Abstract
The objective of this study was to evaluate the physiological importance of the mitochondrial fatty acid synthesis pathway in mammalian cells using the RNA interference strategy. Transfection of HEK293T cells with small interfering RNAs targeting the acyl carrier protein (ACP) component reduced ACP mRNA and protein levels by >85% within 24 h. The earliest phenotypic changes observed were a marked decrease in the proportion of post-translationally lipoylated mitochondrial proteins recognized by anti-lipoate antibodies and a reduction in their catalytic activity, and a slowing of the cell growth rate. Later effects observed included a reduction in the specific activity of respiratory complex I, lowered mitochondrial membrane potential, the development of cytoplasmic membrane blebs containing high levels of reactive oxygen species and ultimately, cell death. Supplementation of the culture medium with lipoic acid offered some protection against oxidative damage but did not reverse the protein lipoylation defect. These observations are consistent with a dual role for ACP in mammalian mitochondrial function. First, as a key component of the mitochondrial fatty acid biosynthetic pathway, ACP plays an essential role in providing the octanoyl-ACP precursor required for the protein lipoylation pathway. Second, as one of the subunits of complex I, ACP is required for the efficient functioning of the electron transport chain and maintenance of normal mitochondrial membrane potential.
Eukaryotes employ two distinct systems for the synthesis of fatty acids de novo. The bulk of fatty acids destined for membrane biogenesis and energy storage are synthesized in the cytosolic compartment by megasynthases in which the component enzymes are covalently linked in very large polypeptides; this system is referred to as the type I fatty acid synthase (FAS)2 (1, 2). A second system localized in mitochondria is composed of a suite of discrete, freestanding enzymes that closely resemble their counterparts in prokaryotes (3–10), which are characterized as type II FASs (11). Most of the constituent enzymes of the mitochondrial fatty acid biosynthetic system have been identified and characterized in fungi and animals; all are nuclear-encoded proteins that are transported to the matrix compartment of mitochondria. Fungi with deleted mitochondrial FAS genes fail to grow on non-fermentable carbon sources, have low levels of lipoic acid and elevated levels of mitochondrial lysophospholipids (12, 13). These observations indicate that the mitochondrial FAS may serve to provide the octanoyl precursor required for the biosynthesis of lipoyl moieties de novo, as well as providing fatty acids that are utilized in remodeling of mitochondrial membrane phospholipids (14). The mitochondrial FAS system in animals is less well characterized. However, kinetic analysis of the β-ketoacyl synthase enzyme responsible for catalysis of the chain extension reaction in human mitochondria suggested that this system is uniquely engineered to produce mainly octanoyl moieties and has limited ability to form long-chain products (9). Indeed, studies with a reconstituted system from bovine heart mitochondrial matrix extracts confirmed that octanoyl moieties are the main product and are utilized for the synthesis of lipoyl moieties (15). One of the key components of the prokaryotic and mitochondrial FAS systems is a small molecular mass, freestanding protein, the ACP, that shuttles substrates and pathway intermediates to each of the component enzymes. The mitochondrial ACP is localized primarily in the matrix compartment (16), but a small fraction is integrated into complex I of the electron transport chain (17–23). As is the case with many of the other 45 subunits of complex I, the role of the ACP subunit is unclear (24). To clarify the physiological importance of the mitochondrial FAS, and the mitochondrial ACP in particular, in mammalian mitochondrial function we have utilized an RNA interference strategy to knockdown the mitochondrial ACP in cultured HEK293T cells.
EXPERIMENTAL PROCEDURES
Culturing and Transfection of HEK293T Cells—Human embryonic kidney (HEK) 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C in a 5% CO2 atmosphere. On-Target Plus siRNA reagents, as well as nonspecific siRNAs, were purchased from Dharmacon. The former consist of pooled siRNA species that are chemically modified to reduce off-target effects and favor antisense strand entry into the RNA-induced Silencing Complex. Transfection was performed using DharmaFECT 1 lipid transfection reagent. HEK293T cells were seeded at a density of 6 × 104 cells per 0.5 ml in each well of 24-well plates. DharmaFECT 1 (0.8 μl) was diluted in 50 μl of Opti-MEM I medium (Invitrogen), the mixture was incubated at 20 °C for 5 min, siRNA was added, and incubation continued for another 20 min. The final mixture was added to culture plate wells, resulting in a final siRNA concentration in the 10–100 nm range. In earlier experiments 100 nm was used until we subsequently established that 10 nm was equally effective in down-regulation of the ACP. In some control experiments, no siRNA was included with the transfection reagent. Cells were collected, counted, and assayed at various times after transfection. Some cell cultures were supplemented with dl-α-lipoic acid (Sigma) as either Tris or sodium salt.
Quantitative Real-time PCR—Total RNA was isolated using the RNeasy Mini kit (Qiagen) and treated with DNase (Qiagen) to avoid genomic DNA contamination. Total RNA yield was determined using a Nano-drop® ND-1000 Spectrophotometer. Total RNA (1 μg) was reverse transcribed using SuperScript® III Platinum® according to the manufacturer's instructions. All samples within an experiment were reverse transcribed at the same time, the resulting cDNA was diluted 1:5 in nuclease-free water and stored in aliquots at -80 °C. Real-time PCR was performed with intron-spanning target-specific primers and SYBR Green detection in 384-well plates, using an ABI Prism 7900 Fast Real-time PCR system (Applied Biosystems). Sequence Detector System software was used for data analysis.
Oligonucleotide Sequences—The On-Target Plus Smart Pool siRNA sequences were: for ACP, GGACCGUGUUCUUUACGUA, GGCCAUGGAAGACGAAUUU, CCAGAGAAGCUUUCAGUAA, and UGGACCAAGUGGAGAUUAU; for NDUFB8, GUAUGCAGCUCUUCGGUUUUU, GCAAAGGGCAUCCCGGAACUU, CCGCCAAGAAGUAUAAUAUUU, and GAGAGAGAUCCAUGGUAUAUU. The primer sequences for Real-time PCR were: for ACP, CTATGACAAGATTGACCCAGAGAAG and CAGCATCTATATCAGGAATTTCAAACC; for pyruvate dehydrogenase E2 subunit, AGAAGTTTTGTTGGTACGGAAAGAAC and AGTACTGACCGCAACACTGACAT; for β-actin, CATGTACGTTGCTATCCAGGC and CTCCTTAATGTCACGCACGAT.
MTT Assay—Cell viability was assessed using the MTT assay in HEK293T cells (5 × 103) seeded in a 96-well plate in 100-μl volumes. siRNA transfection was done at the time cells were seeded. MTT (10 μl of 5 mg/ml) was added at various times, and the cells were incubated for 4 h in a tissue culture incubator. Formazan produced in the cells was dissolved in 50 μl of lysis buffer (20% SDS, 0.2 m HCl), and the mixture incubated overnight in a humidified atmosphere. Absorbance of the formazan product was measured at 590 nm and the reference wavelength was set at 700 nm. The absorbance was directly proportional to cell number over the range 5 × 103 to 4 × 104 cells (A590 nm range 0.01–0.6). A unit of formazan production corresponds to A590 nm–A700 nm.
Fluorescence Microscopy—Loss of mitochondrial transmembrane potential and accumulation of ROS in cultured HEK293T cells were assessed at various times following siRNA treatment using the JC-1 mitochondrial potential detection kit (Cell Technology, Inc.) and CM-H2DCFDA dye (Invitrogen), respectively, according to the manufacturer's instructions. Labeled cells were examined using a Zeiss Axiovert 25 light fluorescence inverted microscope with mercury illumination interfaced to a computer equipped with basic spot processing software.
