Abstract
Although parathyroid hormone (PTH) induces 25-hydroxyvitamin D3 (25(OH)D3) 1α-hydroxylase (1α(OH)ase) under hypocalcemic conditions, previous studies showed that calcitonin, not PTH, has an important role in the maintenance of serum 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) under normocalcemic conditions. In this study we report that 1α(OH)ase transcription is strongly induced by calcitonin in kidney cells and indicate mechanisms that underlie this regulation. The transcription factor C/EBPβ is up-regulated by calcitonin in kidney cells and results in a significant enhancement of calcitonin induction of 1α(OH)ase transcription and protein expression. Mutation constructs of the 1α(OH)ase promoter demonstrate the importance of the C/EBPβ binding site at –79/–73 for activation of the 1α(OH)ase promoter by calcitonin. The SWI/SNF chromatin remodeling complex was found to cooperate with calcitonin in the regulation of 1α(OH)ase. Chromatin immunoprecipitation analysis showed that calcitonin recruits C/EBPβ to the 1α(OH)ase promoter, and Re-chromatin immunoprecipitation analysis (sequential chromatin immunoprecipitations using different antibodies) showed that C/EBPβ and BRG1, an ATPase that is a component of the SWI/SNF complex, bind simultaneously to the 1α(OH)ase promoter. These findings are the first to address the dynamics between calcitonin, C/EBPβ, and SWI/SNF in the regulation of 1α(OH)ase and provide a mechanism, for the first time, for calcitonin induction of 1α(OH)ase. Because plasma calcitonin as well as 1,25(OH)2D3 have been reported to be increased during pregnancy and lactation and in early development, these findings suggest a mechanism that may account, at least in part, for the increase in plasma 1,25(OH)2D3 during these times of increased calcium requirement.
Vitamin D is a principal factor required for maintaining normal calcium homeostasis (1). The active form of vitamin D, 1,25-dihydroxyvitamin D3 (1,25(OH)2D3)3 is generated by two successive hydroxylations; 25-hydroxylation in the liver and 1α-hydroxylation in the kidney (1–3). Inactivating mutations in the 25-hydroxvitamin D3 1α-hydroxylase (1α(OH)ase) gene result in vitamin D dependent rickets type I despite normal intake of vitamin D, indicating the importance of the 1α(OH)ase enzyme (4, 5). Elevated parathyroid hormone (PTH) resulting from hypocalcemia is a primary signal mediating the renal synthesis of 1,25(OH)2D3 (6). PTH stimulates 1α(OH)ase transcription, resulting in increased 1,25(OH)2D3 synthesis (7–9). However, under normocalcemic conditions, PTH fails to stimulate 1α(OH)ase expression (10). Previous studies showed that calcitonin can enhance renal conversion of 25(OH)D3 to 1,25(OH)2D3 and that calcitonin, not PTH, is the major regulator of 1α(OH)ase in the normocalcemic state (10–13). In early development and during pregnancy and lactation calcitonin levels are increased under normocalcemic conditions and correlated to an increase in serum 1,25(OH)2D3 levels (14–17). The stimulation of 1,25(OH)2D3 under normocalcemic conditions by calcitonin may have biological significance to control calcium homeostasis during the perinatal period and during pregnancy and lactation when the need for calcium is increased. However, the mechanisms involved in the stimulation of 1α(OH)ase by calcitonin have not been examined.
Calcitonin, a 32-amino acid peptide hormone generated from the thyroid C cells, has been reported to have diverse physiological actions that include inhibition of osteoclastic bone resorption (18), inhibition of prolactin secretion (19), effects on the growth of prostate and breast cancer cells (20–22), and effects on maternal-fetal calcium exchange (23). The calcitonin receptor, a G-protein coupled receptor, is widely expressed in numerous tissues including osteoclasts (18), kidney (12), placenta (23), and breast and prostate cancer cells (20–22). Thus, calcitonin has both skeletal and extra-skeletal effects. Although it had been thought that a major function of calcitonin is to lower serum calcium, it has been shown that patients with medullary thyroid carcinoma, a neoplasm of C cells, have high calcitonin levels but normal serum calcium (24). In addition, in the absence of calcitonin, serum calcium is unaffected (25). The elevation in plasma 1,25(OH)2D3 in rats treated with calcitonin (11) and the increased serum 1,25(OH)2D3 levels in patients with medullary thyroid carcinoma as well as the decrease in 1,25(OH)2D3 levels after surgical cure of medullary thyroid carcinoma and hypercalcitoninemia (24, 26) further suggest multiple roles of calcitonin and specifically a direct effect of calcitonin on renal 1α(OH)ase.
Although 1α(OH)ase, a mitochondrial P450 enzyme, is regulated by many factors including PTH, calcitonin, and 1,25(OH)2D3, the mechanisms involved in the regulation of 1α(OH)ase expression are only now beginning to be defined. In the human 1α(OH)ase promoter, an SP1 site involved in basal expression and a negative vitamin D response element, which associates with VDR/retinoid X receptor as well as histone deacetylase complex in the presence of 1,25(OH)2D3, have been reported (27, 28). Regions of the mouse 1α(OH)ase promoter that contribute to its basal activity have also been characterized (29). Differences between the mouse and human promoter have been noted, suggesting different regulation of the two genes (27, 29, 30). Although responsiveness to PTH has been noted for both the mouse and human promoter (7–9, 30), a direct inhibition of the activity of the mouse 1α(OH)ase promoter by 1,25(OH)2D3 has not been observed (7).
The results of this investigation indicate that the mouse promoter, similar to the human 1α(OH)ase promoter (9), confers positive responsiveness to calcitonin. Maximal calcitonin activation is observed using the minimal promoter region (–85/+22). Mutation of a C/EBPβ binding site at –79/–73 resulted in marked attenuation of activation of 1α(OH)ase transcription by calcitonin. SWI/SNF, which remodels chromatin using the energy of ATP hydrolysis, is recruited by C/EBPβ to the 1α(OH)ase promoter and also mediates calcitonin regulation of 1α(OH)ase transcription. Chromatin immunoprecipitation (ChIP) analysis also showed an increase in acetylated histone 4 in response to calcitonin, suggesting cooperation between acetylation and chromatin remodeling. These findings are the first to address the dynamics between calcitonin, C/EBPβ, and SWI/SNF in the regulation of 1α(OH)ase and provide a mechanism for the first time for calcitonin induction of 1α(OH)ase.
EXPERIMENTAL PROCEDURES
Materials—Deoxy-[γ-32P]ATP (3000 Ci/mmol) was obtained from PerkinElmer Life Sciences. A Random Primers DNA labeling kit was purchased from Invitrogen. The 1,25(OH)2D3 RIA kit was purchased from Immunodiagnostics Systems Inc. (Fountain Hills, AZ). Prestained protein molecular weight markers and an electrochemiluminescent detection system were obtained from PerkinElmer Life Sciences. Salmon calcitonin was obtained from Sigma. C/EBPβ, BRG1, Brm, and β-actin antisera were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Culture—AOK-B50 porcine renal proximal tubular cells (LLCPK1 cells that express PTH/PTHrP type I receptors as well as calcitonin receptors (31, 32)) and MCT mouse renal proximal tubular cells (33) that also express both PTH and calcitonin receptors (9) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 7% heat-inactivated fetal bovine serum (Gemini Biological Products, Calabasas, CA) in a humidified atmosphere of 95% air, 5%CO2 at 37 °C. Cells were grown to 70–80% confluence and changed to medium supplemented with 2% charcoal-dextran-treated fetal bovine serum before treatment. C33A cells (from ATCC) that lack Brm and BRG1 were used in some studies and were similarly maintained. Cells were treated with the vehicle or the compounds noted at the indicated times and concentrations.
