Abstract
We describe novel, cell-permeable, and bioavailable salicylic acid derivatives that are potent and selective inhibitors of GLEPP1/protein-tyrosine phosphatase φ. Two previously described GLEPP1 substrates, paxillin and Syk, are both required for cytoskeletal rearrangement and cellular motility of leukocytes in chemotaxis. We show here that GLEPP1 inhibitors prevent dephosphorylation of Syk1 and paxillin in resting cells and block primary human monocyte and mouse bone marrow-derived macrophage chemotaxis in a gradient of monocyte chemotactic protein-1. In mice, the GLEPP1 inhibitors also reduce thioglycolate-induced peritoneal chemotaxis of neutrophils, lymphocytes, and macrophages. In murine disease models, the GLEPP1 inhibitors significantly reduce severity of contact hypersensitivity, a model for allergic dermatitis, and dextran sulfate sodium-induced ulcerative colitis, a model for inflammatory bowel disease. Taken together, our data provide confirmation that GLEPP1 plays an important role in controlling chemotaxis of multiple types of leukocytes and that pharmacological inhibition of this phosphatase may have therapeutic use.
The protein-tyrosine phosphatases (PTPs)4 constitute a mammalian gene family of about 100 members, roughly similar in number to the 90 tyrosine kinases (1, 2). By exerting negative control on kinase signaling cascades (3), PTPs, themselves tightly regulated, play important roles in human disease, including cancer (4), neurodegenerative diseases (5), and especially inflammatory diseases (6–9). For instance, a common missense SNP (single nucleotide polymorphism) in PTPN22 is a major risk factor for a long list of human autoimmune diseases (10). Since PTPs dephosphorylate and “reset” cytokine and growth factor receptors and their downstream effectors, their inhibition or genetic deletion may result in enhanced receptor signaling. For example, deletion of PTP-1B sensitizes insulin receptor signaling, making this PTP a clinical target for diabetes (11, 12), and mice that lack PTP-H1, a PTP with substrate selectivity for the growth hormone receptor (13), display enhanced systemic growth (14). Despite this gene family's clear potential as drug targets, few PTP inhibitors have reached the clinic (15, 16). One hurdle toward understanding PTP function and target validation is that these enzymes are usually involved in multiple signaling pathways. Also, the medicinal chemistry for developing selective inhibitors is challenging, with little guidance for clinically tolerable selectivity profiles. The discovery of orally available PTP inhibitors with defined selectivity is therefore critical to validate PTPs both conceptually and as drugable targets. We describe here the serendipitous discovery of two orally bioavailable inhibitors with selectivity for GLEPP1 (glomerular epithelial protein-1) and use these to explore GLEPP1 function in inflammatory disease.
GLEPP1 is also known as glomerular epithelial protein-1, PTPRO, PTP-U2, PTP-φ, PTP-oc, CRYP2, or PTP-BK. Like most PTPs, GLEPP1 is well conserved among mammals, forming an unambiguous orthologous gene cluster (17). GLEPP1 is a receptor PTP with an extracellular region that contains eight fibronectin FN III repeats and a single intracellular catalytic domain (18). In addition to this “full-length” GLEPP1, which is expressed in kidney and brain, a shorter variant exists that lacks the eight FN III repeats in the extracellular domain, called PTPROt, PTPφ, or PTP-oc, and is expressed in the hematopoietic lineage only, including osteoclasts (19–21). Both variants encode the catalytic domain, and it is now clear that they are produced from distinct promoters rather than by differential splicing (19, 20, 22, 23). In addition, both transcripts undergo differential splicing as well (20). Mice that cannot produce the full-length form of GLEPP1 (only) display mild kidney abnormalities (24). Throughout this study, we shall refer with “GLEPP1” to both the short and long variants, since enzymatic inhibitors presumably affect the catalytic domain present in both forms.
Apart from a role in the kidney, GLEPP1 has been associated with four other physiological processes. (i) Using EphA and -B receptors as substrate, GLEPP1 plays a role in axonal guidance (25, 26). (ii) GLEPP/PTPRO was characterized as a tumor suppressor gene, with CpG island promoter hypermethylation and reduced expression seen in primary tumors and tumor cell lines (27–29). (iii) GLEPP1 promotes osteoclastic bone resorption, which can be prevented using GLEPP1 antisense (30) or targeted gene deletion (31). (iv) Finally, GLEPP1 and its substrates mediate chemotaxis. It was shown that treatment of macrophages with chemoattractant CSF-1 (colony-stimulating factor-1) induces expression of GLEPP1, which dephosphorylates paxillin and controls cell motility (20, 32, 33). More recently, Syk was identified as another GLEPP1 substrate in B-cells. Since Syk, too, is essential for chemotaxis (34) and cell motility (35), we decided to use the two GLEPP1 inhibitors as pharmacological tools and investigate GLEPP1 function in chemotaxis under different settings, both in vitro and in vivo.
We demonstrate here that the two GLEPP1 inhibitors are cell-permeable and block GLEPP1-mediated dephosphorylation of Syk and paxillin, both identified earlier as GLEPP1 substrates, in cells. Since both substrates have been shown earlier to mediate cytoskeletal rearrangement and chemotaxis, we tested the compounds and discovered that they block chemotaxis of mouse bone marrow-derived macrophages and human monocytes in vitro. We also show that the compounds partially prevent thioglycolate-induced, intraperitoneal chemotaxis in mice. Based on these observations, we decided to test GLEPP1 inhibitors in disease models that strongly rely on lymphocyte infiltration in diseased tissues. We found that the compounds protect animals from delayed type contact hypersensitivity and dextran sodium sulfate (DSS)-induced ulcerative colitis. We conclude from our data that GLEPP1 may play a controlling role in chemotaxis through dephosphorylation of Syk and/or paxillin substrates and that GLEPP1 inhibitors prevent chemotaxis in vitro and in vivo with potential clinical use in inflammatory diseases that are sustained by infiltration of immune cells.
EXPERIMENTAL PROCEDURES
Synthesis of Compounds 1 and 2—The synthesis of compounds 1 and 2 uses a common intermediate 3, as depicted in supplemental Scheme 1. Then a series of reductive alkylation, hydrolysis, or amidification was used successfully to prepare these novel compounds as shown in the following detailed protocol. Compounds 1 and 2 were prepared in eight and six steps, respectively, from commercially available reagents. The synthesis is described in detail in the supplemental material.
PTP Selectivity Analysis of Inhibitors—Unless otherwise stated, chemicals were purchased from Sigma. 6,8-Difluoro-4-methyumbelliferyl phosphate was from Molecular Probes/Invitrogen. 96-well plates were from Corning Glass. The glutathione S-transferase-tagged catalytic domains of human phosphatases were cloned, expressed in Escherichia coli, and affinity-purified on a GSH column, as described (13, 36). Selectivity phosphatase assays were carried out using a 6,8-difluoro-4-methyumbelliferyl phosphate concentration corresponding to the Km value of the enzyme studied (37). In 96-well plates containing 5 μl of diluted compound or solvent (100% DMSO) in each well, 55 μl of 6,8-difluoro-4-methyumbelliferyl phosphate diluted in PTP buffer (20 mm BisTris-HCl, pH 7.5, 0.1% Brij 35, 1 mm dl-dithiothreitol) was added, followed by 40 μl of recombinant enzyme diluted in PTP buffer in order to start the reaction. After 45 min at room temperature, fluorescence intensity (FI) was measured (excitation at 355 nm and emission at 460 nm for 0.2 s) on a PerkinElmer Life Sciences fusion spectrofluorimeter. Negative controls were performed in the absence of enzyme, and positive controls were carried out in the presence of enzyme without compound. The percentage of activity was calculated according to the formula, % activity = 100 × (FIcompound - FIlow control)/(FIhigh control - FIlow control). IC50 values were determined in triplicate in two independent experiments.