Preparation of Mitochondrial Extracts—All operations were carried out at 4 °C. HEK293T cells were washed with cold phosphate-buffered saline, pH 7, scraped from the wells, suspended in 250 mm sucrose buffered with 20 mm imidazole-HCl, pH 7, containing 1 mm EDTA and protease inhibitors (leupeptin (5 μg/ml), antitrypsin (5 μg/ml), pepstatin (1 μg/ml) trans-epoxysuccinyleucylamido(4-guanidino)butane (3.6 μg/ml), and phenylmethanesulfonyl fluoride (1 mm)) and homogenized with 30 strokes in a Dounce homogenizer. Homogenates were centrifuged at 1,000 × g for 10 min. The pellet was homogenized again with a new portion of the buffer and centrifuged. Both supernatants were combined and centrifuged at 12,000 × g for 30 min at 4 °C. The resulting pellet was used for preparation of mitochondrial extract or stored at -80 °C. Mitochondria were extracted by incubation for 30 min, with occasional mixing, in a solution (20 μl/21 mg wet cells) containing 2% lauryl maltoside, 50 mm sodium chloride, 50 mm imidazole-HCl, pH 7, 2 mm 6-aminohexanoic acid, 1 mm EDTA and protease inhibitors. The extract was centrifuged at 12,000 × g for 15 min, the supernatant was collected, and the extraction repeated. Both supernatants were combined and centrifuged at 100,000 × g for 20 min. The extract, which contained both matrix and solubilized membrane proteins, was aliquoted, flash-frozen in liquid nitrogen, and stored at -80 °C. Protein concentration in the extracts was measured with bicinchoninic acid reagent (Pierce) using bovine serum albumin as standard.
SDS-PAGE and Western Blotting—The procedures used were based on the methods described earlier (25, 26) except that the denaturation solution also contained 3 m urea, the transfer solution for Western blotting contained 20% methanol, and the nitrocellulose-membrane blocking solution was 5% fat-free dried milk powder dissolved in Tris-buffered saline containing 0.1% Tween 20. After exposure to the primary antibody, the membranes were washed in Tris-buffered saline containing 0.1% Tween 20 and incubated with peroxidase-conjugated secondary antibody. Enhanced Chemiluminescent or SuperSignal West Pico Chemiluminescent Reagent (Thermo Scientific Pierce) was used as substrate for peroxidase, according to manufacturer's recommendations.
The procedure used for SDS-PAGE of mitochondrial extract proteins was modified as follows. Denatured proteins (32–40 μg) were separated by Tricine SDS-PAGE using 10% acrylamide gels (27). Separated proteins were transferred to a polyvinylidene fluoride membrane in a semi-dry transfer system using CAPS buffer, pH 11, containing 10% methanol.
Enzyme Assays—Cell extracts initially were prepared either by lauryl maltoside extraction of the isolated mitochondrial fraction or by hypotonic treatment (freeze-thawing in 10 mm Tris-HCl, pH 7.8). For all complexes but complex III, the highest specific activities were observed with the mitochondria swollen by hypotonic treatment and this method was employed for preparation of mitochondria in subsequent assays. All assays were performed at 30 °C.
Respiratory complex I (NADH dehydrogenase) was assayed spectrophotometrically at 600 nm, in the presence of antimycin A, using 2,6-dichloroindophenol as the acceptor of electrons from decylubiquinone, which is reduced after oxidation of NADH by complex I (28). Rotenone-sensitive activity was defined as complex I activity. Complex II (succinate dehydrogenase) was assayed, in the presence of antimycin A, potassium cyanide, and rotenone, by measuring the rate of malonate-sensitive and succinate-dependent reduction of 2,6-dichloroindophenol in the presence of decylubiquinone (28, 29). Complex III activity (ubiquinol-cytochrome c reductase) was assayed by monitoring at 550 nm the antimycin A-sensitive reduction of ferri- to ferrocytochrome c by decylubiquinol, in the presence of Tween 20, albumin, and sodium azide (30). Complex IV activity (cytochrome c oxidase) was assayed by monitoring the azide-sensitive decrease in absorbance at 550 nm that accompanies the oxidation of ferro- to ferricytochrome c (31, 32). This assay was not linear and the first-order rate constant was used to calculate complex IV activities. Complex V (ATP synthase) was measured in the reverse direction by coupling the formation of ADP with the pyruvate kinase and lactic dehydrogenase reactions; pyruvate formed in the kinase reaction is converted to lactate resulting in the oxidation of NADH and a decrease in absorbance at 340 nm (33). Oligomycin-sensitive ATPase activity, catalyzed by F0 subunit, is reported as complex V activity.
Pyruvate and oxoglutarate dehydrogenase complex activities in hypotonically swollen mitochondria were monitored spectrophotometrically at 500 nm (34). β-Fluoropyruvate sensitivity (pyruvate dehydrogenase) and CoASH dependence (oxoglutarate dehydrogenase) were used to verify specificity. Prior to the assay pyruvate dehydrogenase complex was activated by incubation of hypotonically swollen mitochondria, at 1 mg protein/ml, with dichloroacetate and magnesium and calcium ions at 37 °C for 10 min (35).
Antibodies—Human mitochondrial ACP was expressed and purified as described previously (36) and polyclonal rabbit antibodies prepared by Antibodies Inc., Davis, CA. Polyclonal rabbit antibodies recognizing 4-hydroxy-2-nonenal and the E2 subunit of oxoglutarate dehydrogenase were gifts from Dr. Luke Szweda (Oklahoma Medical Research Foundation). Rabbit polyclonal antibodies raised against lipoic acid were purchased from Calbiochem. Individual mouse monoclonal antibodies against five subunits of human complex I and against the E2 subunit of human pyruvate dehydrogenase were obtained from Mitosciences (Eugene, OR). Mouse monoclonal antibody against branched-chain oxoacid dehydrogenase was purchased from Abcam (Cambridge, MA) and those recognizing β-actin from Sigma.
Blue Native PAGE and Complex I Activity—Mitochondrial complexes were separated on 1-mm thick 4–13% acrylamide gel gradients, using a 3.5% stacking gel, prepared in a Bio-Rad Mini Protean II system (37). Each sample, usually 32 μg of protein, contained 5% glycerol and 0.5% G-250 suspension in 0.5 m 6-aminohexanoic acid to achieve a mass ratio between lauryl maltoside and G-250 equal to 30. Electrophoresis was performed at 4 °C, 60 V for the first 15 min, then at 5 mA constant current. Deep blue buffer was replaced with light blue buffer (37) when the front reached 2/3 of the gel length. Complex I activity was measured directly in the gel using nitro blue tetrazolium (38). For two-dimensional analysis, Blue Native gel electrophoresis was performed in the first dimension, 3-cm sections of the gel were cut out, soaked for 1 h at 20 °C in Tricine SDS-PAGE sample buffer containing 1% mercaptoethanol and 10 mm tris(2-carboxyethyl)phosphine (39), and sealed into a 4% acrylamide stacking gel poured over a 10% acrylamide SDS-Tricine running gel. Electrophoresis was performed first at 15 V for 2.5 h and then at constant current, 16 mA/plate, for 2.5 to 3 h. Western blotting, in a semi-dry system, was performed as described above. Integrated density measurements were done with ImageJ version 1.38x software (National Institutes of Health).
