Abstract
ABCA1 plays a major role in cholesterol homeostasis and high density lipoprotein (HDL) metabolism. ABCA1 contains disulfide bond(s) between its N- and C-terminal halves, but it remains unclear whether disulfide bond formation is important for the functions of ABCA1 and which cysteines are involved in disulfide bond formation. To answer these questions, we constructed >30 ABCA1 mutants in which 16 extracellular domain (ECD) cysteines were replaced with serines and examined disulfide bond formation, apoA-I binding, and HDL formation in these mutants. From the single cysteine replacements, two cysteines (Cys75 and Cys309) in ECD1 were found to be essential for apoA-I binding. In contrast, in ECD2, only Cys1477 was found to be essential for HDL formation, and no single cysteine replacement impaired apoA-I binding. The concurrent replacement of two cysteines, Cys1463 and Cys1465, impaired apoA-I binding and HDL formation, suggesting that four of five extracellular cysteines (Cys75, Cys309, Cys1463, Cys1465, and Cys1477) are involved in these functions of ABCA1. Trypsin digestion experiments suggested that one disulfide bond is not sufficient and that two intramolecular disulfide bonds (between Cys75 and Cys309 in ECD1 and either Cys1463 or Cys1465 and Cys1477 in ECD2) are required for ABCA1 to be fully functional.
Maintenance of cellular cholesterol homeostasis is important for normal human physiology; its disruption can lead to a variety of pathological conditions, including cardiovascular disease (1). ABCA1 (ATP-binding cassette protein A1), a key factor in cholesterol homeostasis, mediates the secretion of cellular free cholesterol and phospholipids to an extracellular acceptor, apoA-I, to form high density lipoprotein (HDL)2 (2, 3). HDL formation is the only known pathway for the elimination of excess cholesterol from peripheral cells. Defects in ABCA1 cause Tangier disease (4–6), in which patients have a near absence of circulating HDL, prominent cholesterol ester accumulation in tissue macrophages, and premature atherosclerotic vascular disease (1, 7).
ABCA1 is a member of the ABCA subclass of ABC transporters, which contain the basic architecture of the “full-length” ABC transporters organized into two tandemly arranged halves. Each half contains several transmembrane α-helices (TMs), which provide a translocation pathway, followed by a cytoplasmic nucleotide-binding domain, which hydrolyze ATP. In the case of “half-size” ABC transporters, such as ABCG1, ABCD1, TAP1/TAP2 (transporter associated with antigen processing), and the bacterial homolog Sav1866, they dimerize to form the full transporter. Crystallographic analysis of the bacterial homolog Sav1866 revealed that the TMs of one subunit are closely related to the TMs of the other subunit, forming two “wings” in an outward-facing conformation (8).
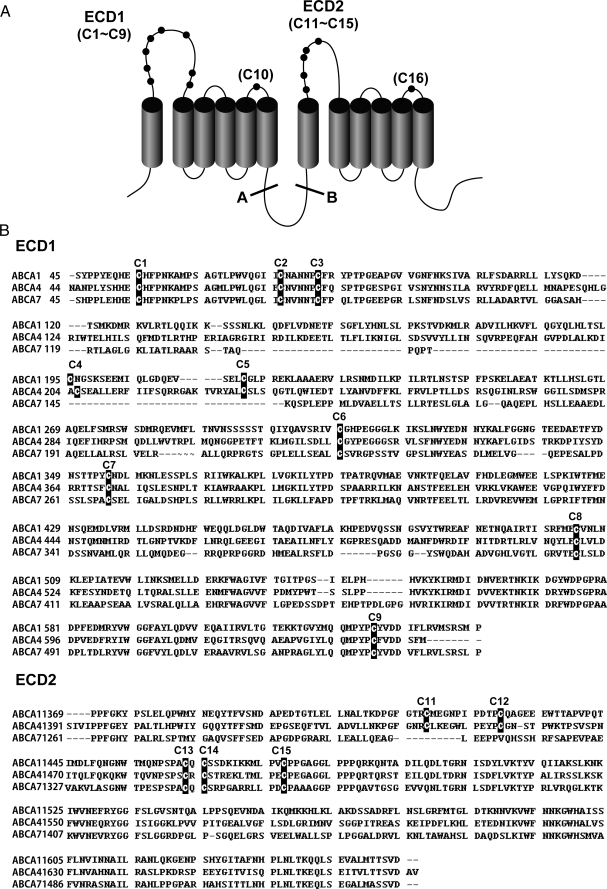
When ABCA1 is partially digested by trypsin, ABCA1 is cleaved at site A, just C-terminal to TM6, and at site B, just N-terminal to TM7, to produce four fragments of 170 and 150 kDa and subsequently of 125 and 110 kDa (Fig. 1A) (9). When these fragments are analyzed by SDS-PAGE under nonreducing conditions, they co-migrate with undigested ABCA1. These results suggest that the N- and C-terminal halves of ABCA1 are connected by disulfide bond(s), as reported for ABCA4 (ABCR) (10). The ABCA subclass is distinguished from other ABC transporter subclasses by the presence of large extracellular domains (ECDs) (Fig. 1A) (11, 12). ECD1 and ECD2 of ABCA1 contain nine and five cysteine residues, respectively, and each connecting loop between TM5 and TM6 and between TM11 and TM12 contains a cysteine residue. These cysteine residues were assigned numbers, C1 to C16, based on their distance from the N terminus (Fig. 1A). These cysteine residues are well conserved among ABCA1, ABCA4, and ABCA7 (Fig. 1B). All of the cysteine residues in ECD1 are conserved between ABCA1 and ABCA4, and seven cysteine residues (except C4 and C5) are conserved in ABCA7. All five of the cysteine residues in ECD2 are conserved between ABCA1 and ABCA4, and three cysteine residues (except C11 and C12) are conserved in ABCA7. Because ABCA7, like ABCA1, mediates apoA-I-dependent lipid efflux (13, 14), conserved cysteine residues might be important for its function. Indeed, the Tangier disease mutation C1477R has been reported to abolish apoA-I binding and HDL formation (15–17), and several missense mutations in cysteine residues within ECD1 (C54Y, C75G) and ECD2 (C1488R, C1490Y) of ABCA4 have been linked to Stargardt disease (18–21). It remains unclear, however, whether disulfide bond formation is important for the proper folding and/or the functions of ABCA subclass proteins.