Real Time PCR Procedure—Human and mouse leukocytes were isolated from healthy donors using MACS® reagents, the AutoMacs® equipment, and protocols from Miltenyi Biotec GmbH. Total RNA was prepared using TRIzol reagent (catalog number 15596-026; Invitrogen) using the manufacturer's protocol. 1 μg of total RNA was used for cDNA synthesis (RT-PCR Advantage kit; Clontech), and qPCRs were performed in triplicates, using the manufacturer's instructions (QuantiFast® SYBR Green PCR kit; Qiagen®) with 2.5 ng of the total cDNA and the respective QuantiTect® primer assays, in an Applied Biosystems 7900HT Fast Real-Time PCR system, using the manufacturer's instructions (13).
qPCR Primers—PCR primers for human PTPs were designed using the Primer Express software from PerkinElmer Life Sciences based on the published sequences: GLEPP1 (mouse, reverse), TCG GGC CAG GCT GTG TAG TTA AA and CCA GTA GAC ACT TCC GGA TCA AC; GLEPP1 (human, reverse), TCA GGC CAT GCA GTG TAG TTA AA and CCT GTA GAC ACT TCC GGA TCA AC; GAPDH (mouse, reverse), GGA GAC AAC CTG GTC CTC AGT G and ACC TGC CAA GTA TGA TGA CAT CA; GAPDH (human, reverse), GAT GGG ATT TCC ATT GAT GAC A and CCA CCC ATG GCA AAT TCC; GAPDH intron (negative control for DNA contamination), TGG AAA TTG TGA GGG AGA TGC and TCG GTT GCT GAT GAG TC (mouse) and CCT AGT CCC AGG GCT TTG ATT and CTG TGC TCC CAC TCC TGA TTT C (human).
Interference of GLEPP1 Inhibitors with Signaling—PT-PROt-WT tetracycline-inducible DHL10 cells were cultured with or without 1 μg/ml doxycycline and subsequently serum-starved. The cells were then treated with 3 μm inhibitor (Compound 1 or Compound 2) or vehicle alone at 37 °C for 1 h. Thereafter, the cells were stimulated with goat anti-human IgG plus IgM (10 μg/ml) for 5 min or left untreated, lysed, and analyzed for Syk phosphorylation, as described previously, using anti-phosphotyrosine 4G10 from Upstate Biotechnology, Inc. (Lake Placid, NY) (39).
BAC1.2F5 macrophages were starved of CSF-1 overnight, treated with 20 μm of inhibitor (Compound 1 or Compound 2) or vehicle alone at 37 °C for 1 h. The cells were subsequently stimulated with CSF-1 (120 ng/ml) for either 0 or 15 min prior to lysis. Paxillin immunoprecipitations (BD Transduction Laboratories) were carried out as previously described (32) and analyzed for paxillin phosphorylation with PY100 antibody (Cell Signaling, Danvers, MA).
Chemotaxis—Monocytes from healthy human donors were isolated from buffy coats by negative depletion using the autoMACS® magnetic cell sorting kit (Miltenyi Biotec; catalog number 130-091-153) for preparation of “untouched” monocytes, according to the manufacturer's instructions. Cells (106 cells/ml) were incubated with test compounds to chosen final concentration(s) for 20 min at 37 °C. Chemotaxis plates (96-well filter plates from Neuro Probe Inc.; ChemoTX; catalog number 101-5; 5-μm pore size) were filled with medium (RPMI 1640 without phenol red, containing l-glutamine, 0.3% DMSO, and 2% fetal calf serum), MCP-1 (monocyte chemotactic protein-1; 1 μm; R&D Systems) in the lower compartment, and test compound in both compartments. The monocytes (2 × 104 cells/well in a total volume of 20 μl) were layered on top of the plate filter, taking care to avoid air bubbles, the lid was replaced, and plates were incubated at 37 °C for 2 h. Following disassembly, filters were gently rinsed with 10 ml of phosphate-buffered saline, to remove adhering cells, and the migrated cells were transferred to a black 96-well reader plate (Costar; catalog number 3915) with the aid of a 96-well funnel assembly (Neuro Probe Inc.; catalog number FP1) and by centrifuging at 2,000 rpm for 2 min. The plates, covered with an adhesive sheet, were frozen overnight at -80 °C and thawed, and cell mass was stained using a CyQUANT cell proliferation assay kit (Molecular Probes, Inc.; catalog number C7026) and Wallac Victor Fluorescent Counter (excitation at 480 nm, emission at 520 nm). Chemotaxis of mouse bone marrow-derived macrophages and THP-1 cells was tested as described previously (40).
Thioglycolate-induced Peritoneal Recruitment of Leukocytes—Female C3H mice (6 animals/group, 8 weeks old; Janvier Breeding) received thioglycolate (1.5%, 40 ml/kg, intraperitoneally) after oral administration of test compounds and a second administration of the test compounds 24 h later. Compound 2 and Compound 1 were administered at doses of 1, 3, and 10 mg/kg and 3, 10, 30, 60 mg/kg, respectively. Forty-eight hours after the challenge, the animals were sacrificed, and lavage of the peritoneal cavity was conducted using 2 × 5 ml of phosphate-buffered saline, 1 mm EDTA (4 °C). After centrifugation (10 min at 3,000 rpm), the pellet was resuspended in 1 ml of phosphate-buffered saline. The peritoneal cells were counted using a Beckman/Coulter counter. Dexamethasone (1 mg/kg, per os), was used as reference compound.
Contact Hypersensitivity—Procedures were mostly as described (41). Briefly, female BALB/c mice (8 weeks old) were primed on day 0 on the shaved back with antigen (0.5% dinitrofluorobenzene (DNFB)). On day 5, an ear was challenged with 0.2% DNFB, and swelling measured 24 h after challenge. Test compound was administered per os 60 min before the challenge. Methotrexate was used as positive control. “No priming” refers to animals that did not receive the initial DNFB application (but were subsequently challenged on ear skin).
Ulcerative Colitis—Ulcerative colitis was induced in female BALB/c mice (6 animals/group, 20–22 g; Janvier Breeding) by 4% DSS (molecular mass 36–50 kDa; ICN Biomedicals) administered in drinking water. The mice had free access to DSS during 7 days. Body weight was determined daily. The severity of the ulcerative colitis was assessed by a clinical score estimating the constituency of the stool (0 = firm, 1 = loose, 2 = diarrhea) and the presence of blood (0 = no blood, 1 = occult blood, 2 = gross rectal bleeding). Seven days after the induction of the disease, the animals were sacrificed. The length of the colon and the weight of spleen were determined. The test compounds were orally administered at days 3, 4, 5, and 6 after the induction of the ulcerative colitis at doses of 3, 10, and 30 mg/kg (Compound 2) or 30 mg/kg (Compound 1), twice daily. Control mice received the vehicle (0.5% Methocel, 0.25% Tween). Sulfasalazine (200 mg/kg, per os) was used as a reference compound.