RESULTS
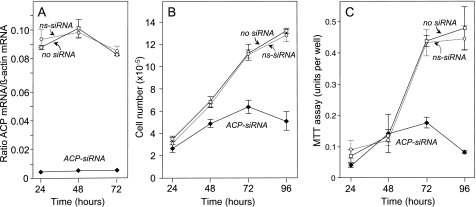
Effect of ACP siRNA on ACP mRNA Level and Cell Growth—Transfection of HEK293T cells with siRNAs targeting mitochondrial ACP was extremely effective in down-regulating ACP expression. Within 24 h, the ACP mRNA level was reduced by 95% and this low level was maintained for at least 72 h, whereas that in control cells, treated with either nonspecific siRNA or vehicle only, were relatively unaffected (Fig. 1A). Cell growth was slowed significantly by 48 h and by 72 h cells began to die and detach from the culture plate (Fig. 1B). The reduction in the viable cell population was also reflected in the decreased ability of the ACP siRNA-treated cells to reduce 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide to the insoluble formazan dye in the MTT assay (Fig. 1C) and the appearance of cells that stained with 7-amino-actinomycin D (details not shown).
FIGURE 1.
Effect of mitochondrial ACP down-regulation on growth of HEK293T cells. HEK293T cells were treated with 100 nm mitochondrial ACP siRNA (diamonds), nonspecific siRNA (circles, dotted lines), or no siRNA (squares). A, ACP mRNA level, normalized to β-actin mRNA. B, cell number. C, MTT assay.
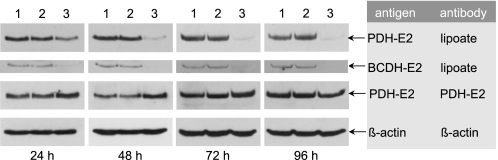
Effect of ACP Down-regulation on the Protein Lipoylation Profile—Our previous finding that the fatty acid biosynthetic and protein lipoylation pathways are directly linked in mammalian mitochondria prompted us to examine the effect of ACP down-regulation on the lipoylation status of mitochondrial proteins, using antibodies that recognize lipoyl moieties. Three lipoylated proteins were recognized by the antibodies in HEK293T cells. The species giving the strongest anti-lipoyl signal was identified as pyruvate dehydrogenase, using antibodies that recognize specifically the E2 subunits of this complex. The level of pyruvate dehydrogenase E2 protein was not decreased by treatment of cells with ACP siRNA (Fig. 2). However, the ability of anti-lipoyl antibodies to recognize the E2 subunits was dramatically curtailed as early as 24 h post-transfection; this reduced reactivity toward anti-lipoyl antibodies was maintained for at least 96 h post-transfection. Cells treated with nonspecific siRNA, or vehicle alone, did not exhibit altered lipoylation profiles. Recognition of a second, faster-moving lipoylated protein was also dramatically reduced by treatment of the cells with ACP siRNA. This species was identified as the E2 subunit of branched-chain oxoacid dehydrogenase using antibodies specific for that protein (details not shown). Lipoylation of a third protein, identified as the H-protein of the glycine cleavage system based on its molecular mass of ∼14 kDa, was also dramatically reduced by transfection of HEK293T cells with ACP siRNAs (details not shown).
FIGURE 2.
Effect of siRNA treatment on the lipoylation profile of HEK293T cells. Cells were treated with either vehicle alone (lanes 1), 100 nm nonspecific siRNA (lanes 2), or 100 nm ACP siRNA (lanes 3) and cultured for up to 96 h. Whole cell extracts were prepared and the proteins from 30,000 cells were subjected to SDS-PAGE/Western blotting using antibodies specific for lipoic acid, the E2 subunit of pyruvate dehydrogenase (PDH) or β-actin. The electrophoretic mobilities of PDH-E2 and branched chain oxoacid dehydrogenase E2 (BCDH-E2) corresponded to molecular masses of ∼60 and 47 kDa, respectively.
Direct spectrophotometric assay of pyruvate and oxoglutarate dehydrogenases revealed that 48 h following transfection with ACP siRNA, the activities were lowered by ∼40% (Table 1). These results suggested that down-regulation of mitochondrial ACP may reduce the availability of octanoyl moieties required for the synthesis of lipoyl moieties de novo, thus compromising the post-translational modification reactions.
TABLE 1.
Effect of down-regulation of mitochondrial ACP on activities of oxoacid dehydrogenases and respiratory complexes HEK293T cells were treated with 10 nm nonspecific siRNA (control) or ACP siRNA, cultured for 48 h, and swollen mitochondria were prepared for assay. Values are means (± S.D.) for two to three determinations. Specificity of the assays was evaluated using specific inhibitors: approximately 70% activity measured in the pyruvate dehydrogenase assay was inhibited by β -fluoropyruvate, 90% activity measured in oxoglutarate assay was CoASH dependent, 75% of the activity measured in the complex I assay was inhibited by rotenone; 95% of the activity measured in the complex II assay was inhibited by malonate; 75% of the activity in the complex III assay was inhibited by antimycin A; 95% of activity in the complex IV assay was inhibited by sodium azide; 50% of activity in the complex V assay was inhibited by oligomycin. 100% values, in nanomole min–1 mg protein–1, were: for pyruvate dehydrogenase, 27.7; oxoglutarate dehydrogenase, 43.5; complex I. 153; complex II, 212, complex III, 377, complex V, 108. The first-order rate constant of complex IV was 1,480 nmol–1 min–1 mg–1.
| Enzyme | Nonspecific siRNA | No treatment | +ACP siRNA | ||
|---|---|---|---|---|---|
| activity, % control | |||||
| Pyruvate dehydrogenase | 100 ± 5.5 | 97.3 ± 4.4 | 51.9 ± 0.3a | ||
| Oxoglutarate dehydrogenase | 100 ± 7.8 | 110 ± 0.8 | 51.6 ± 2.5a | ||
| Complex I | 100 ± 5.1 | 124 ± 0.9a | 61.6 ± 5.7a | ||
| Complex II | 100 ± 1.2 | 111 ± 4.1b | 75.3 ± 0.1a | ||
| Complex III | 100 ± 1.3 | 96.4 ± 13.7 | 94.2 ± 4.6 | ||
| Complex IV | 100 ± 3.9 | 104 ± 2.8 | 108 ± 3.4 | ||
| Complex V | 100 ± 10.6 | 122 ± 0.8 | 112 ± 13.5 | ||
p values indicate activities statistically different from the nonspecific siRNA control: <0.02
p values indicate activities statistically different from the nonspecific siRNA control: <0.05
Effect of Down-regulation of Mitochondrial ACP on Integrity of Respiratory Chain Complexes—Because mitochondrial ACP has been implicated as one of the many subunits of respiratory complex I, we evaluated the effect of ACP siRNA treatment on the integrity of this complex. For comparison, HEK293T cells were also transfected with siRNAs targeting NDUFB8, another of the nuclear-encoded complex I subunits. The mitochondrial content of ACP and NDUFB8 were both significantly lowered by treatment with their respective siRNAs (Fig. 3A). In contrast, the mitochondrial content of NDUFS3, another of the complex I subunits, was unaffected by treatment with siRNAs targeting either ACP or NDUFB8. Complex I was separated from other respiratory complexes by Blue Native gel electrophoresis, localized by Western blotting with antibodies directed against the NDUFA9 subunit, and its activity was assessed directly in the gel (Fig. 3B). Although ACP protein levels in HEK293T cell mitochondria were significantly reduced 24 h after transfection with ACP siRNA, no decrease in complex I activity was apparent until 48 h post-transfection. Activity at this time was estimated as ∼47% of that of untreated control cells. Transfection of cells with siRNAs targeting the NDUFB8 subunit resulted in a similar decrease in complex I activity by 48 h post-transfection; activity was reduced to ∼45% of the control.
FIGURE 3.