Transient Transfection and Luciferase Assay—For transfection studies, the mouse 1α(OH)ase promoter –1651/+22 placed upstream of a luciferase reporter gene in the pGL2b vector and the deletion constructs –144/+22, –85/+22, and –74/+22 were kindly provided by Dr. H. F. DeLuca (University of Wisconsin at Madison, Madison, WI). pMex-C/EBPβ was a gift of Dr. Simon Willimas, Texas Tech University (Lubbock, TX). A-C/EBP, a dominant negative that heterodimerizes with C/EBP family members and blocks DNA binding, was provided by Dr. Charles Vinson (NIH, Bethesda, MD) (34). The C/EBPβ promoter luciferase construct (–1400/+16) was provided by Dr. Christian Trautwein (35). pCMV-mutant Brm (with the ATPase site mutated that acts as a dominant negative inhibitor) was obtained from M. Yaniv (Institut Pasteur, Paris). PBJ5 BRG1 (K785R) with the ATPase site mutated was from J. DiRenzo and M. Brown (36). Mutant Brm and BRG1 (with the ATPase sites mutated) have previously been characterized and shown to be incorporated into the SWI/SNF complex resulting in interference in transcriptional activation (37). LIP, the dominant negative isoform of C/EBPβ, originally described by Descombes and Schibler (38), was provided by A. Dusso (Washington University School of Medicine, St. Louis, MO). Cells were seeded in a 24-well culture dish 24 h before transfection at 70% confluence. Empty vectors were transfected to keep the total DNA concentration equal. Cells in each well were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Efficiency of transfection, as assessed by green fluorescent protein cotransfection and subsequent visualization, was estimated at 75–85% for AOK-B50 cells. Efficiency of transfection of MCT cells, similarly analyzed, was 25–35%. Maximal induction of 1α(OH)ase transcription (using transfected promoter constructs) by calcitonin in MCT cells was 2.4 ± 0.3-fold (compared with 9–10-fold for AOK-B50 cells). Thus, for most studies examining transcriptional regulation of 1α(OH)ase using transfected cells, AOK-B50 cells were used. Cells were treated 24-h post-transfection for another 24 h. Time course studies with calcitonin indicated a peak of activation at 6 h (1 nm calcitonin resulted in a 20–21-fold induction in transcription using either the –1651/+22 or the –85/+22 promoter construct). Studies were done at 24 h (suboptimal conditions) similar to previous studies with calcitonin (100 nm) (9). Cells were washed twice with phosphate-buffered saline and harvested by incubating with 1× passive lysis buffer, supplied by the dual-luciferase reporter assay kit (Promega, Madison, Wisconsin). The luciferase activity assay was performed according to the protocol of the manufacturer and normalized based on protein content of the cell lysates. Protein levels were determined by the Bradford assay (39).
Site-directed Mutagenesis—The C/EBPβ binding site at –79/–73 was mutated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotides used to generate the mutated site at –79/–73 were as follows: 5′-GGA GTC TGG GAG ACT CTG AAG AGC-3′ (top strand) and 5′-GCT CTT CAG AGT CTC CCA GAC TCC-3′ (lower strand). The mutated constructs were confirmed by DNA sequencing.
Western Blot Analysis—For Western blot analysis of 1α(OH)ase, 50 μg of protein from total cell extracts was loaded onto a 15% SDS-polyacrylamide gel, separated by electrophoresis, and transferred onto a polyvinylidene difluoride membrane (Bio-Rad). Membranes were blocked with 5% milk, Tris-buffered saline for 30 min and incubated at room temperature with 1α(OH)ase antibody (from Armbrecht et al. laboratory (40); rabbit antiserum was raised against a synthetic peptide consisting of the last 12 amino acids of the mouse/pig 1α(OH)ase sequence) at a dilution of 1:1000 in 1% milk, Tris-buffered saline for 60 min. Membranes were then rinsed with Tris-buffered saline (TBS) and incubated at room temperature with goat anti-rabbit IgG-horseradish peroxidase antibody (sc-2004; Santa Cruz Biotechnology) at 1:10,000 in 1% milk, TBS for 30 min. The antigen-antibody complex was detected by the electrochemiluminescent detection system. For analysis of C/EBPβ protein, nuclear extracts were prepared, and Western blot analysis was performed as previously described (41).
ChIP Assay—Cells were cultured to 95% confluence before the experiment and then treated with calcitonin for the different times to perform the ChIP assay as previously described (42). Briefly, cells were first washed with phosphate-buffered saline and subjected to a cross-link reaction with 1% formaldehyde for 15 min. The cross-link reaction was stopped by adding glycine to a final concentration of 0.125 m. Cells were washed with phosphate-buffered saline twice, collected by scraping, and lysed sequentially in 5 mm Pipes, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40, and then in 1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1, for 20 min individually. The chromatin pellets were sonicated to an average DNA size of 500 bp using a Fisher model 100 sonic dismembrator at a power setting of 2. The sonicated extract was centrifuged for 10 min at maximum speed and then diluted into ChIP dilution buffer (16.7 mm Tris-HCl, pH 8.1, 150 mm NaCl, 0.01% SDS, 1.1% Triton X-100, and 1.2 mm EDTA). Immunoprecipitations were performed at 4 °C overnight with the indicated antibody. After a 1-h incubation with salmon sperm DNA and bovine serum albumin-pretreated Zysorbin (Zymed Laboratories Inc., San Francisco, CA), the precipitates were collected by centrifugation. Precipitates were washed sequentially in buffer I (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 150 mm NaCl), buffer II (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 500 mm NaCl), buffer III (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.1), and TE buffer (10 mm Tris, 1 mm EDTA) twice. The protein-DNA was eluted by using 1% SDS and 0.1 m NaHCO3 for 15 min twice. Cross-links were reversed by incubating at 65 °C overnight in elution buffer with 0.2 m NaCl. DNA fragments were purified using Qiagen QIAquick PCR purification kits (Valencia, CA) and subjected to PCR using the primers designed to amplify the fragment containing the C/EBP binding site (upper, 5′-CTA CAC AGA CCA CTT GCA AA-3′; lower, 5′-TCA TGT CTG TGT TTG GGG AG-′). PCR products were resolved in 1% agarose gel and visualized using ethidium bromide staining. PCR was carried out in the linear range of DNA amplification. DNA acquired before precipitation was collected and used as the input. 10% of input was used for PCR evaluation. PCR using the primers designed to amplify the upstream region of 1α(OH)ase promoter (–1300/–980) were used as a negative control. DNA acquired from immunoprecipitates performed with IgG was subjected to PCR using the primers designed to amplify the fragment containing the C/EBP binding site to exclude nonspecific binding.
Re-ChIP experiments were also done using sequential chromatin immunoprecipitations and two different antibodies (α-C/EBPβ and α-BRG1) to assay for the simultaneous presence of these two factors at the same site in the 1α(OH)ase promoter. In Re-ChIP experiments, on the second day of the ChIP experiment complexes were eluted in 60 μl of elution buffer containing 10 mm dithiothreitol for 30 min at 37 °C. The eluted samples were diluted 50 times with ChIP dilution buffer and subjected again to the ChIP procedure using the second antibody (α-BRG1) (42).
Electrophoretic Mobility Shift Assay—22-Mer complementary oligonucleotides spanning the C/EBP binding site at –79/–73 of the mouse 1α(OH)ase promoter or mutated C/EBP binding site were used for the gel shift assays. The sequences of the oligo nucleotides were 5′-CTT CAG CCA ATC CCA GAC GCG-3′ and 5′-CGC GTC TGG GAT TGG CTG AAG-3′ for the wild type and 5′-CTT CAG AGT CTC CCA GAC GCG-3′ and 5′-CGC GTC TGG GAG ACT CTG AAG-3′ for the mutant construct. Overlapping oligonucleotide strands were heat-denatured and annealed overnight. Fifty nanograms of duplex oligonucleotides were 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (Invitrogen) and purified using a Micro Bio-Spin P-30 column (Bio-Rad). Aliquots of nuclear preparations from calcitonin-treated AOK-B50 cells (5 μg of protein) were incubated for 20 min at 27 °C with 2 μg of poly(dI-dC) with or without unlabeled wild type or mutant DNA competitor or C/EBPβ antibody in binding buffer (4 mm Tris-HCl, pH 7.9, 1 mm EDTA, pH 8.0, 60 mm KCl, 12% glycerol, 12 mm HEPES, 1 mm dithiothreitol) followed by the addition of 0.3–0.5 ng of the labeled oligonucleotide probe (∼100,000 cpm) and incubation for 30 min at 27 °C. The samples were separated by electrophoresis on a 6% nondenaturing polyacrylamide which had been pre-electrophoresed for 30 min at 100 V/cm at 4 °C in 45 mm Tris, 45 mm boric acid, 1 mm EDTA for 2.5 h under identical conditions. The dried gels were exposed to x-ray film at –80 °C with an intensifying screen.