Authorization for Animal Experiments—The DSS and in vivo chemotaxis models and animal handling procedures have been approved and authorized by the local public authorities (Office Vétérinaire Cantonal de Genève; Permission 3116/2 for the DSS model and 3121/1 for in vivo chemotaxis). The delayed-type hypersensitivity experiment (at the Istituto di Ricerche Biomediche) was in accordance with EC Directive 86/609/EEC as implemented by the Italian Decketo Legge. 116 of January 27, 1992. Physical facilities and equipment for accommodation and care of animals were in accordance with the provisions of EEC Council Directive 86/609. The Institute is fully authorized by competent veterinary health authorities. All parts of this protocol concerning animal care have been approved by the official Istituto di Ricerche Biomediche veterinarian. The delayed-type hypersensitivity protocol was authorized by the Italian Ministry of Health (Decree 15/02-B).
RESULTS
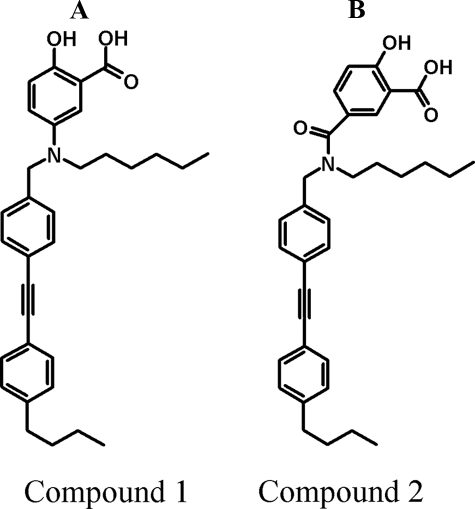
Discovery of Selective, Cell-permeable Inhibitors of Protein-tyrosine Phosphatase GLEPP1—A screening program for PTP-1B inhibitors, PTP family selectivity assays, and in-house medicinal chemistry efforts resulted in the serendipitous discovery of two PTP inhibitors with selectivity for GLEPP1. The compounds are novel salicylic acid derivatives whose hydroxyl-benzoic acid moiety may mimic phosphorylated tyrosine in a phosphoprotein side chain. Compound 1 is a tertiary amine, and Compound 2 is the corresponding amide at the same position (Fig. 1, A and B). The compounds behave as kinetically competitive, nanomolar inhibitors in enzyme assays with Ki values of 0.47 and 0.36 μm, respectively (supplemental material). Both compounds displayed relative selectivity toward GLEPP1, with an in vitro IC50 of 100–300 nm, among a set of 10 human PTPs tested (Table 1). When tested on a reference set of 34 non-PTP enzymes, receptors, and channels, neither compound displayed significant inhibition when tested at 10 μm (data not shown).
FIGURE 1.
Structure and selectivity of GLEPP1 inhibitors Compound 1 and Compound 2 used in this study. Shown are the structures of Compound 1 (A) and Compound 2 (B).
TABLE 1.
Selectivity of inhibitors among protein-tyrosine phosphatases Shown are Compound 1 and 2 IC50 values (nm) for 10 PTPs, representing different PTP family subclasses. S.D. values represent at least three independent experiments.
| PTP | Compound 1 | Compound 2 |
|---|---|---|
| nm | nm | |
| Glepp1 | 123 ± 35 | 340 ± 407 |
| PTP1B | 589 ± 88 | 611 ± 238 |
| Pac1 | 5,630 ± 1,520 | 7,500 ± 1,252 |
| PTP-β | 8,050 ± 1,399 | 5,866 ± 2,804 |
| PTPH1 | 5,438 ± 2,417 | 3,240 ± 2,894 |
| PTP-μ | 5,610 ± 914 | 20,903 ± 2,975 |
| SHP-1 | 673 ± 188 | 1,008 ± 499 |
| SHP-2 | 393 ± 81 | 652 ± 506 |
| TC-PTP | 553 ± 419 | 973 ± 460 |
| VHR | 650 ± 101 | 703 ± 286 |
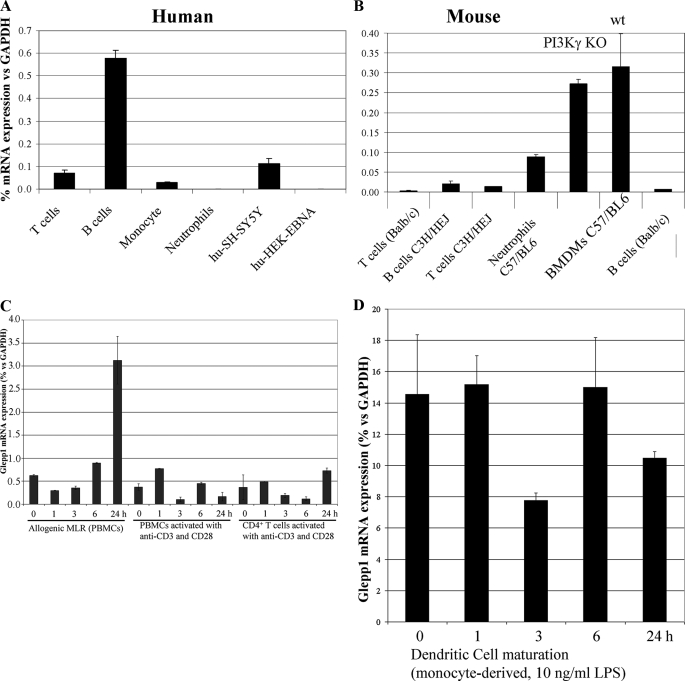
Since we were interested in investigating GLEPP1 inhibitors in immune cell chemotaxis (see Introduction), we evaluated the expression of the enzyme in human and mouse leukocytes. We used qPCR to detect mRNA that expresses the enzyme's catalytic domain, which is present in both the full-length and truncated GLEPP1 protein variants as well as in all described splice forms. As shown in Fig. 2, GLEPP1 mRNA was readily detectable in both human (Fig. 2A) and mouse leukocytes (Fig. 2B), except for human neutrophils. Expression in human B cells was higher than in T cells, whereas expression in mouse B and T cells was similar and did not vary by mouse strain (including phosphoinositide kinase-3γ (PI3Kγ) knockouts; see also Ref. 42 for complete expression data in C57BL/6). Relatively high expression was seen in mouse neutrophils and bone marrow-derived macrophages. Apart from the mixed lymphocyte reaction, stimulation of peripheral blood mononuclear cells, T cells, or dendrocytes did not induce Glepp1 mRNA expression in human cells (Fig. 2, C and D).
FIGURE 2.