Effect of siRNA treatment on the integrity of respiratory complex I. HEK293T cells were treated with 10 nm siRNA targeting either the ACP or NDUFB8 subunits of complex I and cultured up to 48 h. Mitochondria were isolated and a 2% lauryl maltoside extract prepared. A, the content of three complex I subunits was assessed by SDS-PAGE and Western blotting with specific antibody probes for ACP, NDUFB8, and NDUFS3. In the control experiment (c) no siRNA treatment was performed; si, siRNA-treated. B, respiratory complexes were fractionated by Blue Native gel electrophoresis and complex I activity was assayed directly in the gel. The location of complex I in the gel (arrow) was confirmed by blotting onto a polyvinylidene fluoride membrane and probing with antibodies specific for antibodies of the NDUFA9 subunit (far right lane). C and D, two-dimensional electrophoretic analysis of complex I. After Blue Native gel electrophoresis in the direction indicated by the arrows, SDS-PAGE was performed in the second dimension and selected subunits of complex I were detected by Western blotting using a mixture of five specific antibodies. Integrated density measurements for all detected subunits indicate that the amount of complex I in ACP and NDUFB8 siRNA-treated cells was 122 ± 41 and 40 ± 19%, respectively, of the control values.
To distinguish between the possibilities that the decreased activity in complex I induced by siRNA treatment could reflect either a decrease in the total amount of complex or a decrease in specific activity of the complex, electrophoresis was performed in a second dimension, using SDS-PAGE, and the amounts of five subunits present was assessed immunochemically (Fig. 3, C and D). The results revealed that in the NDUFB8 siRNA-treated cells the amount of these subunits present in complex I was ∼40% of that of control cells, of similar magnitude to the decrease in activity (∼45%); thus the total amount of complex I, but not the specific activity was reduced. In contrast, cells treated with ACP siRNA actually had 122% of the five subunits present in complex I compared with controls, even though the activity of complex I was only 47% of that of the control cell; thus the specific activity of complex I was decreased by treatment with ACP siRNA. Unfortunately, the sensitivity of the Western blotting procedure was insufficient to detect the ACP subunit in complex I using rabbit anti-ACP antibodies.
The inference that treatment with ACP siRNA caused a decrease in the specific activity of complex I was supported by direct spectrophotometric assay in mitochondria swollen by hypotonic treatment (Table 1). The specific activity of complex I in mitochondria from ACP siRNA-treated HEK293T cells was reduced to ∼60% of that of mitochondria from cells treated with nonspecific siRNAs. In contrast, the specific activities of complexes III, IV, and V were not significantly reduced by treatment with ACP siRNA; the specific activity of complex II was reduced, but to a lesser extent than that of complex I.
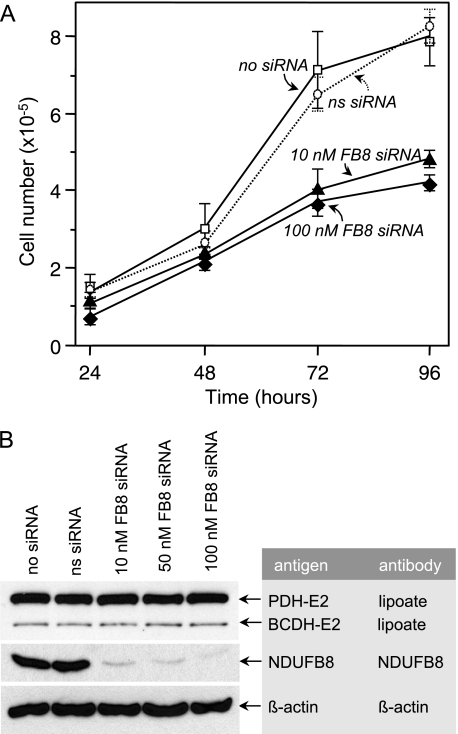
To verify that the altered lipoylation profile induced by ACP siRNA was unrelated to any effect on complex I, we also monitored the effects of treatment with siRNAs targeting the NDUFB8 subunit of complex I (Fig. 4). Treatment with NDUFB8 siRNA, even at only 10 nm, effectively lowered the cellular content of the NDUFB8 protein (Fig. 4B) and slowed the rate of growth of the HEK293T cells (Fig. 4A). However, no effect on the lipoylation profile was observed, even at 100 nm siRNA (Fig. 4B).
FIGURE 4.
Effect of down-regulation of NDUFB8 on cell growth and lipoylation profile. HEK293T cells were treated with various concentrations of siRNAs targeting the NDUFB8 subunit of respiratory complex I for up to 96 h. A, cell growth. B, lipoylation profile, monitored 96 h post-transfection. Control cells were treated with either vehicle alone or nonspecific siRNAs. The electrophoretic mobility of NDUFB8 corresponds to a molecular mass of ∼19 kDa.
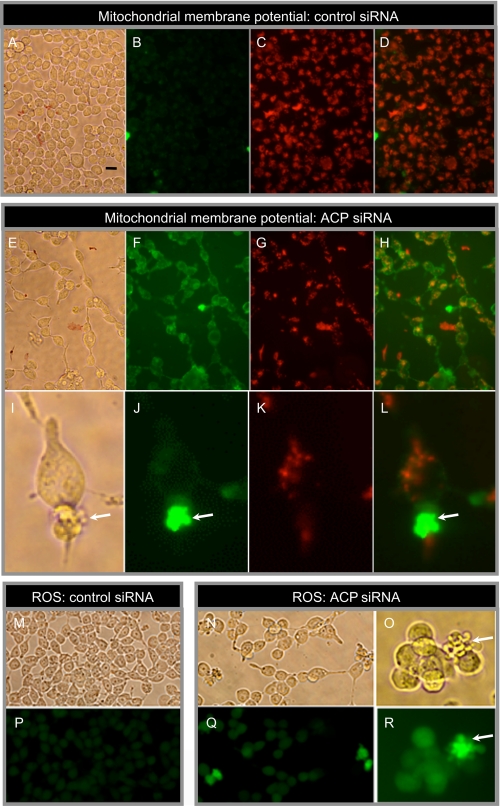
Effect of siRNA Treatment on Cell Morphology, Mitochondrial Membrane Potential, and ROS Levels—By about 72 h post-transfection with ACP siRNA, HEK293T cells began to detach from the plastic dish, so that cell density was significantly lower than in control cells treated with nonspecific siRNA (Fig. 5E and A, respectively). We assessed mitochondrial membrane potential of siRNA-treated cells using the lipophilic cation probe JC-1. This green fluorescent probe is selectively imported into mitochondria where it forms red fluorescent aggregates; depolarization of mitochondrial membranes results in lower uptake and retention of the green fluorescent form of the probe in the cytosolic compartment (40). Compared with controls, cells treated with ACP siRNA exhibited markedly reduced mitochondrial uptake of the probe by 80 h post-transfection (Fig. 5, B–D and F–H). This difference is best illustrated in panels D and H, which show a substantial shift in the red/green balance in cells treated with ACP siRNA. We also examined the levels of ROS using the probe CM-H2DCFDA, which emits an intense green fluorescence only after deacylation and subsequent oxidation by ROS (Fig. 5, M–R). By 96 h post-transfection, cells treated with ACP siRNA showed only a slightly increased green fluorescence, compared with controls (Fig. 5, compare P and Q). This finding was confirmed by quantitative fluorescence-activated cell sorter analysis (details not shown). However, some of the cells treated with ACP siRNA developed cytoplasmic membrane extrusions and these blebs lacked functional mitochondria, as revealed by the lowered membrane potential (Fig. 5, I–L) and were highly enriched in ROS, as evidenced by their intense green fluorescence (Fig. 5, O and R). Neither the loss of membrane potential nor the appearance of ROS-enriched cytoplasmic blebs was detectable prior to 72 h post-transfection.