FIGURE 1.
Structural features of ABCA1. A, topological model for human ABCA1. ABCA1 consists of 12 transmembrane α-helices (TM1–TM12) and two large ECDs. ECD1 and ECD2 contain nine and five cysteine residues, respectively, and each connecting loop between TM5 and TM6 and between TM11 and TM12 contains a cysteine residue. These cysteine residues were assigned numbers, from C1 to C16, based on their distance from the N terminus. ABCA1 is cleaved at sites A and B by limited trypsin digestion. B, amino acid sequence alignment of ECDs of human ABCA1, ABCA4, and ABCA7. Conserved cysteine residues are indicated in black boxes.
In this study, we analyzed which cysteine residues are involved in disulfide bond formation and examined whether disulfide bond formation is necessary for the functions of ABCA1. Cysteine substitution experiments suggested that two disulfide bonds are formed between C2 and C6 in ECD1 and between either C13 or C14 and C15 in ECD2 and that this two-disulfide bond formation is necessary for apoA-I-dependent cholesterol efflux by ABCA1.
EXPERIMENTAL PROCEDURES
Materials—N-Ethylmaleimide was purchased from Nacalai Tesque; dithiothreitol (DTT) was from Wako Pure Chemical Industries. Collagen solution and trypsin (tosylphenylalanyl chloromethyl ketone-treated, from bovine pancreas) were from Sigma.
Antibodies—Rat anti-human ABCA1 monoclonal antibody KM3073 was generated against the first ECD (amino acids 45–639) of human ABCA1 as described previously (3).
Cell Culture and Transfection—HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. Cells in 100-mm culture dishes were transfected with 8 μg of DNA using Lipofectamine 2000 (Invitrogen). Cells in 12-well culture plates were transfected with 3 μg of DNA using calcium phosphate buffer (2.5 m CaCl2, 1 mm Tris-HCl, 0.1 mm EDTA, pH 7.5, and 2× BES (280 mm NaCl, 50 mm BES, and 1.5 mm Na2HPO4, pH 6.95)).
ApoA-I Binding Assay—HEK293 cells were subcultured on collagen-coated glass coverslips in 12-well dishes at a density of 2 × 104 cells in DMEM containing 10% fetal bovine serum. After 18 h of incubation, cells were transfected with green fluorescent protein (GFP)-tagged ABCA1 using calcium phosphate buffer. Cells were washed with DMEM 28 h after transfection and then incubated with DMEM containing 5 μg/ml Alexa 546-labeled apoA-I as described previously (22).
Constructions of Mutants—Site-directed mutations were introduced into ABCA1; in individual mutants, each cysteine residue was replaced with serine. Mutations were generated using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) with the appropriate synthetic oligonucleotides (supplemental Table I). The integrity of the mutated DNA was confirmed by sequencing the entire DNA.
Biotinylation of Cell-surface Proteins—HEK293 cells were transfected with GFP-tagged ABCA1. 28 h after transfection, cells were washed with ice-cold phosphate-buffered saline containing 0.1 g/liter CaCl2 and MgCl2·6H2O (PBS+) and incubated with 0.5 mg/ml sulfo-NHS-biotin solubilized in PBS+ for 30 min on ice in the dark. Cells were washed with PBS+ to remove unbound sulfo-NHS-biotin and lysed in radioimmune precipitation assay buffer (20 mm Tris-Cl, pH 7.5, 1% Triton X-100, 0.1% SDS, and 1% sodium deoxycholate) containing protease inhibitors (100 μg/ml (p-amidinophenyl)methanesulfonyl fluoride, 2 μg/ml leupeptin, and 2 μg/ml aprotinin). ImmunoPure immobilized monomeric avidin gel (Pierce) was added to the cell lysate to precipitate the biotinylated proteins. The biotinylated proteins were electrophoresed on a 7% SDS-polyacrylamide gel and detected by immunoblotting (23).
Limited Trypsin Digestion of ABCA1—HEK293 cells were transfected with GFP-tagged ABCA1. 24 h after transfection, cells were lysed with suspension buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 250 mm sucrose) containing 1% n-dodecyl-β-d-maltoside. Protein (20 μg) was digested by 5 μg/ml trypsin on ice for 15 or 40 min. The digested proteins were denatured with or without DTT, electrophoresed on a 5% PAGEL (ATTO), and detected by immunoblotting. Two fragments of ABCA1, 170 and 150 kDa, were recognized by anti-ABCA1 antibody KM3073.
