Abstract
Curcuminoids found in the rhizome of turmeric, Curcuma longa, possess various biological activities. Despite much attention regarding the biosynthesis of curcuminoids because of their pharmaceutically important properties and biosynthetically intriguing structures, no enzyme systems have been elucidated. Here we propose a pathway for curcuminoid biosynthesis in the herb C. longa, which includes two novel type III polyketide synthases. One of the type III polyketide synthases, named diketide-CoA synthase (DCS), catalyzed the formation of feruloyldiketide-CoA by condensing feruloyl-CoA and malonyl-CoA. The other, named curcumin synthase (CURS), catalyzed the in vitro formation of curcuminoids from cinnamoyldiketide-N-acetylcysteamine (a mimic of the CoA ester) and feruloyl-CoA. Co-incubation of DCS and CURS in the presence of feruloyl-CoA and malonyl-CoA yielded curcumin at high efficiency, although CURS itself possessed low activity for the synthesis of curcumin from feruloyl-CoA and malonyl-CoA. These findings thus revealed the curcumin biosynthetic route in turmeric, in which DCS synthesizes feruloyldiketide-CoA, and CURS then converts the diketide-CoA esters into a curcuminoid scaffold.
The rhizome of turmeric, Curcuma longa, has been used as a food additive, especially as a spice in curry, as a coloring agent, and in cosmetics (1, 2). In addition to its aromatic, stimulant, and coloring properties, the powdered rhizome of turmeric possesses anti-inflammatory, antioxidant, and antitumor activities and has been taken orally to treat dyspepsia, flatulence, and liver and urinary tract diseases (3–6). Most of these pharmaceutical properties of turmeric depend on the curcuminoids, including curcumin (3e), demethoxycurcumin (4e), and bisdemethoxycurcumin (2e) (see Fig. 1A). The amounts (wild type t%) of these curcuminoids were ∼2, 0.7, and 0.6% in powdered turmeric (7).
FIGURE 1.
Curcuminoid biosynthesis in C. longa, as proposed by the present study (A) and the enzyme activities of various type III PKSs, including DCS and CURS (B). A, the starter substrates, cinnamoyl-CoA (1a), p-coumaroyl-CoA (2a), or feruloyl-CoA (3a), are synthesized from phenylalanine by the activity of phenylalanine ammonia-lyase (PAL), 4-coumarate:CoA ligase (4CL), cinnamate-4-hydroxylase (C4H), hydroxycinnamoyl transferase (HCT), cinnamate-3-hydroxylase (C3H), and O-methyltransferase (OMT). DCS catalyzes formation of diketide-CoAs by condensing p-coumaroyl-CoA and feruloyl-CoA with malonyl-CoA. CURS catalyzes formation of curcuminoids by condensing diketide-CoAs with starter substrates. B, the reactions catalyzed by DCS, CURS, benzalacetone synthase (BAS) from Rheum palmatum, CUS from O. sativa, and CHS are shown.
Type III polyketide synthases (PKSs),4 consisting of a homodimer of ketosynthase, play an important role in the biosynthesis of most plant polyketides (8). Chalcone synthase (CHS), a typical type III PKS, catalyzes the formation of naringenin chalcone by the following mechanism (see Fig. 1B) (9, 10). The reaction of CHS is primed by a transfer of the acyl moiety of p-coumaroyl-CoA, which is called a starter substrate, to the catalytic cysteine of CHS. Subsequent iterative decarboxylative condensation of three malonyl-CoAs, which are called extender substrates, yields a tetraketide intermediate. These iterative condensations are catalyzed by a Cys-Asn-His catalytic triad (8, 9). The resultant tetraketide intermediate is then cyclized by Claisen condensation, followed by aromatization resulting in naringenin chalcone formation. Like CHS, most type III PKSs catalyze condensation of one starter substrate and several extender substrates.
Due to the pharmaceutically important properties of curcuminoids, the biosynthetic pathway has been the subject of much attention, and radio tracer studies were performed decades ago (11). Based on those results, Schröder (12) proposed that curcuminoids are derived from the phenylpropanoid pathway and that one or more type III PKSs were responsible for the formation of its scaffold. Curcuminoids, consisting of two phenylpropanoid units, are chemically derived from phenylalanine connected by a central carbon unit derived from malonyl-CoA. The 13C feeding experiment carried out by Kita et al. (13) showed that the curcuminoid scaffolds originate from two phenylalanines. One possible pathway from phenylalanine to curcumin is presented in Fig. 1A. However, it is not known when the methyl ethers at the 3′ position of curcumin are incorporated, i.e. before or after curcumin scaffold formation, or how the curcumin scaffold is synthesized, i.e. by a single type III PKS enzyme or a multiple enzyme system.
On the basis of these findings, several groups attempted to identify one or more type III PKSs responsible for the formation of the curcuminoid scaffold. Brand et al. (14) characterized a type III PKS in Wachendorfia thyrsiflora, a plant that produces phenylphenalenones derived from a curcuminoid scaffold. This type III PKS, named WtPKS1, synthesizes a benzalacetone scaffold and triketide pyrones from various starter substrates (see Fig. 1B). Therefore, they predicted that benzalacetone synthesis might be included in a step for the formation of the curcuminoid scaffold. This hypothesis was also mentioned by Abe et al. (15). Ramirez-Ahumada et al. (16) detected curcuminoid synthetic activity in an extract from the leaves of turmeric. Their attempts to purify a type III PKS responsible for this reaction by anion-exchange column chromatography failed.
We recently discovered a type III PKS, named curcuminoid synthase (CUS), in the rice Oryza sativa, that catalyzes the formation of bisdemethoxycurcumin by condensing two p-coumaroyl-CoAs and one malonyl-CoA by the mechanism shown in Fig. 1B (17). CUS synthesizes p-coumaroyldiketide-CoA (2b) by condensing p-coumaroyl-CoA with malonyl-CoA. The resultant diketide-CoA (2b) is hydrolyzed to yield a β-keto acid (2c), which then serves as the second extender substrate and is condensed with another p-coumaroyl-CoA, resulting finally in the formation of bisdemethoxycurcumin (2e). CUS is the first type III PKS that catalyzes condensation of two starter substrates with one extender substrate. CUS also produces triketide pyrones from one p-coumaroyl-CoA and two malonyl-CoAs (Fig. 1B). O. sativa produces no detectable curcuminoids, and therefore the in vivo function of CUS still remains unknown. Although the discovery of CUS supported the hypothesis proposed by Schröder (12), the type III PKS catalyzing the curcuminoid synthesis in turmeric still remained undiscovered.
In this report, we describe the cloning and characterization of two type III PKSs from turmeric, which were named diketide-CoA synthase (DCS) and curcumin synthase (CURS) on the basis of their activities. DCS catalyzes the formation of a diketide intermediate via condensation of feruloyl-CoA with malonyl-CoA. CURS catalyzes the formation of curcumin from the diketide intermediate and feruloyl-CoA. CURS also catalyzes the formation of a diketide intermediate from feruloyl-CoA and malonyl-CoA, but at a very low rate. We thus concluded that DCS and CURS participate in curcumin synthesis using the abundantly present CoA esters in turmeric.