Immunoprecipitation—Immunoprecipitation experiments were done to examine the interaction of C/EBPβ and BRG1. Nuclear extracts were prepared from MCT cells, and protein concentration was determined by the Bradford method (39). 500 μg of each preparation was used for immunoprecipitation with the addition of 4 μg of C/EBPβ antiserum or 4 μg of BRG1 antiserum for 24 h at 4 °C. 30 μl of protein A-Sepharose 4 Fast Flow Beads (Amersham Biosciences) were added to each sample and, after further incubation by rotating at 4 °C for 3 h, the immunoprecipitated complex was collected by centrifuging at 3000 rpm for 5 min. The complex was separated by 7.5 or 15% SDS-PAGE and probed with BRG1 antibody or C/EBPβ antibody (42).
1,25(OH)2D3 Production—To determine that 1α(OH)ase protein (examined by Western blot) was catalytically active, 1,25(OH)2D3 production in AOK-B50 cells in response to calcitonin was measured. Cells were cultured 60–70% confluent in T25 flasks before serum deprivation overnight and then treated with vehicle or calcitonin for different doses for 12 h. After treatment, cells were incubated with 100 nm 25(OH)D3 for 4 h. Cellular 1,25(OH)2D3 production was determined by radioimmunoassay using the 1,25(OH)2D3 radioimmunoassay kit according to the protocol of the manufacturer (IDS, Inc., Fountain Hills, AZ). Samples of medium containing 100 nm 25(OH)D3 were incubated without cells to control for cross-reactivity of 25(OH)D3 in the assay. Cell pellets were lysed, delipidated, and immunoextracted using an immobilized monoclonal antibody to the 1α-hydroxyl group. The extracted samples were incubated with primary antibody overnight at 4 °C, and then 125I-1,25(OH)2D3 was added and incubated for another 2 h at room temperature. Primary antibody-bound or free 1,25(OH)2D3 was separated using an immobilized second antibody. Bound radioactivity, quantified with a gamma counter, was inversely proportional to 1,25(OH)2D3 production in the samples (40).
Statistical Analysis—Results are expressed as the mean ± S.E., and significance was determined by analysis with Students' t test for two-group comparison or analysis of variance for multiple group comparisons.
RESULTS
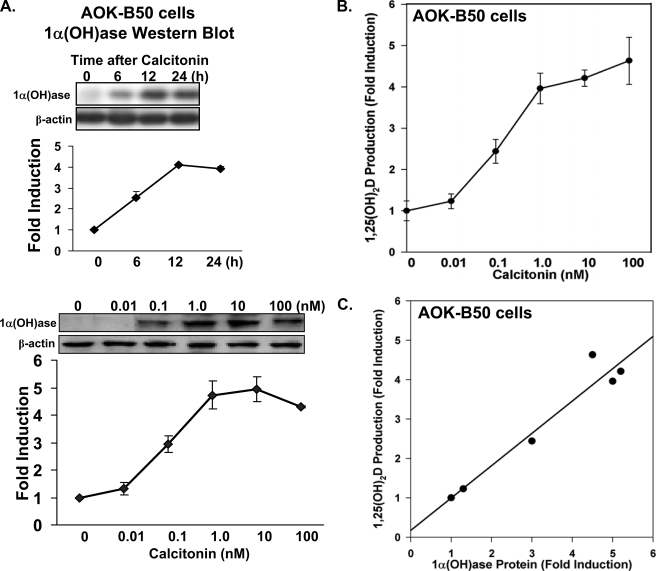
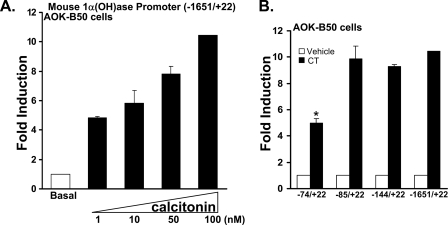
1α(OH)ase Expression and Transcription Are Induced by Calcitonin in Kidney Cells—Previous studies have shown increased 1α(OH)ase mRNA and enzymatic activity upon calcitonin administration in normocalcemic animals (10). AOK-B50 porcine renal proximal tubule cells (LLCPK1 cells that express PTH/PTH-related protein (PTHrP) type I receptors as well as calcitonin receptors (31, 32)) have previously been used to study the regulation of 1α(OH)ase by 1,25(OH)2D3 and PTH (7, 8) and represent a good in vitro model of hormonal regulation of renal 1α(OH)ase in proximal tubules. Therefore, these cells were used to investigate the mechanisms involved in the calcitonin-mediated 1α(OH)ase activation. We found by Western blot that calcitonin induced 1α(OH)ase protein levels (Fig. 1A). The 1α(OH)ase protein stimulated by calcitonin in AOK-B50 cells effectively converted 25(OH)D3 to 1,25(OH)2D3 (Fig. 1B). Fig. 1C indicates the correlation between 1α(OH)ase protein and 1,25(OH)2D3 production in these cells in response to calcitonin (r = 0.97, p < 0.01). To examine the mechanism of activation of 1α(OH)ase by calcitonin, AOK-B50 cells were transfected with the mouse 1α(OH)ase promoter (–1651/+22) as well as different deletion constructs (Fig. 2). The enhancement of luciferase activity using the –1651/+22 construct by calcitonin was concentration-dependent (Fig. 2A). A 9.3–10.4-fold induction in response to calcitonin (100 nm) was observed using the –1651/+22, –144/+22, and –85/+22 constructs, with a significant decrease in calcitonin-induced transcription observed using the –74/+22 construct (4.9 ± 0.4-fold induction; p < 0.05 compared with other constructs) (Fig. 2B). The enhancement of luciferase activity of the –85/+22 construct by calcitonin was also concentration-dependent and similar to the response observed using the –1651/+22 1α(OH)ase promoter; Fig. 2A (data not shown). Thus, although the –74/+22 region contributes to the calcitonin responsiveness, the decrease in transcription observed using the –74/+22 construct suggests the presence of a calcitoninresponsive region between –74 and –85 that is required for maximal activation of the 1α(OH)ase promoter by calcitonin.
FIGURE 1.
Regulation of 1α(OH)ase protein and catalytic activity by calcitonin in AOK-B50 cells. A, Western blot was performed using total extracts from AOK-B50 porcine kidney proximal tubule cells (LLCPK1 cells that express PTH/PTHrP Type 1 receptors as well as calcitonin receptors (31, 32)). Upper panel, cells were treated with vehicle (0) or with calcitonin (100 nm) for 6, 12, or 24 h. Lower panel, AOK-B50 cells were treated with vehicle (0) or increasing concentrations of calcitonin (0.01–100 nm). Results represent the mean ± S.E. of three separate experiments. For all times of calcitonin treatment (A) and for all concentrations of calcitonin (lower panel), 1α(OH)ase levels were significantly induced compared with vehicle (p < 0. 05). B, 1,25(OH)2D3 production in AOK-B50 cells in response to increasing concentrations of calcitonin. Cells were treated with vehicle (0) or with calcitonin (0.01, 0.1, 1, 10, 100 nm) for 12 h and then incubated with 100 nm 25(OH)D3 for 4 h. Cellular 1,25(OH)2D3 production was determined by radioimmunoassay as described under “Experimental Procedures.” Results are reported as the mean ± S.E. (n = 4). Calcitonin treatment (0.1–100 nm) resulted in a significant increase in 1,25(OH)2D3 production (p < 0.05). C, correlation between 1α(OH)ase protein (A, lower panel) and 1,25(OH)2D3 production (B) in AOK-B50 cells in response to calcitonin (r = 0.97, p < 0.01).
