Abstract
Recent studies have shown that cytoplasmic proteins are exported efficiently in Escherichia coli only if they are attached to signal peptides that are recognized by the signal recognition particle and are thereby targeted to the SecYEG complex cotranslationally. The evidence suggests that the entry of these proteins into the secretory pathway at an early stage of translation is necessary to prevent them from folding into a translocation-incompetent conformation. We found, however, that several glycolytic enzymes attached to signal peptides that are recognized by the signal recognition particle were exported inefficiently. Based on previous studies of post-translational export, we hypothesized that the export block was due to the presence of basic residues at the extreme N terminus of each enzyme. Consistent with our hypothesis, we found that the introduction of negatively charged residues into this segment increased the efficiency of export. Export efficiency was sensitive to the number, position, and sequence context of charged residues. The importance of charge for efficient export was underscored by an in silico analysis that revealed a conserved negative charge bias at the N terminus of the mature region of bacterial presecretory proteins. Our results demonstrate that cotranslational targeting of a protein to the E. coli SecYEG complex does not ensure its export but that export also depends on a subsequent event (most likely the initiation of translocation) that involves sequences both within and just beyond the signal peptide.
Since the “signal hypothesis” was proposed over 30 years ago (1), it has become clear that signal sequences are not simply generic hydrophobic peptides that earmark proteins for secretion. In bacteria, the features of a signal peptide determine the mechanism by which a given presecretory protein is targeted to the SecYEG translocation complex in the inner membrane (IM).2 Whereas most or all signal peptides are recognized by the signal recognition particle (SRP) in mammalian cells, only a small fraction of Escherichia coli signal peptides are recognized by SRP. These signal peptides are typically extremely hydrophobic (2, 3), but SRP apparently can also recognize slightly less hydrophobic signal peptides that contain a highly basic N terminus (4). SRP recognizes signal peptides as they emerge from translating ribosomes and then targets ribosome-nascent chain complexes to the IM cotranslationally (5). The binding of SRP to its receptor (FtsY), which interacts with the SecYEG complex (6), leads to the release of the nascent chain in the immediate vicinity of the translocation machinery. By targeting nascent polypeptides to the SecYEG complex at an early stage of translation, SRP prevents its substrates from folding into a conformation that is incompatible with translocation through the narrow channel formed by the SecYEG complex (7). Because most signal peptides are not recognized by E. coli SRP, the majority of presecretory proteins are fully synthesized and targeted post-translationally to the IM. These proteins are maintained in a translocation-competent conformation by molecular chaperones such as SecB that keep them unfolded (or loosely folded) (8). Signal peptides themselves also appear to play a role in maintaining translocation competence (9, 10). After mediating the targeting reaction, signal peptides likely play a role in gating open the SecYEG complex to initiate translocation.
Interestingly, although signal sequences are the most salient feature of presecretory proteins, they are neither completely necessary nor sufficient to mediate protein export in E. coli (11–13). A version of alkaline phosphatase that lacks a signal peptide is still exported, albeit very inefficiently (11). The export of the leaderless protein, unlike the export of wild-type alkaline phosphatase, is strictly dependent on SecB (11). Conversely, the attachment of signal peptides to cytoplasmic proteins often does not promote their export (14). In light of evidence that folding and export are competing events, these observations led to the proposal that exported proteins tend to fold slowly (or are prevented from folding by chaperones) and therefore remain translocation-competent even without a signal peptide, whereas cytoplasmic proteins fold rapidly into a conformation that is incompatible with export. Recent studies that used thioredoxin as a model protein have validated this hypothesis. Whereas the wild-type protein attached to a typical signal peptide remained trapped in the cytoplasm, four of five slow folding mutants were exported efficiently (15). Furthermore, attachment of a signal peptide that is recognized by SRP to thioredoxin led to efficient export (16). This idea was further confirmed by a report in which various DARPins (designed ankyrin Repeat proteins) were attached to different signal peptides. Most of the DARPins were exported efficiently when they were fused to signal peptides that mediate cotranslational targeting but remained in the cytoplasm when they were attached to signal peptides that are bypassed by SRP (17).
Despite these observations, there are several lines of evidence suggesting that export efficiency is not simply dictated by the ability of a protein to reach the SecYEG complex before folding into a translocation-incompetent conformation. For reasons that are unclear, some DARPins are secreted inefficiently even when they are routed into the SRP pathway (17). In addition, numerous reports have indicated that the amino acid composition of the segment of post-translationally targeted presecretory proteins that lies just beyond the signal peptide cleavage site has a dramatic effect on export efficiency. Statistical analysis has shown that the first ∼5–15 residues of the mature region of most presecretory proteins produced by Gram-negative bacteria is neutral or has a net negative charge (18). Consistent with the observed sequence bias, the presence of multiple basic residues at the N terminus of the mature region often leads to accumulation of the secretory precursor, whereas conversion of the basic residues to acidic residues restores export (19–22). Because different combinations of proteins and signal peptides were used in these studies, the exact number and location of charged residues that impinge on the efficiency of export is unclear. In any case, the effect of the net charge in the region distal to the signal peptide on protein export has never been explained. Although basic residues might conceivably promote premature folding of presecretory proteins or block the cleavage of signal peptides by leader peptidase, it is also possible that they inhibit an uncharacterized post-targeting event. Even if effects on signal peptide cleavage could have been ruled out in the aforementioned studies, however, it would not have been possible to distinguish between effects on protein folding and effects on a hypothetical post-targeting step because only proteins that are targeted post-translationally were monitored.