FIGURE 5.
Effect of siRNA treatment on cell morphology, mitochondrial membrane potential, and ROS. HEK293T cells were treated with nonspecific control siRNA or ACP siRNA (10 nm), cultured for 80 h, and exposed to the fluorescent dye JC-1 to monitor mitochondrial membrane potential. Healthy cells with normal mitochondrial membrane potential translocate the green fluorescent dye from the cytosol into mitochondria, where it forms red aggregates. A, E, and I, phase-contrast images. Bar in panel A, 20 μm. B, F, and J, green fluorescence. C, G, and K, red fluorescence. D, H, and L, superimposed green and red fluorescence. Panels I–L are higher magnification images illustrating cytoplasmic membrane blebs (arrows). In M–R, HEK293T cells, cultured with either nonspecific control siRNA or ACP siRNA for 96 h, were exposed to CM-H2DCFDA, which is taken up by mitochondria and converted to a green fluorescent product in the presence of ROS. M–O, phase-contrast images. P—R, green fluorescence. Panels O and R are higher magnification images illustrating ROS accumulation in cytoplasmic membrane blebs (arrows).
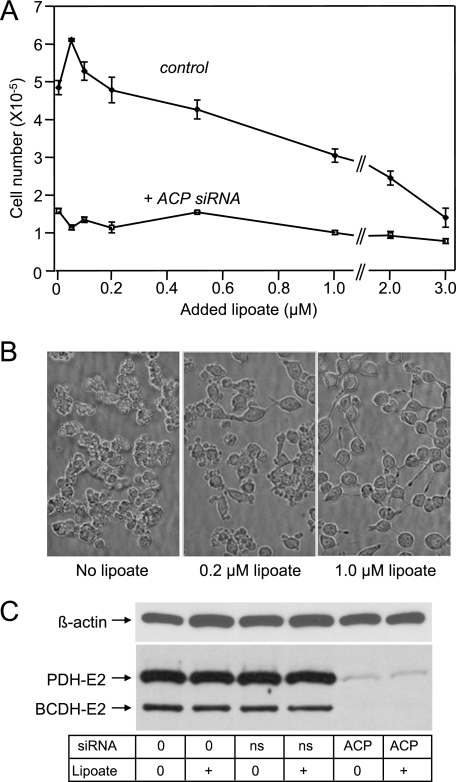
Effect of Exogenous Lipoate on HEK293T Cells—To determine whether increased availability of exogenous lipoic acid might relieve any of the effects of ACP down-regulation, we supplemented the culture medium with lipoate. HEK293T cells were extremely sensitive to added lipoate so that 50% inhibition of cell growth was observed with ∼2 μm added lipoate, in the absence of ACP siRNA (Fig. 6A). This observation was surprising, because lipoic acid is typically well tolerated by normal cells. However, there are reports in the literature indicating that lipoic acid can induce apoptosis in some cancer and transformed cell lines; the concentration of lipoic acid required to induce this effect appears to vary considerably, depending on the particular cell line (41–46). In the presence of ACP siRNAs, cell growth was less affected by added lipoate than in their absence (Fig. 6A), overall cell morphology was improved and blebbing was reduced (Fig. 6B). Nevertheless, lipoate supplementation neither increased cell growth (Fig. 6A), nor reversed the lipoylation defect induced by ACP siRNA (Fig. 6C). This finding suggests that, whereas exogenous free lipoate may improve the morphology of cells treated with ACP siRNA through its action as an antioxidant, it is unable to reverse the protein lipoylation defect.
FIGURE 6.
Effect of exogenous lipoate on siRNA-treated HEK293T cells. A, cells were treated with either ACP siRNA or nonspecific siRNA (10 nm), cultured in medium supplemented with various concentrations of Tris-lipoate for 96 h, and cell number monitored. B, morphology of cells treated with ACP siRNA supplemented with lipoate. C, lipoylation profile of cells grown in medium supplemented with 1 μm lipoate. Lipoylation was monitored by Western blotting using anti-lipoyl antibodies as probe. Ns, nonspecific siRNA. Similar results were obtained using the sodium salt of lipoic acid.
DISCUSSION
The human mitochondrial fatty acid biosynthetic system appears uniquely adapted for the synthesis of octanoyl-ACP as the major product and our previous studies had shown that these octanoyl moieties can be directly translocated to apoproteins, where they presumably are substrates for the insertion of sulfur atoms by lipoic acid synthase (9, 15). Consistent with this hypothesis, the earliest effect of mitochondrial ACP down-regulation that we observed was a change in the lipoylation profile of covalently modified mitochondrial enzymes, as monitored by Western blotting using anti-lipoyl antibodies. We were surprised to find that anti-lipoyl antibodies recognized only pyruvate dehydrogenase E2, branched chain oxoacid dehydrogenase E2, and the glycine cleavage H-protein in HEK293T cells. The pyruvate dehydrogenase E3bp subunit and oxoglutarate dehydrogenase E2 subunits were not detected, although the latter clearly are present, as we confirmed with antibodies recognizing the protein moiety (details not shown). In addition, immunocapture and analysis of the pyruvate dehydrogenase complex from HEK293T cells confirmed by protein staining that the 48-kDa E3bp subunits were present but not recognized by the anti-lipoyl antibodies (details not shown). The inability of the antibodies to recognize these proteins is not unique to HEK293T cells, as we were unable to detect these lipoylated proteins in mitochondria isolated from bovine heart as well as mouse heart, liver, kidney, and skeletal muscle (details not shown). It seems highly unlikely that these proteins would be present entirely in the apo-form, so the reason for this apparent selectivity of the anti-lipoyl antibodies, which are raised against lipoic acid conjugated to keyhole limpet hemocyanin, is unclear.
The ACP siRNA-induced change in reactivity of pyruvate dehydrogenase E2, branched chain oxoacid dehydrogenase E2, and glycine cleavage H-protein toward the anti-lipoyl antibodies could reflect either the absence of lipoyl moieties on the protein, or the presence of modified lipoyl moieties that are not recognized by the antibodies. There are reports in the literature indicating that oxidatively damaged lipoyl moieties are no longer recognized by anti-lipoyl antibodies (47, 48). However, we did not detect any signs of elevated ROS until several days following transfection of cells with ACP siRNA. Earlier studies have shown that protein-linked lipoyl moieties modified by 4-hydroxy-2-nonenal, a well characterized product of free radical damage to mitochondrial lipids, are reactive toward antibodies that recognize the 4-hydroxy-2-nonenal lipoyl moiety (49). However, neither of the E2 proteins from ACP siRNA-treated HEK293T cells exhibited any reactivity toward these antibodies (details not shown). These observations suggest that the change in the lipoylation profile observed as a consequence of ACP down-regulation more likely reflects the accumulation of apo-proteins lacking lipoyl moieties.