FIGURE 2.
A regulatory region for calcitonin stimulation of 1α(OH)ase transcription is localized within –85/+22. A, AOK-B50 cells were plated in a 24-well culture dish, and cells in each well were transfected with 0.3 μg of the mouse 1α(OH)ase promoter construct (–1651/+22). After 24 h, cells were treated with vehicle (Basal) or 1–100 nm calcitonin for another 24 h and harvested, and luciferase activity was determined and normalized based on protein contents of cell lysates. 1α(OH)ase promoter activity is represented as -fold induction (mean ± S.E.; n = 3–6 observations per group) by comparison to basal levels. Calcitonin treatment (1–100 nm) resulted in a significant increase in 1α(OH)ase promoter activity compared with basal levels (p < 0.05). For all transcription experiments, empty vectors were used to keep the total DNA concentration the same. B, AOK-B50 cells were plated in a 24-well culture dish, and cells in each well were transfected with 0.3 μg of mouse 1α(OH)ase promoter –1651/+22 and deletion constructs (–144/+22, –85/+22, –74/+22). After 24 h, cells were treated with vehicle or 100 nm calcitonin (CT) for another 24 h. Results represent the mean ± S.E. of 4–8 observations/group. *, p < 0.05 compared with the activity of the –1651/+22 promoter and the two deletion constructs in response to calcitonin.
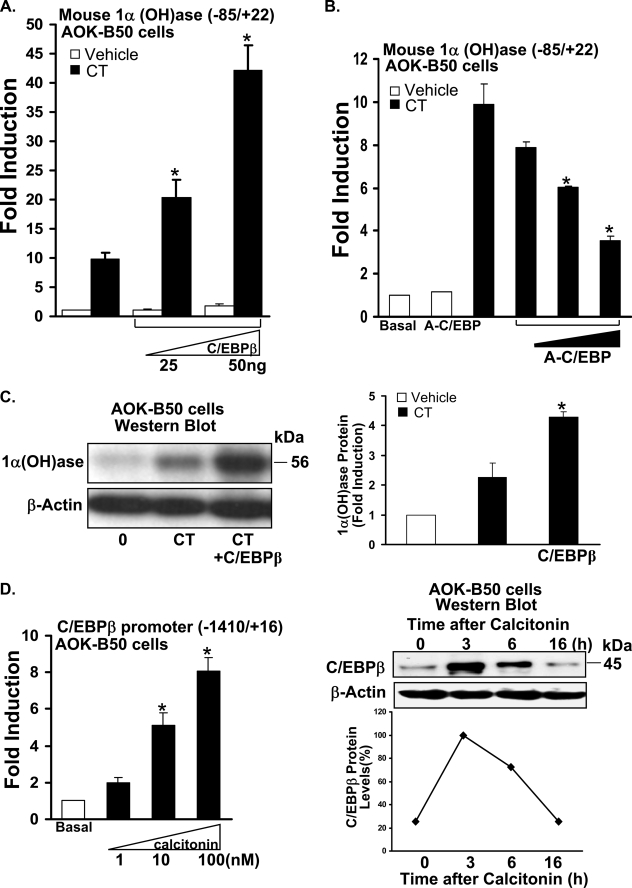
C/EBPβ Can Enhance the Calcitonin-mediated Induction of 1α(OH)ase and Calcitonin Induces C/EBPβ in Kidney Cells—Because examination of the mouse 1α(OH)ase promoter indicated by sequence homology a putative C/EBP binding site at –79/–73, we investigated the possibility that C/EBPβ may be involved in the regulation of 1α(OH)ase by calcitonin. C/EBPβ (0.025, 0.05 μg) significantly enhanced calcitonin induction of 1α(OH)ase transcription 2.0–4.2-fold (p < 0.05 compared with calcitonin alone; maximal stimulation of transcription in the presence of C/EBPβ (0.05 μg) and calcitonin was 42.0-fold; Fig. 3A). Using the –1651/+22 construct as well as the –144/+22 construct, C/EBPβ (0.025 and 0.05 μg) similarly significantly enhanced calcitonin induced transcription (1.7-4.0-fold; p < 0.05 compared with calcitonin alone (1, 10, and 100 nm)). Using the –74/+22 promoter construct, significant enhancement of calcitonin induction of transcription by C/EBPβ (0.01–0.05 μg) was not observed (p > 0.1 compared with calcitonin alone). However, using the –1651/+22 and the –144/+22 constructs and 0.2 μg of C/EBPβ, a significant inhibition of calcitonin induced transcription was observed (10.0 ± 1-fold versus 4.0 ± 0.5-fold and 9.1 ± 0.2-fold versus 4.9 ± 0.3-fold induction in 1α(OH)ase transcription, calcitonin (100 nm) versus calcitonin + 0.2 μg of C/EBPβ (p < 0.05 compared with calcitonin alone) with each promoter construct, respectively). This inhibition of calcitonin induced transcription was not observed using the –85/+22 promoter construct and higher concentrations of C/EBPβ (0.1–0.5 μg), suggesting an upstream regulatory region of inhibition of 1α(OH)ase transcription in the presence of high concentrations of C/EBPβ. A-C/EBP, which functions as a dominant negative inhibitor for C/EBPs, however, inhibited calcitonin induction of 1α(OH)ase transcription dose-dependently (Fig. 3B). This finding suggests that endogenous C/EBP is required for calcitonin induction of transcription, further supporting a predominant positive, cooperative role of C/EBPβ with calcitonin in the induction of 1α(OH)ase transcription. Note there was no effect of A-C/EBP on basal levels of 1α(OH)ase transcription even at high concentrations (1 μg; open bar A-C/EBP; Fig. 3B). LIP also decreased calcitonin induction of 1α(OH)ase transcription; however, basal levels of 1α(OH)ase transcription were similarly decreased (not shown). A similar dose-dependent inhibition of calcitonin-induced 1α(OH)ase transcription by A-C/EBP was observed using the –1651/+22 promoter construct (not shown). Western blot analysis also indicated enhancement of calcitonin induction of 1α(OH)ase protein levels by C/EBPβ (Fig. 3C). In addition, calcitonin was found to induce the transcription of C/EBPβ as well as C/EBPβ protein expression (Fig. 3D). Note in Fig. 3D, right panel, that the induction of C/EBPβ protein by calcitonin precedes the induction of 1α(OH)ase protein by calcitonin (see Fig. 1A), consistent with a role for C/EBPβ in calcitonin induction of 1α(OH)ase.
FIGURE 3.
Cooperative role of C/EBPβ in the regulation of 1α(OH)ase by calcitonin and calcitonin induction of C/EBPβ protein and transcription in kidney cells. A and B, AOK-B50 cells were plated in a 24-well culture dish, and cells in each well were co-transfected with 0.3 μg of mouse 1α(OH)ase promoter construct (–85/+22) and C/EBPβ expression vector (0.025 and 0.050 μg) (A) or A-C/EBP (0.25, 0.5, 1.0 μg) (B). After 24 h, cells were treated with vehicle or 100 nm calcitonin (CT) for another 24 h. C, C/EBPβ enhances calcitonin induced 1α(OH)ase protein levels. AOK-B50 cells in 100-mm tissue culture dishes were transfected with pMEX-C/EBPβ for 24 h and treated with vehicle (0) or with calcitonin (100 nm) for 6 h (a time when the level of 1α(OH)ase protein induced by calcitonin is suboptimal (see Fig. 1A)). Left panel, representative Western blot. Right panel, graphic representation of densitometric scans of Western blots from three separate experiments (mean ± S.E.). D, calcitonin induces C/EBPβ transcription and protein in AOK B-50 cells. Left panel, AOK-B50 cells were transfected with 0.3 μg of C/EBPβ promoter construct (–1400/+16). After 24 h, cells were treated with vehicle (Basal) or 1–100 nm calcitonin for another 24 h. Calcitonin (10 and 100 nm) significantly induced C/EBPβ promoter activity (p < 0.05). Right panel, top, representative Western blot of C/EBPβ expression in nuclear extracts from AOK-B50 cells treated with vehicle or with calcitonin (100 nm) for 3–16 h. Bottom, graphic representation of densitometric scans of Western blots from three separate experiments (mean ± S.E.). Western blot analysis of nuclear extracts from MCT cells also showed low levels of C/EBPβ at 0 time and induction of C/EBPβ at 3 and 6 h (not shown). 1α(OH)ase (A and B) or C/EBPβ (D, left panel) promoter activity was measured by firefly luciferase activity/protein concentration and represented as -fold induction (mean ± S.E.; n = 3 or more experiments) by comparison to basal levels. *, p < 0.05 compared with calcitonin alone (A, B, and C). *, p < 0.05 compared with basal (D, left panel).