GLEPP1 expression in human and murine leukocytes. Expression of mRNA encoding the GLEPP1 catalytic domain was measured by qPCR, as a percentage of GAPDH mRNA, starting with RNA from human (A) or mouse (B) cells. Human cells for RNA isolation were from two or more donors; mouse cells were from the indicated strains. BMDMs were either from PI3Kγ knock-out (KO) (40) or wild type (wt) mice, as indicated. C and D, Glepp1 mRNA expression in variously activated human peripheral blood mononuclear cells, T cells, and dendritic cells. MLR, mixed lymphocyte reaction.
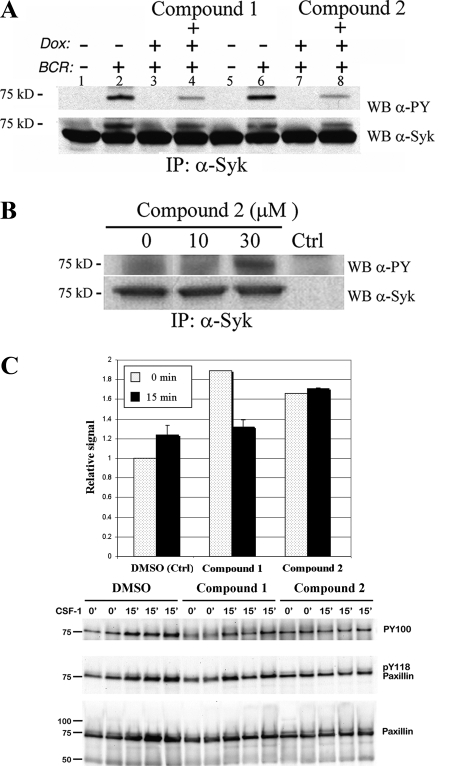
In order to evaluate cell permeability and activity of the inhibitors in cells, we utilized a recently described cellular assay in which doxycycline-induced expression of PTPROt (truncated GLEPP1) in B-cell lymphoma cell lines inhibited the phosphorylation of GLEPP1 substrate Syk (39). After inducing PTPROt with doxycycline, we treated the human lymphoma cell line with one of two different GLEPP1 inhibitors (Compound 1 or Compound 2) or vehicle alone. Thereafter, the cells were stimulated with anti-human immunoglobulin (activating Syk) and lysed for biochemical analysis of Syk. After Syk was immunoprecipitated with a pan-Syk antibody, phospho-Syk was detected by immunoblotting with the anti-phosphotyrosine antibody 4G10. BCR-induced Syk tyrosine phosphorylation was significantly inhibited by PTPROt expression, as previously described (Fig. 3A, compare lane 2 with lane 3, and compare lane 6 with lane 7). In contrast, BCR-induced Syk tyrosine phosphorylation was partially restored by treatment with either Compound 1 or Compound 2 (compare lanes 3 and 4 and lanes 7 and 8). Similarly, treatment of human monocytes with Compound 2 resulted in increased steady-state phosphorylation of Syk (Fig. 3B). We next examined the phosphorylation state of paxillin in BAC1.2F5 macrophages. In this system, it has been shown earlier that overexpression of GLEPP1 wild-type protein enhances cell motility, whereas a dominant mutant form of the enzyme promotes adhesion and results in paxillin hyperphosphorylation (32). In BAC1.2F5 macrophages, both GLEPP1 inhibitors induced paxillin phosphorylation, although only transiently for Compound 1 (Fig. 3C, last lane). We conclude that both GLEPP1 inhibitors are cell-permeable, inhibit GLEPP1 in cells, and induce increased steady-state phosphorylation of previously identified GLEPP1 substrates Syk and paxillin in human and mouse leukocytes.
FIGURE 3.
GLEPP1 inhibitors prevent cellular dephosphorylation of Syk and paxillin. A, evaluation of GLEPP1 inhibitors in human DHL10 lymphoma cells (39). Dox, doxycycline, used to induce expression of GLEPP1. BCR, B-cell receptor cross-linking using IgG and IgM to activate Syk kinase autophosphorylation. IP, immunoprecipitation. WB, Western blot immunostained using antibodies against Syk (α-Syk) or phosphotyrosine (α-PY). B, same as A but using human THP1 monocytes to measure steady-state, Compound 2-induced Syk phosphorylation. C, phosphorylation state of paxillin in BAC1.2F5 macrophages treated with GLEPP1 inhibitors (0 min) or following treatments with CSF1 for 15 min; averaged relative band density (top) and Western blot (bottom).
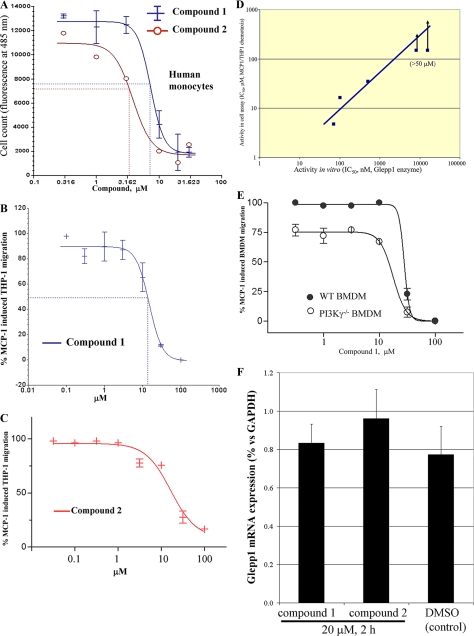
GLEPP1 Inhibitors Block Chemotaxis in Vitro and in Vivo—We next evaluated the two GLEPP1 inhibitors in several cellular assays for chemotaxis. Although the inhibitors were discovered using human GLEPP1 enzyme, we assume, given the close sequence similarity between human and mouse enzymes (>96% identity between the two catalytic domains) that the inhibitors should affect enzymes from both species equally. As shown in Fig. 4A, both compounds showed dose-dependent inhibition of MCP-1-directed chemotaxis of primary human monocytes, resulting in nearly complete inhibition at the highest inhibitor concentration. Similar activity was seen using the human monocytic leukemia cell line THP-1 (Fig. 4, B and C). In order to correlate cellular (THP1) and enzymatic activity, we compared five structurally related compounds. As Fig. 4D shows, in vitro inhibitory activity correlated well with cellular inhibition, further implicating GLEPP1 as the relevant target of the inhibitors. Two compounds that inhibited the GLEPP1 enzyme in the 10–20 μm range failed to show cellular activity in our assay (IC50 >50 μm; arrows). Finally, we evaluated Compound 1 in murine bone marrow-derived macrophage (BMDM) chemotaxis, used here to model primary, naive monocytes. As shown in Fig. 4E, the GLEPP1 inhibitor displayed dose-dependent inhibition in this assay as well. Neither inhibitor affected Glepp1 mRNA expression in THP1 cells (Fig. 4F).
FIGURE 4.
GLEPP1 inhibitors block cellular chemotaxis. Compound 1 and Compound 2 were tested in cellular assays for MCP-1-directed chemotaxis of primary human monocytes (A), the monocytic human THP1 cell line (B and C), and BMDM cells (E). D, correlation between the IC50 values for GLEPP1 inhibitors in vitro (horizontal) with activity in the THP1 cell assay (vertical). Two of the tested compounds failed to show activity in the cell assay at the highest dose tested (50 μm; indicated by arrows). F, Glepp1 mRNA expression analysis (qPCR) following a 2-h exposure of THP-1 cells to 20 μm Compound 1 or 2 or to diluted solvent (DMSO; all biological triplicates).