The loss of immunoreactivity of pyruvate dehydrogenase E2 subunits toward anti-lipoyl antibodies was not accompanied by a proportional decrease in overall pyruvate dehydrogenase activity. Thus, 48 h following treatment of HEK293T cells with ACP siRNA, immunoreactivity was decreased ∼90% but activity was decreased only ∼50%. The most likely explanation is that each active site of the E1 component is serviced by at least two different lipoyl moieties. Indeed at least 40% of the E2 domains of mammalian pyruvate dehydrogenase can be removed without reducing overall catalytic activity (50). A similar observation was made with the pyruvate dehydrogenase complex from Escherichia coli, from which two of the three lipoyl domains on each E2 subunit can be removed without loss in activity (51). Direct enzyme assays also revealed that the activity of oxoglutarate dehydrogenase was reduced ∼50% in HEK283T cells treated with ACP siRNA for 48 h (Table 1). Approximately 50% inhibition of oxoglutarate dehydrogenase in PC12 cells, induced by α-keto-β-methyl-n-valerate, is sufficient to cause necrotic cell death, even in the absence of changes in mitochondrial membrane potential or in the level of ROS (52). It seems likely therefore, that in cells compromised in their ability to synthesize lipoyl moieties, lowered activity of the oxoacid dehydrogenases may be responsible, at least in part, for the decreased growth rate and eventual death.
We did not observe any significant change in the amounts of pyruvate dehydrogenase E2 protein (Fig. 2) or its mRNA (details not shown) in response to ACP siRNA exposure, as monitored by Western blotting and quantitative PCR, respectively. This finding was hardly surprising, because mitochondrial proteins typically turn over slowly and the half-life for pyruvate dehydrogenase in normal rat liver has been estimated to be more than 1 week (53). Thus, the rapid accumulation of the apoE2 subunits observed on down-regulation of ACP strongly suggests that the lipoyl moieties turn over independently of the protein moiety. Because lipoyl moieties are sensitive to oxidative damage, possibly their rapid turnover ensures that the lipoyl moieties are maintained in a fully functional state without the necessity of replacing the entire protein moieties.
The inability of exogenous lipoate to rescue the lipoylation defect was surprising, because it is known to enter cells and mitochondria quite readily. Indeed, we did observe effects of lipoate supplementation that were consistent with protection from the oxidative damage observed as a relatively late consequence of siRNA treatment, so that it is highly unlikely that limited permeability of the cells to lipoate was an issue. Free lipoate is thought to be utilized in the protein lipoylation pathway in prokaryotes and eukaryotes in a two-step reaction sequence. Initially ATP is used to form a lipoyl-AMP intermediate and the lipoyl moiety is subsequently translocated from AMP to specific lysine residues on the acceptor proteins. In prokaryotes, both reactions are catalyzed by the same enzyme (54). However, in animals two discrete enzymes have been implicated: a ligase believed to be identical with a medium-chain fatty acyl-CoA synthetase (55) and a lipoyltransferase (56–58). The failure of this alternative pathway to rescue the siRNA-induced defect in the pathway for formation and utilization of lipoyl moieties raises questions as to how widely the exogenous pathway is utilized in mammalian cells.
Chronologically, the next observable siRNA-induced phenotypic change was a decrease in respiratory complex I activity, first detected 48 h after transfection. The exact role of ACP in complex I assembly and/or activity is unclear. Only a small fraction of the mitochondrial ACP is sequestered in complex I, most being free in the matrix compartment (16). It has been reported that the mass of the ACP subunit of bovine complex I is higher than anticipated, most likely because the phosphopantetheinyl moiety is in thioester linkage with an acyl chain corresponding to a free acid having a mass of 241 Da (19). This is fairly close to the theoretical mass for 3-hydroxytetradecanoic acid (244 Da), which has been implicated as the acyl moiety present on complex I in Neurospora crassa (59). However, saturated fatty acids with 6–18 carbon atoms have also been detected attached to the ACP subunit of N. crassa complex I (17, 60), so it is unclear whether the presence of acyl chains on the phosphopantetheine has any functional significance. What is clear from our results is that the specific activity of the mammalian complex I is compromised when ACP availability is limited.
It is well established that inhibition of respiratory complex I can result in elevation of mitochondrial superoxide formed as a biproduct of the oxidative phosphorylation process (61, 62). Superoxide, in turn, may release ferrous iron from iron-sulfur center-containing enzymes, which then can react with hydrogen peroxide to form the highly reactive hydroxyl radical that can initiate lipid peroxidation. Several days following transfection of HEK293T cells with ACP siRNA, we did see evidence of oxidative damage, primarily localized in protrusions from the cytoplasmic membranes. Because the specific activity of complex I was reduced by treatment with ACP siRNA, it appears likely that this event was the primary cause of the oxidative damage and blebbing observed as a late phenotypic change. Interestingly, we did not observe cytoplasmic membrane blebbing in cells treated with NDUFB8 siRNA, even 96 h following transfection (data not shown). Presumably, because in this case the specific activity of complex I was unaltered, there was no increase in the production of ROS biproducts. Although certain forms of cytoplasmic protrusions can occur reversibly in normal cells in response to various physiological stimuli (63, 64), membrane blebbing often is a terminal event associated with necrotic and apoptotic cell death (65, 66). The membrane blebs observed on transfection of cells with ACP siRNA contain elevated levels of ROS and non-functional mitochondria, thus almost certainly formed as a result of oxidative damage. Parenthetically, we also observed that treatment of HEK293T cells with hydrogen peroxide produced a membrane blebbing response very similar to that induced by ACP siRNA exposure (data not shown). Furthermore, the extent of blebbing could be reduced by exogenous free lipoate, presumably acting as an antioxidant. The appearance of the cytoplasmic membrane blebs and the loss of membrane potential as late responses to ACP siRNA treatment strongly suggest that these changes represent a prelude to cell death that becomes most notable 3–4 days post-transfection. Although collapse of mitochondrial membrane potential is often associated with the onset of apoptosis, it can also lead to caspase-independent necrosis (67–69), so the precise details of the cause of death induced by down-regulation of mitochondrial ACP remain to be ironed out. Our studies on down-regulation of the mitochondrial fatty acid synthesis pathway thus far have been limited to HEK293T cells. We chose these cells primarily because they are easy to culture and transfect, they have been used as a model system for the study of complex I assembly (70), and they are derived from kidney cells, which express significant levels of the mitochondrial FAS proteins (9). Similar studies now need to be performed on different cell types and ideally a knock-out mouse model should be engineered to assess the overall importance of the pathway in the organism as a whole.
Previous studies have established that the mitochondrial pathway for fatty acid biosynthesis de novo is required for the normal function of plant (14, 48), fungal (13), and protozoan (9, 71, 72) cells. Disruption of the genes encoding yeast mitochondrial FAS proteins leads to a respiratory-deficient phenotype that is accompanied by lowered cellular levels of lipoate. Neither the lowered lipoate nor the respiratory-deficient phenotype could be corrected by exogenous lipoate (13). In N. crassa, which unlike yeast contains a large multisubunit respiratory complex I that includes an ACP subunit, deletion of the ACP gene also results in improper assembly of this complex (12). In protozoa, dependence on the mitochondrial lipoylation pathway appears to vary in different species. Thus, in Toxoplasma gondii, mitochondrial lipoate appears to be derived primarily from the host cell, even though the secondary endosymbiotic organ present in this organism, the apicoplast, is capable of lipoate synthesis using a type II FAS system (71). In contrast, Trypanosoma brucei, which does not possess an apicoplast, has a functional type II mitochondrial FAS system and down-regulation of the ACP component results in defective cytochrome-mediated respiration. However, it is unclear whether this organism actually contains a multisubunit complex I that includes an ACP and down-regulation of the protein lipoylation pathway does not appear to be the primary cause of the respiratory defect. Rather, this mitochondrial system appears to be important for the production of long-chain fatty acids that are essential for maintaining phospholipid levels essential for the activity of respiratory complexes II, III, and IV (72). The appearance of ROS-enriched membrane blebs and loss of mitochondrial potential in HEK293T cells following exposure to ACP siRNA does not appear attributable to generalized loss of membrane phospholipid integrity, because only complex I, and to a lesser extent complex II, is affected. Although the ability of the human mitochondrial FAS system to synthesize long-chain fatty acids appears to be severely limited (9, 15), the possibility that longer term down-regulation of this pathway may also lead to abnormalities in phospholipid metabolism cannot be entirely discounted at this time.