EXPERIMENTAL PROCEDURES
Materials—Escherichia coli JM109, pUC19, restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were purchased from Takara Biochemicals (Shiga, Japan). pET16b was used for expression of the His-tagged proteins in E. coli BL21(DE3) (Novagen, Darmstadt, Germany). Bisdemethoxycurcumin (2f) was purchased from Chromadex (Santa Ana, CA). Curcumin (3f) was purchased from Sigma (Steinheim, Germany). trans-Cinnamic acid, trans-4-hydroxycinnamic acid (p-coumaric acid), and trans-4-hydroxy-3-methoxycinnamic acid (ferulic acid) were purchased from Wako (Tokyo, Japan). Benzalacetone (1d) and dehydrozingerone (3d) were purchased from Tokyo Chemical Industry (Tokyo, Japan). 4-Hydroxybenzalacetone (2d) was purchased from Sigma-Aldrich. p-Coumaroyl-CoA (2a), cinnamoyl-CoA (1a), and feruloyl-CoA (3a) were synthesized according to the procedures reported by Blecher (18). Cinnamoyldiketide-N-acetylcysteamine (4b) was synthesized according to the reported method (19). trans-3-Oxo-5-phenylpent-4-enoic acid (1c) was synthesized by a modification of the procedure reported by Matsumura et al. (20). Cinnamoyl-p-coumaroylmethane (5e), dicinnamoylmethane (1e), and cinnamoylferuloylmethane (6e) were synthesized according to previously reported methods (21). trans-5-(4-Hydroxy-3-methoxyphenyl)-3-oxopent-4-enoic acid methyl ester was synthesized by a modification of a published procedure (22).
cDNA Preparation—Curcuma longa was grown under standard conditions in a field in Chiba, Japan, for 4 months before rhizomes and leaves were collected. Total RNA was prepared from 80 mg each of rhizomes and leaves using the RNeasy– Plant Mini kit (Qiagen, Valencia, CA). The DNA was digested using the TURBO DNA-free kit (Applied Biosystems, Foster City, CA), and total RNA was concentrated using the RNeasy– MinElute– Cleanup kit (Qiagen), yielding total RNAs of 1010 ng from the leaf and 360 ng from the rhizome. cDNA was synthesized from these RNAs using the SMART RACE cDNA Amplification kit (Clontech, Palo Alto, CA) and PrimeScript– Reverse Transcriptase (Takara).
Amplification of Full-length cDNAs—The full-length cDNA of CURS was amplified by the primers 5′-CTCTGCGACTGCGAGAAGAAG-3′ and 5′-GTCCTATAACAGATAGACAGC-3′ from a cDNA library. These primers were designed on the basis of previously reported expressed sequence tag sequences (NCBI accession numbers DY384950 and DY386934). A 1.3-kb cDNA fragment encoding CURS was obtained. The full-length cDNA of DCS was amplified as follows. A BLAST search using CUS from O. sativa as a query established conserved sequences of type III PKSs that would be appropriate for the primers to amplify cDNAs of type III PKS genes. The primers thus designed were 5′-TCATGGCCATCGGCACNGCNAMNCC-3′ (M = A or C; N = A, T, G, or C) and 5′-CCTGTTGTTCTCGGCCADNTCYTTNGC-3′ (D = G, A, or T; Y = T or C). A partial sequence of the cDNA encoding the conserved regions of type III PKSs was amplified from the cDNA library using these primers, and the fragment obtained was sequenced. Using this sequence, a gene-specific primer, 5′-GACATCACGCACCTGGTCTTCTG-3′, was designed. 5′-Rapid amplification of cDNA ends (RACE) was carried out using the SMART RACE cDNA amplification kit (Clontech). The amplified 5′-RACE fragment was purified and sequenced. The primer 5′-GGTAACACGCGTCATTCTTTGC-3′ was designed to carry out 3′-RACE using the SMART RACE cDNA amplification kit. The amplified 3′-RACE fragment was purified and sequenced, yielding a 1.4-kb cDNA fragment encoding DCS.
Relative Quantification by Quantitative Real-time PCR—cDNA was prepared and purified as described above. Quantitative real-time PCR was carried out on a Smart Cycler II (Takara) using SYBER Premix Ex Taq polymerase and 0.2 μm of gene-specific forward and reverse primers (supplemental Table S1) in a total volume of 25 μl. The conditions were as follows. The initial incubation at 95 °C was carried out for 10 s, followed by 40 cycles of 95 °C for 10 s and 60 °C for 20 s. For quantification of the PCR products, 18 S rRNA was used as a control (DNA accession no. AB047718). The ratio of gene-specific expression to 18 S rRNA signals was defined as the relative expression.
Production and Purification of DCS—Using the DCS cDNA as a template, the 1.1-kb DNA fragment containing the DCS-coding region was amplified by PCR with the primers 5′-CCGAATTCCATATGGAAGCGAACGGCTACCG-3′ (the bold letters indicate an EcoRI site, the underline indicates an NdeI site, and the italics indicate the start codon of DCS) and 5′-CGCGGATCCCTAGTTCAGTCTGCAACTAT-3′ (the bold letters indicate a BamHI site, and italics indicate the stop codon of DCS). The amplified fragment was cloned between the EcoRI and BamHI sites of pUC19, resulting in pUC19-DCS. The DCS sequence was excised as an NdeI-BamHI fragment from pUC19-DCS and cloned between the NdeI and BamHI sites of pET16b, resulting in pET16b-DCS.
For the production of histidine-tagged DCS, E. coli BL21(DE3) harboring pET16b-DCS was grown overnight in Luria Broth containing 100 μg/ml ampicillin. Cells were harvested by centrifugation and resuspended in buffer containing 50 mm potassium phosphate buffer (pH 8.0) and 10% glycerol. After sonication, cell debris was removed by centrifugation, and the cleared lysate was applied to a column containing His-bind metal chelation resin (Qiagen). The purified histidine-tagged protein was dialyzed against 50 mm potassium phosphate buffer (pH 8.0), 1 mm dithiothreitol, and 10% glycerol.
DCS Assay—The standard reaction mixture contained 100 μm starter substrate (cinnamoyl-CoA, p-coumaroyl-CoA, or feruloyl-CoA), 100 μm extender substrate (malonyl-CoA), 100 mm potassium phosphate buffer (pH 7.5), and 4.0 μg of DCS in a total volume of 100 μl. Reactions were incubated at 37 °C for 1 h before being quenched with 20 μl of 6 m HCl or 5 μl of 10 m NaOH. The solutions, to which NaOH was added at a final concentration of 0.5 m, were incubated at 65 °C for 10 min for alkaline hydrolysis. The solutions obtained were applied directly to LC-ESI MS/MS analysis. LC-ESI MS/MS analysis was carried out using the Esquire High-capacity Trap (HCT) plus system (Bruker Daltonics, Bremen, Germany). The HPLC was equipped with a Pegasil-B C4 reversed-phase HPLC column (4.6 × 250 mm, Senshu Scientific, Tokyo, Japan) and eluted with a linear acetonitrile gradient (5% for 5 min, 5 to 40% over 35 min, and 40–80% over 5 min) in water containing 25 mm ammonium acetate at a flow rate of 1.0 ml/min. UV spectra were detected on an Agilent G1315B system.
Identification of Feruloyldiketide-CoA (3b)—Feruloyldiketide-CoA was purified by HPLC. HPLC on a Waters 600 system equipped with a Pegasil-B C4 reversed-phase HPLC column (4.6 × 250 mm) was carried out by elution with a linear acetonitrile gradient (10–40% over 40 min and 40–70% over 5 min) in water containing 25 mm KH2PO4 at a flow rate of 1.0 ml/min. The purified feruloyldiketide-CoA was dialyzed by HPLC. The feruloyldiketide-CoA thus obtained was hydrolyzed in 1 m KOH at 65 °C for 10 min. The resultant solution was acidified and kept on ice until directly applied to LC-ESI MS analysis. LC-ESI MS/MS analysis was carried out using the Esquire HCT system. The HPLC was equipped with a Pegasil-B ODS reversed-phase HPLC column (2 × 250 mm, Senshu) and eluted with a linear acetonitrile gradient (5% for 5 min, 5–40% over 35 min, and 40–80% over 5 min) in water containing 25 mm ammonium acetate at a flow rate of 0.3 ml/min. The compound derived as a result of hydrolysis was identified as trans-5-(4-hydroxy-3-methoxyphenyl)-3-oxopent-4-enoic acid by comparison to the sample synthesized by hydrolysis of trans-5-(4-hydroxy-3-methoxyphenyl)-3-oxopent-4-enoic acid methyl ester.