Earlier studies have identified the importance of phosphatidylinositol 3,4,5-trisphosphate gradients in chemotaxis, which are generated by the action of PI3K, as one of several semiredundant ways of guiding cells (reviewed in Ref. 43). Accordingly, it has been shown that deletion of PI3Kγ in BMDMs results in partial blockade of chemotaxis (40). Similarly, ablation of SHIP1 (SH2-containing inositol phosphatase-1) interferes partially with chemotaxis (43). As shown in Fig. 4E, inhibition of GLEPP1 completely inhibited chemotaxis both in wild-type and PI3Kγ-/- BMDMs with indistinguishable IC50, making it unlikely that Glepp1 affects the phosphatidylinositol 3,4,5-trisphosphate gradient. Our data are consistent with a proposed role for PI3Kγ in early chemotaxis receptor signaling (40), whereas GLEPP1, by interacting with paxillin and/or Syk, may be involved in later events that involve the cytoskeletal actin, integrin, and tubulin rearrangements required for cell motility and locomotion (20, 32–35).
We also tested GLEPP1 inhibitors in an unrelated immune response assay, namely a mixed lymphocyte reaction, using human cells from HLA-mismatched donors. In this 3-day assay, GLEPP1 inhibitors displayed no activity. Furthermore, this assay, which depends on cellular proliferation as readout, demonstrated a lack of cytostatic activity (toxicity) of our GLEPP1 inhibitors at the concentrations tested earlier (data not shown).
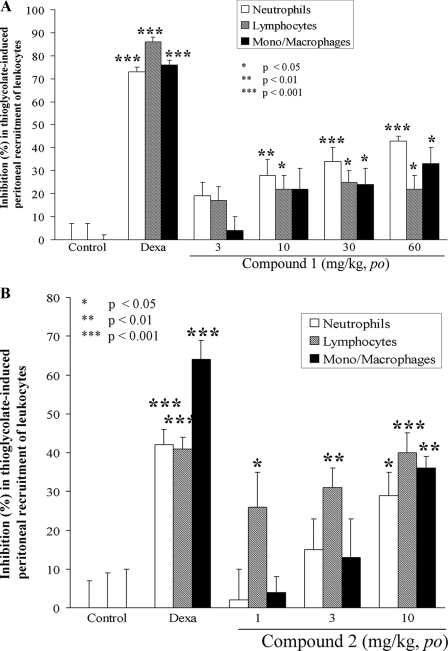
We next tested inhibitor functionality in vivo, following our observation that they showed good oral bioavailability in rodents (data not shown). We chose a model for peritonitis induced by intraperitoneal injection of thioglycolate, an indirect chemoattractant for a wide variety of leukocytes (44). Chemotaxis in this model is known to depend on signaling to paxillin (45), a substrate of GLEPP1 (20, 32, 33). Full recruitment using thioglycolate results in 3–4 × 103/μl neutrophils, 8–11 × 103/μl lymphocytes, and 4–6 × 103/μl mono-/macrophages (see supplemental material for complete data). We found that Compound 1 (Fig. 5A) and Compound 2 (Fig. 5B) each reduced thioglycolate-induced peritoneal recruitment of leukocytes (neutrophils, lymphocytes, and monocytes/macrophages) in a dose-dependent manner.
FIGURE 5.
GLEPP1 inhibitors prevent thioglycolate-induced peritoneal recruitment of leukocytes. Compound 1 (A), Compound 2 (B), or vehicle (control) were orally administered 15 min prior to and 24 h after intraperitoneal injection of thioglycolate in C3H mice. Peritoneal leukocytes were counted 48 h later. The percentage inhibition was calculated against control (vehicle-treated) animals and corrected for background (from animals not injected with thioglycolate; see supplemental material for calculation of raw data). Statistics were calculated using analysis of variance and Dunnett's test, and p value cut-offs are indicated. Dexamethasone was used as a reference compound for inducing inhibition.
In a separate experimental setting, we also found that Compound 1 did not interfere with lipopolysaccharide-induced tumor necrosis factor-α release in vivo when tested at the same doses (data not shown), suggesting that GLEPP1 inhibitors do not interfere with cytokine release but block cell motility only.
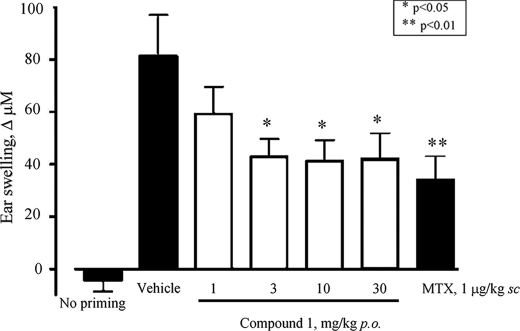
Activity of GLEPP1 Inhibitors in Disease Models—Based on our results so far, we decided to test GLEPP1 inhibitors in disease models presumed to depend on ongoing immune cell migration and infiltration. The first model corresponds to delayed type contact hypersensitivity (41), which approximates allergic dermatitis in humans and which is mediated by interferon-γ-producing Th1/Tc1 cells. Briefly, mice are sensitized (on the shaven back) with dinitrofluorobenzene (DNFB), and 5 days later (challenge), the same compound is applied externally to the pinna (ear flap), resulting in infiltration of neutrophils and macrophages (46) and causing significant swelling. As shown in Fig. 6, animals that received Compound 1 1 h before the DNFB challenge showed significantly reduced swelling (thickness) of the ear, roughly similar to the reduction seen for methotrexate, used as positive control. Good activity was seen at low doses (1–3 mg/kg), presumably because the contact hypersensitivity model allows timing of peak plasma inhibitor levels (∼1 h after compound administration) to coincide with the DNFB challenge and induction of chemotaxis. We next chose a more complex disease model, namely DSS-induced ulcerative colitis, considered to represent a good translational model for therapeutics in human inflammatory bowel disease (47).
FIGURE 6.
Compound 1 reduces contact hypersensitivity in vivo.Compound 1 was administered orally at the indicated dose 1 h prior to the topical ear challenge with dinitrofluorobenzene, followed by measurement of ear skin swelling (indicated as increments, in μm, on the y axis). MTX, methotrexate.
