955. doi: 10.1074/jbc.M308317200. [DOI] [PubMed] [Google Scholar]
- 18.DiMarco AA, Bobik TA, Wolfe RS. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 19.Reszka KJ, Matuszak Z, Chignell CF, Dillon J. Oxidation of biological electron donors and antioxidants by a reactive lactoperoxidase metabolite from nitrite (NO2−): An EPR and spin trapping study. Free Radic Biol Med. 1999;26:669–678. doi: 10.1016/s0891-5849(98)00244-5. [DOI] [PubMed] [Google Scholar]
- 20.Choi KP, Bair TB, Bae YM, Daniels L. Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F420 biosynthesis by Mycobacterium bovis BCG. J Bacteriol. 2001;183:7058–7066. doi: 10.1128/JB.183.24.7058-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakshmi VM, Hsu FF, Zenser TV. Nitric oxide-mediated nitrosation of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline potentiated by hemin and myeloperoxidase. Chem Res Toxicol. 2005;18:1038–1047. doi: 10.1021/tx0500070. [DOI] [PubMed] [Google Scholar]
- 22.Ricoux R, Boucher JL, Mansuy D, Mahy JP. Microperoxidase 8 catalyzed nitration of phenol by nitrogen dioxide radicals. Eur J Biochem. 2001;268:3783–3788. doi: 10.1046/j.1432-1327.2001.02288.x. [DOI] [PubMed] [Google Scholar]
- 23.Yu K, et al. Toxicity of nitrogen oxides and related oxidants on mycobacteria: M. tuberculosis is resistant to peroxynitrite anion. Tuber Lung Dis. 1999;79:191–198. doi: 10.1054/tuld.1998.0203. [DOI] [PubMed] [Google Scholar]
- 24.Purwantini E, Daniels L. Purification of a novel coenzyme F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J Bacteriol. 1996;178:2861–2866. doi: 10.1128/jb.178.10.2861-2866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooney RV, et al. γ-Tocopherol detoxification of nitrogen dioxide: Superiority to α-tocopherol. Proc Natl Acad Sci USA. 1993;90:1771–1775. doi: 10.1073/pnas.90.5.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stover CK, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 27.Singh R, et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacMicking JD, et al. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nathan C. Microbiology: An antibiotic mimics immunity. Science. 2008;322:1337–1338. doi: 10.1126/science.1167452. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay B, Daniels L. Aerobic purification of N5,N10-methylenetetrahydromethanopterin dehydrogenase, separated from N5,N10-methylenetetrahydromethanopterin cyclohydrolase, from Methanobacterium thermoautotrophicum strain Marburg. Can J Microbiol. 1989;35:499–507. doi: 10.1139/m89-077. [DOI] [PubMed] [Google Scholar]
- 31.Purwantini E, Mukhopadhyay B, Spencer RW, Daniels L. Effect of temperature on the spectral properties of coenzyme F420 and related compounds. Anal Biochem. 1992;205:342–350. doi: 10.1016/0003-2697(92)90446-e. [DOI] [PubMed] [Google Scholar]
- 32.Mattson BM, Lannan J. Microscale gas chemistry. Part 5. Experiments with nitrogen oxides. J. Chem13 News. 1997:255. [Google Scholar]
- 33.Malaga W, Perez E, Guilhot C. Production of unmarked mutations in mycobacteria using site-specific recombination. FEMS Microbiol Lett. 2003;219:261–268. doi: 10.1016/S0378-1097(03)00003-X. [DOI] [PubMed] [Google Scholar]
- 34.Pelicic V, Reyrat JM, Gicquel B. Generation of unmarked directed mutations in mycobacteria, using sucrose counterselectable suicide vectors. Mol Microbiol. 1996;20:919–925. doi: 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 35.Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 36.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 37.Purwantini E. Microbiology. Iowa City: University of Iowa; 1997. [Google Scholar]
- 38.Garbe TR, et al. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 39.Guerra-Lopez D, Daniels L, Rawat M. Mycobacterium smegmatis mc2 155 fbiC and MSMEG_2392 are involved in triphenylmethane dye decolorization and coenzyme F420 biosynthesis. Microbiology. 2007;153:2724–2732. doi: 10.1099/mic.0.2006/009241-0. [DOI] [PubMed] [Google Scholar]
- 40.Liley PE, Reid RC, Buck E. In: Perry's Chemical Engineers' Handbook. Perry RH, Green DW, Maloney JO, editors. New York: McGraw–Hill; 1984. pp. 1–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.