Determination of the Kinetic Parameters of DCS—A standard reaction contained 100 mm potassium phosphate buffer (pH 7.5), 200 μm malonyl-CoA, feruloyl-CoA (2.5–100 μm), and 4.0 μg of DCS in a total volume of 100 μl for determination of kinetic values for feruloyl-CoA, and contained 100 mm potassium phosphate buffer (pH 7.5), malonyl-CoA (2.5–50 μm), 100 μm feruloyl-CoA, and 4.0 μg of DCS in a total volume of 100 μl for determination of kinetic values for malonyl-CoA. After the reaction mixture had been preincubated at 37 °C for 2 min, reactions were initiated by adding the substrate and continued for 5 min. We observed that the product formation was linear throughout this period. The reactions were stopped with 5 μl of 10 m NaOH, and the solution was incubated at 65 °C for 15 min to hydrolyze the products. The solution was acidified by adding 10 μl of 6 n HCl, and the material in the mixture was extracted with ethyl acetate. The organic layer was collected and evaporated. The residual materials were dissolved in 20 μl of methanol for HPLC analysis. The velocity of the reaction was measured by monitoring the formation of dehydrozingerone that was synthesized via nonenzymatic hydrolysis and decarboxylation (3d) of feruloyldiketide-CoA (3b). HPLC analysis was carried out using a Waters 600 HPLC apparatus equipped with a COSMOSIL 5C18MS-II reversed-phase HPLC column (4.6 × 250 mm, Nacalai Tesque, Kyoto, Japan) and isocratically eluted with acetonitrile in water containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min. UV was detected by the Waters 996 photodiode array detector. Dehydrozingerone (3d) was used to generate the standard curve for quantification of the products. Steady-state parameters were determined by fitting the curve to v = Vmax[S]n/(S50n + [S]n) (n indicates the slope value) and v = Vmax[S]/(Km + [S]).
Production and Purification of CURS—Using CURS cDNA as a template, the 1.1-kb DNA fragment containing the CURS-coding region was amplified by PCR with the primers 5′-CCGAATTCCATATGGCCAACCTCCACGCGTT-3′ (the bold letters indicate an EcoRI site, the underline indicates an NdeI site, and the italics indicate the start codon of CURS) and 5′-CGCGGATCCCTACAGTGGCATACTGCGCA-3′ (the bold letters indicate a BamHI site, and italics indicate the stop codon of CURS). The amplified fragment was cloned between the EcoRI and BamHI sites of pUC19, resulting in pUC19-CURS. The CURS sequence was excised as an NdeI-BamHI fragment from pUC19-CURS and cloned between the NdeI and BamHI sites of pET16b, resulting in pET16b-CURS.
For production of histidine-tagged CURS, E. coli BL21(DE3) harboring pET16b-CURS was grown overnight in Luria Broth containing 100 μg/ml ampicillin. Cells were harvested by centrifugation and resuspended in buffer containing 50 mm potassium phosphate buffer (pH 8.0), 10 mm β-mercaptoethanol, and 10% glycerol. After sonication, cell debris was removed by centrifugation, and the cleared lysate was applied to a column containing His-bind metal chelation resin (Qiagen). The purified histidine-tagged protein was dialyzed against 50 mm potassium phosphate buffer (pH 8.0), 1 mm dithiothreitol, and 10% glycerol.
CURS Assay—The standard reaction mixture contained 100 μm starter substrate (cinnamoyl-CoA, p-coumaroyl-CoA, or feruloyl-CoA), 100 μm extender substrate (malonyl-CoA or cinnamoyldiketide-NAC), 100 mm potassium phosphate buffer (pH 8.0), and 4.0 μg of CURS in a total volume of 100 μl. Reactions were incubated at 37 °C for 1 h before being quenched with 20 μl of 6 m HCl. The products were extracted with ethyl acetate, and the organic layer was evaporated to dryness. The residual material was dissolved in 20 μl of DMSO for LC-APCI MS/MS analysis, which was carried out using the esquire HCT plus system. The HPLC was equipped with a Pegasil-B C4 reversed-phase HPLC column (4.6 × 250 mm) and eluted with a linear acetonitrile gradient (10–100% over 45 min) in water containing 0.1% acetic acid at a flow rate of 1.0 ml/min. UV spectra were detected on an Agilent G1315B system.
Determination of the Kinetic Parameters of CURS—The reaction mixture contained 100 mm potassium phosphate buffer (pH 8.0), 2–100 μm each of feruloyl-CoA or p-coumaroyl-CoA, 100 μm cinnamoyl-diketide-NAC, and 4.0 μg of CURS in a total volume of 100 μl. After the reaction mixture had been preincubated at 37 °C for 2 min, reactions were initiated by adding the substrates and continued for 5 min. We confirmed that the product formation was linear throughout this period. The reactions were stopped with 20 μl of 6 m HCl, and the material in the mixture was extracted with ethyl acetate. The organic layer was collected and evaporated. The residual materials were dissolved in 20 μl of methanol for HPLC analysis. HPLC analysis was carried out using the Waters 600 HPLC apparatus equipped with a Pegasil-B C4 reversed-phase HPLC column (4.6 × 250 mm, Senshu) and isocratically eluted with 60% acetonitrile in water containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min. UV absorbance was detected using a Waters 996 photodiode array detector. Cinnamoyl-p-coumaroylmethane (4f) and cinnamoylferuloylmethane (5f) were used to generate the standard curves for quantification of the products. Steady-state parameters were determined by fitting the curve to v = Vmax[S]/(Km + [S]).
Spectroscopic Data of Synthetic Samples—Dicinnamoylmethane (1e): 1H NMR (500 MHz, CHCl3) δ = 7.68 (d, 2H, J = 16 Hz), 7.57 (dd, 4H, J = 2, 7 Hz), 7.39 (m, 6H), 6.65 (d, 2H, J = 16 Hz), 5.87 (s, 1H). LC-APCI MS (positive): MS, m/z 277 [M+H]+, MS/MS (precursor ion at m/z 277), m/z 131 (100) [C9H7O1]+ m/z 193 (61) [M+H-C4H4O2]. UV: λmax 390 nm. Cinnamoyl-p-coumaroylmethane (5e): 1H NMR (500 MHz, CDCl3): δ = 7.67 (d, 1H, J516 Hz), 7.64 (d, 1H, J516 Hz), 7.57 (dd, 2H, J = 2, 7.5 Hz), 7.49 (d, 2H, J = 8.5 Hz), 7.40 (m, 3H), 6.87 (d, 2H, J = 8.5 Hz), 6.63 (d, 1H, J = 16 Hz), 6.52 (d, 1H, J = 16 Hz), 5.83 (s, 1H). LC-APCI MS (positive): MS, m/z 293 [M+H]+, MS/MS (precursor ion at m/z 293), m/z 131 (17) [C9H7O1]+ m/z 147 (26) [C9H7O2]+ m/z 209 (100) [M+H-C4H4O2]+. λmax 404 nm. Cinnamoylferuloylmethane (6e). 1H NMR (500 MHz, CH3COCH3) δ = 7.69 (dd, 1H J = 1, 7.5 Hz), 7.66 (d, 1H, J = 16 Hz), 7.64 (d, 1H, J = 16 Hz), 7.43 (m, 3H) 7.36 (d, 1H, J = 1.5 Hz), 7.20 (dd, 1H, J = 1.5, 8 Hz), 6.89 (d, 1H, J = 8 Hz), 6.86 (d, 1H, J = 16 Hz), 6.75 (d, 1H, J = 16 Hz), 6.06 (s, 1H) 3.93 (s, 3H). LC-APCI MS (positive): MS, m/z 323 [M+H]+. MS/MS (precursor ion at m/z 323), m/z 175 (50) [C10H7O3]+ m/z 177 (100) [C10H9O3]+ m/z 239 (100) [M+H-C4H4O2]+. λmax 404 nm.