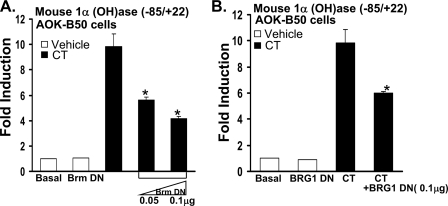
SWI/SNF Chromatin Remodeling Complex Is Involved in the Calcitonin Induction of 1α(OH)ase Transcription—Because C/EBPβ has been reported to recruit the SWI/SNF complex to regulate cell type-specific genes (43, 44), we examined the role of SWI/SNF complex, which contains one of two homologous ATPases, Brahma (Brm), and Brahma/related gene 1 (BRG1), in calcitonin induction of 1α(OH)ase transcription using ATPase site-mutated BRG1 or Brm, which function as dominant-negative inhibitors. In the presence of Brm-DN or BRG1-DN, the calcitonin induction of 1α(OH)ase transcription was significantly reduced, suggesting the involvement of the SWI/SNF complex in the calcitonin effect on 1α(OH)ase transcription (Fig. 4). In C33A cells (which lack endogenous Brm and BRG1), BRG1 induced the activity of the –85/+22 1α(OH)ase promoter construct (1.8 ± 0.1-fold, data not shown), further suggesting a role for BRG1 in activation of 1α(OH)ase transcription. Using MCT cells, BRG1-DN also significantly inhibited the calcitonin induction of transcription (2.4 ± 0.4-fold induction, calcitonin alone; BRG1-DN (0.1 μg) + calcitonin (100 nm), 1.6 ± 0.3-fold induction, p < 0.05 compared with calcitonin alone).
FIGURE 4.
Mutant Brm or BRG1, which act as dominant negative inhibitors, inhibit calcitonin induction of 1α(OH)ase transcription. AOK-B50 cells were co-transfected with 0.3 μg mouse 1α(OH)ase promoter (–85/+22) and Brm-DN expression vectors (0.05, 0.1 μg) (A) or BRG1-DN (0.1 μg) (B). After 24 h, cells were treated with vehicle or 100 nm calcitonin (CT) for another 24 h. 1α(OH)ase promoter activity was measured by firefly luciferase activity/protein concentration and represented as -fold induction (mean ± S.E.; n = at least three observations per group) by comparison to basal levels. Similar results were obtained using the –1651/+22 promoter construct (not shown). Note that there was no effect of 0.1 μg of Brm-DN or 0.1 μg of BRG1-DN on basal levels of 1α(OH)ase transcription (open bar, Brm-DN (A); open bar, BRG1-DN (B)).
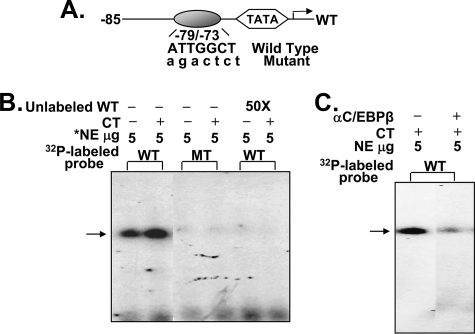
C/EBPβ Binding Site (–79/–73) on the Mouse 1α(OH)ase Promoter Is Detected by Electrophoretic Mobility Shift Assay—Gel shift analysis was performed to determine whether calcitonin can modify the binding of transcription factors involved in the 1α(OH)ase promoter activation. Calcitonin treatment resulted in increased binding of nuclear extracts from AOK-B50 cells to the site at –79/–73. No binding was observed using the mutated sequence or preincubation with the unlabeled wild type oligonucleotide (Fig. 5B). Preincubation with C/EBPβ antibody depleted the binding (Fig. 5C), indicating the ability of C/EBPβ to bind to the element. Gel shift analysis using COS-7 cell extracts transfected with C/EBPβ expression vector also showed binding of C/EBPβ to the site (–79/–73). No binding was observed using COS-7 cell nuclear extracts transfected with vector alone (data not shown).
FIGURE 5.
C/EBPβ binding site in 1α(OH)ase promoter detected by electrophoretic mobility shift assay. A, schematic of the C/EBPβ binding site at –79/–73 in the mouse 1α(OH)ase promoter. Oligonucleotides corresponding to wild type or mutated C/EBPβ binding site were used. B, calcitonin (CT) treatment (100 nm, 6 h) resulted in increased binding to the C/EBPβ binding site. No binding was observed using the mutated sequence (MT) or preincubation with the unlabeled wild type oligonucleotide (WT). C, preincubation with C/EBPβ antibody depleted the binding. Results are representative of four separate experiments. *, NE, AOK-B50 cell nuclear extract.
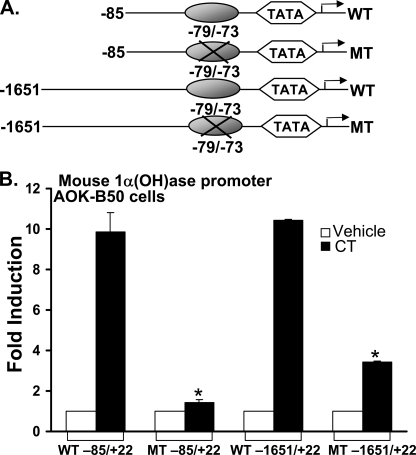
Mutation of the C/EBPβ Binding Site (–79/–73) Inhibits the Activation of 1α(OH)ase Transcription Mediated by Calcitonin—Mutation of the C/EBP binding site at –79/–73 within the –85/+22 construct inhibited the response to calcitonin (Fig. 6). Mutation of this site within the –1651/+22 promoter construct also markedly reduced the response to calcitonin (Fig. 6). These findings indicate that the C/EBPβ binding site at –79/–73 plays an important role in the calcitonin effect on 1α(OH)ase transcription.
FIGURE 6.
Mutation of the C/EBPβ binding site (–79/–73) inhibits the activation of 1α(OH)ase transcription mediated by CT. AOK-B50 cells were transfected with 0.3 μg of mouse 1α(OH)ase (–85/+22) promoter (wild type (WT) or mutant (MT)) or 1α(OH)ase (–1651/+22) promoter (wild type or mutant). After 24 h, cells were treated with vehicle or 100 nm calcitonin (CT) for another 24 h. 1α(OH)ase promoter activity was measured by firefly luciferase activity/protein concentration and represented as -fold induction (mean ± S.E.) by comparison to basal levels (3–5 observations/group). *, p < 0.05 compared with wild type, calcitonin-treated.