Cellular Lipid Release Assay—HEK293 cells subcultured on 100-mm dishes were transfected with GFP-tagged ABCA1 using Lipofectamine 2000. 12 h after transfection, cells were trypsinized and subcultured in 6-well dishes at a density of 6 × 105 cells in DMEM with 10% fetal bovine serum. After 12 h of incubation, the culture medium was removed, and cells were washed with DMEM containing 0.02% bovine serum albumin. Next, cells were incubated with DMEM containing 0.02% bovine serum albumin and 5 μg/ml apoA-I for 24 h at 37 °C. The lipid content of the medium was determined as described previously (23).
Fluorescence Microscopy—Cells were cultured on glass coverslips and fixed with 3.7% paraformaldehyde in PBS+ for 30 min in the dark at room temperature. Cells were viewed using a Zeiss confocal microscope (LSM 5 Pascal).
RESULTS
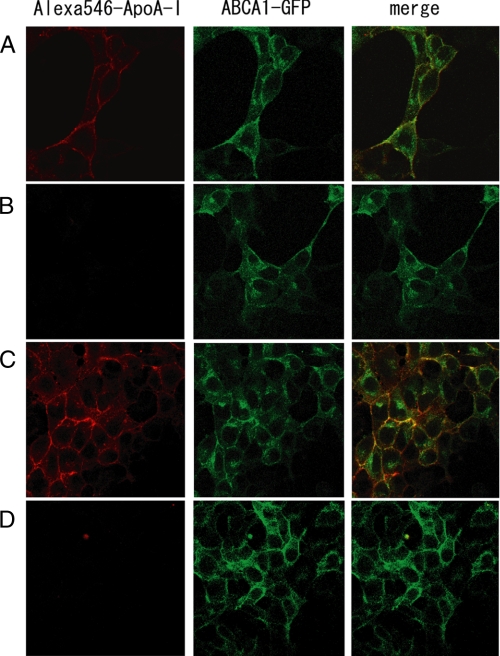
Effect of DTT Treatment of Cells on ApoA-I Binding—ABCA1 contains disulfide bond(s) between the N- and C-terminal halves, but it is unclear whether disulfide bond formation is important for the functions of ABCA1. To explore the roles of disulfide bonding in ABCA1, we first examined the effects of DTT treatment of cells on apoA-I binding. When Alexa 546-conjugated apoA-I was added to cells expressing ABCA1-GFP, apoA-I binding to the cell surface was clearly observed after 15 min (Fig. 2A). ApoA-I colocalized with ABCA1 and did not bind to untransfected HEK293 host cells (supplemental Fig. 2), suggesting that apoA-I binding is ABCA1-dependent. When 2 mm DTT was subsequently added to the medium and cells were incubated for 5 min in the presence of DTT, apoA-I bound to the cell surface dissociated, and no apoA-I binding was detected (Fig. 2B). Next, cells were washed with fresh medium, and Alexa 546-conjugated apoA-I was added again (Fig. 2C); apoA-I bound to cells just as it had before the DTT treatment, indicating that the inhibitory effect of DTT on apoA-I binding is reversible. When cells were treated with DTT and subsequently with 3 mm N-ethylmaleimide, which covalently modifies free thiol groups, apoA-I did not bind to cells even after washing with fresh medium (Fig. 2D), although subcellular localization of ABCA1 was not much affected. ApoA-I treated with DTT and N-ethylmaleimide bound to cells expressing ABCA1-GFP (data not shown). These results suggest that disulfide bonding is important for ABCA1-dependent apoA-I binding to cells.
FIGURE 2.
Effect of DTT treatment of the cell surface on apoA-I binding. A, HEK293 cells stably expressing ABCA1-GFP were incubated with Alexa 546-conjugated ApoA-I for 15 min at 37 °C. B, 2 mm DTT was added to the medium, and cells were incubated for 5 min. C, Alexa 546-conjugated apoA-I was added again after washing cells with fresh medium, followed by incubation for 15 min at 37 °C. D, 3 mm N-ethylmaleimide was added to the medium after DTT treatment, and then Alexa 546-conjugated apoA-I was added again after washing cells with fresh medium, followed by incubation for 15 min at 37 °C.
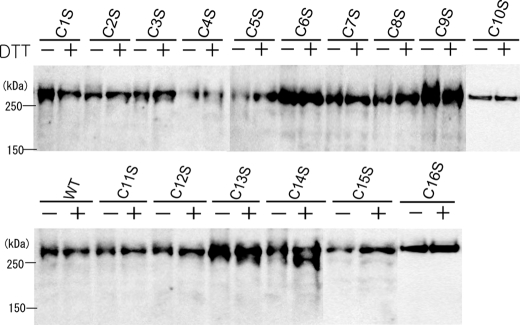
Construction and Expression of Cysteine Mutants—To investigate the role of ECD cysteine residues in the structure and functions of ABCA1, each of the 16 extracellular cysteine residues was replaced with serine using site-directed mutagenesis. Each mutant was transiently expressed in HEK293 cells as a GFP fusion protein; expression was validated by immunoblotting. Wild-type ABCA1 and all 16 single cysteine mutants yielded a product with an apparent mass of 280 kDa as the major product, although some mutants, such as C13S and C14S also yielded significant levels of a 250-kDa product, which we speculate contains an immature oligosaccharide. Although each mutant showed considerable variation in expression (Fig. 3), the variation was not due to the mutation but due to the transient expression because C4S and C5S showed higher expression than the wild type in separate experiments (data not shown). The electrophoretic mobility of wild-type and mutant ABCA1 did not differ between reducing and nonreducing conditions. Subcellular localization of most mutants was similar to that of wild-type ABCA1-GFP. Some mutants, however, such as C13S and C14S, showed significantly higher accumulation in intracellular compartments (supplemental Fig. 1).