RESULTS
Isolation and Cloning of cDNAs Encoding Type III PKSs—We cloned and sequenced two full-length cDNAs obtained by amplifying the cDNA library from the rhizome and leaf of turmeric (supplemental Fig. S1). One full-length cDNA was obtained via amplification using degenerate oligonucleotide primers designed on the basis of the conserved sequence of type III PKSs (the details of the method are described under “Experimental Procedures”). Another was obtained using primer sequences based on the expressed sequence tag sequences reported by Gang's group (NCBI accession numbers DY384950 and DY386934). Direct sequencing of these DNA fragments predicted that both cDNAs encoded different type III PKSs. These type III PKSs were named DCS and CURS on the basis of their enzyme activities (see below). The entire DCS and CURS cDNAs were cloned using standard DNA manipulation methods. DCS and CURS shared 62% identity in their amino acid sequences and contained a conserved Cys-Asn-His catalytic triad (supplemental Fig. S2) (9).
Expression of DCS and CURS in turmeric was checked by quantitative PCR. Total RNA was extracted from the rhizome and leaf of turmeric grown for 4 months, and cDNAs were prepared using the SMART RACE cDNA amplification kit (Clontech). Quantitative PCR showed that DCS and CURS were expressed in both the leaf and rhizome (supplemental Fig. S3), consistent with our observation that the leaves of turmeric contain dihydrocurcuminoids probably synthesized via reduction of the double bond of curcuminoids.5 The expression levels of both enzyme genes were higher in the rhizome than in the leaf (supplemental Fig. S3), which is also in agreement with the amounts of curcuminoids; the total amount of curcuminoids is higher in the rhizome than in the leaf.
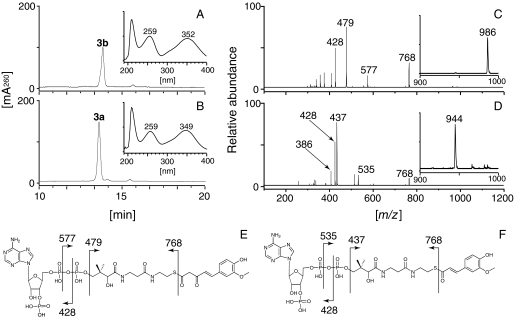
Characterization of DCS—DCS fused with a His tag at its N terminus was produced in E. coli BL21(DE3) using a pET system and purified on an nickel-nitrilotriacetic acid affinity column (supplemental Fig. S4). Recombinant DCS, giving a major protein band on SDS-PAGE, was incubated with cinnamoyl-CoA (1a), p-coumaroyl-CoA (2a), or feruloyl-CoA (3a) as starter substrates and malonyl-CoA as an extender substrate. The reaction was quenched by addition of HCl, and the solution was extracted with ethyl acetate. Liquid chromatography-atmospheric pressure chemical ionization mass spectrometry (LC-APCI MS) analysis of the organic layer revealed the presence of a trace amount of dehydrozingerone (3d) in the feruloyl-CoA-primed reaction (Fig. 2). 4-Hydroxybenzalacetone (2d) was also produced, when examined after scaling up the reaction (data not shown). These compounds were characterized by comparing their retention times, UV, MS, and MS/MS spectra with those of commercially available authentic samples. The low benzalacetone synthesis activity of DCS and a plausible curcumin biosynthesis route via diketide-CoA formation suggested that DCS acted as a diketide-CoA synthase to produce feruloyldiketide-CoA (3b), and the dehydrozingerone detected was a byproduct formed via artificial or nonenzymatic hydrolysis and subsequent decarboxylation of the diketide-CoA. Acidification of the solution might result in hydrolysis of a thioester bond. It is known that β-keto acids are easily decarboxylated at room temperature and converted to 2-ketones (23). In addition, some β-keto acids are decarboxylated rapidly under acidic conditions (24). To prove this prediction, we applied the reaction mixture directly to the LC-ESI MS without acidification or ethyl acetate extraction. As expected, no production of dehydrozingerone or β-keto acids was detected, and, instead, most of the feruloyl-CoA (3a) was consumed and a new compound whose molecular weight was [M + H]+ = 986 was observed (Fig. 3). This compound was predicted to be a diketide-CoA derived from feruloyl-CoA (3a), because it possessed a λmax near 260 nm and because its MS/MS spectra exhibited several fragments predicted to be derived from CoA fragmentation (Fig. 3). Furthermore, this compound was converted to trans-5-(4-hydroxy-3-methoxyphenyl)-3-oxopent-4-enoic acid (3c) by alkaline hydrolysis (supplemental Fig. S5). This compound was characterized by comparing its retention time, UV, MS, and MS/MS spectra with those of the synthetic sample. All of these findings suggested that DCS was a diketide-CoA synthase, producing feruloyldiketide-CoA (3b) from feruloyl-CoA (3a) and malonyl-CoA.
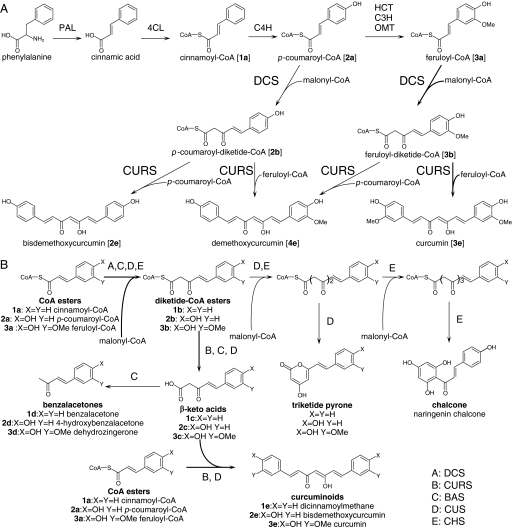
FIGURE 2.
Analysis of the ethyl acetate extracts of reactions containing DCS. 4-Hydroxybenzalacetone (2d) was not detected in the extract of the reaction containing p-coumaroyl-CoA (2a), malonyl-CoA, and DCS (A) but appeared in the extract after alkaline hydrolysis (C). Dehydrozingerone (3d) in a low amount was detected in the extract of the reaction containing feruloyl-CoA (3a), malonyl-CoA, and DCS (B). The yield of dehydrozingerone (3d) increased after alkaline hydrolysis of the reaction mixture containing feruloyl-CoA (3d), malonyl-CoA, and DCS (D). 4-Hydroxybenzalacetone (E) and dehydrozingerone (F) were identified by comparing their retention times, MS, MS/MS, and UV spectra with those of the authentic samples.
