Abstract
Many alkylated peptide amphiphiles have been reported to self-assemble into cylindrical nanofibers with diameters on the order of a few nanometers and micrometer scale lengths; these nanostructures can be highly bioactive and are of great interest in many biomedical applications. We have discovered the sequences for these molecules that can eliminate all curvature from the nanostructures they form in water and generate completely flat nanobelts with giant dimensions relative to previously reported systems. The nanobelts have fairly monodisperse widths on the order of 150 nm and lengths of up to 0.1 millimeters. The sequences have an alternating sequence with hydrophobic and hydrophilic side chains and variations in monomer concentration generate a “broom” morphology with twisted ribbons that reveals the mechanism through which giant nanobelts form. Interestingly, a variation in pH generates reversibly periodic 2nm grooves on the surfaces of the nanobelts. With proper functionalization, these nanostructures offer a novel architecture to present epitopes to cells for therapeutic applications.
One-dimensional (1D) nanostructures have attracted extensive research interest over the past decade due to the beneficial influence of their dimensionality on electronic and optical materials properties1–4 and very recently bioactivity combined with ability to crosslink into gel networks.5–11 Interesting examples include the vapor deposition synthesis of carbon nanotubes1, semiconductor nanowires2, 3 and nanobelts4, and on the soft matter side the self-assembly of cylindrical micelles12, 13, ribbons14 and peptide nanofibers5–7, 10, 11. The key factor in formation of these 1D nanostructures is control of preferential growth in only one dimension. While both cylinders (one-dimensional growth) and membranes (two-dimensional growth) have been frequently reported through self-assembly of molecular building units12, 15–17, flat 1D nanostructures, namely nanobelts, are less common because of the difficulty of maintaining two significantly different growth rates along two different dimensions.
Peptides or proteins with one or more β-sheet strands exhibit the extraordinary ability to assemble into long, fibrillar nanostructures via intermolecular hydrogen bonding.5, 6, 18–36 Over the past few decades, extensive research efforts have been devoted to the structure and the formation mechanisms of amyloid fibers due to their links to neurodegenerative diseases21–23. In other work, there has been interest in 1D nanostructures formed by self-assembly of de novo designed peptides and peptidomimetics for their potential applications as biomaterials.5, 6, 24–31, 37–41 Due to the intrinsic chirality of natural amino acids, the observed peptidic 1D nanostructures are often twisted or helical, and occasionally, they can further interwind into supramolecular filamentous bundles or sheets27, 37, 42–44. Yet untwisted belt-like structures are rare and have only been observed with limited dimension in width under specific solution conditions6, 28, 45–47 such as highly concentrated peptide solutions6, in impure peptide systems45, in the presence of salts47, or through mixing of enantiomers46. In these systems it is likely that strong forces driving lateral growth of peptide nanostructures to significant dimensions are effectively absent.
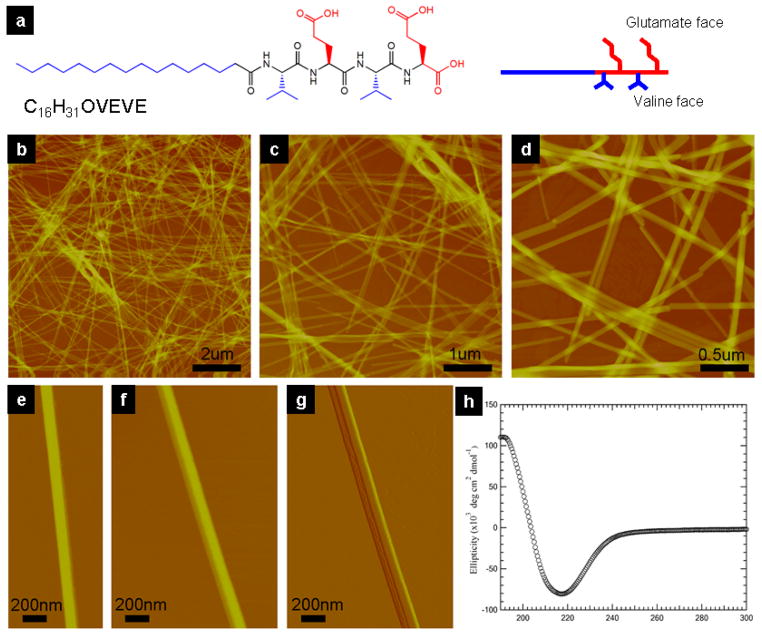