To gain further insight into the factors that govern the efficiency of protein export, we sought an explanation for the observation that the cotranslational targeting of at least some cytoplasmic proteins is insufficient to guarantee their translocation across the IM. We found that the export of several different endogenous E. coli cytoplasmic proteins required not only the attachment of a signal peptide that is recognized by SRP but also a net negative charge just past the signal peptide cleavage site. Taken together with previous results, our data show that the charge of the segment just beyond the signal peptide influences export efficiency irrespective of the mechanism by which a protein is targeted to the IM. Because proteins that are targeted cotranslationally reach the IM before they have a chance to fold, our results imply the existence of a post-targeting step (most likely the initiation of translocation) that is facilitated by acidic residues distal to the signal peptide and inhibited or delayed by basic residues. These results help to resolve a long-standing puzzle about the influence of the mature region of presecretory proteins on protein export and have significant implications for optimizing the export of cytosolic and heterologous proteins in E. coli.
EXPERIMENTAL PROCEDURES
Reagents, Bacterial Strains, and Media—A rabbit polyclonal antiserum against the influenza hemagglutinin (HA) epitope HA1.1 was obtained from Covance. A polyclonal antiserum against Ffh was described previously (23). All of the experiments were conducted using the E. coli strains MC4100 (F-araD139 Δ(argF-lac)U169 rpsL150 relA1 thi fib5301 deoC1 ptsF25 rbsR) and MRE600 (24). Media preparation and basic bacterial manipulations were performed by standard methods (25). Selective media contained 100 μg/ml ampicillin.
Plasmid Construction—Plasmid pHL33, which encodes an HA-tagged version of MBPSP-Pgk under the control of the tac promoter, has been described (26). Plasmids pHL40 and pHL41, which encode MBPSP-GapA and MBPSP-Eno, respectively, were constructed in an analogous manner. Initially the gapA and eno genes were amplified and HA-tagged by PCR using the oligonucleotides 5′-ACACGATTCCTCTAGACGCTGCGTAAGGT-3′ and 5′-CCCAAGCTTACAGGCTCGCATAATCCGGCACATCGTACGGATAACATTTGGAGATGTGAGCGATCAGGTCC-3′ (for gapA) and 5′-GTACGCGTTGTTTGTCTAGAGTTTCAGTTTAA-3′ and 5′-CCCAAGCTTACAGGCTCGCATAATCCGGCACATCGTACGGATAACATGCCTGGCCTTTGATCTCTTTACGA-3′ (for eno) and genomic DNA from strain LE392 as a template. The amplified DNA was then digested with XbaI and HindIII and cloned into pTRC99a (GE Healthcare) to construct pHL38 and pHL39. Next, the tagged versions of gapA and eno were reamplified using the oligonucleotides 5′-CTGGTGGAATATAAGATCTTCAAAGTAGGTAT-3′ (for gapA), 5′-CCTAATGTCCAAGATCTTAAAAATCATCGGTC-3′ (for eno), and 5′-CCCAAGCTTACAGGCTCGCATAATC-3′ (for both genes) and plasmids pHL38 and pHL39 as templates. The resulting PCR products were then digested with BglII and HindIII and cloned into a derivative of pMal-p2X (New England Biolabs) containing a BglII site at nucleotide 1607 to generate pHL40 and pHL41. The MBP signal peptide in pHL33, pHL40, and pHL41 was converted to the MBP*1 signal peptide (to make pHL42-pHL44), and additional changes were introduced into the signal peptide and the enzymes by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene). To produce templates for coupled transcription-translation reactions, a BamHI site was introduced upstream of the MBP or MBP*1 signal peptide using the oligonucleotide 5′-CCAGGACAAGCTGGATCCGTTTACCTGGGATG-3′, and an internal HindIII site was introduced into pgk, gapA, and eno. BamHI-HindIII fragments encoding the N terminus of the signal peptide-enzyme fusions were then cloned into pGEM-4z (Promega) under the control of the T7 promoter.
Pulse-Chase Labeling and Proteinase K Digestion—The cells were grown at 37 °C in M9 medium containing 0.2% glucose. Overnight cultures were washed and diluted into fresh medium at A550 = 0.02. When cultures reached A550 = 0.2, isopropyl β-d-thiogalactopyranoside (100 μm) was added to induce the expression of plasmid-borne derivatives of Pgk, GapA, and Eno, and cultures were incubated for an additional 30 min. The cells were then subjected to pulse-chase labeling by the addition of 30 μCi/ml Tran35S-label (MP Biomedical) for 30 s followed by the addition of 1 mm l-methionine and l-cysteine. The aliquots were removed at various time points, and the proteins were precipitated with cold 10% trichloroacetic acid. In experiments that involved proteinase K digestion, radiolabeled cells were pelleted by centrifugation at 6,800 × g for 8 min and resuspended in 1 ml of cold 40% sucrose/33 mm Tris, pH 8.0. The samples were then mixed with 100 μg/ml lysozyme, 2 mm EDTA and incubated on ice for 20 min. One half of each sample was trichloroacetic acid-precipitated, and the other half was treated with proteinase K (200 μg/ml) for 20 min on ice. Protease digestions were stopped by the addition of 2 mm phenylmethanesulfonyl fluoride, and the proteins were trichloroacetic acid-precipitated. Immunoprecipitations were performed as described previously (27). The proteins were resolved by SDS-PAGE on 8–16% Tris-glycine minigels (Invitrogen).