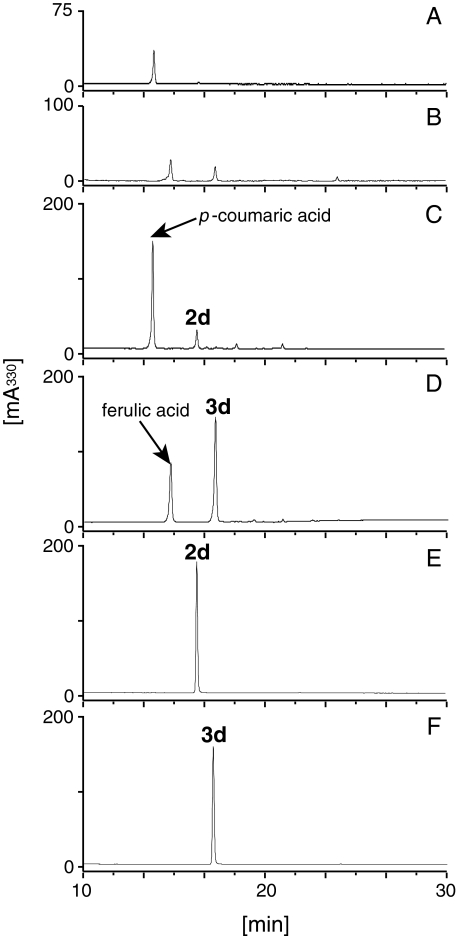
FIGURE 3.
HPLC analysis of the reaction products of DCS. Incubation of DCS in the presence of feruloyl-CoA (3a) and malonyl-CoA yielded feruloyldiketide-CoA (3b) (A), whereas the reaction with boiled DCS, as a negative control, yielded no products (B). The UV spectra of feruloyldiketide-CoA (3b) and feruloyl-CoA (3a) are shown in the insets in A and B, respectively. The comparison of MS/MS spectra of feruloyldiketide-CoA (3b) (C) and feruloyl-CoA (3a) (D) showed that the product was feruloyldiketide-CoA. The parent ion peaks of feruloyldiketide-CoA (3b) and feruloyl-CoA (3a) are shown in the insets in C and D, respectively. The predicted fragmentation patterns of feruloyldiketide-CoA (E) and feruloyl-CoA (F) are shown.
When the reaction mixture was acidified before application to LC-MS, a trace amount of dehydrozingerone (3d) was observed, probably due to hydrolysis induced by acidification of the solution (data not shown). In this analysis, production of β-keto acids would not be observed because β-keto acids are readily decarboxylated under acidic conditions; the acidification of the reaction mixture might convert most of the β-keto acids to dehydrozingerone (3d) (23, 24). Therefore, we concluded that the dehydrozingerone (3d) observed in the initial assay was formed by nonenzymatic hydrolysis and decarboxylation of feruloyldiketide-CoA. Consistent with this idea, the yield of dehydrozingerone (3d) increased dramatically when alkaline hydrolysis was performed before acidification and ethyl acetate extraction (Fig. 2, B and D).
When p-coumaroyl-CoA (2a) or cinnamoyl-CoA (1a) was added instead of feruloyl-CoA (3a), no diketide-CoA (1b or 2b) was detected, probably due to the low preference against these two CoA esters as a starter substrate of DCS. We next performed alkaline hydrolysis and ethyl acetate extraction of the reaction mixture, because benzalacetones (1d or 2d) could be detected more easily than diketide-CoAs. As a result, 4-hydroxybenzalacetone (2d) was detected, but benzalacetone (1d) was not (data not shown). From these results, we concluded that DCS preferred feruloyl-CoA as a starter substrate and could accept p-coumaroyl-CoA (2a) but at a lower efficiency. Cinnamoyl-CoA (1a) was not used as a starter substrate by DCS at any detectable level.
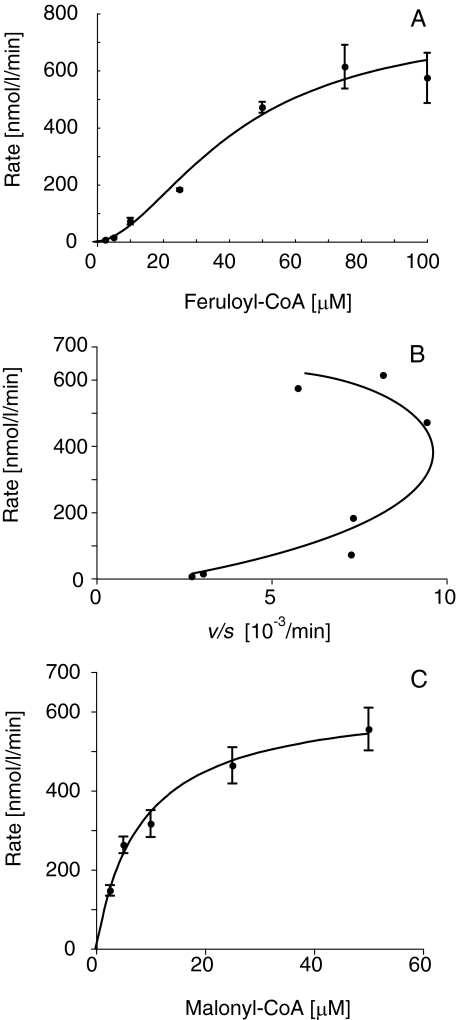
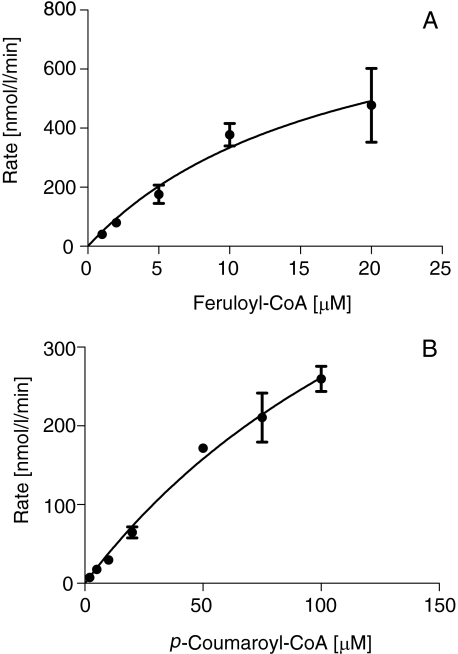
The temperature and pH optima of DCS were 25–35 °C and 6.5–7.5, respectively, when determined for the diketide-CoA formation. We then investigated the kinetic parameters of DCS under these conditions. We confirmed that the products formation was liner throughout the assay. An unexpected finding was that the rate-feruloyl-CoA concentration profile exhibited a sigmoidal curve, which is characteristic of allosteric enzymes and homotropic activation properties (Fig. 4A) (25, 26). Eadie-Hofstee plot also exhibited curves characteristic of allosteric enzymes (Fig. 4B). Therefore, we determined the kinetic parameters of DCS for feruloyl-CoA (3a) according to its allosteric properties. The kcat, S50, and Hill slope values were 1.2 ± 0.2 min–1, 46 ± 9 μm, and 1.8 ± 0.2, respectively. The kinetic parameters for p-coumaroyl-CoA or cinnamoyl-CoA could not be determined due to low activity. Instead, the rate-malonyl-CoA concentration profile exhibited a hyperbolic curve (Fig. 4C). The kcat and Km values of DCS for malonyl-CoA were 0.67 ± 0.1 min–1 and 8.4 ± 2.0 μm, respectively.
FIGURE 4.
Kinetic analysis of DCS. The rate-feruloyl-CoA concentration profile (A), Eadie-Hofstee plot (B), and the rate-malonyl-CoA concentration profile (C) are shown. The data, obtained from three independent experiments, suggest that DCS is an allosteric enzyme.