Our previous work showed that a great diversity of peptide amphiphiles, molecules that contain peptide sequences covalently grafted to alkyl segments, self-assemble into cylindrical nanofibers as a result of hydrogen bonding among peptide segments and hydrophobic collapse of alkyl tails.11, 18, 19, 48 We show here that alternating tetrapeptide sequences in these molecules with hydrophobic and negatively charged residues (V and E) and alkyl segments with 16 carbons self-assemble into 1D nanostructures that lose all curvature and grow laterally to create nanobelts (Fig. 1a, molecular synthesis, purification and characterization are provided in supporting material). Tapping-mode atomic force microscopy (AFM) imaging (Fig. 1b–g) reveals the flat, belt-like morphology of the nanostructures formed in aqueous solution at a concentration of 0.1 wt% over the course of two weeks (Nanobelts are observed after 2 days). These nanobelt assemblies exhibit lengths well over tens of micrometers (up to 0.1mm) and widths on the order of 150 nm. AFM also reveals a variation in height (between 10nm and 20nm) among the nanobelts which exceeds the expected thickness of a peptide bilayer (2×3.3nm). There is a consistent, quantized difference in nanostructure height of approximately 4.3nm, a distance reasonably close to that of an interdigitated peptide bilayer (4.7nm). Whereas most have a flat surface, some reveal a terraced structure in AFM micrographs revealing their multilayered structure (See supplemental info, Fig. S4). Stacking of bilayers along the z-direction (thickness) is most likely due to protonation of the peptide C termini at low pH that weakens electrostatic repulsions between different nanobelt layers. Further growth of the nanobelts in solution over a period of two months causes them to eventually settle from aqueous solution.
Figure 1.
Giant (ultralong and wide) nanobelts assembled from a peptide amphiphile containing four amino acids and an alkyl tail. a, chemical structure of the peptide amphiphile. b–d, AFM images of peptide nanobelts at different scanning sizes. The assembled nanobelts are the dominant structures in the assembly system (almost artifact free). e and f, AFM images of a single-layer and a double-layer nanobelt morphology. g, AFM amplitude image of f. h, CD spectrum of the peptide nanobelt solution at a concentration of 0.05 wt % proves the existence of β-sheet secondary structure in the supramolecular assemblies.
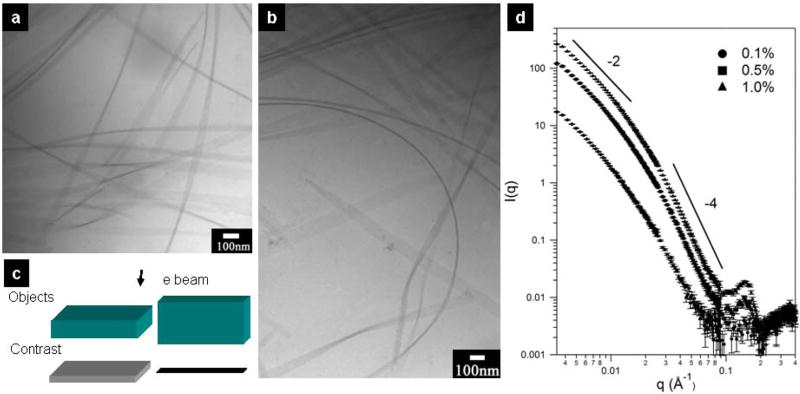
To further characterize the nanobelt morphology, cryogenic transmission electron microscope (cryo-TEM) was carried out on the nanobelt stock solutions after two weeks of incubation at room temperature (cryo TEM offers the possibility of characterizing structure in solution49). Fig. 2a and 2b show that the nanobelt morphology is indeed the dominant structure in solution. Additionally, it can be seen from the cryo-TEM images that nanobelts exhibit great flexibility in the solution state (Fig. 2b). This observation indicates that the rigid, well-extended nanobelt morphology seen by AFM is possibly a result of specimen preparation. The cryo-TEM image contrast is associated with different tilt angles of the nanobelts as depicted in Fig. 2c. The darkest thin lines in Fig. 2b originate from the greater mass-thickness contrast of the nanobelts with approximately 90° tilt angle (the nanobelt surfaces are parallel to the direction of the electron beam). A direct measurement from cryo-TEM images reveals that the height of the nanobelt in Fig. 2b is approximately 12 nm, suggesting a stacking of three peptide amphiphile bilayers.
Figure 2.
Cryo-TEM images of nanobelt morphology and small-angle neutron scattering profiles of nanobelt solutions. a, Cryo-TEM image shows that the nanobelts can flip, tilt and entangle with other nanobelts in solution. b, Cryo-TEM image reveals the mechanical flexibility of nanobelts. c, Schematic representation of the origin of contrast in cryo-TEM images. In the current system, the dominant contrast mechanism arises from the mass-thickness contrast associated with different tilt angles of the nanobelt morphology. When the nanobelt is tilted at a right angle, electrons travel the longest distance inside the nanobelt, and thus have the highest possibility to be scattered. d, SANS profiles at different nanobelt concentrations. The similar shape of three scattering profiles suggests a stable nanobelt morphology over the concentration range of 0.1 wt %, 0.5 wt % and 1.0 wt %.