In summary, the ACP appears to turn over unusually rapidly for a mitochondrial protein, exhibiting a half-life of less than 24 h (Fig. 3A). Down-regulation of ACP quickly induces multiple effects, ranging from compromised function of the citric acid cycle, through accumulation of the apoE2 forms of pyruvate and oxoglutarate dehydrogenases, compromised ability to catabolize branched chain amino acids and glycine, through effects on branched chain oxoacid dehydrogenase and the glycine cleavage H-protein, as well as reduced functionality of the respiratory chain through its role in complex I.
Mammalian mitochondria contain more than 1100 proteins that are involved in many biochemical pathways. Surprisingly, the precise function of many of these proteins remains to be elucidated and only recently have the enzymes involved in the mitochondrial FAS pathway been identified and characterized. Defective mitochondrial function has been linked to more than 50 identified diseases and most likely is implicated in many other diseases of uncertain etiology. Almost half of all known mitochondrial disorders involve complex I abnormalities (73). So, in view of the important role played by the ACP in this protein and in supporting the post-translational lipoylation pathways, it might be anticipated that lack of normal ACP function could cause serious metabolic problems. To date, only a small group of patients with complex I deficiency have been screened for possible mutations in the ACP subunit and none were found (18, 22). The results of our study indicate that the hypomorphic ACP phenotype may be more complex than originally anticipated and suggest some abnormal phenotypes that could serve as possible indicators of ACP defects.
Acknowledgments
We thank Dr. Luke Szweda of the Oklahoma Medical Research Foundation for the generous gift of rabbit polyclonal antibodies recognizing 4-hydroxy-2-nonenal and the E2 subunit of oxoglutarate dehydrogenase.
This work was supported, in whole or in part, by National Institutes of Health Grant GM069717.
Footnotes
The abbreviations used are: FAS, fatty acid synthase; ACP, acyl carrier protein; ROS, reactive oxygen species; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; HEK, human embryonic kidney; siRNA, small interfering RNA; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]-glycine; CAPS, 3-(cyclohexylamino)propanesulfonic acid.
References
- 1.Smith, S., and Tsai, S. C. (2007) Nat. Prod. Rep. 24, 1041-1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schweizer, E., and Hofmann, J. (2004) Microbiol. Mol. Biol. Rev. 68, 501-517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harington, A., Herbert, C. J., Tung, B., Getz, G. S., and Slonimski, P. P. (1993) Mol. Microbiol. 9, 545-555 [DOI] [PubMed] [Google Scholar]
- 4.Schneider, R., Brors, B., Burger, F., Camrath, S., and Weiss, H. (1997) Curr. Genet. 32, 384-388 [DOI] [PubMed] [Google Scholar]
- 5.Torkko, J. M., Koivuranta, K. T., Miinalainen, I. J., Yagi, A. I., Schmitz, W., Kastaniotis, A. J., Airenne, T. T., Gurvitz, A., and Hiltunen, K. J. (2001) Mol. Cell. Biol. 21, 6243-6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miinalainen, I. J., Chen, Z. J., Torkko, J. M., Pirila, P. L., Sormunen, R. T., Bergmann, U., Qin, Y. M., and Hiltunen, J. K. (2003) J. Biol. Chem. 278, 20154-20161 [DOI] [PubMed] [Google Scholar]
- 7.Zhang, L., Joshi, A. K., and Smith, S. (2003) J. Biol. Chem. 278, 40067-40074 [DOI] [PubMed] [Google Scholar]
- 8.Kastaniotis, A. J., Autio, K. J., Sormunen, R. T., and Hiltunen, J. K. (2004) Mol. Microbiol. 53, 1407-1421 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, L., Joshi, A. K., Hofmann, J., Schweizer, E., and Smith, S. (2005) J. Biol. Chem. 280, 12422-12429 [DOI] [PubMed] [Google Scholar]
- 10.Autio, K. J., Kastaniotis, A. J., Pospiech, H., Miinalainen, I. J., Schonauer, M. S., Dieckmann, C. L., and Hiltunen, J. K. (2008) FASEB J. 22, 569-578 [DOI] [PubMed] [Google Scholar]
- 11.White, S. W., Zheng, J., Zhang, Y.-M., and Rock, C. O. (2005) Annu. Rev. Biochem. 74, 791-831 [DOI] [PubMed] [Google Scholar]
- 12.Schneider, R., Massow, M., Lisowsky, T., and Weiss, H. (1995) Curr. Genet. 29, 10-17 [DOI] [PubMed] [Google Scholar]
- 13.Brody, S., Oh, C., Hoja, U., and Schweizer, E. (1997) FEBS Lett. 408, 217-220 [DOI] [PubMed] [Google Scholar]
- 14.Wada, H., Shintani, D., and Ohlrogge, J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94, 1591-1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witkowski, A., Joshi, A. K., and Smith, S. (2007) J. Biol. Chem. 282, 14178-14185 [DOI] [PubMed] [Google Scholar]
- 16.Cronan, J. E., Fearnley, I. M., and Walker, J. E. (2005) FEBS Lett. 579, 4892-4896 [DOI] [PubMed] [Google Scholar]
- 17.Zensen, R., Husmann, H., Schneider, R., Peine, T., and Weiss, H. (1992) FEBS Lett. 310, 179-181 [DOI] [PubMed] [Google Scholar]
- 18.Triepels, R., Smeitink, J., Loeffen, J., Smeets, R., Buskens, C., Trijbels, F., and van den Heuvel, L. (1999) J. Inherit. Metab. Dis. 22, 163-173 [DOI] [PubMed] [Google Scholar]
- 19.Carroll, J., Fearnley, I. M., Shannon, R. J., Hirst, J., and Walker, J. E. (2003) Mol. Cell. Proteomics 2, 117-126 [DOI] [PubMed] [Google Scholar]
- 20.Carroll, J., Fearnley, I. M., Skehel, J. M., Runswick, M. J., Shannon, R. J., Hirst, J., and Walker, J. E. (2005) Mol. Cell. Proteomics 4, 693-699 [DOI] [PubMed] [Google Scholar]
- 21.Schilling, B., Bharath, M. M. S., Row, R. H., Murray, J., Cusack, M. P., Capaldi, R. A., Freed, C. R., Prasad, K. N., Andersen, J. K., and Gibson, B. W. (2005) Mol. Cell. Proteomics 4, 84-96 [DOI] [PubMed] [Google Scholar]
- 22.Hinttala, R., Uusimaa, J., Remes, A. M., Rantala, H., Hassinen, I. E., and Majamaa, K. (2005) J. Mol. Med. 83, 786-794 [DOI] [PubMed] [Google Scholar]
- 23.Brandt, U. (2006) Annu. Rev. Biochem. 75, 69-92 [DOI] [PubMed] [Google Scholar]