Coupled in Vitro Transcription-Translation Reactions and Chemical Cross-linking—S-30 extracts were prepared from strain MRE600, and coupled transcription-translation reactions were performed essentially as described (28). Linear DNA templates encoding the first 86 amino acids of each protein were synthesized by PCR using the oligonucleotide 5′-GAGCGGATAACAATTTCACACAGGAAACAGCTATGACC-3′ and an appropriate downstream oligonucleotide. For each 100-μl reaction, 1–2 μg of purified DNA template was added. The reactions were performed at 37 °C for 30 min and chilled for 5 min on ice. A portion (10 μl) of each sample was then removed, and the proteins were acetone-precipitated. Bis(sulfosuccinimidyl) suberate (BS3; 2 mm) was added to the remainder of each sample, and cross-linking was conducted at room temperature for 30 min. The cross-linking reactions were then quenched by incubating samples with 30 mm Tris, pH 8.0, at room temperature for 15 min. An aliquot (25 μl) of each sample was acetone-precipitated, and the remainder (70 μl) was trichloroacetic acid-precipitated and used for immunoprecipitations. Finally, the proteins were resolved on 10–20% Tricine minigels (Invitrogen).
Sequence Analysis—The complete set of proteins predicted to be produced by each of the organisms we analyzed was extracted from GenBank™ (except human proteins, which were extracted from BaCeILo data sets (29)). The proteins predicted with a 99.9% confidence level by SignalP (30) to contain signal peptides were defined as presecretory proteins. The set of cytoplasmic proteins was obtained by removing integral membrane proteins (predicted by TMHMM (31)) and low confidence presecretory proteins (proteins predicted to contain a signal peptide with a confidence level of 10–99.9%). For bacterial genomes, proteins predicted to be exported via the TAT pathway were also removed from the sets of presecretory and cytoplasmic proteins using TatP (32).
RESULTS
The Presence of a Highly Hydrophobic Signal Peptide Is Not Sufficient to Promote Efficient Export of Three Glycolytic Enzymes—In our initial experiments we wished to examine the efficiency with which different signal peptides direct the translocation of three endogenous glycolytic enzymes, phosphoglycerate kinase (Pgk), glyceraldehyde-3-phosphate dehydrogenase (GapA), and enolase (Eno), across the E. coli IM. To this end we attached either the native maltose-binding protein (MBP) signal peptide (MBPSP) or a more hydrophobic derivative (MBP*1SP) to each protein (Fig. 1A). We also attached a C-terminal HA epitope tag to facilitate detection. During the cloning process the first 2–3 amino acids of each protein were modified (Fig. 1B). Whereas the MBP signal peptide is bypassed by SRP and targets proteins to the SecYEG complex post-translationally, the MBP*1 signal peptide is recognized by SRP and promotes cotranslational targeting (2). MC4100 transformed with plasmids that encode the signal sequence-bearing proteins under the control of the tac promoter were grown in minimal medium. After the addition of isopropyl β-d-thiogalactopyranoside to induce synthesis of the modified enzymes, the cells were subjected to pulse-chase labeling. Each enzyme was then immunoprecipitated with an anti-HA antiserum. Translocation efficiency was monitored by following signal peptide cleavage and examining the accessibility of each protein to proteinase K digestion following conversion of the cells to spheroplasts. In experiments that involved proteinase K treatment, the protein component of SRP (Ffh) was also immunoprecipitated and served as a control for the integrity of the IM.
FIGURE 1.
N-terminal sequences of the constructs used in this study. In this study, the MBP signal peptide (MBPSP) or a derivative shown in A was fused to the E. coli glycolytic enzymes Pgk, GapA, or Eno or a variant that begins with one of the N-terminal sequences shown in B. Point mutations and insertions are underlined. To facilitate the attachment of signal peptides to each enzyme, the first 2–3 residues of Pgk, GapA, and Eno were mutated and differ from those found in the native enzyme.
Consistent with previous results (26), we found that <20% of the MBPSP-Pgk was exported (Fig. 2A, lanes 1–4). Because the fraction of the precursor that was converted to the mature form did not increase over time, it appears that post-translational targeting of Pgk leads to a rapid loss of translocation competence. Furthermore, essentially no export of MBPSP-GapA or MBPSP-Eno was observed (Fig. 2, B, lanes 1 and 2, and C, lanes 1–4). Surprisingly, attaching MBP*1SP to the three enzymes did not significantly enhance export efficiency (Fig. 2, A, lanes 5–8; B, lanes 3–7; and C, lanes 5–8). Although MBP*1SP-GapA migrated slightly faster than MBPSP-GapA, the resistance of the protein to proteinase K digestion shows that the immunoprecipitated band corresponds to the precursor form.
FIGURE 2.
Neither post-translational nor cotranslational signal peptides direct efficient secretion of glycolytic enzymes. MC4100 were transformed with a plasmid that encodes Pgk (A), GapA (B), or Eno (C) fused to the indicated signal sequence and subjected to pulse-chase labeling. A C-terminal HA tag was attached to each protein during cloning. Because of the difficulty of resolving the precursor (p) and mature (m) forms of the proteins, in some cases cells were converted to spheroplasts after a 1-min chase and treated with proteinase K (PK). Precursor and mature forms of each enzyme were immunoprecipitated with an anti-HA antiserum. In one case the cytoplasmic protein Ffh was immunoprecipitated to demonstrate the integrity of the IM.
We next conducted chemical cross-linking experiments to confirm that the enzymes were targeted to the SecYEG complex by the expected pathway. Coupled transcription-translation reactions were programmed with linear DNA templates that encode the first 86 amino acids of each presecretory protein. Because the DNA templates lack a stop codon, nascent polypeptide chains synthesized in these reactions remain attached to ribosomes. Following polypeptide synthesis, the homobifunctional cross-linker BS3 was added to each reaction, and SRP binding was assessed by immunoprecipitating Ffh-containing adducts from the cross-linking reactions. A significant amount of a ∼58-kDa radiolabeled adduct (“X”) was observed when nascent chains containing MBP*1SP were synthesized (Fig. 3, lanes 2, 8, and 14). This adduct was the size of Ffh and the nascent chain combined and could be immunoprecipitated with an anti-Ffh antiserum (Fig. 3, lanes 20, 23, and 26). Although a very slightly larger product was observed when nascent chains containing MBPSP were synthesized (Fig. 3, lanes 1, 7, and 13, asterisk), this product was not effectively immunoprecipitated by anti-Ffh (Fig. 3, lanes 19, 22, and 25). These results provide direct evidence that SRP specifically recognizes MBP*1SP-Pgk, MBP*1SP-Eno, and MBP*1SP-GapA and rule out the possibility that sequences derived from the cytoplasmic enzymes interfere with SRP binding. Taken together, the data strongly suggest that the targeting of the cytoplasmic enzymes to the SecYEG complex by the cotranslational SRP pathway is insufficient to ensure their export.