We also carried out small angle neutron scattering (SANS) experiments to characterize the bulk solution assembly behavior at different concentrations. Fig. 2d displays absolute scattering intensities from 0.1%, 0.5% and 1.0% peptide solutions. First, the flat nanobelt morphology was indicated by the −2 scaling for flat particles at low q and the −4 Porod scaling for scattering from an infinite interface at high q50 (Fig. S5). The scaling of approximately −4 in the 2 wt % and 1 wt % scattering curves suggests a well-defined, sharp interface between the self-assembled nanobelts and the D2O solvent. In the 0.1 wt % solution, the high q slope is −3.54, implying more hydrated, fractal nanobelt surfaces. Secondly, the peak at 0.145Å−1 does not shift with variations in solution concentration and was therefore identified to be the diffraction peak corresponding to the periodic stacking of multiple bilayers of the peptide molecules in the nanostructures. The calculated d-spacing is 4.3nm, given by d=2π/q. This value is consistent with the AFM measurement. Third, over the measured q range, the three SANS profiles assume very similar shapes, indicating that nanobelts are stable and that the interactions between nanobelts are negligible in the concentration range from 0.1 wt % to 1 wt %.
The key structural feature for nanobelt as opposed to the very common cylindrical nanostructure is the alternating hydrophobic and hydrophilic amino acid sequence. Peptides with alternating hydrophobic and hydrophilic amino acids are known to have a strong propensity to form β-sheet structures.5, 7, 26, 27, 40, 51–54 Circular dichroism (CD) spectroscopy confirms that the peptide amphiphile studied here forms a β-sheet secondary structure (Fig. 1h), as indicated by a minimum at 217 nm. The structural motif of VEVE, when adopting an extended β-strand conformation, flips the hydrophilic and hydrophobic side chains to the opposite sides of the peptide backbone. We hypothesize that in aqueous solution, the valine surfaces have a strong tendency to associate with each other in order to minimize exposure to water, resulting in an attractive dimerization of two peptide segments. The existence of the peptide amphiphile dimers was inferred from the presence of the corresponding peaks in mass spectrometry of samples (Fig. S3). It is not clear what is the nature of the interaction among glutamic acid side chains, however, these alternating sequences seem to allow close packing among peptide segments and therefore loss of curvature in the aggregates, leading to the nanobelt morphology. Indeed, when we disrupted the alternating hydrophobic and hydrophilic amino acid sequence by replacing the VEVE peptide segment with a structural motif of VVEE, the resulting nanostructures regain their interfacial curvature, forming cylindrical nanofibers under the exact same conditions (Fig. S6). It is therefore likely that nanobelt flat architecture is associated with highly effective packing among β sheets with peptides containing alternating sequences. The detailed molecular explanation remains to be identified in the future, it may be the result of dimerization of the sequences or even the nature of hydration between β sheets with alternating hydrophobic-hydrophilic side chains.
The growth of these nanostructures along each dimension is obviously regulated by different forces. One dimensionality is clearly linked to fast growth along the direction of hydrogen bonding in β-sheets relative to a much slower lateral adhesion among them. However, it is not clear why these nanostructures exhibit a relatively uniform width (Fig. S7). A single nucleation event followed by uniform growth in the fast and slow directions, in analogy to living polymerization, is a possible explanation. Lateral growth of the nanobelt should depend not only on the hydrophobic collapse of alkyl tails and amino acid side chain interactions, but also on the elastic penalty of untwisting the natural shape of peptide β-sheets.44 Within a cylindrical aggregate the natural twisting of β-sheets can be easily accommodated18, however, this would not be the case within a flat structure of significant width like the nanobelt.