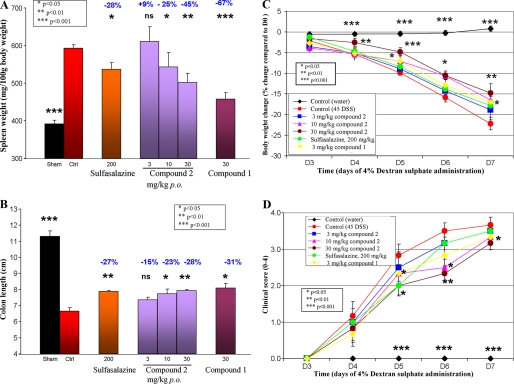
We found that Compound 2 (10 or 30 mg/kg) significantly reduced (25 and 45%, p < 0.05 and p < 0.01, respectively) DSS-induced splenomegaly in a dose-dependent manner (Fig. 7A). Compound 1 (30 mg/kg) and sulfasalazine (200 mg/kg) also showed protective effect (67% (p < 0.001) and 28% (p < 0.05), respectively; Fig. 7A). Compound 2 (10 or 30 mg/kg) significantly prevented (23 and 28%, p < 0.05 and p < 0.01, respectively) DSS-induced colon reduction in a dose-dependent manner (Fig. 7B). Compound 1 (30 mg/kg) and sulfasalazine (200 mg/kg) showed similar efficacy (31% (p < 0.05) and 27% (p < 0.01), respectively; Fig. 7B). In addition, Compounds 1 and 2 (30 mg/kg) reduced disease-related loss of body weight (Fig. 7C). Finally, Compound 2 (10 or 30 mg/kg), Compound 1 (30 mg/kg), and sulfasalazine (200 mg/kg) reduced weakly but significantly (p < 0.05/0.01) the clinical score evaluating stool constituency and presence of blood (Fig. 7D). Overall, Compound 1 tended to show higher efficacy due to its superior bioavailability and longer half-life (data not shown). In order to address the possibility that GLEPP1 inhibitors might somehow interfere with intestinal absorption and local activity of DSS, we administered Compound 1 also by subcutaneous injection and observed similar protection from disease as seen for equivalent plasma exposure following oral administration (data not shown). We therefore do not consider it likely that the GLEPP1 inhibitors somehow interfered with disease induction by DSS per se.
FIGURE 7.
Glepp1 inhibitors reduce DSS-induced ulcerative colitis. Ulcerative colitis was induced in mice through administration of DSS in drinking water over 7 days. The test compounds were orally administered at days 3, 4, 5, and 6 after the induction of the ulcerative colitis at doses of 3, 10, and 30 mg/kg (Compound 2) or 30 mg/kg (Compound 1) twice daily. Sham, healthy mice that did not receive DSS. Sulfasalazine. ns, nonsignificant (i.e. p > 0.05). Readouts for spleen weight (A), colon length (B), percentage body weight change (C), and clinical score (D) are described under “Experimental Procedures.” Percentages refer to readout improvement over controls.
DISCUSSION
We describe here the synthesis of novel inhibitors for GLEPP1, a protein-tyrosine phosphatase for which no inhibitors have so far been described, and use these to evaluate function and drugability of GLEPP1, partially guided by published substrates that have been discovered for this enzyme, and its reported role in chemotaxis. Earlier work had identified that overexpression of GLEPP1 wild-type or dominant negative enzyme “freezes” cells in adhesive or motile states (32). Induction in macrophages of CSF-1-induced chemotaxis results in Pyk2-mediated phosphorylation of paxillin, the establishment of actin bundles, and formation of lamellipodia. These cellular locomotive components are then recycled through GLEPP1-mediated dephosphorylation of paxillin, actin disruption, and the formation of dorsal ruffles in a cyclic process (32). Microscopic imaging of resting macrophages shows that they maintain a highly dynamic state with constant cytoskeletal reorganization and (apparently) random pseudopodia formation and retraction until they initiate controlled migration in the direction of a linear chemotactic gradient. Although signaling, the overall chemotactic process, and cellular motility are poorly understood, it is clear that cyclical phosphorylation and dephosphorylation events accompany the associated cytoskeletal remodeling. Our data suggest that pharmacological inhibition of GLEPP1 may “freeze” cells in an adhesive, nonmotile state, as was previously shown by overexpression of dominant negative GLEPP1 enzyme (32), thus preventing cells from responding to chemotactic stimuli. Our results also suggest that GLEPP1 does not affect early, redundant receptor signaling events mediated by PI3Ks and other (uncharacterized) factors but probably controls phosphorylation of the structural cytoskeletal components. Interference with these “late,” downstream events apparently results in chemotactic paralysis to an extent not seen for complete (genetic) PI3K or SHIP1 inhibition, and Glepp1 may therefore represent a more sensitive target to block chemotaxis than PI3K or SHIP1.
Recent data indicate that (at least) in B cells, GLEPP1 also dephosphorylates Syk, a kinase that is critical in signal transduction of BCR and other immunoreceptor tyrosine-based activation motif-containing receptors (39). Constitutively active Syk, disconnected from ligand-induced BCR stimulation, is known to drive B-lymphoma proliferation (e.g. see Ref. 48). The demonstration that overexpression of GLEPP1 limits lymphoma cell line proliferation (39) explains how PTPRO may behave as a tumor suppressor gene, at least in B cell lymphomas. However, both GLEPP1 and Syk are expressed in all hematopoietic cells, not only in B cells. Multiple lines of evidence point to an additional role of Syk in chemotaxis; Syk autophosphorylation is seen in CCL4/CCR5-mediated chemotaxis in B-cells and primary T-cells (49) and also in MonoMac6 cells as they respond to CX3CL1 (50) or MCP-1 (51). In addition, down-regulation of Syk disrupts CX3CL1-induced cytoskeletal rearrangement in the murine monocyte/macrophage RAW cell line (34). How Syk controls cytoskeletal reorganization is unclear, but, as for paxillin, it is probably a late event downstream of chemokine receptor signal transduction (34). A successful clinical Phase II study was recently completed for a Syk inhibitor (52), one of the very few kinase inhibitors demonstrating clinical safety and efficacy in patients for inflammatory disease, suggesting that Syk is perhaps not only involved in BCR signaling but in chemotaxis as well, thus explaining its success as a drug target.
We confirm here that inhibition of GLEPP1 enzymatic activity in cells blocks Syk phosphorylation in the cellular assay shown in Fig. 3A, and we also show that GLEPP1 inhibitors interfere with steady-state phosphorylation of Syk and paxillin in resting cells. It is not clear whether this dephosphorylation is a regulated process or represents a GLEPP1 “housekeeping” function. Either way, our data indicate that interference with the paxillin or Syk phosphorylation state (or both) negatively affects the roles these enzymes play in cytoskeletal reorganization as induced by chemotactic agents.
In order to evaluate our inhibitors in vivo, we selected disease models suspected to require sustained infiltration of leukocytes, and indeed, we found that our compounds were very effective in models for contact hypersensitivity and DSS-induced ulcerative colitis. However, GLEPP1 inhibitors were less protective (although not totally without efficacy) in collagen-induced arthritis and experimental allergic encephalitis, murine models for human arthritis and multiple sclerosis, respectively (data not shown). We believe that the difference in observed inhibitor efficacy between the two sets of autoimmune diseases (delayed-type hypersensitivity and DSS-colitis versus experimental allergic encephalitis and collagen-induced arthritis) may be explained by the fact that the hypersensitivity and colitis models involve leukocyte infiltration into skin and colon epithelial tissues (bordering the external environment), whereas arthritis and multiple sclerosis represent an “endogenous” autoimmune attack on cartilaginous tissue and myelin, respectively. Possibly, the latter diseases rely less on sustained leukocyte recruitment. Thus, better disease-protective efficacy in those models might require GLEPP1 inhibitor treatment lasting longer or with more frequent administration than used in our disease models.