FIGURE 3.
MBP*1SP-enzyme chimeras are recognized effectively by SRP. The N-terminal 86 residues of the indicated MBPSP-enzyme or MBP*1SP-enzyme (including the signal peptide) were synthesized in coupled transcription-translation reactions. Portions of each reaction that were treated with the cross-linker BS3 (lanes 1–3, 7–9, and 13–15) or untreated (lanes 4–6, 10–12, and 16–18) were analyzed by SDS-PAGE. T, major translation products; X, ∼58-kDa cross-linked product; *, ∼60-kDa cross-linked product. Portions of each sample that were treated with the cross-linker were also subjected to immunoprecipitation with an anti-Ffh antiserum (lanes 19–27).
Mutation of N-terminal Basic Residues to Acidic Residues Increases the Efficiency of Glycolytic Enzyme Export—In examining the sequences of the three glycolytic enzymes that we chose as model cytoplasmic proteins, we noticed that the net charge of the first 15 residues ranged from +1 (for Pgk) to +4 (for GapA) (Fig. 1B). Although N-terminal basic residues have only been shown to inhibit post-translational export, we hypothesized that a net positive charge in the segment just beyond the signal peptide might also interfere with cotranslational export. To test this idea, we introduced a variety of charge mutations into the N terminus of the three glycolytic enzymes and examined their effect on post-translational and cotranslational export. The mutant constructs were designated enzyme(-N) (e.g. Pgk(-2)) to indicate the net alteration in charge. MC4100 were transformed with the mutant plasmids, and translocation across the IM was assessed by monitoring signal peptide cleavage and the accessibility of presecretory proteins to proteinase K added to spheroplasts.
Interestingly, we found that converting basic and uncharged residues to acidic residues not only markedly enhanced the cotranslational export of Pgk but also stimulated its post-translational export to a lesser degree. Pulse-chase analysis showed that ∼50% of MBP*1SP-Pgk(-2) and ∼25% of MBPSP-Pgk(-2), which both contain a glutamate in place of the N-terminal lysine, was exported within the pulse-labeling period (Fig. 4A, top panel). No significant export was observed during the chase period. Pgk(-3) derivatives that contain a second charge mutation (L3E) were exported even more efficiently. Essentially all of the MBP*1SP-Pgk(-3) was exported within the pulse-labeling period (Fig. 4A, bottom panel, lanes 3–5). Treatment of spheroplasts with proteinase K confirmed that the single band observed at all time points corresponds to the mature form of the protein (Fig. 4B). Chemical cross-linking experiments indicated that SRP binds to MBP*1SP-Pgk and MBP*1SP-Pgk(-3) with similar affinities (Fig. 3, lanes 20 and 21) and thereby strongly suggest that the acidic residues dramatically increase export efficiency by facilitating a post-targeting event rather than by affecting protein targeting per se. Furthermore, whereas only ∼25% of MBPSP-Pgk(-3) was exported within the pulse-labeling period, more than half of the protein was exported by 2 min (Fig. 4A, bottom panel, lanes 1 and 2). Although it is possible that the acidic residues increase the translocation competence of the fully synthesized presecretory protein simply by altering its folding, in all probability they enhance post-translational and cotranslational export by acting at a step (most likely the initiation of translocation) where both targeting pathways converge.
FIGURE 4.
Charge mutations at the N terminus of glycolytic enzymes increase the efficiency of cotranslational export. MC4100 were transformed with a plasmid that encodes the indicated signal peptide-enzyme chimera and subjected to pulse-chase labeling. Modified versions of Pgk (A and B), GapA (C and D), and Eno (E and F) were examined. In B, D, and F, cells were converted to spheroplasts after a 1-min chase and treated with proteinase K (PK). Precursor (p) and mature (m) forms of each enzyme were immunoprecipitated with an anti-HA antiserum. The cytoplasmic protein Ffh was also immunoprecipitated to demonstrate the integrity of the IM.
The conversion of basic and neutral residues to acidic residues had a similar effect on the export of GapA and Eno. Because the precursor and mature forms of several of the derivatives (especially those containing the MBP*1SP signal peptide) were difficult to resolve, it was necessary to assess export by treating spheroplasts with proteinase K. The conversion of two lysines to glutamate (which produced a charge change of -4) led to the cotranslational export of nearly half of GapA and Eno and a charge alteration of -6 led to quantitative export (Fig. 4, C and E, lanes 3–5, and D and F, top and middle panels). Chemical cross-linking confirmed that MBP*1SP-GapA, MBP*1SP-GapA(-6), MBP*1SP-Eno, and MBP*1SP-Eno(-6) are all recognized effectively by SRP (Fig. 3, lanes 24 and 27; moderate differences in the level of cross-linked products appear to reflect differences in translation efficiency). These data provide further evidence that a net negative charge just past the signal peptide stimulates cotranslational export by promoting a post-targeting reaction. Although more mutations were required to achieve efficient export of GapA and Eno than Pgk, in each case a net negative charge of -2 to -3 within the first 15 residues was sufficient. Furthermore, it is noteworthy that altering the charge of GapA and Eno improved post-translational export only very modestly (Fig. 4, C and E, lanes 1 and 2, and F, bottom panel). Presumably the acceleration of post-targeting events caused by the charge alteration is insufficient to overcome the tendency of fully synthesized GapA and Eno to become translocation-incompetent before they reach the SecYEG complex.