FIGURE 3.
Western blot analysis of single cysteine mutants of ABCA1 under reducing and nonreducing conditions. Each mutant fused with GFP was transiently expressed in HEK293 cells. Cells were lysed 28 h after transfection, and expressed proteins were denatured under reducing (+DTT) or nonreducing (-DTT) conditions. Western blotting was performed with anti-ABCA1 antibody KM3073. We confirmed in three separate experiments that the variation of expression was not due to the mutation but due to the transient expression. WT, wild type.
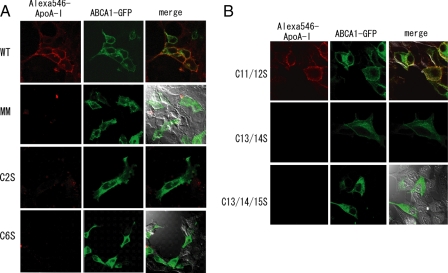
Effect of Mutations on ApoA-I Binding—ApoA-I binding to cells expressing the wild type or any one of the 16 single cysteine mutants was examined. ApoA-I bound to cells expressing wild-type ABCA1 but not to cells expressing ABCA1-MM (ABCA1(K939M/K1952M)), a mutant in which the Walker A lysines in both nucleotide-binding domains are replaced by methionines, although the expression level (data not shown) and surface expression (Fig. 4A) of the mutant were comparable with those of the wild-type protein, as reported (24). ApoA-I bound to cells expressing almost all of the cysteine mutants as efficiently as to cells expressing the wild type (Fig. 4A, Table 1, and supplemental Fig. 2). However, apoA-I did not bind to cells expressing mutant C2S or C6S. These results suggested that only two cysteine residues in ECD1 (C2 and C6) are essential for apoA-I binding.
FIGURE 4.
Effect of cysteine mutations on apoA-I binding. Single cysteine mutants (A) or double or triple cysteine mutants (B) were transiently expressed as GFP fusion proteins in HEK293 cells. Cells were incubated, 28 h after transfection, with Alexa 546-conjugated apoA-I for 15 min. WT, wild type.
TABLE 1.
Cysteine mutants of ABCA1
| Protein | ApoA-I binding | Cholesterol efflux | Mutations |
|---|---|---|---|
| Wild type | + | + | None |
| C1S | + | + | C54S |
| C2S | – | – | C75S |
| C3S | + | + | C81S |
| C4S | + | + | C195S |
| C5S | + | + | C215S |
| C6S | – | – | C309S |
| C7S | + | + | C355S |
| C8S | + | + | C504S |
| C9S | + | + | C626S |
| C10S | + | + | C791S |
| C11S | + | + | C1418S |
| C12S | + | + | C1429S |
| C13S | + | + | C1463S |
| C14S | + | + | C1465S |
| C15S | + | – | C1477S |
| C16S | + | + | C1814S |
| C2/6S | – | – | C75S/C309S |
| C11/12S | + | + | C1418S/C1429S |
| C13/14S | – | – | C1463S/C1465S |
| C13/14/15S | – | – | C1463S/C1465S/C1477S |
| C11/12/13/14S | – | – | C1418S/C1429S/C1463S/C1465S |
| C11/12/13/15S | – | – | C1418S/C1429S/C1463S/C1477S |
| C11/12/14/15S | – | – | C1418S/C1429S/C1465S/C1477S |
| C11/13/14/15S | – | – | C1418S/C1463S/C1465S/C1477S |
| C12/13/14/15S | – | – | C1429S/C1463S/C1465S/C1477S |
| C11/12/13/14/15S | – | – | C1418S/C1429S/C1463S/C1465S/C1477S |
| C2/6/13/14/15S | – | – | C75S/C309S/C1463S/C1465S/C1477S |
| Cys4#1 | + | + | C54S/C81S/C195S/C215S/C355S/C504S/C626S/C791S/C1418S/C1429S/C1465S/1814S |
| Cys4#2 | + | + | C54S/C81S/C195S/C215S/C355S/C504S/C626S/C791S/C1418S/C1429S/C1463S/1814S |
| Cys5 | + | + | C54S/C81S/C195S/C215S/C355S/C504S/C626S/C791S/C1418S/C1429S/C1814S |
Although previous reports have suggested that ABCA1 contains disulfide bond(s) between the N- and C-terminal halves, no cysteine in ECD2 has been shown to be important for apoA-I binding. To analyze the roles of cysteine residues in ECD2, we replaced all of the five cysteines (C11–C15) in ECD2 with serines but found that mutant C11/12/13/14/15S was localized mainly in the intracellular compartment and was scarcely expressed on the plasma membrane (supplemental Fig. 3). Next, we constructed the quad cysteine mutants C11/12/13/14S, C11/12/13/15S, C11/12/14/15S, C11/13/14/15S, and C12/13/14/15S. ApoA-I did not bind to cells expressing any of these mutants, although at least a fraction of the mutant proteins was observed on the cell surface (supplemental Fig. 3). These results suggested that more than one cysteine residue in ECD2 could be required for the function. Therefore, we next examined the effects of double and triple cysteine substitutions and found that apoA-I bound to cells expressing the double cysteine mutant C11/12S but not to cells expressing mutant C13/14S or C13/14/15S, even though the surface expression of these mutants was comparable with that of the wild-type protein (Fig. 4B). These results suggested that C13 and C14 are important for apoA-I binding.