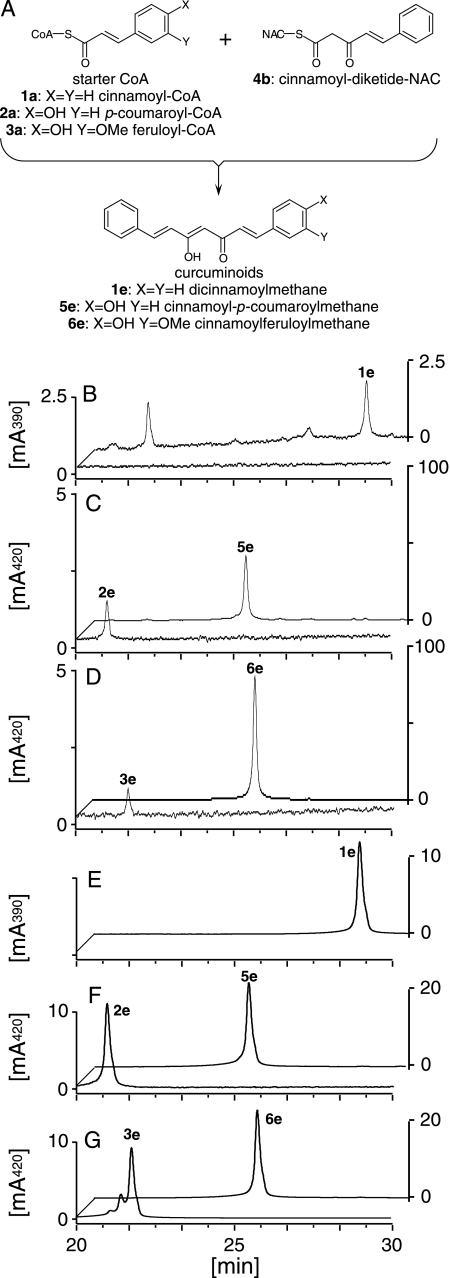
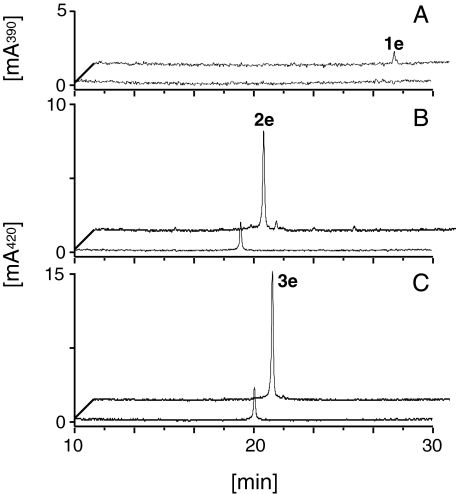
Characterization of CURS—CURS fused with a His tag at the N terminus was similarly produced in and purified from E. coli (supplemental Fig. S4). Incubation of the purified recombinant CURS, giving a major protein band on SDS-PAGE, with cinnamoyl-CoA (1a), p-coumaroyl-CoA (2a), or feruloyl-CoA (3a) as starter substrates and malonyl-CoA as an extender gave trace amounts of curcuminoids (2e and 3e) from p-coumaroyl-CoA (2a) and feruloyl-CoA (3a) (Fig. 5). These compounds were characterized by comparing their retention times, UV, MS, and MS/MS spectra with those of commercially available authentic samples (Fig. 5). Triketide pyrones or benzalacetones were not produced. The diketide-CoA-forming activity of DCS prompted us to examine the reaction of CURS in the presence of diketide-CoAs as a probable extender. We synthesized cinnamoyldiketide-N-acetylcysteamine (NAC) (4b), which is a mimic of diketide-CoAs (Fig. 5). We chose this analog derived from cinnamoyl-CoA (1a) because of the ease of chemical synthesis. As expected, incubation of starter substrates (1a, 2a, and 3a) with cinnamoyl-diketide-NAC (4b) yielded curcuminoids (1e, 5e, and 6e) (Fig. 5). These compounds were characterized by comparing their retention times, UV, MS, and MS/MS spectra with those of the synthetic samples (Fig. 5). The reaction containing feruloyl-CoA (3a) exhibited the highest activity, showing the preference of CURS for feruloyl-CoA (3a) as a starter substrate. Incubation of CURS with feruloyl-CoA (3a) and cinnamoyl-diketide-NAC (4b) produced ∼100-fold more curcuminoids over that with feruloyl-CoA and malonyl-CoA. This is in vivid contrast to the curcuminoid synthesis by CUS from O. sativa; the yields of curcuminoids produced from p-coumaroyl-CoA with either malonyl-CoA or cinnamoyldiketide-NAC (4e) by CUS were similar.5 In addition, only a trace amount of feruloyldiketide-CoA (3b) was detected in the reaction mixture containing CURS, malonyl-CoA, and feruloyl-CoA, which suggested that the malonyl-CoA condensation activity of CURS is much lower than DCS. We therefore assumed that CURS catalyzed formation of curcuminoids from starter substrates and diketide-CoAs. CURS produced no curcuminoids when incubated with feruloyl-CoA and dehydrozingerone (3d). Instead, the reaction with feruloyl-CoA and trans-5-phenyl-3-oxopent-4-enoic acid (1c) yielded a curcuminoid (5f) (data not shown). These results suggested that CURS condensed diketide-CoAs with a starter substrate in a manner similar to CUS from O. sativa.
FIGURE 5.
HPLC analysis of the reaction products of CURS. The scheme of the CURS reaction starting with CoA esters and cinnamoyl-diketide-NAC (4b) (A) is illustrated. In chromatograms B–D, the front chromatograms show the CURS reactions beginning with a starter substrate and malonyl-CoA and the back chromatograms show the CURS reaction beginning with a starter substrate and cinnamoyl-diketide-NAC (4b). Incubation of CURS in the presence of p-coumaroyl-CoA (2a) (C, front) and feruloyl-CoA (3a) (D, front), in addition to malonyl-CoA, yielded bisdemethoxycurcumin (2e) and curcumin (3e), respectively. No products were detected in the reactions starting with cinnamoyl-CoA (1a) and malonyl-CoA (B, front). When cinnamoyl-CoA (1a) (B, back), p-coumaroyl-CoA (2a) (C, back), or feruloyl-CoA (3a) (D, back) were added to a reaction containing cinnamoyl-diketide-NAC (4b), dicinnamoylmethane (1e), cinnamoyl-p-coumaroylmethane (5e), or cinnamoylferuloylmethane (6e), respectively, were produced. The dicinnamoylmethane (E, back), bisdemethoxycurcumin (F, front), cinnamoyl-p-coumaroylmethane (F, back), curcumin (G, front), and cinnamoylferuloylmethane (G, back) were identified by comparing their retention times, MS, MS/MS, and UV spectra with those of the authentic or synthetic samples.
The optimum pH and temperature of the CURS reaction for production of cinnamoylferuloylmethane (6e) were 9 and 50 °C, respectively, both of which are similar to CUS. We calculated the kinetic parameters by monitoring the production rates of curcuminoid (5e or 6e) from starter substrates (2a or 3a) and cinnamoyl-diketide-NAC (4b). In contrast to DCS, the rate-substrate concentration profile exhibited a hyperbolic curve, which is a feature of the Michaelis-Menten model (Fig. 6). The kcat and Km values for cinnamoylferuloylmethane (6e) formation from feruloyl-CoA (3a) were 1.1 ± 0.4 min–1 and 18 ± 5 μm, respectively. The kcat and Km values for cinnamoyl-p-coumaroylmethane (5e) formation from p-coumaroyl-CoA (2a) were 0.85 ± 0.04 min–1 and 189 ± 6 μm, respectively. We could not determine the kinetic parameters for dicinnamoylmethane (1e) formation due to the low activity toward cinnamoyl-CoA (1a). Thus, CURS preferred feruloyl-CoA (3a) as a starter substrate.
FIGURE 6.