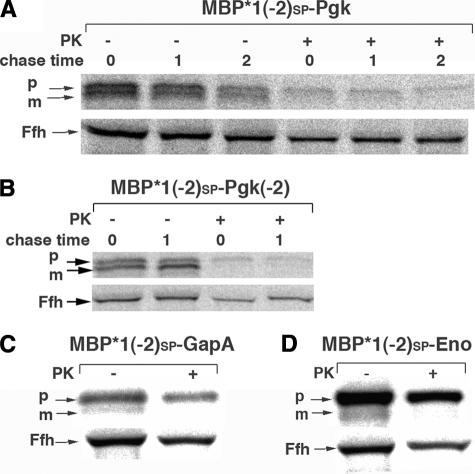
We next wished to determine whether the position of acidic residues at the N terminus of the glycolytic enzymes influences their export. To this end we examined the effect of inserting one or more aspartate residues immediately adjacent to the signal peptide. The addition of a single aspartate to the N terminus of Pgk (to create D-Pgk; Fig. 1B) led to the cotranslational export of ∼30% of the protein (Fig. 5A, top panel). The addition of two aspartates (to create DD-Pgk; Fig. 1B) led to the cotranslational export of nearly all of the protein and the post-translational export of ∼75% of the protein after a 2-min chase (Fig. 5A, bottom panel). In contrast, only about 50% of MBP*1SP-Pgk(-2) and <30% of MBPSP-Pgk(-2), which both have the same charge change as MBP*1ss-DD-Pgk and MBPSP-DD-Pgk, was exported (Fig. 4A, top panel). Likewise, the addition of two aspartates to the N terminus of GapA (to create DD-GapA) led to the cotranslational export of ∼50% of the protein (Fig. 5, B and C). Thus MBP*1SP-DD-GapA was exported with about the same efficiency as MBP*1SP-GapA(-4) (Fig. 4, C and D, top panels) despite the fact that the N terminus of the enzyme was less acidic. Curiously, the insertion of up to eight aspartates at the extreme N terminus of GapA had essentially the same effect as the insertion of two aspartates (data not shown). Taken together, these results suggest that both the magnitude of the charge as well as the position of charged residues within the N terminus of the mature region of a cotranslationally targeted protein can influence export efficiency.
FIGURE 5.
The position of charged residues influences the efficiency of glycolytic enzyme secretion. MC4100 were transformed with a plasmid that encodes the indicated signal peptide-enzyme chimera and subjected to pulse-chase labeling. Modified versions of Pgk (A) and GapA (B and C) were examined. In C the cells were converted to spheroplasts after a 1-min chase and treated with proteinase K (PK). Precursor (p) and mature (m) forms of each enzyme were immunoprecipitated with an anti-HA antiserum. The cytoplasmic protein Ffh was also immunoprecipitated to demonstrate the integrity of the IM.
Acidic Residues Are Overrepresented at the N Terminus of the Mature Region of Eubacterial Presecretory Proteins—Taken together with previous studies, our results suggest that acidic residues at the N terminus of E. coli presecretory proteins play a key role in promoting protein translocation irrespective of the pathway by which a protein is targeted to the SecYEG complex. The data predict that the N terminus of the mature region of most presecretory proteins in E. coli as well as other organisms in which translocation is initiated by the same mechanism should be negatively charged. Indeed a previous in silico analysis performed on a limited data set found a negative charge bias at the N terminus of the mature region of presecretory proteins produced by Gram-negative bacteria but not those produced by Gram-positive bacteria or humans (18). To perform a more complete analysis, we examined the entire set of proteins produced by a variety of organisms that are predicted with a confidence level of 99.9% to contain signal peptides by the latest version of SignalP (30). We found that the first ∼10–15 residues of the mature region of the vast majority of predicted presecretory proteins produced not only by E. coli and ten other arbitrarily selected Gram-negative bacteria but also those produced by Bacillus subtilis and six other Gram-positive bacteria all have a neutral or net negative charge (Fig. 6, A and B). As the distance from the signal peptide cleavage site increases, the skewed charge distribution disappears. In contrast, no charge bias was detected at the N terminus of cytoplasmic proteins produced by any organism. The identification of a charge bias in presecretory proteins produced by Gram-positive bacteria is striking because their signal peptides are especially hydrophobic (33, 34), and they therefore might utilize the SRP pathway more extensively than Gram-negative organisms. Curiously, the N terminus of a significant number of E. coli, Bacillus subtilis, and other bacterial presecretory proteins is basic (Fig. 6, F and G, and data not shown). This observation suggests that although a net negative charge is generally of functional significance, it is not essential for the export of all proteins. Furthermore, the finding that presecretory proteins produced by yeast, roundworms, and humans do not exhibit a charge bias (Fig. 6, C–E) suggests that the mechanism of translocation initiation might differ in prokaryotes and eukaryotes.
FIGURE 6.