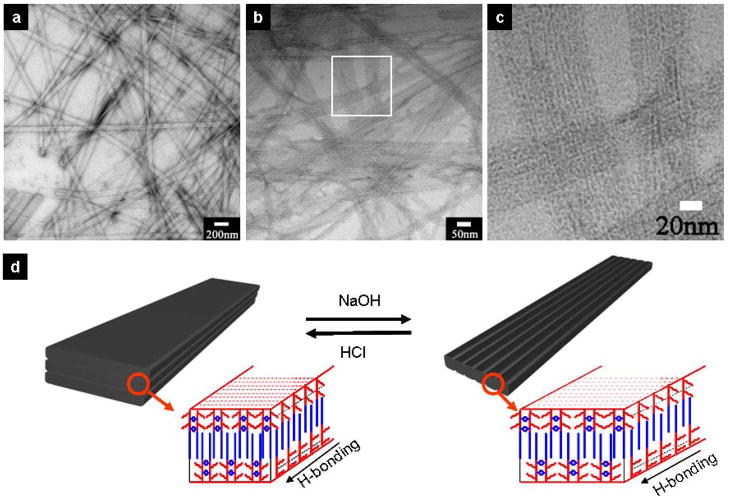
Interestingly, the flat nanobelts (Fig. 3a) can transform into “grooved” nanobelts (Fig. 3b and 3c) when the solution pH is increased. Addition of a small amount of basic solution (2mM final concentration of NaOH in 0.1 wt % peptide solution) can separate the multilayered aggregates into nanobelts with single bilayer (Fig. S8). In addition, nanogrooves parallel to the nanobelt long axis were formed as a result of electrostatic repulsion among surfaces with glutamate side chains at higher pH. The parallel dark lines, with a periodic spacing of approximately 4.5nm, are the result of uranyl acetate deposition used to negatively stain the TEM samples (Fig. 3b and Fig. 3c). The expected width of the peptide amphiphile dimer with fully extended side chains is approximately 2.5 nm. Therefore, these parallel nanochannels within the nanostructures must be approximately 2 nm wide (4.5nm–2.5nm). Although the nanobelts can retain their belt-like shape due to hydrophobic attraction among alkyl chains, the electrostatic repulsions of partially deprotonated glutamate surfaces are able to cause moderate lateral dissociation (Fig. 3d). In the presence of 2mM NaOH, the width of nanobelts was observed to decrease from ~150nm to ~50nm, and the process was found to be completely reversible. When the pH was restored with HCl, nanobelts with flat, seamless surfaces can be formed again within a few hours.
Figure 3.
Grooved nanobelt morphology produced at high pH. a, TEM image of nanobelts in a 0.1 wt % solution. b, Grooved nanobelts in a 0.1 wt % solution containing 2 mM NaOH. The parallel nanochannels can be clearly seen in the image. c, A closer view of the grooved nanobelts in b. d, Schematic representation of the molecular packing inside the nanobelts and the grooved nanobelts. All the TEM samples were negatively stained with 2% (w/v) uranyl acetate aqueous solution.
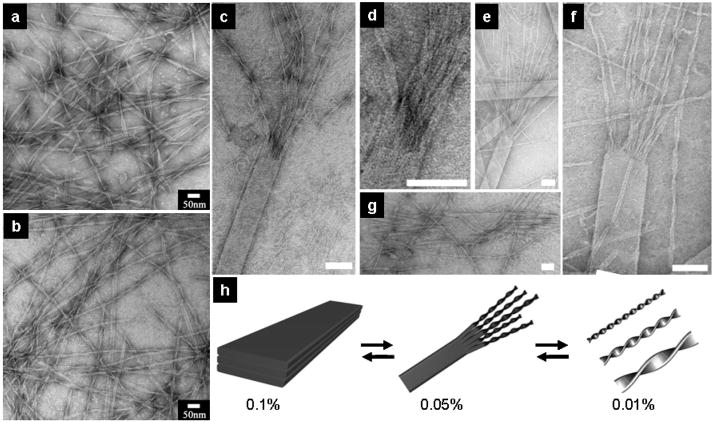
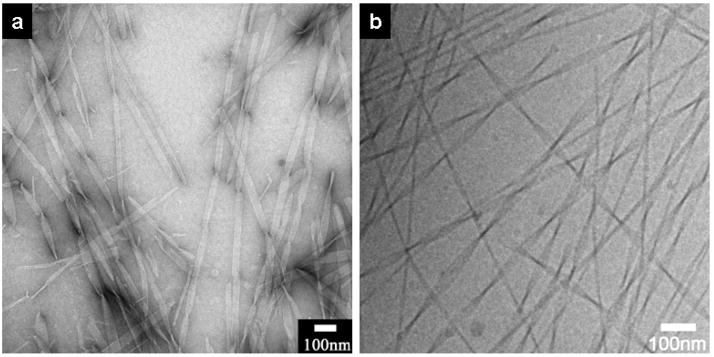
We observed that lateral adhesion among β-sheets is also concentration dependant. Narrower nanobelts and twisted nanoribbons are revealed by TEM when 0.1 wt % peptide amphiphile solutions were diluted to 0.01 wt %. Fig. 4a and 4b show a mixture of twisted nanoribbons with different pitches, and it is evident that the twist pitch varies with ribbon width. As expected, all the twisted nanoribbons here are left-handed because of the intrinsic L-chirality of the amino acids55. Measurement on the narrowest part of each single twisted nanoribbon reveals that they all share the same thickness of approximately 4.3nm, which is the expected value of one single interdigitated bilayer. This suggests that, in dilute solution, the twisted nanoribbons tend to exist as single bilayers of dimerized peptide amphiphiles. The transition from nanobelts to nanoribbons can be directly observed when 0.1 wt % solutions containing nanobelts are diluted to a final peptide amphiphile concentration of 0.05 wt %. The coexistence of twisted nanoribbons and flat nanobelts can be seen after aging the solution for 24 hours. Fig. 4c–g show a clear transition from nanobelts to twisted ribbons starting from the ends of these nanobelts, generating a “broom” morphology. As shown in Fig. 4, the nanobelt ends have a tendency to split into narrow nanobelts. These structures could be either flat or twisted depending on their width and distance from the wide and flat nanobelt they sprout from. One can see in Fig. 4c–f a gradual transition from a flat nanobelt to the separated twisted nanoribbons of the broom morphology. Fig. 4g shows a nanobelt with both ends splitting into narrow nanoribbons. To the best of our knowledge, observation of the “broom” morphology by TEM in this work offers the first direct mechanistic evidence for the transition from a flat and wide aggregate of β-sheet assemblies to twisted one-dimensional nanostructures.