Kinetic analysis of CURS. The rate-feruloyl-CoA concentration profile (A) for cinnamoylferuloylmethane (6e) synthesis from feruloyl-CoA (3a) and cinnamoyldiketide-NAC (4b) are shown. Similarly, the rate-p-coumaroyl-CoA concentration profile (B) for cinnamoyl-p-coumaroylmethane (5e) synthesis from p-coumaroyl-CoA (2a) and cinnamoyldiketide-NAC (4b) are shown. The data were obtained from three independent experiments.
Production of Curcuminoids by Co-incubation of CURS and DCS—To prove our working hypothesis that CURS produces curcuminoids by condensing the diketide-CoAs produced from starter substrates by the action of DCS, we co-incubated CURS and DCS in the presence of feruloyl-CoA (3a) and malonyl-CoA and measured the yield of curcumin, compared with a reference experiment in which CURS alone was used. As a result, the reaction mixture containing the two enzymes produced 1300 ± 230 nmol/liter/h (n = 3) of curcumin (3e), whereas the control reaction containing only CURS produced 85 ± 8 nmol/liter/h (n = 3) (Fig. 7). A trace amount of dehydrozingerone (3d) was simultaneously detected (data not shown), which was probably derived from nonenzymatic hydrolysis of feruloyldiketide-CoA (3b) and decarboxylation of the β-keto acid in the reaction containing the two enzymes. The yields of bisdemethoxycurcumin (2e) and dicinnamoylmethane (1e) were also increased by co-incubating the enzymes with the corresponding starter substrates (Fig. 7). These observations clearly showed that CURS synthesized curcuminoids in collaboration with DCS. When two starter substrates, feruloyl-CoA (3a) and p-coumaroyl-CoA (2a), were present, demethoxycurcumin (4e) and a trace amount of bisdemethoxycurcumin (2e) were also produced, in addition to curcumin (3e) (supplemental Fig. S6A). However, the product profile of the curcuminoids of the in vitro reaction was different from that of an ethyl acetate extract of the rhizome of turmeric (supplemental Fig. S6B). The observed difference probably resulted from the difference of the abundance of two starter substrates; the concentrations of p-coumaroyl-CoA and feruloyl-CoA are not equal in vivo.
FIGURE 7.
HPLC analysis of the reaction products of DCS/CURS co-incubation. The front chromatograms show control reactions containing only CURS, and the back chromatograms indicate the reactions containing both DCS and CURS. Co-incubation of DCS and CURS in the presence of cinnamoyl-CoA (1a) and malonyl-CoA yielded a trace amount of dicinnamoylmethane (1e), whereas the control reaction containing only CURS did not (A). Co-incubation of DCS/CURS in the presence of p-coumaroyl-CoA (2a) (B) or feruloyl-CoA (3a) (C), in addition to malonyl-CoA, yielded a larger amount of bisdemethoxycurcumin (2e) or curcumin (3e), respectively, than the control incubation of only CURS (B and C).
DISCUSSION
In vitro analysis of the two type III PKSs, DCS and CURS, isolated from the herb C. longa revealed that DCS catalyzed the formation of feruloyldiketide-CoA (3b) from feruloyl-CoA (3a) and malonyl-CoA, and CURS catalyzed the formation of curcuminoids from feruloyl-CoA (3a) and cinnamoyl-diketide-NAC (4b). CURS also catalyzed the formation of curcumin from feruloyl-CoA (3a) and malonyl-CoA, but at a low rate. Co-incubation with DCS accelerated the curcumin synthetic rate catalyzed by CURS ∼15-fold by supplying diketide-CoA as an extender. These results implied that curcumin (3e) is synthesized by these two type III PKSs in turmeric. In addition, both enzymes preferred feruloyl-CoA (3a) as a starter substrate, suggesting that curcumin (3e) is synthesized via feruloyl-CoA (3a). Therefore, as shown in Fig. 1A, the hydroxylation and methylation at the 3′ position occurs before the polyketide formation during curcumin (3e) biosynthesis. DCS and CURS also accept p-coumaroyl-CoA (2a) as a starter substrate. When p-coumaroyl-CoA (1a) and feruloyl-CoA (3a) were simultaneously used for the reaction, demethoxycurcumin (4e) and bisdemethoxycurcumin (2e) were produced in addition to curcumin (3e). These results indicate that demethoxycurcumin (4e) and bisdemethoxycurcumin (2e) are also synthesized by these two enzymes. Dihydrocurcuminoids detected in the leaf of C. longa are presumably synthesized by these type III PKSs.
The kinetic analysis of DCS suggested that DCS is an allosteric enzyme. Type III PKSs are homodimers of ketosynthase; therefore, one dimer of type III PKSs possesses two active sites (8). The binding of a starter substrate to the active site in one subunit may affect the structure of the active site in the other subunit during the DCS reaction. The homotropic activation might be important for the synthesis of curcumin in turmeric. DCS and CURS use the same starter substrate, but CURS uses the feruloyldiketide-CoA (3b) synthesized and supplied by DCS. Therefore, the initial stage of reaction is catalyzed by DCS. In enzymes such as DCS, homotropic activation decreases their activity as the substrate concentration becomes lower more quickly than those exhibiting Michaelis-Menten-type kinetics. In curcumin synthetic reactions, the DCS reaction precedes the CURS reaction. Therefore, feruloyl-CoA must not be fully consumed by DCS. The homotropic activation property of DCS will leave feruloyl-CoA to be consumed by CURS, whose activity increases after diketide-CoAs are synthesized by DCS. Although continuous synthesis of the starter substrate may make this property unnecessary, this property probably possesses some role in the curcuminoid biosynthesis. For a full understanding of the allosteric properties of DCS, further study, including x-ray crystallography, is necessary.
There are several type III PKSs that catalyze a single condensation of an extender substrate (15, 27, 28). Because PstrCHS2 from Pinus strobus and germicidin synthase from Streptomyces coelicolor A3(2) catalyze formation of a triketide from an diketide intermediate and an extender substrate, they are obviously different from DCS. Benzalacetone synthase catalyzes a single condensation of malonyl-CoA with p-coumaroyl-CoA, followed by hydrolysis of the thioester bond and decarboxylation catalyzed by the enzyme. Because DCS releases the diketide intermediate without cleaving the thioester bond, it is also distinct from benzalacetone synthase. DCS is a unique type III PKS that releases the product as a CoA-bound form. There is only an example of type III PKSs thus far reported that releases the product as a CoA ester (29, 30).
Although CUS itself from O. sativa synthesizes curcuminoids in vitro from malonyl-CoA via diketide intermediate formation (16), in turmeric DCS and CURS are allotted respective steps in curcuminoid synthesis. CUS synthesizes curcuminoids accompanied by triketide pyrone formation. On the other hand, the curcumin synthesis catalyzed by CURS and DCS is accompanied by the formation of only a trace amount of dehydrozingerone (3d) as a by-product, which is derived by nonenzymatic hydrolysis and decarboxylation of feruloyldiketide-CoA (3b). Therefore, the DCS/CURS-coupled reaction synthesizes only curcumin and its intermediate, feruloyldiketide-CoA. The curcumin synthetic system in turmeric avoids production of a triketide pyrone as a by-product by dividing the pathway into two parts: one including DCS and the other including CURS. The substrate specificity of CUS and CURS/DCS toward starter substrates was also different. CUS prefers p-coumaroyl-CoA (2a) as a starter substrate, whereas both CURS and DCS prefer feruloyl-CoA (3a).