The N terminus of the mature region of bacterial presecretory proteins shows a distinct charge bias. The average charge per residue (Q) in predicted presecretory proteins (green) and cytoplasmic proteins (red) in Gram-negative bacteria (A), Gram-positive bacteria (B), Homo sapiens (C), Caenorhabditis elegans (D), and Saccharomyces cerevisiae (E) was plotted for the first N residues from the N terminus of each set of proteins. The Gram-negative organisms analyzed were: E. coli K-12; Chlamydophila pneumoniae; Pseudomonas syringae pv. tomato str. DC3000; Lawsonia intracellularis PHE/MN1–00; Synechococcus elongatus PCC 6301; Rhodopseudomonas palustris CGA009; Myxococcus xanthus DK 1622; Anaeromyxobacter dehalogenans 2CP-C; Rhodoferax ferrireducens T118; Yersinia pestis Antiqua; Ralstonia metallidurans CH34; and Sphingopyxis alaskensis RB2256. The Gram-positive organisms analyzed were: Bacillus anthracis Sterne; Bacillus subtilis 168; Mycobacterium tuberculosis H37Rv; Staphylococcus saprophyticus ATCC 15305; Mycobacterium bovis AF2122/97; Staphylococcus aureus MRSA252; Bacillus licheniformis ATCC 14580; Staphylococcus aureus COL; and Rubrobacter xylanophilus DSM 9941. The percentage of E. coli and B. subtilis presecretory proteins that have a given net charge within the first 15 residues of their mature regions is depicted in F and G.
Elimination of Basic Residues within the Signal Peptide Enhances Pgk Secretion—Because the N terminus of signal sequences is usually positively charged (35), we hypothesized that acidic residues at the start of the mature region of most bacterial presecretory proteins might facilitate the formation of a structure (e.g. a hairpin) that accelerates the initiation of translocation. As a corollary, we conjectured that the presence of basic residues just past the signal peptide might delay the onset of translocation by causing charge repulsion and preventing the formation of the appropriate structure. One prediction of this hypothesis is that translocation might be enhanced not only by eliminating positive charges at the start of the mature region of a presecretory protein but also by reducing the positive charge of the N terminus of the signal peptide. To test this idea, we mutated both of the lysine residues of MBP*1SP to asparagine to create MBP*1SP(-2) and examined the effect of the modified signal peptide on the export of Pgk, GapA, and Eno. Consistent with our hypothesis, we found that the MBP*1SP(-2) signal peptide significantly enhanced Pgk export (Fig. 7A). Interestingly, the -2 charge change in the signal peptide had essentially the same effect on Pgk export as a charge alteration of the same magnitude at the N terminus of the enzyme (Fig. 4A). Attaching the MBP*1SP(-2) signal peptide to Pgk(-2) only subtly increased export efficiency, suggesting that charge alterations at the N terminus of the signal peptide and the N terminus of Pgk act redundantly (compare Fig. 7B to Fig. 4A, top panel, lanes 3–5). Fusion of the MBP*1SP(-2) signal peptide to GapA and Eno did not promote significant export (Fig. 7, C and D), but this result was not surprising because translocation of these proteins across the IM appeared to require a larger charge alteration than Pgk. Taken together, the data support the idea that a charge balance between the signal peptide and the N terminus of the mature region of a bacterial presecretory protein stimulates a key post-targeting reaction.
FIGURE 7.
The charge of the signal peptide influences the efficiency of Pgk secretion. MC4100 were transformed with a plasmid that encodes the indicated signal peptide-enzyme chimera and subjected to pulse-chase labeling. Modified versions of Pgk (A) and GapA (B) and Eno (C) were examined. In B and C cells were converted to spheroplasts after a 1-min chase and treated with proteinase K (PK). Precursor (p) and mature (m) forms of each enzyme were immunoprecipitated with an anti-HA antiserum. The cytoplasmic protein Ffh was also immunoprecipitated to demonstrate the integrity of the IM.
DISCUSSION
In this study we obtained evidence that cotranslational targeting of cytoplasmic proteins to the E. coli SecYEG complex is necessary but not sufficient for their translocation across the IM. We found that efficient export of three E. coli glycolytic enzymes required both the attachment of a signal peptide that is recognized by SRP and the introduction of mutations that change the net charge of the first 15 residues from basic to acidic. Both the magnitude of the charge and the position of acidic residues influenced the efficiency of translocation. The requirement for an acidic N terminus was not detected in previous studies on cotranslational export of cytoplasmic proteins because those studies fortuitously employed a model protein that has an acidic N terminus (thioredoxin) or cassette cloning in which the signal peptide is followed by an acidic epitope tag (FLAG tag) (16, 17). The simplest interpretation of our data is that cotranslational export requires a post-targeting step, most likely the initiation of translocation, that is mediated by a peptide encompassing both the signal peptide and the N terminus of the mature region and that is optimized by the presence of N-terminal acidic residues. Presumably the binding of SRP to a signal peptide directs ribosome-nascent chain complexes to the translocation machinery at an early stage of translation, but a significant delay in the post-targeting reaction results in a loss of translocation competence caused by ongoing polypeptide synthesis. The SRPs of Gram-negative bacteria lack the “translation arrest” domain found in other SRPs and probably do not slow translation upon recognition of a signal peptide, so it is likely that the time window before the onset of translocation is limited even during cotranslational targeting. In any case, our data show that the docking of the ribosome on the SecYEG complex (which only occurs during cotranslational targeting) does not guarantee the success of subsequent steps in the export process.
The inhibitory effect of N-terminal basic residues on post-translational export has been noted in many different studies but has never been explained. In light of our experimental results and the results of our in silico analysis, which indicate that the N-terminal ∼10–15 residues of the mature region of most eubacterial proteins is acidic, it is likely that N-terminal basic residues inhibit a key step in the export process that is independent of the mechanism by which a protein reaches the IM. Consistent with this interpretation, the conversion of basic residues at the N terminus of Pgk to acidic residues increased the efficiency of export even when the protein was targeted post-translationally. Presumably by accelerating the initiation of translocation, the acidic residues compensated for the relatively slow targeting of the protein and promoted export of a subset of molecules that reached the IM prior to folding into a transport-incompetent conformation. Because the N terminus of most signal peptides is basic, it is conceivable that salt bridges between the signal peptide and the mature region of a presecretory protein promote the formation of a transient structure that facilitates downstream events. Indeed the observation that charge mutations in the signal peptide and the N terminus of Pgk exert similar effects on export is consistent with this idea. Interestingly, efficient post-translational export also appears to require opposite charges flanking the hydrophobic core of a signal peptide (22). The significance of basic residues at the N terminus of signal peptides may be more complex, however, because an excess of signal peptide charge can retard export without obviously affecting the formation of a putative hairpin (4). Perhaps these residues mediate an essential interaction with acidic phospholipids, as has been previously proposed (36). Furthermore, it is curious that the basic nature of the signal peptide is generally conserved in cases where the N terminus of the mature region of a bacterial presecretory protein is also positively charged.3 In these cases the formation of an appropriate N-terminal structure might be promoted by as yet unidentified sequence elements. Alternatively, these proteins might have evolved to exit the cytoplasm slowly or to remain translocation-competent for a prolonged period.