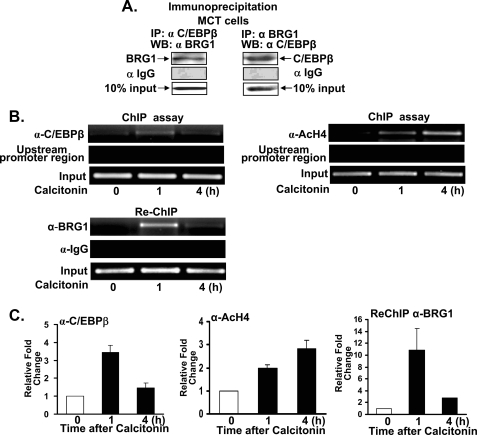
Calcitonin Modulates Binding of Transcription Factors to the Mouse 1α(OH)ase Promoter—To understand mechanisms involved in the calcitonin induction of mouse 1α(OH)ase in vivo, we first determined whether C/EBPβ and SWI/SNF interact within the nuclei of kidney cells, and then we examined the recruitment of C/EBPβ and the binding of SWI/SNF as well as acetylated histone H4 to the 1α(OH)ase promoter using the ChIP assay. Calcitonin was reported to induce 1α(OH)ase mRNA expression in MCT cells (mouse proximal tubular cell line) (9), and our studies using MCT cells showed an enhancement by C/EBPβ of calcitonin induced 1α(OH)ase transcription (2.1 ± 0.2-fold; p < 0.05 compared with calcitonin alone). We used this cell line in our ChIP assays because the mouse 1α(OH)ase promoter sequence, unlike the porcine sequence, is well defined. Using nuclear extracts prepared from MCT cells and immunoprecipitation using BRG1 antibody and Western blot with C/EBPβ or using C/EBPβ antibody for immunoprecipitation and BRG1 antibody for Western blot, C/EBPβ and BRG1 were found to be components of the same nuclear complex in MCT cells (Fig. 7A). ChIP analysis shows that calcitonin recruits C/EBPβ to the 1α(OH)ase promoter in vivo (Fig. 7, B and C). Re-ChIP analysis shows that C/EBPβ and BRG1 bind simultaneously to the 1α(OH)ase promoter (Fig. 7, B and C). These factors do not interact with an upstream sequence of the 1α(OH)ase promoter (–1300/–980), indicating specificity of the C/EBPβ/BRG1 interaction within the context of the proximal promoter. ChIP analysis also indicated an increase in acetylated histone 4 in response to calcitonin (Fig. 7, B and C).
FIGURE 7.
C/EBPβ and BRG1 are components of the same nuclear complex and calcitonin modulates C/EBPβ and BRG1 recruitment to the 1α(OH)ase promoter. A, nuclear extracts were prepared from MCT cells and used for immunoprecipitation (IP) with C/EBPβ antibody, BRG1 antibody, or control rabbit IgG. WB, Western blot. B, ChIP analysis of CEBPβ, acetylated histone H4 (AcH4), and Re-ChIP analysis of BRG1 binding to the 1α(OH)ase promoter. MCT cells were treated with vehicle or calcitonin for 1 and 4 h and cross-linked by 1% formaldehyde for 15 min. Cross-linked cell lysates were subjected to immunoprecipitation first with C/EBPβ antibody (α-C/EBPβ) and then with BRG1 antibody (α-BRG1). DNA precipitates were isolated and then subjected to PCR using specific primers designed according to the C/EBPβ site on the mouse 1α(OH)ase promoter (see “Experimental Procedures”). Analysis of input DNA (0.2%) was taken before precipitation (Input). Recruitment of Brm to the 1α(OH)ase promoter was not observed (it should be noted that, although Brm and BRG1 were detected in AOK-B50 cell nuclear extracts, BRG1 but not Brm was detected by Western blot analysis of nuclear extracts of MCT cells). Using a distal 1α(OH)ase promoter region (–1300/–980) binding of C/EBPβ and BRG1 was not observed. C, quantitation of ChIP analyses (±S.E.).
DISCUSSION
Although the synthesis of 1,25(OH)2D3 is induced in a hypocalcemic state by secondary hyperparathyroidism, under normocalcemic conditions, calcitonin, not PTH, has been reported to play a major role in 1,25(OH)2D3 synthesis (10). The data here provide a mechanism, for the first time that accounts at least in part for the calcitonin induction of 1α(OH)ase. C/EBPβ and SWI/SNF were found to mediate the calcitonin induction of 1α(OH)ase transcription. A positive C/EBPβ site at –79/–73 on the 1α(OH)ase promoter was identified.
C/EBPs, and in particular C/EBPβ, have been implicated in numerous different processes including hormonal control of nutrient metabolism, differentiation, and regulation of cell type-specific gene expression (45). The levels of C/EBP proteins have previously been reported to differentially modulate gene transcription (46, 47). With regard to vitamin D metabolism, C/EBPβ has previously been shown to be induced by 1,25(OH)2D3 in kidney and osteoblasts and to act as an enhancer of negative vitamin D-mediated transcription of 24(OH)ase (41). γ-interferon induction of C/EBPβ expression has been shown to contribute to γ-interferon transcriptional control of 1α(OH)ase expression in monocytes/macrophages (48, 49). It was concluded that C/EBPβ is the essential transcription factor controlling immune-mediated 1α(OH)ase transcription (48, 49).
Recent studies examining mechanisms involved in the PTH regulation of mouse 1α(OH)ase transcription in kidney cells noted that the orphan receptor nuclear receptor 4A2 (NR4A2 or Nurr 1) and not C/EBPβ is a key factor involved in the induction of 1α(OH)ase transcription by PTH (8). NR4A2 was previously shown to have an important role in brain in normal dopamine cell functions (50) in the regulation of osteopontin in osteoblasts (51) and in the regulation of key cytokines in T cells (52). In AOK-B50 cells PTH induces NF4A2, and C/EBPβ was found to inhibit the NF4A2 induction of 1α(OH)ase transcription (8). We found that the 1α(OH)ase promoter was more sensitive to calcitonin stimulation than to PTH (maximal induction by PTH of 1α(OH)ase transcription in AOK-B50 cells is ∼2-fold (8)4, further suggesting that different factors are involved in the regulation of 1α(OH)ase by CT and PTH. Our study is the first demonstration of the induction of C/EPBβ by calcitonin in kidney cells. It is of interest that PTH, which induces C/EBPβ in osteoblastic cells, does not induce C/EBPβ in AOK-B50 cells or other kidney cells (unlike calcitonin) (41). Thus, different mechanisms are involved in the calcitonin induction and PTH induction of 1α(OH)ase transcription.
SWI/SNF chromatin remodeling complex has been shown to participate in cell cycle control, gene regulation, development, and differentiation (53). Although SWI/SNF has been found to associate with several transcription activators including steroid receptors, erythroid Kruppel-like factors, and heat shock factor 1 (54), only a few transcription factors have the capacity to recruit SWI/SNF to the promoter region. Among them is C/EBPβ, which has been reported to cooperate with SWI/SNF to regulate the expression of myeloid genes (43), the osteocalcin gene in osteoblastic cells (44), and mammary-specific casein genes (55). Similar to our study of 1α(OH)ase gene transcription in kidney cells, BRG1-DN inhibited osteocalcin gene transcription in osteoblastic cells and β and γ casein transcription in EpH4 cells (epithelial cells derived from normal mouse mammary gland) (44, 55), and ChIP/Re-ChIP analysis indicated that BRG1 and C/EBPβ interact within the context of the osteocalcin promoter (44). Also, extracellular matrix protein was found to cooperate with prolactin to induce the recruitment of BRG1 and C/EBPβ to the β and γ casein promoters (55). ChIP analysis also demonstrated enhanced histone acetylation after activation in the β casein promoter (55). In the regulation of osteocalcin it has been suggested that SWI/SNF and histone acetylation cooperate in mediating changes in chromatin structure that facilitate osteocalcin transcription (44). In our studies ChIP analysis indicated an increase in acetylated histone H4 in response to calcitonin, similarly suggesting cooperation between acetylation and chromatin remodeling. It is possible that acetylation may allow SWI/SNF to remain stably bound to the promoter, thus facilitating remodeling of nucleosomes. It has previously been reported that acetylation of histone H4 results in firm association of SWI/SNF through BRG1 (56). Thus, increased calcitonin levels would result in enhanced C/EBPβ binding to the 1α(OH)ase promoter, recruitment of SWI/SNF, and cooperation between acetylation and chromatin remodeling, allowing for efficient initiation of transcription. In future studies it will be of interest to examine additional coactivators that may be involved in the C/EBPβ-SWI/SNF mediated calcitonin induction of transcription. Because CBP, a histone acetyltransferase, has been reported to interact with C/EBPβ and is involved in C/EBP activation of transcription (41, 57), it is possible that CBP may be involved in the calcitonin regulation of 1α(OH)ase and in the cooperation between acetylation and chromatin remodeling.