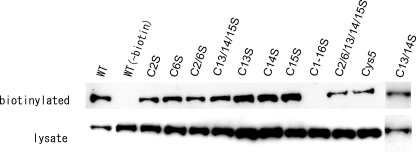
To confirm that replacement of these cysteines by serines did not affect the surface expression of ABCA1, we performed biotinylation experiments to identify surface proteins. Cysteine mutants C2S, C6S, C13S, C14S, C15S, C2/6S, C13/14S, C13/14/15S, and C2/6/13/14/15S were expressed at a level comparable with that of wild-type ABCA1 and also exhibited considerable surface expression (Fig. 5). ABCA1(Cl–16S)-GFP, in which all of the cysteines (C1–C16) in ECDs were replaced by serines, showed a comparable expression level but no surface expression. However, ABCA1(Cys5)-GFP, which contains only the five cysteines C2, C6, C13, C14, and C15 but in which the other 11 extracellular cysteines are replaced by serine residues, showed considerable surface expression (Fig. 5). These results support the importance of C13 and C14 in apoA-I binding.
FIGURE 5.
Cell-surface expression of ABCA1 mutants. Each mutant expressed as a GFP fusion protein was transiently expressed in HEK293 cells. Cells were washed with ice-cold PBS+ 28 h after transfection and incubated with 0.5 mg/ml sulfo-NHS-biotin for 30 min on ice in the dark. Biotinylated proteins were precipitated with avidin gel. Western blotting was performed with anti-ABCA1 antibody KM3073. WT, wild type.
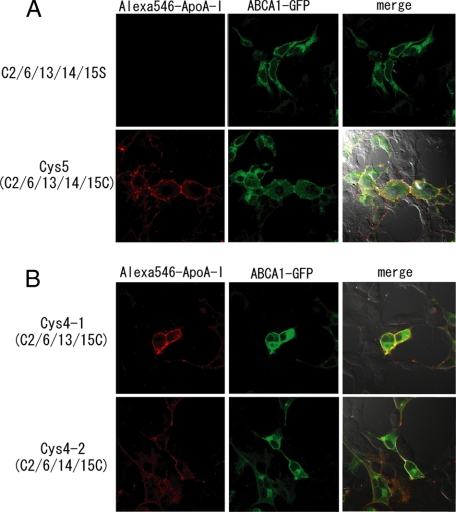
Four of Five Cysteine Residues (C2, C6, C13, C14, and C15) in ECDs of ABCA1 Are Involved in ApoA-I-dependent Cholesterol Efflux—We examined whether five cysteine residues (C2, C6, C13, C14, and C15) in ECD1 and ECD2 are involved in the function of ABCA1. ApoA-I bound to cells expressing ABCA1(Cys5)-GFP as efficiently as to cells expressing the wild-type protein (Fig. 6A). However, apoA-I did not bind to cells expressing C2/6/13/14/15S even though it located on the cell surface comparable with ABCA1(Cys5)-GFP (Fig. 5). When cells expressing ABCA1-(Cys5)-GFP were treated with DTT, bound apoA-I dissociated from cells; apoA-I re-bound to cells again after subsequent washing with fresh medium. DTT treatment of cells in the presence of N-ethylmaleimide irreversibly abolished apoA-I binding to cells expressing ABCA1(Cys5)-GFP (supplemental Fig. 4).
FIGURE 6.
Involvement of five cysteine residues (C2, C6, C13, C14, and C15) in apoA-I binding. A, ABCA1-GFP containing the C2/6/13/14/15S mutation and ABCA1(Cys5)-GFP, containing only the five extracellular cysteines (C2, C6, C13, C14, and C15), were transiently expressed in HEK293 cells. B, ABCA1(Cys4#1)-GFP or ABCA1(Cys4#2)-GFP, containing only four extracellular cysteine residues (C2/C6/C13/C15 and C2/C6/C14/C15, respectively), were transiently expressed in HEK293 cells. Cells were incubated with Alexa 546-conjugated apoA-I for 15 min.
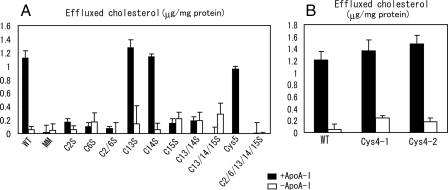
We next examined apoA-I-dependent cholesterol efflux from cells expressing the cysteine mutants (Fig. 7). Like ABCA1-MM, cysteine mutants C2S, C6S, C2/6S, and C13/14S did not show apoA-I-dependent cholesterol efflux, whereas C13S and C14S showed apoA-I-dependent cholesterol efflux as efficiently as the wild type. Interestingly, C15S did not show apoA-I-dependent cholesterol efflux, although this mutation did not affect apoA-I binding. ABCA1(Cys5) showed apoA-I-dependent cholesterol efflux as efficiently as the wild type, whereas C2/6/13/14/15S did not show any activity. These results suggest that four cysteine residues (C2, C6, C15, and either C13 or C14) in ECD1 and ECD2 of ABCA1 are required for apoA-I-dependent cholesterol efflux.
FIGURE 7.
Effects of cysteine mutations on apoA-I-dependent cholesterol efflux. HEK293 cells were transfected with each mutant fused with GFP. The efflux of cellular free cholesterol in the presence (black bars) or absence (white bars) of apoA-I was analyzed. Experiments were performed in triplicate, and the means ± S.E. are represented. WT, wild type.