Although our data imply the existence of at least one hitherto uncharacterized post-targeting reaction, they do not rule out the possibility that the initiation of translocation involves multiple steps. It is almost certain that translocation initiation involves the displacement of the “plug” (segment TM2a of SecY) that closes the SecYEG channel (37), but additional conformational changes, such as the opening of the lateral gate to allow the escape of the signal peptide into the lipid bilayer, may also be required. Moreover, the observation that gating of the translocation channel occurs relatively slowly (at least during membrane protein integration) suggests that gating may actually require multiple well coordinated conformational changes (38). In addition, recent evidence that the functional form of SecYEG is a dimer of heterotrimers in which one heterotrimer serves as a transport channel, whereas the other functions primarily as a docking site for SecA (39) raises the possibility that translocation initiation requires coordination between the two heterotrimers. Interestingly, we found that the SecY prlA4 mutation, which is thought to destabilize the closed form of the channel (7), does not significantly enhance the export of MBP*1SP-Pgk, MBP*1SP-GapA or MBP*1SP-Eno.3 This observation strongly suggests that the post-targeting event we have identified is distinct from plug displacement and that the segment comprised of the signal peptide plus the N terminus of a presecretory protein promotes translocation by mediating a different biochemical reaction.
Although post-targeting events have not previously been described for cotranslational export in bacteria, such events have been identified in studies of protein import into the endoplasmic reticulum in eukaryotic cells. There is considerable evidence that cotranslational import of proteins into the endoplasmic reticulum in mammalian cells requires the recognition of signal peptides by both SRP and the translocation machinery in two separate steps (40, 41). Furthermore, a detailed analysis using site-specific photocross-linking suggested that signal sequences and the adjoining seven residues interact with the translocon in a loop configuration in yeast (42). The significance of the first few residues of the mature region of presecretory proteins in the translocation process in eukaryotic cells has not been investigated, however. Nevertheless, our finding that most eukaryotic presecretory proteins lack the N-terminal acidic residues that enhance translocation in bacteria suggests that either the mechanics of translocation initiation differ in eukaryotes and prokaryotes or that the function mediated by acidic residues in prokaryotes is fulfilled by nonacidic residues in eukaryotic cells. Indeed the identification of multiple eukaryotic-specific factors that participate in translocation (e.g. TRAM, Sec62p complex, and TRAP complex; see Ref. 43) suggests that transport initiation may proceed by a somewhat different pathway.
Besides providing evidence for a post-targeting step that is required for effective cotranslational export in E. coli, our study may have significant practical implications. E. coli is widely used as a host for the expression of both homologous and heterologous proteins (44). Although the localization of these proteins in the periplasm is sometimes desirable, the attachment of signal peptides has not always led to efficient export. Our results suggest that it is necessary to optimize both the signal peptide and the contiguous sequence and that the use of a cotranslational targeting signal followed by a negatively charged peptide should provide a very general strategy for achieving effective export of a wide variety of proteins.
Acknowledgments
We thank Hin Lee for constructing some of the plasmids used in this study, Georg Koch for technical advice, and Yihong Ye for critical reading of the manuscript.
This work was supported, in whole or in part, by the National Institutes of Health.
Footnotes
The abbreviations used are: IM, inner membrane; BS3, bis(sulfosuccinimidyl) suberate; Eno, enolase; GapA, glyceraldehylde-3-phosphate dehydrogenase; HA, influenza hemagglutinin; MBP, maltose-binding protein; Pgk, phosphoglycerate kinase; SRP, signal recognition particle; SP, signal peptide; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