Figure 4.
Twisted nanoribbons at 0.01 wt % aqueous solution and intermediate structures (broom morphology) of nanobelts transforming into twisted nanoribbons at 0.05 wt % solution. a–b, Narrower nanobelts and twisted nanoribbons are observed at a concentration of 0.01 wt %. The twist pitch increases with an increase of nanoribbon width. c–f, Twisted nanoribbons sprouting from one nanobelt end. d, A closer view of c. There is a gradual transition from flat nanobelt to twisted nanoribbons. The longer the distance from the wide nanobelt, the more likely the nanoribbons will twist in their natural states. g, Nanobelts split from both ends into narrower nanobelts. The split ribbons are too short to be twisted because they are too close to the wide nanobelt. g, Schematic representation of the morphological transitions with a change in concentration. Scale bars of Fig c–g: 100nm. All the TEM samples were negatively stained with 2% (w/v) uranyl acetate aqueous solution.
In the hierarchical model developed by Nyrkova and coworkers6, 44, 56, twisted β-sheets could associate laterally into a variety of twisted linear structures with multiple β-sheet stacks. The stacking number is dependant on the peptide concentration and the strength of favorable interactions among the side chains of the peptide residues. Significant lateral growth must untwist the β-sheet to accommodate for close packing of additional sheets. If the lateral adhesion energies are sufficiently large to offset the entropy loss of untwisting β-sheets from their natural states, a flat nanobelt morphology with unlimited β-sheet stacks would be expected. However, the lack of well-defined lateral adhesion forces often causes the peptides to roll into bundles due to the attractive interaction between N-termini and C-termini.42 Often times, aggregates are formed as narrow filaments due to the unfavorable interactions between hydrophilic peptide side chains.5, 26, 53 In the system reported here, we hypothesize that lateral assembly of the nanobelt morphology is due to the hydrophobic collapse of alkyl tails which provides sufficient energy to compensate for the elastic penalty of untwisting peptide sheets. If favorable interactions occur among the glutamate side chains at low pH, for example hydrogen bonding, then these would synergistically contribute to lateral growth of the nanobelts. However, if these glutamate interactions are repulsive because of sterics or electrostatics, then hydrophobic collapse of the alkyl tails would also compensate for them in allowing nanobelt formation. Clearly, at low pH, multilayered nanobelts are produced because most of the COOH groups are protonated. The energies of hydrophobic alkyl attraction are adequate to untwist the β-sheets and overcome any repulsive interactions. At high pH (in the presence of 2mM NaOH), the deprotonation of COOH groups leads to nanobelt dissociation in response to electrostatic repulsion among glutamate side chains. Electrostatic repulsion causes the multilayered nanobelts to break into single bilayer nanobelts and open up the observed 2 nm nanochannels on both sides of the nanobelt surfaces. Nyrkova’s model also predicts that peptides tend to form smaller aggregates in more dilute solution and the observed morphology should be less laminated, and thus, more twisted. This prediction is in good agreement with our observations on the twisted ribbons at 0.01 wt % and the “broom” morphology at 0.05%.