Establishment of the function of protein-tyrosine phosphatases is challenging for a number of reasons. They are generally involved in complex, negative control of kinase-driven phosphorylation cascades, and their own activity is often tightly controlled at many expression levels as well. For instance, GLEPP1 is transcriptionally regulated by phorbol ester (53) and CSF-1 (20). In addition, PTPs may be involved in multiple, completely unrelated physiological processes (again illustrated by the proposed roles of GLEPP1 in axonal guidance, osteoclastic resorption, chemotaxis, and tumorigenesis). Pharmacological inhibitors, along with genetic models, are essential tools to help understand PTP function in health and disease, with one approach complementing the other (38). The difficulty in unambiguously validating PTPs as drug targets using biological approaches conspires with the paucity of efforts to identify selective inhibitors (as contrasted with the massive efforts spent on kinase inhibitor discovery), resulting in turn in limited availability of quality pharmacological research tools that can be used for target validation. Our description of orally bioavailable GLEPP1 inhibitors and demonstration of their use in inhibiting chemotaxis should help further define the molecular role(s) of this enzyme in the events associated with leukocyte mobilization under various pathological conditions, as well as the additionally proposed roles of GLEPP1 in health and disease.
Supplementary Material
Acknowledgments
We thank the Hôpital Universitaire de Genève for the generous donation of blood for monocyte preparation and thank S. Alouani, J. Challier, P. de Lys, and A. Rolland for RNA samples.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Scheme 1, Table 1, and Figs. 1 and 2.
Footnotes
The abbreviations used are: PTP, protein-tyrosine phosphatase; BCR, B-cell receptor; BMDM, bone marrow-derived macrophages; DNFB, dinitrofluorobenzene; DSS, dextran sodium sulfate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PI3K, phosphoinositide kinase-3; qPCR, quantitative (or real time) PCR; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)-propane-1,3-diol; FI, fluorescence intensity.
References
- 1.Tonks, N. K. (2006) Nat. Rev. Mol. Cell. Biol. 7 833-846 [DOI] [PubMed] [Google Scholar]
- 2.Alonso, A., Sasin, J., Bottini, N., Friedberg, I., Friedberg, I., Osterman, A., Godzik, A., Hunter, T., Dixon, J., and Mustelin, T. (2004) Cell 117 699-711 [DOI] [PubMed] [Google Scholar]
- 3.Stoker, A. W. (2005) J. Endocrinol. 185 19-33 [DOI] [PubMed] [Google Scholar]
- 4.Östman, A., Hellberg, C., and Bohmer, F. D. (2006) Nat. Rev. Cancer. 6 307-320 [DOI] [PubMed] [Google Scholar]
- 5.Gee, C. E., and Mansuy, I. M. (2005) Cell. Mol. Life. Sci. 62 1120-1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mustelin, T., Vang, T., and Bottini, N. (2005) Nat. Rev. Immunol. 5 43-57 [DOI] [PubMed] [Google Scholar]
- 7.Dolton, G. M., Sathish, J. G., and Matthews, R. J. (2006) Biochem. Soc. Trans. 34 1041-1045 [DOI] [PubMed] [Google Scholar]
- 8.Mustelin, T., Rahmouni, S., Bottini, N., and Alonso, A. (2003) Immunol. Rev. 191 139-147 [DOI] [PubMed] [Google Scholar]
- 9.Vang, T., Miletic, A. V., Arimura, Y., Tautz, L., Rickert, R. C., and Mustelin, T. (2008) Annu. Rev. Immunol. 26 29-55 [DOI] [PubMed] [Google Scholar]
- 10.Gregersen, P. K., Lee, H. S., Batliwalla, F., and Begovich, A. B. (2006) Semin. Immunol. 18 214-223 [DOI] [PubMed] [Google Scholar]
- 11.Hooft van Huijsduijnen, R., Sauer, W., Bombrun, A., and Swinnen, D. (2004) J. Med. Chem. 47 4142-4146 [DOI] [PubMed] [Google Scholar]
- 12.Hooft van Huijsduijnen, R., Bombrun, A., and Swinnen, D. (2002) Drug. Discov. Today 7 1013-1019 [DOI] [PubMed] [Google Scholar]
- 13.Pasquali, C., Curchod, M. L., Walchli, S., Espanel, X., Guerrier, M., Arigoni, F., Strous, G., and Hooft van Huijsduijnen, R. (2003) Mol. Endocrinol. 17 2228-2239 [DOI] [PubMed] [Google Scholar]
- 14.Pilecka, I., Patrignani, C., Pescini, R., Curchod, M.-L., Perrin, D., Xue, Y., Yasenchak, J., Clark, A., Magnone, M. C., Zaratin, P., Valenzuela, D., Rommel, C., and Hooft van Huijsduijnen, R. (2007) J. Biol. Chem. 282 35405-35415 [DOI] [PubMed] [Google Scholar]
- 15.Erbe, D. V., Wang, S., Zhang, Y. L., Harding, K., Kung, L., Tam, M., Stolz, L., Xing, Y., Furey, S., Qadri, A., Klaman, L. D., and Tobin, J. F. (2005) Mol. Pharmacol. 67 69-77 [DOI] [PubMed] [Google Scholar]
- 16.Zinker, B. A., Rondinone, C. M., Trevillyan, J. M., Gum, R. J., Clampit, J. E., Waring, J. F., Xie, N., Wilcox, D., Jacobson, P., Frost, L., Kroeger, P. E., Reilly, R. M., Koterski, S., Opgenorth, T. J., Ulrich, R. G., Crosby, S., Butler, M., Murray, S. F., McKay, R. A., Bhanot, S., Monia, B. P., and Jirousek, M. R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11357-11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen, J. N., Jansen, P. G., Echwald, S. M., Mortensen, O. H., Fukada, T., Del Vecchio, R., Tonks, N. K., and Møller, N. P. (2004) FASEB J. 18 8-30 [DOI] [PubMed] [Google Scholar]
- 18.Thomas, P. E., Wharram, B. L., Goyal, M., Wiggins, J. E., Holzman, L. B., and Wiggins, R. C. (1994) J. Biol. Chem. 269 19953-19962 [PubMed] [Google Scholar]
- 19.Aguiar, R. C., Yakushijin, Y., Kharbanda, S., Tiwari, S., Freeman, G. J., and Shipp, M. A. (1999) Blood 94 2403-2413 [PubMed] [Google Scholar]
- 20.Pixley, F. J., Lee, P. S., Dominguez, M. G., Einstein, D. B., and Stanley, E. R. (1995) J. Biol. Chem. 270 27339-27347 [DOI] [PubMed] [Google Scholar]
- 21.Wu, L. W., Baylink, D. J., and Lau, K. H. (1996) Biochem. J. 316 515-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amoui, M., Baylink, D. J., Tillman, J. B., and Lau, K. H. (2003) J. Biol. Chem. 278 44273-44280 [DOI] [PubMed] [Google Scholar]