P. Tian, unpublished results.
References
- 1.Blobel, G., and Dobberstein, B. (1975) J. Cell Biol. 67, 835-851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee, H. C., and Bernstein, H. D. (2001) Proc. Natl. Acad. Sci. U. S. A. 98, 3471-3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowers, C. W., Lau, F., and Silhavy, T. J. (2003) J. Bacteriol. 185, 5697-5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson, J. H., Woolhead, C. A., and Bernstein, H. D. (2003) J. Biol. Chem. 278, 46155-46162 [DOI] [PubMed] [Google Scholar]
- 5.Keenan, R. J., Freymann, D. M., Stroud, R. M., and Walter, P. (2001) Annu. Rev. Biochem. 70, 755-775 [DOI] [PubMed] [Google Scholar]
- 6.Angelini, S., Deitermann, S., and Koch, H. G. (2005) EMBO Rep. 6, 476-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Berg, B., Clemons, W. M., Jr., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C., and Rapoport, T. A. (2004) Nature 427, 36-44 [DOI] [PubMed] [Google Scholar]
- 8.Collier, D. N., Bankaitis, V. A., Weiss, J. B., and Bassford, P. J., Jr. (1988) Cell 53, 273-283 [DOI] [PubMed] [Google Scholar]
- 9.Park, S., Liu, G., Topping, T. B., Cover, W. H., and Randall, L. L. (1988) Science 239, 1033-1035 [DOI] [PubMed] [Google Scholar]
- 10.Beena, K., Udgaonkar, J. B., and Varadarajan, R. (2004) Biochemistry 43, 3608-3619 [DOI] [PubMed] [Google Scholar]
- 11.Derman, A. I., Puziss, J. W., Bassford, P. J., Jr., and Beckwith, J. (1993) EMBO J., 12, 879-888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frate, M. C., Lietz, E. J., Santos, J., Rossi, J. P., Fink, A. L., and Ermacora, M. R. (2000) Eur. J. Biochem. 267, 3836-3847 [DOI] [PubMed] [Google Scholar]
- 13.Moreno, F., Fowler, A. V., Hall, M. Silhavy, T. J., Zabin, I., and Schwartz, M. (1980) Nature 286, 356-359 [DOI] [PubMed] [Google Scholar]
- 14.Mallik, I., Smith, M. A., and Flower, A. M. (2002) BMC Microbiol. 2, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber, D., Cha, M. I., Debarbieux, L., Planson, A. G., Cruz, N., Lopez, G., Tasayco, M. L., Chaffotte, A., and Beckwith, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102, 18872-18877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schierle, C. F., Berkman, M., Huber, D., Kumamoto, C., Boyd, D., and Beckwith, J. (2003) J. Bacteriol. 185, 5706-5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiner, D., Forrer, P., Stumpp, M. P., and Pluckthün, A. (2006) Nat. Biotechnol. 24, 823-831 [DOI] [PubMed] [Google Scholar]
- 18.Kajava, A. V., Zolov, S. N., Kalinin, A. E., and Nesmeyanova, M. A. (2000) J. Bacteriol, 182, 2163-2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, P., Beckwith, J., and Inouye, H. (1988) Proc. Natl. Acad. Sci. U. S. A. 85, 7685-7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamane, K., and Mizushima, S. (1988) J. Biol. Chem. 263, 19690-19696 [PubMed] [Google Scholar]
- 21.Summers, R. G., and Knowles, J. R. (1989) J. Biol. Chem. 264, 20074-20081 [PubMed] [Google Scholar]
- 22.Puziss, J. W., Strobel, S. M., and Bassford, P. J., Jr. (1992) J. Bacteriol. 174, 92-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein, H. D., Zopf, D., Freymann, D., and Walter, P. (1993) Proc. Natl. Acad. Sci. U. S. A. 90, 5229-5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cammack, K. A., and Wade, H. E. (1965) Biochem. J. 96, 671-680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, J. H. (1992) A Short Course in Bacterial Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 26.Lee, H. C., and Bernstein, H. D. (2002) J. Biol. Chem. 277, 43527-43535 [DOI] [PubMed] [Google Scholar]
- 27.Newitt, J. A., and Bernstein, H. D. (1998) J. Biol. Chem. 273, 12451-12456 [DOI] [PubMed] [Google Scholar]
- 28.Moser, M., Panahandeh, S., Holzapfel, E., and Muller, M. (2007) Methods Mol. Biol. 390, 63-79 [DOI] [PubMed] [Google Scholar]
- 29.Pierleoni, A., Martelli, P.L., Fariselli, P., and Casadio, R. (2006) Bioinformatics 22, e408-e416 [DOI] [PubMed] [Google Scholar]
- 30.Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007) Nat. Prot. 2, 953-971 [DOI] [PubMed] [Google Scholar]
- 31.Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001) J. Mol. Biol. 305, 567-580 [DOI] [PubMed] [Google Scholar]
- 32.Bendtsen, J. D., Nielsen, H., Widdick, D., Palmer, T., and Brunak, S. (2005) BMC Bioinformatics 6, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Heijne, G., and L. Abrahmsen (1989) FEBS Lett. 244, 439-446 [DOI] [PubMed] [Google Scholar]
- 34.Tjalsma, H., Bolhuis, A., Jongbloed, J. D., Bron, S., and van Dijl, J. M. (2000) Microbiol. Mol. Biol. Rev. 64, 515-547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Heijne, G. (1985) J. Mol. Biol. 184, 99-105 [DOI] [PubMed] [Google Scholar]
- 36.Nesmeyanova, M. A., Karamyshev, A. L., Karamysheva, Z. N., Kalinin, A. E., Ksenzenko, V. N., and Kajava, A. V. (1997) FEBS Lett. 403, 203-207 [DOI] [PubMed] [Google Scholar]
- 37.Smith, M. A., Clemons, W. M., Jr., DeMars, C. J., and Flower, A. M. (2005) J. Bacteriol. 187, 6454-6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng, Z., and Gilmore, R. (2006) Nat. Struct. Mol. Biol. 13, 930-936 [DOI] [PubMed] [Google Scholar]
- 39.Osborne, A. R., and Rapoport, T. A. (2007) Cell 129, 97-110 [DOI] [PubMed] [Google Scholar]
- 40.Jungnickel, B., and Rapoport, T. A. (1995) Cell 82, 261-270 [DOI] [PubMed] [Google Scholar]
- 41.Belin, D., Bost, S., Vassalli, J. D., and Strub, K. (1996) EMBO J. 15, 469-478 [PMC free article] [PubMed] [Google Scholar]
- 42.Plath, K., Mothes, W., Wilkinson, B. M., Stirling, C. J., and Rapoport, T. A. (1998) Cell 94, 795-807 [DOI] [PubMed] [Google Scholar]
- 43.Osborne, A. R., Rapoport, T. A., and van den Berg, B. (2005) Annu. Rev. Cell Dev. Biol. 21, 529-550 [DOI] [PubMed] [Google Scholar]
- 44.Baneyx, F. (1999) Curr. Opin. Biotechnol. 10, 411-421 [DOI] [PubMed] [Google Scholar]