To assess the possibility of using the flat nanobelts for cell signaling, we introduced the bioactive cell adhesion epitope Arginine-Glycine-Aspartic acid (RGD) at the terminus of the peptide segment.57 The RGD epitope, a small peptide sequence from extracellular matrix proteins, has been widely used to create bioactive nanostructures.9 In our design, one glycine (G) residue was placed as a spacer between the VEVE peptide sequence and the RGD epitope to maintain the alternating hydrophobic and hydrophilic structural motif. In the RGD-bearing peptide, TEM examination reveals a nanobelt morphology with a left-handed twist (Fig. 5). The observation of twisted and narrower belt morphology formed by molecules with the extended peptide sequence can be rooted in the greater entropic penalty for untwisting the peptide region and possibly repulsive interactions among side chains, both limiting lateral growth. This observation is consistent with Nyrkova’s model44 and other previous reports6, 52, 56 that peptidic fibrillar nanostructures tend to be twisted when having limited growth in width. Indeed, the widths of these twisted nanobelts are on average narrower by a factor of 3 relative to those without the epitope, on the order of ~50nm. In contrast to the well known cylindrical nanostructures based on peptide amphiphiles, the wide nanobelt architecture offers the possibility of cell signaling using a flat, SAM-like presentation of signals that can still wrap around cells in three dimensions. Fibroblast cells (NIH/3T3), which are known to interact strongly with the RGD epitope, were used to test their adhesion behavior with the nanostructures. Our preliminary data show that suspensions of these fibroblasts in cell media at a concentration of 300,000 cells/ml lead to the formation of a self-supporting gel when combined with 0.5 wt % RGD nanobelt solutions. This observation suggests a high efficiency of interaction between cell receptors and the nanobelts.
Figure 5.
TEM images of twisted nanoribbons of C16H31O-VEVEGRGD at a 0.1 wt % concentration. a, Negatively stained TEM image. b, Cryo-TEM image. The nanoribbons exhibit a uniform width of approximately 50nm.
We have demonstrated that a short hydrophobic-hydrophilic alternating peptide sequence can trigger self-assembly of peptide amphiphiles into wide nanobelts with completely flat architecture. This is in great contrast to the very common 1D cylindrical morphology observed in bioactive nanostructures designed with these molecules. Furthermore, we have shown modifications are possible to generate twisted nanoribbons as opposed to cylinders bearing a bioactive epitope. The system studied here also shows that electrostatic repulsions can reversibly open 2 nm grooves in the nanobelts through changes in pH, and changes in concentration break up the nanobelts into twisted ribbons. These observations have provided mechanisitic insight on giant nanobelt formation. The literature suggests that at least six residues are required to form stable β-sheet structures.6, 58 The designed peptide amphiphile studied here generates stable β-sheet structures with only four amino acids. With proper functionalization, these nanostructures offer a novel architecture to present epitopes to cells for therapeutic applications.
Supplementary Material
Acknowledgments
Funding of this work was supported by the U.S. Department of Energy (grant No. DE-FG02-00ER45810), by National Science Foundation (NSF grant No. DMR-0605427, MRSEC at Northwestern University Grant No. NSF DMR-0520513, and NSEC at Northwestern University Grant No. NSEC EEC-0118025), and by National Institute of Health (grant number NIH/NIBIB R01EB003806). The authors are grateful to Biological Imaging Facility (BIF), Integrated Molecular Structure Education and Research Center (IMSERC) and Keck Biophysics Facility at Northwestern University. The authors also thank Liam Palmer and Ronit Bitton from the authors’ laboratory for insightful discussions.