Finally, we examined apoA-I binding to and apoA-I-dependent cholesterol efflux from cells expressing ABCA1(Cys4#1), containing four extracellular cysteine residues (C2, C6, C13, and C15) or ABCA1(Cys4#2), containing four extracellular cysteine residues (C2, C6, C14, and C15). ApoA-I bound to the cells expressing both of these mutants as efficiently as to cells expressing the wild type (Fig. 6B). ApoA-I-dependent cholesterol efflux from these cells was as high as that from cells expressing wild-type ABCA1 (Fig. 7B). These results suggest that four cysteine residues in ECD1 and ECD2 of ABCA1 are sufficient for apoA-I binding and apoA-I-dependent cholesterol efflux.
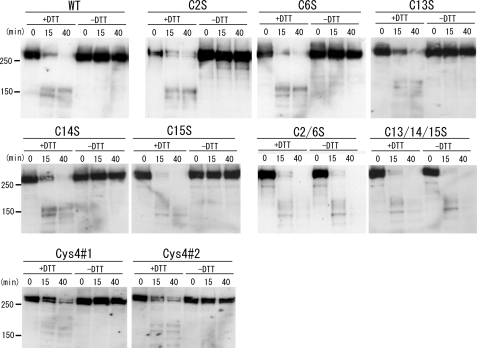
Analysis of Trypsin-digested Wild-type and Mutant ABCA1 under Reducing and Nonreducing Conditions—To determine whether the five cysteine residues are involved in intramolecular disulfide bond formation, the migration behavior of ABCA1 upon SDS-PAGE was analyzed under disulfide reducing and nonreducing conditions (Fig. 8 and supplemental Fig. 5). The 280-kDa band greatly decreased when wild-type ABCA1 was digested with trypsin for 15 min and separated on the gel under reducing conditions, and the band disappeared when digested for 40 min. Instead, faster migrating bands (∼150 kDa) appeared after trypsin digestion, as reported (9). However, under nonreducing conditions, the trypsin-digested ABCA1 co-migrated with undigested ABCA1, indicating that the N- and C-terminal halves of ABCA1 were connected with disulfide bonds.
FIGURE 8.
Analysis of wild-type and mutant ABCA1 after limited trypsin digestion under reducing or nonreducing conditions. HEK293 cells were transfected with wild-type (WT) or mutant ABCA1. Cells were lysed 28 h after transfection, and limited trypsin digestion was performed. The digested proteins were denatured with or without DTT and electrophoresed on 5% PAGEL. Western blotting was performed with anti-ABCA1 antibody KM3073.
Among the single cysteine mutants, C2S, C6S, C13S, C14S, and C15S retained disulfide bond(s) between the N- and C-terminal halves. However, mutants C2/6S and C13/14/15S retained no disulfide bond between the N- and C-terminal halves. Finally, we analyzed the electrophoretic mobility of ABCA1(Cys4#1), ABCA1(Cys4#2), and ABCA1(Cys5) under disulfide reducing and nonreducing conditions and found it to be identical to that of the wild type.
DISCUSSION
ABCA1 contains disulfide bond(s) between the N- and C-terminal halves, and cysteine residues in the two characteristic ECDs are predicted to be involved in disulfide bond formation. However, it remains unclear which cysteines are involved in disulfide bond formation, how many disulfide bonds are formed, and most importantly, whether disulfide bond formation is important for the functions of ABCA1.
To identify the cysteine residues involved in disulfide bond formation in ABCA1, we constructed >30 mutants in which extracellular cysteine residues were replaced by serines and analyzed their function in assays of apoA-I binding and apoA-I-dependent cholesterol efflux. Two cysteine residues, C2 and C6 in ECD1, were found by single cysteine replacement to be essential for apoA-I binding and apoA-I-dependent cholesterol efflux. In contrast, in ECD2, only one cysteine residue, C15, was found to be essential for apoA-I-dependent cholesterol efflux, and no single cysteine replacement in ECD2 impaired apoA-I binding. Instead, the concurrent replacement of two cysteine residues, C13 and C14, by serines impaired apoA-I binding and apoA-I-dependent cholesterol efflux. These results can be interpreted as follows: three cysteine residues (C2, C6, and C15) and either C13 or C14 are essential for the functions of ABCA1. Indeed, ABCA1(Cys4#1) and ABCA1(Cys4#2), containing only four extracellular cysteines (C2/C6/C13/C15 and C2/C6/C14/C15, respectively) were capable of binding apoA-I and participating in apoA-I-dependent cholesterol efflux as efficiently as wild-type ABCA1. These results suggest that four cysteine residues in ECD1 and ECD2 are necessary for the functions of ABCA1 expressed in HEK293 cells.
Trypsin digestion experiments suggested that five cysteine residues (C2, C6, C13, C14, and C15) are involved in disulfide bond formation between ECD1 and ECD2. Replacement of any one of these residues could not abolish disulfide bond formation between ECD1 and ECD2, but no disulfide bonds were formed in ABCA1 containing either the C2/6S or C13/14/15S substitution. These results suggest that more than one disulfide bond can be formed between ECD1 and ECD2. Because C13 and C14 can substitute for each other functionally, two distinct disulfide bonds can be formed between ECD1 and ECD2. These results suggest that the formation of one disulfide bond between two halves is not sufficient to support ABCA1 function; two intramolecular disulfide bonds between ECD1 and ECD2 are necessary for the functions of ABCA1.