Although we identified a positive C/EBPβ binding site within the –85/+22 region of the mouse 1α(OH)ase promoter, it has been noted that C/EBPβ has both activation and repression functions, depending on the promoter context and co-factor interaction (46). C/EBPβ has been reported to activate genes by recruiting chromatin remodeling complexes and by cooperating with transcription factors such as Myb and CBP/P300 (43, 44, 57, 58). C/EBPβ also possesses the capacity to suppress gene expression directly through its repression domains or indirectly through other co-factors. Studies from the Wahli laboratory (59) have shown inhibition of peroxisome proliferator-activated receptor βexpression by C/EBPβ and its association with histone deacetylase in the control of differentiation and proliferation of keratinocytes. It can also bind directly to the C/EBP element on the osteoblast-specific Runx2 promoter independent of deacetylase activity to repress gene expression, resulting in an inhibition of retinoic acid-induced osteoblast differentiation (60). In our studies, C/EBPβ at low concentrations enhances calcitonin-induced 1α(OH)ase transcription, but at high concentrations repression of 1α(OH)ase transcription was observed. Thus, although the predominant effect of C/EPBβ is enhancement, as indicated by increased expression of 1α(OH)ase protein in C/EBPβ-transfected cells and inhibition of calcitonin-induced 1α(OH)ase transcription by A-C/EBP as well as by LIP, it is possible that C/EBPβ may have a dual role depending on the level of C/EBPβ, the hormonal context, the specific intracellular environment, and the level of 1α(OH)ase expression.
In summary, calcitonin is a hormone that has diverse physiological actions, including a role in the maintenance of 1,25(OH)2D3 levels. Previous evidence indicated that the stimulation of 1,25(OH)2D3 under normocalcemic conditions by calcitonin has physiological importance during pregnancy, lactation, and early development (14–16). Our findings provide a mechanism for the first time for calcitonin induction of 1α(OH)ase and identify key regulators involved in the maintenance of 1,25(OH)2D3 levels.
Acknowledgments
M. A. Boltz (Armbrecht laboratory) provided technical assistance in Western blot analysis of 1α(OH)ase and radioimmunoassay of 1,25(OH)2D3. We appreciate the advice and help of Dr. Adriana Dusso (Washington University School of Medicine, St. Louis, MO) with experiments using MCT cells.
This work was supported, in whole or in part, by National Institutes of Health Grant DK-38961 (to S. C.).
This manuscript is dedicated to the memory of Iain MacIntyre (1924–2008), a leader in the field of bone and calcium metabolism, who first discovered the effect of calcitonin on 1α(OH)ase (13) and encouraged us in this investigation.
Footnotes
The abbreviations used are: 1,25(OH)2D3, 1,25-dihydroxyvitamin D3; 1α(OH)ase, 25-hydroxvitamin D3 1α-hydroxylase; PTH, parathyroid hormone; 25(OH)D3, 25-hydroxyvitamin D3; CBP, cAMP-response element-binding protein (CREB)-binding protein; Pipes, 1,4-piperazinediethanesulfonic acid; ChIP, chromatin immunoprecipitation; DN, dominant negative.
Y. Zhong and S. Christakos, unpublished observation.
References
- 1.Bikle, D., Adams, J., and Christakos, S. (2008) in Primer on Metabolic Bone Diseases and Disorder of Mineral Metabolism (Rosen, C., ed) pp. 141–149, American Society for Bone and Mineral Research, Washington DC
- 2.Omdahl, J. L., Bobrovnikova, E. V., Annalora, A., Chen, P., and Serda, R. (2003) J. Cell. Biochem. 88, 356–362 [DOI] [PubMed] [Google Scholar]
- 3.Henry, H. (2005) in Vitamin D (Feldman, D., Glorieux, F. H., and Pike, J. W., ed) pp. 69–83, Academic Press, Inc., San Diego, CA
- 4.St-Arnaud, R., Messerlian, S., Moir, J. M., Omdahl, J. L., and Glorieux, F. H. (1997) J. Bone Miner. Res. 12, 1552–1559 [DOI] [PubMed] [Google Scholar]
- 5.Kitanaka, S., Takeyama, K., Murayama, A., Sato, T., Okumura, K., Nogami, M., Hasegawa, Y., Niimi, H., Yanagisawa, J., Tanaka, T., and Kato, S. (1998) N. Engl. J. Med. 338, 653–661 [DOI] [PubMed] [Google Scholar]
- 6.Garabedian, M., Holick, M. F., Deluca, H. F., and Boyle, I. T. (1972) Proc. Natl. Acad. Sci. U. S. A. 69, 1673–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenza, H. L., Kimmel-Jehan, C., Jehan, F., Shinki, T., Wakino, S., Anazawa, H., Suda, T., and DeLuca, H. F. (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 1387–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zierold, C., Nehring, J. A., and DeLuca, H. F. (2007) Arch. Biochem. Biophys. 460, 233–239 [DOI] [PubMed] [Google Scholar]
- 9.Murayama, A., Takeyama, K., Kitanaka, S., Kodera, Y., Hosoya, T., and Kato, S. (1998) Biochem. Biophys. Res. Commun. 249, 11–16 [DOI] [PubMed] [Google Scholar]
- 10.Shinki, T., Ueno, Y., DeLuca, H. F., and Suda, T. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 8253–8258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaeger, P., Jones, W., Clemens, T. L., and Hayslett, J. P. (1986) J. Clin. Investig. 78, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawashima, H., Torikai, S., and Kurokawa, K. (1981) Nature 291, 327–329 [DOI] [PubMed] [Google Scholar]
- 13.Galante, L., Colston, K. W., MacAuley, S. J., and MacIntyre, I. (1972) Nature 238, 271–273 [DOI] [PubMed] [Google Scholar]
- 14.Nishioka, T., Yasuda, T., Niimi, H., and Nakajima, H. (1988) Eur. J. Pediatr. 147, 148–152 [DOI] [PubMed] [Google Scholar]
- 15.Cooper, C. W., Obie, J. F., Toverud, S. U., and Munson, P. L. (1977) Endocrinology 101, 1657–1664 [DOI] [PubMed] [Google Scholar]
- 16.Stevenson, J. C., Hillyard, C. J., MacIntyre, I., Cooper, H., and Whitehead, M. I. (1979) Lancet 2, 769–770 [DOI] [PubMed] [Google Scholar]
- 17.Kumar, R., Cohen, W. R., Silva, P., and Epstein, F. H. (1979) J. Clin. Investig. 63, 342–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaidi, M., Inzerillo, A. M., Moonga, B. S., Bevis, P. J., and Huang, C. L. (2002) Bone (NY) 30, 655–663 [DOI] [PubMed] [Google Scholar]
- 19.Wang, Y. Q., Yuan, R., Sun, Y. P., Lee, T. J., and Shah, G. V. (2003) Endocrinology 144, 2164–2171 [DOI] [PubMed] [Google Scholar]
- 20.Thomas, S., Chigurupati, S., Anbalagan, M., and Shah, G. (2006) Mol. Endocrinol. 20, 1894–1911 [DOI] [PubMed] [Google Scholar]
- 21.Segawa, N., Nakamura, M., Nakamura, Y., Mori, I., Katsuoka, Y., and Kakudo, K. (2001) Cancer Res. 61, 6060–6063 [PubMed] [Google Scholar]