References
- 1.Ren ZF, Huang ZP, Xu JW, Wang JH, Bush P, Siegal MP, Provencio PN. Science. 1998;282(5391):1105–1107. doi: 10.1126/science.282.5391.1105. [DOI] [PubMed] [Google Scholar]
- 2.Duan XF, Huang Y, Cui Y, Wang JF, Lieber CM. Nature. 2001;409(6816):66–69. doi: 10.1038/35051047. [DOI] [PubMed] [Google Scholar]
- 3.Alivisatos AP. Science. 1996;271(5251):933–937. [Google Scholar]
- 4.Pan ZW, Dai ZR, Wang ZL. Science. 2001;291(5510):1947–1949. doi: 10.1126/science.1058120. [DOI] [PubMed] [Google Scholar]
- 5.Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. Journal of the American Chemical Society. 2002;124(50):15030–15037. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 6.Aggeli A, Bell M, Boden N, Keen JN, McLeish TCB, Nyrkova I, Radford SE, Semenov A. Journal of Materials Chemistry. 1997;7(7):1135–1145. [Google Scholar]
- 7.Caplan MR, Schwartzfarb EM, Zhang SG, Kamm RD, Lauffenburger DA. Biomaterials. 2002;23(1):219–227. doi: 10.1016/s0142-9612(01)00099-0. [DOI] [PubMed] [Google Scholar]
- 8.Hartgerink JD, Zubarev ER, Stupp SI. Current Opinion in Solid State & Materials Science. 2001;5(4):355–361. [Google Scholar]
- 9.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, Stupp SI. Biomaterials. 2007;28(31):4608–4618. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 10.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303(5662):1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 11.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294(5547):1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 12.Won YY, Davis HT, Bates FS. Science. 1999;283(5404):960–963. doi: 10.1126/science.283.5404.960. [DOI] [PubMed] [Google Scholar]
- 13.Cui HG, Chen ZY, Zhong S, Wooley KL, Pochan DJ. Science. 2007;317(5838):647–650. doi: 10.1126/science.1141768. [DOI] [PubMed] [Google Scholar]
- 14.Zubarev ER, Sone ED, Stupp SI. Chemistry-a European Journal. 2006;12(28):7313–7327. doi: 10.1002/chem.200600619. [DOI] [PubMed] [Google Scholar]
- 15.Sackmann E. Science. 1996;271(5245):43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 16.Li LS, Stupp SI. Macromolecules. 1997;30(18):5313–5320. [Google Scholar]
- 17.Stupp SI, Son S, Lin HC, Li LS. Science. 1993;259(5091):59–63. doi: 10.1126/science.259.5091.59. [DOI] [PubMed] [Google Scholar]
- 18.Jiang HZ, Guler MO, Stupp SI. Soft Matter. 2007;3(4):454–462. doi: 10.1039/b614426h. [DOI] [PubMed] [Google Scholar]
- 19.Guler MO, Hsu L, Soukasene S, Harrington DA, Hulvat JF, Stupp SI. Biomacromolecules. 2006;7(6):1855–1863. doi: 10.1021/bm060161g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajangam K, Behanna HA, Hui MJ, Han XQ, Hulvat JF, Lomasney JW, Stupp SI. Nano Letters. 2006;6(9):2086–2090. doi: 10.1021/nl0613555. [DOI] [PubMed] [Google Scholar]
- 21.Serpell LC. Biochimica ET Biophsica ACTA-Molecular Basis of Disease. 2000;1502(1):16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 22.Sipe JD, Cohen AS. Journal of Structural Biology. 2000;130(2–3):88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 23.Wetzel R. Accounts of Chemical Research. 2006;39(9):671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 24.Lu K, Guo L, Mehta AK, Childers WS, Dublin SN, Skanthakumar S, Conticello VP, Thiyagarajan P, Apkarian RP, Lynn DG. Chemical Communications. 2007;(26):2729–2731. doi: 10.1039/b701029j. [DOI] [PubMed] [Google Scholar]
- 25.Hendler N, Sidelman N, Reches M, Gazit E, Rosenberg Y, Richter S. Advanced Materials. 2007;19(11):1485–1488. [Google Scholar]
- 26.Yokoi H, Kinoshita T, Zhang SG. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(24):8414–8419. doi: 10.1073/pnas.0407843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lashuel HA, LaBrenz SR, Woo L, Serpell LC, Kelly JW. Journal of the American Chemical Society. 2000;122(22):5262–5277. doi: 10.1021/ja9937831. [DOI] [PubMed] [Google Scholar]
- 28.Matsumura S, Uemura S, Mihara H. Chemistry - A European Journal. 2004;10(11):2789–2794. doi: 10.1002/chem.200305735. [DOI] [PubMed] [Google Scholar]
- 29.Yu YC, Berndt P, Tirrell M, Fields GB. Journal of the American Chemical Society. 1996;118(50):12515–12520. [Google Scholar]
- 30.Hamley IW. Angewandte Chemie-International Edition. 2007;46:8128–8147. doi: 10.1002/anie.200700861. [DOI] [PubMed] [Google Scholar]