Some type III PKSs, which appear to possess activity similar to DCS and CURS, have been reported in other plants. WtPKS1 and WtPKS2 (DNA data base accession number AAW50922) in W. thyrsiflora accumulate phenylphenalenone via curcuminoid formation (14, 31–34). WtPKS1 shows 63% identity in amino acid sequence to DCS, and WtPKS2 possesses 62% identity to CURS. These two enzymes may synthesize a curcuminoid scaffold in the same manner as the pairing of DCS and CURS. In vitro analysis by Brand et al. (14) revealed that WtPKS1 catalyzes formation of benzalacetones and triketide pyrones. Our present study with DCS and CURS prompts us to assume that WtPKS1 may produce a curcuminoid scaffold when incubated with WtPKS2. We suppose that a curcuminoid scaffold is synthesized by an enzyme system similar to a pairing of DCS and CURS in various plants, including ginger, which synthesizes diarylheptanoids and phenylphenalenones (31–34).
Supplementary Material
This work was supported in part by the Research Grant Program 2005 of the New Energy and Industrial Technology Development Organization of Japan and a Grant-in-Aid for Scientific Research on Priority Area “Applied Genomics” from Monkasho.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
Footnotes
The abbreviations used are: PKS, polyketide synthase; CHS, chalcone synthase; CUS, curcuminoid synthase; HPLC, high-performance liquid chromatography; NAC, N-acetylcysteamine; CURS, curcumin synthase; DCS, diketide-CoA synthase; LC-ESI MS, liquid chromatography-electro spray ionization mass spectrometry; APCI, atmospheric pressure chemical ionization; MS/MS, tandem mass spectrometry.
Y. Katsuyama, T. Kita, N. Funa, and S. Horinouchi, unpublished data.
References
- 1.Sharma, R. A., Gescher, A. J., and Steward, W. P. (2005) Eur. J. Cancer 41 1955–1968 [DOI] [PubMed] [Google Scholar]
- 2.Tayyem, R. F., Heath, D. D., Al-Delaimy, W. K., and Rock, C. L. (2006) Nutr. Cancer 55 126–131 [DOI] [PubMed] [Google Scholar]
- 3.Ringman, J. M., Frautschy, S. A., Cole, G. M., Masterman, D. L., and Cummings, J. L. (2005) Curr. Alzheimer Res. 2 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber, W. M., Hunsaker, L. A., Abcouwer, S. F., Deck, L. M., and Vander Jagt, D. L. (2005) Bioorg. Med. Chem. 13 3811–3820 [DOI] [PubMed] [Google Scholar]
- 5.Cao, H., Sasaki, Y., Fushimi, H., and Komatsu, K. (2001) Biol. Pharm. Bull. 24 1389–1394 [DOI] [PubMed] [Google Scholar]
- 6.Maheshwari, R. K., Singh, A. K., Gaddipati, J., and Srimal, R. C. (2006) Life Sci. 78 2081–2087 [DOI] [PubMed] [Google Scholar]
- 7.Hiserodt, R., Hartman, T. G., Ho, C.-T., and Rosen, R. T. (1996) J. Chromatogr. A 740 51–63 [Google Scholar]
- 8.Austin, M. B., and Noel, J. P. (2003) Nat. Prod. Rep. 20 79–110 [DOI] [PubMed] [Google Scholar]
- 9.Jez, J. M., Ferrer, J.-L., Bowman, M. E., Dixon, R. A., and Noel, J. P. (2000) Biochemistry 39 890–902 [DOI] [PubMed] [Google Scholar]
- 10.Ferrer, J. L., Jez, J. M., Bowman, M. E., Dixon, R. A., and Noel, J. P. (1999) Nat. Struct. Biol. 6 775–784 [DOI] [PubMed] [Google Scholar]
- 11.Roughley, P. J., and Whiting, D. A. (1973) J. Chem. Soc. Perkin Trans. I 1973 2379–2388 [Google Scholar]
- 12.Schröder, J. (1997) Trends Plant Sci. 2 373–378 [Google Scholar]
- 13.Kita, T., Imai, S., Sawada, H., Kumagai, H., and Seto, H. (2008) Biosci. Biotechnol. Biochem. 72 1789–1798 [DOI] [PubMed] [Google Scholar]
- 14.Brand, S., Holscher, D., Schierhorn, A., Svatos, A., Schröder, J., and Schneider, B. (2006) Planta 224 413–428 [DOI] [PubMed] [Google Scholar]
- 15.Abe, I., Takahashi, Y., Morita, H., and Noguchi, H. (2001) Eur. J. Biochem. 268 3354–3359 [DOI] [PubMed] [Google Scholar]
- 16.Ramirez-Ahumada, M. d. C., Timmermann, B. N., and Gang, D. R. (2006) Phytochemistry 67 2017–2029 [DOI] [PubMed] [Google Scholar]
- 17.Katsuyama, Y., Matsuzawa, M., Funa, N., and Horinouchi, S. (2007) J. Biol. Chem. 282 37702–37709 [DOI] [PubMed] [Google Scholar]
- 18.Blecher, M. (1981) Methods Enzymol. 72 404–408 [DOI] [PubMed] [Google Scholar]
- 19.Gilbert, I. H., Ginty, M., O'Neill, J. A., Simpson, T. J., Staunton, J., and Willis, C. L. (1995) Bioorg. Med. Chem. Lett. 5 1587–1590 [Google Scholar]
- 20.Matsumura, S., Asai, N., and Yoneda, S. (1985) Bull. Chem. Soc. Jpn. 3 310–360 [Google Scholar]
- 21.Katsuyama, Y., Matsuzawa, M., Funa, N., and Horinouchi, S. (2008) Microbiology 154 2620–2628 [DOI] [PubMed] [Google Scholar]
- 22.Holtz, E., Alberecht, U., and Langer, P. (2007) Tetrahedron 63 3293–3301 [Google Scholar]
- 23.Guthrie, J. P. (2002) Bioorg. Chem. 30 32–52 [DOI] [PubMed] [Google Scholar]
- 24.Marcus, J. P., and Dekker, E. E. (1993) Biochem. Biophys. Res. Commun. 190 1066–1072 [DOI] [PubMed] [Google Scholar]
- 25.Hammes, G. G., and Wu, C. W. (1974) Annu. Rev. Biophys. Bioeng. 3 1–33 [DOI] [PubMed] [Google Scholar]
- 26.Houston, J. B., and Galetin, A. (2005) Arch. Biochem. Biophys. 433 351–360 [DOI] [PubMed] [Google Scholar]
- 27.Schröder, J., Raiber, S., Berger, T., Schmidt, A., Schmidt, J., Soares-Sello, A. M., Bradshiri, E., Strack, D., Simpson, T. J., Veit, M., and Schröder, G. (1998) Biochemistry 37 8417–8425 [DOI] [PubMed] [Google Scholar]
- 28.Song, L., Barona-Gomez, F., Corre, C., Xiang, L., Udwary, D. W., Austin, M. B., Noel, J. P., Moore, B. S., and Challis, G. L. (2006) J. Am. Chem. Soc. 128 14754–14755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeifer, V., Nicholson, G. J., Ries, J., Recktenwald, J., Schefer, A. B., Shawky, R. M., Schröder, J., Wohlleben, W., and Pelzer, S. (2001) J. Biol. Chem. 276 38370–38377 [DOI] [PubMed] [Google Scholar]
- 30.Chen, H., Tseng, C. C., Hubbard, B. K., and Walsh, C. T. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14901–14906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamo, T., Hirai, N., Tsuda, M., Fujioka, D., and Ohigashi, H. (2000) Biosci. Biotechnol. Biochem. 64 2089–2098 [DOI] [PubMed] [Google Scholar]
- 32.Schmitt, B., Holscher, D., and Schneider, B. (2000) Phytochemistry 53 331–337 [DOI] [PubMed] [Google Scholar]
- 33.Holscher, D., and Schneider, B. (1995) J. Chem. Soc. Chem. Commun. 1995 525–526 [Google Scholar]
- 34.Denniff, P., Macleod, I., and Whiting, D. A. (1980) J. Chem. Soc. Perkin Trans. I 1980 2637–2644 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.