- 22.Nakamura, M., Han, B., Nishishita, T., Bai, Y., and Kakudo, K. (2007) J. Mol. Endocrinol. 39, 375–384 [DOI] [PubMed] [Google Scholar]
- 23.Kovacs, C. S., Chafe, L. L., Woodland, M. L., McDonald, K. R., Fudge, N. J., and Wookey, P. J. (2002) Am. J. Physiol. Endocrinol. Metab. 282, 721–732 [DOI] [PubMed] [Google Scholar]
- 24.Emmertsen, K., Melsen, F., Mosekilde, L., Lund, B., Sorensen, O. H., Nielsen, H. E., Solling, H., and Hansen, H. H. (1982) Metab. Bone Dis. Relat. Res. 4, 17–23 [DOI] [PubMed] [Google Scholar]
- 25.Hoff, A. O., Catala-Lehnen, P., Thomas, P. M., Priemel, M., Rueger, J. M., Nasonkin, I., Bradley, A., Hughes, M. R., Ordonez, N., Cote, G. J., Amling, M., and Gagel, R. F. (2002) J. Clin. Investig. 110, 1849–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emmertsen, K., Melsen, F., Mosekilde, L., Lund, B., Sorensen, O. H., Charles, P., and Moller, J. (1984) Acta Endocrinol. (Copenh) 106, 346–349 [DOI] [PubMed] [Google Scholar]
- 27.Gao, X. H., Dwivedi, P. P., Choe, S., Alba, F., Morris, H. A., Omdahl, J. L., and May, B. K. (2002) Int. J. Biochem. Cell Biol. 34, 921–930 [DOI] [PubMed] [Google Scholar]
- 28.Murayama, A., Kim, M. S., Yanagisawa, J., Takeyama, K., and Kato, S. (2004) EMBO J. 23, 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Brenza, H. L., and DeLuca, H. F. (2001) Arch. Biochem. Biophys. 388, 121–126 [DOI] [PubMed] [Google Scholar]
- 30.Kong, X. F., Zhu, X. H., Pei, Y. L., Jackson, D. M., and Holick, M. F. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 6988–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bringhurst, F. R., Juppner, H., Guo, J., Urena, P., Potts, J. T., Jr., Kronenberg, H. M., Abou-Samra, A. B., and Segre, G. V. (1993) Endocrinology 132, 2090–2098 [DOI] [PubMed] [Google Scholar]
- 32.Lin, H. Y., Harris, T. L., Flannery, M. S., Aruffo, A., Kaji, E. H., Gorn, A., Kolakowski, L. F., Jr., Lodish, H. F., and Goldring, S. R. (1991) Science 254, 1022–1024 [DOI] [PubMed] [Google Scholar]
- 33.Haverty, T. P., Kelly, C. J., Hines, W. H., Amenta, P. S., Watanabe, M., Harper, R. A., Kefalides, N. A., and Neilson, E. G. (1988) J. Cell Biol. 107, 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinson, C., Myakishev, M., Acharya, A., Mir, A. A., Moll, J. R., and Bonovich, M. (2002) Mol. Cell. Biol. 22, 6321–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niehof, M., Manns, M. P., and Trautwein, C. (1997) Mol. Cell. Biol. 17, 3600–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DiRenzo, J., Shang, Y., Phelan, M., Sif, S., Myers, M., Kingston, R., and Brown, M. (2000) Mol. Cell. Biol. 20, 7541–7549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de La Serna, I. L., Carlson, K. A., Hill, D. A., Guidi, C. J., Stephenson, R. O., Sif, S., Kingston, R. E., and Imbalzano, A. N. (2000) Mol. Cell. Biol. 20, 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Descombes, P., and Schibler, U. (1991) Cell 67, 569–579 [DOI] [PubMed] [Google Scholar]
- 39.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 40.Armbrecht, H. J., Boltz, M. A., Ritter, C. S., and Brown, A. J. (2007) J. Steroid Biochem. Mol. Biol. 103, 330–333 [DOI] [PubMed] [Google Scholar]
- 41.Dhawan, P., Peng, X., Sutton, A. L., MacDonald, P. N., Croniger, C. M., Trautwein, C., Centrella, M., McCarthy, T. L., and Christakos, S. (2005) Mol. Cell. Biol. 25, 472–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen, Q., and Christakos, S. (2005) J. Biol. Chem. 280, 40589–40598 [DOI] [PubMed] [Google Scholar]
- 43.Kowenz-Leutz, E., and Leutz, A. (1999) Mol. Cell 4, 735–743 [DOI] [PubMed] [Google Scholar]
- 44.Villagra, A., Cruzat, F., Carvallo, L., Paredes, R., Olate, J., van Wijnen, A. J., Stein, G. S., Lian, J. B., Stein, J. L., Imbalzano, A. N., and Montecino, M. (2006) J. Biol. Chem. 281, 22695–22706 [DOI] [PubMed] [Google Scholar]
- 45.Ramji, D. P., and Foka, P. (2002) Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Screpanti, I., Romani, L., Musiani, P., Modesti, A., Fattori, E., Lazzaro, D., Sellitto, C., Scarpa, S., Bellavia, D., Lattanzio, G., Bistoni, F., Frati, L., Cortese, R., Gulino, A., Ciliberto, G., Costantini, F., and Poli, V. (1995) EMBO J. 14, 1932–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chew, C. H., Chew, G. S., Najimudin, N., and Tengku-Muhammad, T. S. (2007) Int. J. Biochem. Cell Biol. 39, 1975–1986 [DOI] [PubMed] [Google Scholar]
- 48.Stoffels, K., Overbergh, L., Giulietti, A., Verlinden, L., Bouillon, R., and Mathieu, C. (2006) J. Bone Miner. Res. 21, 37–47 [DOI] [PubMed] [Google Scholar]
- 49.Esteban, L., Vidal, M., and Dusso, A. (2004) J. Steroid Biochem. Mol. Biol. 89–90, 131–137 [DOI] [PubMed] [Google Scholar]
- 50.Saucedo-Cardenas, O., Quintana-Hau, J. D., Le, W. D., Smidt, M. P., Cox, J. J., De Mayo, F., Burbach, J. P., and Conneely, O. M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 4013–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lammi, J., Huppunen, J., and Aarnisalo, P. (2004) Mol. Endocrinol. 18, 1546–1557 [DOI] [PubMed] [Google Scholar]
- 52.Doi, Y., Oki, S., Ozawa, T., Hohjoh, H., Miyake, S., and Yamamura, T. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 8381–8386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neely, K. E., and Workman, J. L. (2002) Biochim. Biophys. Acta 1603, 19–29 [DOI] [PubMed] [Google Scholar]
- 54.Narlikar, G. J., Fan, H. Y., and Kingston, R. E. (2002) Cell 108, 475–487 [DOI] [PubMed] [Google Scholar]
- 55.Xu, R., Spencer, V. A., and Bissell, M. J. (2007) J. Biol. Chem. 282, 14992–14999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agalioti, T., Chen, G., and Thanos, D. (2002) Cell 111, 381–392 [DOI] [PubMed] [Google Scholar]
- 57.Cui, T. X., Piwien-Pilipuk, G., Huo, J. S., Kaplani, J., Kwok, R., and Schwartz, J. (2005) Mol. Endocrinol. 19, 2175–2186 [DOI] [PubMed] [Google Scholar]
- 58.Gutierrez, J., Paredes, R., Cruzat, F., Hill, D. A., van Wijnen, A. J., Lian, J. B., Stein, G. S., Stein, J. L., Imbalzano, A. N., and Montecino, M. (2007) J. Biol. Chem. 282, 9445–9457 [DOI] [PubMed] [Google Scholar]
- 59.Di-Poi, N., Desvergne, B., Michalik, L., and Wahli, W. (2005) J. Biol. Chem. 280, 38700–38710 [DOI] [PubMed] [Google Scholar]
- 60.Wiper-Bergeron, N., St-Louis, C., and Lee, J. M. (2007) Mol. Endocrinol. 21, 2124–2135 [DOI] [PubMed] [Google Scholar]