- 23.Yang, J. H., Amoui, M., Strong, D. D., and Lau, K. H. (2007) Arch. Biochem. Biophys. 465 72-81 [DOI] [PubMed] [Google Scholar]
- 24.Wharram, B. L., Goyal, M., Gillespie, P. J., Wiggins, J. E., Kershaw, D. B., Holzman, L. B., Dysko, R. C., Saunders, T. L., Samuelson, L. C., and Wiggins, R. C. (2000) J. Clin. Invest. 106 1281-1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stepanek, L., Stoker, A. W., Stoeckli, E., and Bixby, J. L. (2005) J. Neurosci. 25 3813-3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shintani, T., Ihara, M., Sakuta, H., Takahashi, H., Watakabe, I., and Noda, M. (2006) Nat. Neurosci. 9 761-769 [DOI] [PubMed] [Google Scholar]
- 27.Motiwala, T., Kutay, H., Ghoshal, K., Bai, S., Seimiya, H., Tsuruo, T., Suster, S., Morrison, C., and Jacob, S. T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13844-13849 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Mori, Y., Yin, J., Sato, F., Sterian, A., Simms, L. A., Selaru, F. M., Schulmann, K., Xu, Y., Olaru, A., Wang, S., Deacu, E., Abraham, J. M., Young, J., Leggett, B. A., and Meltzer, S. J. (2004) Cancer. Res. 64 2434-2438 [DOI] [PubMed] [Google Scholar]
- 29.Motiwala, T., Ghoshal, K., Das, A., Majumder, S., Weichenhan, D., Wu, Y. Z., Holman, K., James, S. J., Jacob, S. T., and Plass, C. (2003) Oncogene 22 6319-6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suhr, S. M., Pamula, S., Baylink, D. J., and Lau, K. H. (2001) J. Bone Miner. Res. 16 1795-1803 [DOI] [PubMed] [Google Scholar]
- 31.Yang, J. H., Amoui, M., and Lau, K. H. (2007) FEBS. Lett. 581 2503-2508 [DOI] [PubMed] [Google Scholar]
- 32.Pixley, F. J., Lee, P. S., Condeelis, J. S., and Stanley, E. R. (2001) Mol. Cell. Biol. 21 1795-1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pixley, F. J., and Stanley, E. R. (2004) Trends Cell. Biol. 14 628-638 [DOI] [PubMed] [Google Scholar]
- 34.Gevrey, J. C., Isaac, B. M., and Cox, D. (2005) J. Immunol. 175 3737-3745 [DOI] [PubMed] [Google Scholar]
- 35.Luangdilok, S., Box, C., Patterson, L., Court, W., Harrington, K., Pitkin, L., Rhys-Evans, P., O-charoenrat, P., and Eccles, S. (2007) Cancer. Res. 67 7907-7916 [DOI] [PubMed] [Google Scholar]
- 36.Wälchli, S., Curchod, M.-L., Pescini Gobert, R., Arkinstall, S., and Hooft van Huijsduijnen, R. (2000) J. Biol. Chem. 275 9792-9796 [DOI] [PubMed] [Google Scholar]
- 37.Grundner, C., Ng, H. L., and Alber, T. (2005) Structure (Camb.) 13 1625-1634 [DOI] [PubMed] [Google Scholar]
- 38.Hooft van Huijsduijnen, R., and Rommel, C. (2006) J. Mol. Med. 84 802-813 [DOI] [PubMed] [Google Scholar]
- 39.Chen, L., Juszczynski, P., Takeyama, K., Aguiar, R. C., and Shipp, M. A. (2006) Blood 108 3428-3433 [DOI] [PubMed] [Google Scholar]
- 40.Camps, M., Ruckle, T., Ji, H., Ardissone, V., Rintelen, F., Shaw, J., Ferrandi, C., Chabert, C., Gillieron, C., Francon, B., Martin, T., Gretener, D., Perrin, D., Leroy, D., Vitte, P. A., Hirsch, E., Wymann, M. P., Cirillo, R., Schwarz, M. K., and Rommel, C. (2005) Nat. Med. 11 936-943 [DOI] [PubMed] [Google Scholar]
- 41.Xu, H., DiIulio, N. A., and Fairchild, R. L. (1996) J. Exp. Med. 183 1001-1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su, A. I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K. A., Block, D., Zhang, J., Soden, R., Hayakawa, M., Kreiman, G., Cooke, M. P., Walker, J. R., and Hogenesch, J. B. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6062-6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kay, R. R., Langridge, P., Traynor, D., and Hoeller, O. (2008) Nat. Rev. Mol. Cell. Biol. 9 455-463 [DOI] [PubMed] [Google Scholar]
- 44.Den Otter, W., De Groot, J. W., Van Basten, C. D., Rademakers, L. H., De Weger, R. A., and Pels, E. (1982) Exp. Mol. Pathol. 36 403-413 [DOI] [PubMed] [Google Scholar]
- 45.Feral, C. C., Rose, D. M., Han, J., Fox, N., Silverman, G. J., Kaushansky, K., and Ginsberg, M. H. (2006) J. Clin. Invest. 116 715-723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garaczi, E., Szell, M., Janossy, T., Koreck, A., Pivarcsi, A., Buzas, E., Pos, Z., Falus, A., Dobozy, A., and Kemeny, L. (2004) Int. Immunol. 16 1781-1788 [DOI] [PubMed] [Google Scholar]
- 47.Melgar, S., Karlsson, L., Rehnstrom, E., Karlsson, A., Utkovic, H., Jansson, L., and Michaelsson, E. (2008) Int. Immunopharmacol. 8 836-844 [DOI] [PubMed] [Google Scholar]
- 48.Chen, L., Monti, S., Juszczynski, P., Daley, J., Chen, W., Witzig, T. E., Habermann, T. M., Kutok, J. L., and Shipp, M. A. (2008) Blood 111 2230-2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jinquan, T., Anting, L., Jacobi, H. H., Glue, C., Jing, C., Ryder, L. P., Madsen, H. O., Svejgaard, A., Skov, P. S., Malling, H. J., and Poulsen, L. K. (2001) J. Immunol. 167 4405-4413 [DOI] [PubMed] [Google Scholar]
- 50.Cambien, B., Pomeranz, M., Schmid-Antomarchi, H., Millet, M., Breittmayer, V., Rossi, B., and Schmid-Alliana, A. (2001) Blood 97 2031-2037 [DOI] [PubMed] [Google Scholar]
- 51.Cambien, B., Pomeranz, M., Millet, M. A., Rossi, B., and Schmid-Alliana, A. (2001) Blood 97 359-366 [DOI] [PubMed] [Google Scholar]
- 52.Braselmann, S., Taylor, V., Zhao, H., Wang, S., Sylvain, C., Baluom, M., Qu, K., Herlaar, E., Lau, A., Young, C., Wong, B. R., Lovell, S., Sun, T., Park, G., Argade, A., Jurcevic, S., Pine, P., Singh, R., Grossbard, E. B., Payan, D. G., and Masuda, E. S. (2006) J. Pharmacol. Exp. Ther. 319 998-1008 [DOI] [PubMed] [Google Scholar]
- 53.Seimiya, H., and Tsuruo, T. (1993) Cell Growth & Differ. 4 1033-1039 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.