We tried to determine which pairs of disulfide bonds, C2-C15 and C6-C13/14 or C2-C13/14 and C6-C15, are formed between ECD1 and ECD2. However, we found that both disulfide bonds C2-C14 and C2-C15 could be formed when ECDs contained only two cysteine residues (C2 and C14 or C2 and C15) (supplemental Fig. 5B). Furthermore, both disulfide bonds (C6-C14 and C6-C15) could form when ECDs contained only two cysteine residues (C6 and C14 or C6 and C15). Because C13, C14, and C15 reside within 15 amino acids in the region predicted as a flexible loop, they may be able to substitute for each other in disulfide bond formation. C13 and C14, with one residue between them, seem to be functionally equivalent and can substitute for each other; they cannot functionally substitute for C15, however, because C15S replacement impaired apoA-I-dependent cholesterol efflux. C13 and C14 are conserved among ABCA1, ABCA4, and ABCA7, and mutations of both C13 (C1488R) and C14 (C1490Y) of ABCA4 are linked to Stargardt disease (20), suggesting that these two cysteines are functionally important and cannot substitute for each other in ABCA4. ABCA4 prepared from bovine retina rod outer segment was reported to migrate on an SDS gel more rapidly under nonreducing conditions than under reducing conditions (10). C13 or C14 might be involved in intermolecular disulfide bond formation, and the resulting bond might affect the structure or functions of ABCA4. Because ABCA1 expressed in human fibroblasts forms a covalently linked tetramer (25), either C13 or C14 of ABCA1 might be involved in intermolecular disulfide bond formation in vivo, although it is not necessary for ABCA1 expressed in HEK293 cells.
ApoA-I did not bind to cells expressing ABCA1-MM. The electrophoretic mobility of the trypsin-digested ABCA1-MM was identical to that of the wild type under disulfide reducing and nonreducing conditions (data not shown), suggesting normal disulfide bond formation in ABCA1-MM. Therefore, conformational changes of ABCA1 caused by ATP binding and/or hydrolysis could be required for apoA-I binding. The single cysteine substitutions C2S and C6S impaired apoA-I binding, suggesting that two disulfide bonds between ECD1 and ECD2 are required for the conformation for apoA-I binding. In contrast, the C15S mutation did not impair apoA-I binding. Two disulfide bonds may be formed between C2 and C6 and between C13 and C14 in the C15S mutant, and the resulting structure may mimic the conformation after ATP binding/hydrolysis. However, these disulfide bonds were not able to support cholesterol efflux, probably because they cannot allow the conformational changes coupled with ATP hydrolysis. The Tangier mutation W590S impairs apoA-I-dependent cholesterol efflux but not apoA-I binding (15). The replacement of C15 by serine may cause conformational changes similar to those caused by the W590S mutation. These results also suggest that apoA-I binding and apoA-I-dependent cholesterol efflux are separable because C15S did not impair apoA-I binding but did impair apoA-I-dependent cholesterol efflux.
In summary, our results indicate that two intramolecular disulfide bonds are formed in ABCA1, between C2 and C6 in ECD1 and between either C13 or C14 and C15 in ECD2, and that these two disulfide bonds are necessary for apoA-I-dependent cholesterol efflux by ABCA1 expressed in HEK293 cells. These cysteine residues are conserved among ABCA1, ABCA4, and ABCA7. This study will facilitate our understanding of the mechanism of HDL formation by ABCA1, as well as the function of other ABCA subfamily proteins.
Supplementary Material
This work was supported by a grant-in-aid for scientific research (S) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, and by the World Premier International Research Center Initiative, Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Table I.
Footnotes
The abbreviations used are: HDL, high density lipoprotein; TM, transmembrane α-helix; ECD, extracellular domain; DTT, dithiothreitol; DMEM, Dulbecco's modified Eagle's medium; BES, 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid; GFP, green fluorescent protein; PBS, phosphate-buffered saline.