- 31.Lowik D, van Hest JCM. Chemical Society Reviews. 2004;33(4):234–245. doi: 10.1039/b212638a. [DOI] [PubMed] [Google Scholar]
- 32.Goux WJ, Kopplin L, Nguyen AD, Leak K, Rutkofsky M, Shanmuganandam VD, Sharma D, Inouye H, Kirschner DA. Journal of Biological Chemistry. 2004;279(26):26868–26875. doi: 10.1074/jbc.M402379200. [DOI] [PubMed] [Google Scholar]
- 33.Hecht MH, Das A, Go A, Bradley LH, Wei YN. Protein Science. 2004;13(7):1711–1723. doi: 10.1110/ps.04690804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Nuallain B, Williams AD, Westermark P, Wetzel R. Journal of Biological Chemistry. 2004;279(17):17490–17499. doi: 10.1074/jbc.M311300200. [DOI] [PubMed] [Google Scholar]
- 35.Yang ZM, Xu KM, Guo ZF, Guo ZH, Xu B. Advanced Materials. 2007;19(20):3152–3156. [Google Scholar]
- 36.Deming TJ. Progress in Polymer Science. 2007;32:858–875. [Google Scholar]
- 37.Marini DM, Hwang W, Lauffenburger DA, Zhang SG, Kamm RD. Nano Letters. 2002;2(4):295–299. [Google Scholar]
- 38.Lamm MS, Sharma N, Rajagopal K, Beyer FL, Schneider JP, Pochan DJ. Advanced Materials. 2008;20(3):447–451. [Google Scholar]
- 39.Nagarkar RP, Hule RA, Pochan DJ, Schneider JP. Journal of the American Chemical Society. 2008;130(13):4466–4474. doi: 10.1021/ja710295t. [DOI] [PubMed] [Google Scholar]
- 40.Sakurai T, Koga M, Takafuji M, Ihara H. Chemistry Letters. 2003;32(2):152–153. [Google Scholar]
- 41.Borner HG, Smarsly BM, Hentschel J, Rank A, Schubert R, Geng Y, Discher DE, Hellweg T, Brandt A. Macromolecules. 2008;41(4):1430–1437. [Google Scholar]
- 42.Bauer HH, Aebi U, Haner M, Hermann R, Muller M, Arvinte T, Merkle HP. Journal of Structural Biology. 1995;115(1):1–15. doi: 10.1006/jsbi.1995.1024. [DOI] [PubMed] [Google Scholar]
- 43.Selinger JV, Spector MS, Schnur JM. Journal of Physical Chemistry B. 2001;105(30):7157–7169. [Google Scholar]
- 44.Nyrkova IA, Semenov AN, Aggeli A, Boden N. European Physical Journal B. 2000;17(3):481–497. [Google Scholar]
- 45.Adams DJ, Holtzmann K, Schneider C, Butler MF. Langmuir. 2007;23:12729–12736. doi: 10.1021/la7011183. [DOI] [PubMed] [Google Scholar]
- 46.Brizard A, Aime C, Labrot T, Huc I, Berthier D, Artzner F, Desbat B, Oda R. Journal of the American Chemical Society. 2007;129(12):3754–3762. doi: 10.1021/ja0682172. [DOI] [PubMed] [Google Scholar]
- 47.Smith AM, Williams RJ, Tang C, Coppo P, Collins RF, Turner ML, Saiani A, Ulijn RV. Advanced Materials. 2008:20–37. [Google Scholar]
- 48.Hartgerink JD, Beniash E, Stupp SI. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(8):5133–5138. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui H, Hodgdon TK, Kaler EW, Abezgauz L, Danino D, Lubovsky M, Talmon Y, Pochan DJ. Soft Matter. 2007;3(8):945–955. doi: 10.1039/b704194b. [DOI] [PubMed] [Google Scholar]
- 50.Porte G. Neutrons, X-rays and Light: Scattering Methods Applied to Soft Condensed Matter. Elsevier Science B.V; 2002. pp. 299–315. [Google Scholar]
- 51.Xiong HY, Buckwalter BL, Shieh HM, Hecht MH. Proceedings of the National Acedemy of Sciences of the United States of America. 1995;92(14):6349–6353. doi: 10.1073/pnas.92.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamm MS, Rajagopal K, Schneider JP, Pochan DJ. Journal of the American Chemical Society. 2005;127(47):16692–16700. doi: 10.1021/ja054721f. [DOI] [PubMed] [Google Scholar]
- 53.Dong H, Paramonov SE, Aulisa L, Bakota EL, Hartgerink JD. Journal of the American Chemical Society. 2007;129(41):12468–12472. doi: 10.1021/ja072536r. [DOI] [PubMed] [Google Scholar]
- 54.Broome BM, Hecht MH. Journal of Molecular Biology. 2000;296(4):961–968. doi: 10.1006/jmbi.2000.3514. [DOI] [PubMed] [Google Scholar]
- 55.Chothia C. Journal of Molecular Biology. 1973;75(2):295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- 56.Aggeli A, Nyrkova IA, Bell M, Harding R, Carrick L, McLeish TCB, Semenov AN, Boden N. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(21):11857–11862. doi: 10.1073/pnas.191250198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pierschbacher MD, Ruoslahti E. Nature. 1984;309(5963):30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 58.Krejchi MT, Atkins EDT, Waddon AJ, Fournier MJ, Mason TL, Tirrell DA. Science. 1994;265(5177):1427–1432. doi: 10.1126/science.8073284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.