References
- 1.Oram, J. F., and Vaughan, A. M. (2006) Circ. Res. 99 1031-1043 [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama, S. (2000) Biochim. Biophys. Acta 1529 231-244 [DOI] [PubMed] [Google Scholar]
- 3.Munehira, Y., Ohnishi, T., Kawamoto, S., Furuya, A., Shitara, K., Imamura, M., Yokota, T., Takeda, S., Amachi, T., Matsuo, M., Kioka, N., and Ueda, K. (2004) J. Biol. Chem. 279 15091-15095 [DOI] [PubMed] [Google Scholar]
- 4.Brooks-Wilson, A., Marcil, M., Clee, S., Zhang, L., Roomp, K., van Dam, M., Yu, L., Brewer, C., Collins, J., Molhuizen, H., Loubser, O., Ouelette, B., Fichter, K., Ashbourne-Excoffon, K., Sensen, C., Scherer, S., Mott, S., Denis, M., Martindale, D., Frohlich, J., Morgan, K., Koop, B., Pimstone, S., Kastelein, J., and Hayden, M. (1999) Nat. Genet. 22 336-345 [DOI] [PubMed] [Google Scholar]
- 5.Bodzioch, M., Orso, E., Klucken, J., Langmann, T., Bottcher, A., Diederich, W., Drobnik, W., Barlage, S., Buchler, C., Porsch-Ozcurumez, M., Kaminski, W., Hahmann, H., Oette, K., Rothe, G., Aslanidis, C., Lackner, K., and Schmitz, G. (1999) Nat. Genet. 22 347-351 [DOI] [PubMed] [Google Scholar]
- 6.Rust, S., Rosier, M., Funke, H., Real, J., Amoura, Z., Piette, J., Deleuze, J., Brewer, H., Duverger, N., Denefle, P., and Assmann, G. (1999) Nat. Genet. 22 352-355 [DOI] [PubMed] [Google Scholar]
- 7.Singaraja, R. R., Brunham, L. R., Visscher, H., Kastelein, J. J., and Hayden, M. R. (2003) Arterioscler. Thromb. Vasc. Biol. 23 1322-1332 [DOI] [PubMed] [Google Scholar]
- 8.Dawson, R. J., and Locher, K. P. (2006) Nature 443 180-185 [DOI] [PubMed] [Google Scholar]
- 9.Takahashi, K., Kimura, Y., Kioka, N., Matsuo, M., and Ueda, K. (2006) J. Biol. Chem. 281 10760-10768 [DOI] [PubMed] [Google Scholar]
- 10.Bungert, S., Molday, L. L., and Molday, R. S. (2001) J. Biol. Chem. 276 23539-23546 [DOI] [PubMed] [Google Scholar]
- 11.Peelman, F., Labeur, C., Vanloo, B., Roosbeek, S., Devaud, C., Duverger, N., Denefle, P., Rosier, M., Vandekerckhove, J., and Rosseneu, M. (2003) J. Mol. Biol. 325 259-274 [DOI] [PubMed] [Google Scholar]
- 12.Tanaka, A. R., Ikeda, Y., Abe-Dohmae, S., Arakawa, R., Sadanami, K., Kidera, A., Nakagawa, S., Nagase, T., Aoki, R., Kioka, N., Amachi, T., Yokoyama, S., and Ueda, K. (2001) Biochem. Biophys. Res. Commun. 283 1019-1025 [DOI] [PubMed] [Google Scholar]
- 13.Hayashi, M., Abe-Dohmae, S., Okazaki, M., Ueda, K., and Yokoyama, S. (2005) J. Lipid Res. 46 1703-1711 [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, Y., Abe-Dohmae, S., Munehira, Y., Aoki, R., Kawamoto, S., Furuya, A., Shitara, K., Amachi, T., Kioka, N., Matsuo, M., Yokoyama, S., and Ueda, K. (2003) Biochem. Biophys. Res. Commun. 311 313-318 [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald, M. L., Morris, A. L., Rhee, J. S., Andersson, L. P., Mendez, A. J., and Freeman, M. W. (2002) J. Biol. Chem. 277 33178-33187 [DOI] [PubMed] [Google Scholar]
- 16.Rigot, V., Hamon, Y., Chambenoit, O., Alibert, M., Duverger, N., and Chimini, G. (2002) J. Lipid Res. 43 2077-2086 [DOI] [PubMed] [Google Scholar]
- 17.Singaraja, R. R., Visscher, H., James, E. R., Chroni, A., Coutinho, J. M., Brunham, L. R., Kang, M. H., Zannis, V. I., Chimini, G., and Hayden, M. R. (2006) Circ. Res. 99 389-397 [DOI] [PubMed] [Google Scholar]
- 18.Lewis, R. A., Shroyer, N. F., Singh, N., Allikmets, R., Hutchinson, A., Li, Y., Lupski, J. R., Leppert, M., and Dean, M. (1999) Am. J. Hum. Genet. 64 422-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stone, E. M., Webster, A. R., Vandenburgh, K., Streb, L. M., Hockey, R. R., Lotery, A. J., and Sheffield, V. C. (1998) Nat. Genet. 20 328-329 [DOI] [PubMed] [Google Scholar]
- 20.Webster, A. R., Heon, E., Lotery, A. J., Vandenburgh, K., Casavant, T. L., Oh, K. T., Beck, G., Fishman, G. A., Lam, B. L., Levin, A., Heckenlively, J. R., Jacobson, S. G., Weleber, R. G., Sheffield, V. C., and Stone, E. M. (2001) Investig. Ophthalmol Vis. Sci. 42 1179-1189 [PubMed] [Google Scholar]
- 21.Maugeri, A., van Driel, M. A., van de Pol, D. J., Klevering, B. J., van Haren, F. J., Tijmes, N., Bergen, A. A., Rohrschneider, K., Blankenagel, A., Pinckers, A. J., Dahl, N., Brunner, H. G., Deutman, A. F., Hoyng, C. B., and Cremers, F. P. (1999) Am. J. Hum. Genet. 64 1024-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azuma, Y., Takada, M., Shin, H.-W., Kioka, N., Nakayama, K., and Ueda, K. (2009) Genes Cells 14 191-204 [DOI] [PubMed] [Google Scholar]
- 23.Hozoji, M., Munehira, Y., Ikeda, Y., Makishima, M., Matsuo, M., Kioka, N., and Ueda, K. (2008) J. Biol. Chem. 283 30057-30063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamon, Y., Broccardo, C., Chambenoit, O., Luciani, M., Toti, F., Chaslin, S., Freyssinet, J., Devaux, P., McNeish, J., Marguet, D., and Chimini, G. (2000) Nat. Cell Biol. 2 399-406 [DOI] [PubMed] [Google Scholar]
- 25.Denis, M., Haidar, B., Marcil, M., Bouvier, M., Krimbou, L., and Genest, J. (2004) J. Biol. Chem. 279 41529-41536 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.