- 24.Hirst, J., Carroll, J., Fearnley, I. M., Shannon, R. J., and Walker, J. E. (2003) Biochim. Biophys. Acta 1604, 135-150 [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. (1970) Nature 227, 680-685 [DOI] [PubMed] [Google Scholar]
- 26.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A. 76, 4350-4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schagger, H. (2006) Nat. Protoc. 1, 16-22 [DOI] [PubMed] [Google Scholar]
- 28.Janssen, A. J., Trijbels, F. J., Sengers, R. C., Smeitink, J. A., van den Heuvel, L. P., Wintjes, L. T., Stoltenborg-Hogenkamp, B. J., and Rodenburg, R. J. (2007) Clin. Chem. 53, 729-734 [DOI] [PubMed] [Google Scholar]
- 29.Rustin, P., Chretien, D., Bourgeron, T., Gerard, B., Rotig, A., Saudubray, J. M., and Munnich, A. (1994) Clin. Chim. Acta 228, 35-51 [DOI] [PubMed] [Google Scholar]
- 30.Luo, C., Long, J., and Liu, J. (2008) Clin. Chim. Acta 395, 38-41 [DOI] [PubMed] [Google Scholar]
- 31.Birch-Machin, M. A., and Turnbull, D. M. (2001) Methods Cell Biol. 65, 97-117 [DOI] [PubMed] [Google Scholar]
- 32.Cooperstein, S. J., and Lazarow, A. (1951) J. Biol. Chem. 189, 665-670 [PubMed] [Google Scholar]
- 33.Zheng, J., and Ramirez, V. D. (1999) Biochem. Biophys. Res. Commun. 261, 499-503 [DOI] [PubMed] [Google Scholar]
- 34.Schwab, M. A., Kolker, S., van den Heuvel, L. P., Sauer, S., Wolf, N. I., Rating, D., Hoffmann, G. F., Smeitink, J. A., and Okun, J. G. (2005) Clin. Chem. 51, 151-160 [DOI] [PubMed] [Google Scholar]
- 35.Sheu, K. F., Hu, C. W., and Utter, M. F. (1981) J. Clin. Investig. 67, 1463-1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi, A. K., Zhang, L., Rangan, V. S., and Smith, S. (2003) J. Biol. Chem. 278, 33142-33149 [DOI] [PubMed] [Google Scholar]
- 37.Wittig, I., Braun, H. P., and Schagger, H. (2006) Nat. Protoc. 1, 418-428 [DOI] [PubMed] [Google Scholar]
- 38.Nijtmans, L. G., Henderson, N. S., and Holt, I. J. (2002) Methods 26, 327-334 [DOI] [PubMed] [Google Scholar]
- 39.Schagger, H., and Von Jagow, G. (1987) Anal. Biochem. 166, 368-379 [DOI] [PubMed] [Google Scholar]
- 40.Salvioli, S., Ardizzoni, A., Franceschi, C., and Cossarizza, A. (1997) FEBS Lett. 411, 77-82 [DOI] [PubMed] [Google Scholar]
- 41.Sen, C. K., Sashwati, R., and Packer, L. (1999) Cell Death Differ. 6, 481-491 [DOI] [PubMed] [Google Scholar]
- 42.Casciari, J. J., Riordan, N. H., Schmidt, T. L., Meng, X. L., Jackson, J. A., and Riordan, H. D. (2001) Br. J. Cancer 84, 1544-1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pack, R. A., Hardy, K., Madigan, M. C., and Hunt, N. H. (2002) Mol. Immunol. 38, 733-745 [DOI] [PubMed] [Google Scholar]
- 44.van de Mark, K., Chen, J. S., Steliou, K., Perrine, S. P., and Faller, D. V. (2003) J. Cell Physiol. 194, 325-340 [DOI] [PubMed] [Google Scholar]
- 45.Wenzel, U., Nickel, A., and Daniel, H. (2005) Apoptosis 10, 359-368 [DOI] [PubMed] [Google Scholar]
- 46.Moungjaroen, J., Nimmannit, U., Callery, P. S., Wang, L., Azad, N., Lipipun, V., Chanvorachote, P., and Rojanasakul, Y. (2006) J. Pharmacol. Exp. Ther. 319, 1062-1069 [DOI] [PubMed] [Google Scholar]
- 47.Humphries, K. M., and Szweda, L. I. (1998) Biochemistry 37, 15835-15841 [DOI] [PubMed] [Google Scholar]
- 48.Ewald, R., Kolukisaoglu, U., Bauwe, U., Mikkat, S., and Bauwe, H. (2007) Plant Physiol. 145, 41-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lucas, D. T., and Szweda, L. I. (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 510-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahmatullah, M., Radke, G. A., Andrews, P. C., and Roche, T. E. (1990) J. Biol. Chem. 265, 14512-14517 [PubMed] [Google Scholar]
- 51.Guest, J. R., Lewis, H. M., Graham, L. D., Packman, L. C., and Perham, R. N. (1985) J. Mol. Biol. 185, 743-754 [DOI] [PubMed] [Google Scholar]
- 52.Huang, H. M., Ou, H. C., Xu, H., Chen, H. L., Fowler, C., and Gibson, G. E. (2003) J. Neurosci. Res. 74, 309-317 [DOI] [PubMed] [Google Scholar]
- 53.Weinberg, M. B., and Utter, M. F. (1980) Biochem. J. 188, 601-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morris, T. W., Reed, K. E., and Cronan, J. E., Jr. (1994) J. Biol. Chem. 269, 16091-16100 [PubMed] [Google Scholar]
- 55.Tsunoda, J. N., and Yasunobu, K. T. (1967) Arch. Biochem. Biophys. 118, 395-401 [DOI] [PubMed] [Google Scholar]
- 56.Fujiwara, K., Okamura-Ikeda, K., and Motokawa, Y. (1994) J. Biol. Chem. 269, 16605-16609 [PubMed] [Google Scholar]
- 57.Fujiwara, K., Okamura-Ikeda, K., and Motokawa, Y. (1997) J. Biol. Chem. 272, 31974-31978 [DOI] [PubMed] [Google Scholar]
- 58.Fujiwara, K., Okamura-Ikeda, K., and Motokawa, Y. (1996) J. Biol. Chem. 271, 12932-12936 [DOI] [PubMed] [Google Scholar]
- 59.Sackmann, U., Zensen, R., Rohlen, D., Jahnke, U., and Weiss, H. (1991) Eur. J. Biochem. 200, 463-469 [DOI] [PubMed] [Google Scholar]
- 60.Mikolajczyk, S., and Brody, S. (1990) Eur. J. Biochem. 187, 431-437 [DOI] [PubMed] [Google Scholar]
- 61.Pitkanen, S., and Robinson, B. H. (1996) J. Clin. Investig. 98, 345-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koopman, W. J., Verkaart, S., Visch, H. J., van der Westhuizen, F. H., Murphy, M. P., van den Heuvel, L. W., Smeitink, J. A., and Willems, P. H. (2005) Am. J. Physiol. 288, C1440-C1450 [DOI] [PubMed] [Google Scholar]
- 63.Trinkaus, J. P. (1973) CIBA Found. Symp. 14, 233-249 [DOI] [PubMed] [Google Scholar]
- 64.Charras, G. T., Hu, C. K., Coughlin, M., and Mitchison, T. J. (2006) J. Cell Biol. 175, 477-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarthy, N. J., Whyte, M. K., Gilbert, C. S., and Evan, G. I. (1997) J. Cell Biol. 136, 215-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mills, J. C., Stone, N. L., Erhardt, J., and Pittman, R. N. (1998) J. Cell Biol. 140, 627-636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lieberthal, W., Menza, S. A., and Levine, J. S. (1998) Am. J. Physiol. 274, F315-F327 [DOI] [PubMed] [Google Scholar]
- 68.Finucane, D. M., Waterhouse, N. J., Amarante-Mendes, G. P., Cotter, T. G., and Green, D. R. (1999) Exp. Cell Res. 251, 166-174 [DOI] [PubMed] [Google Scholar]
- 69.Mayer, B., and Oberbauer, R. (2003) News Physiol. Sci. 18, 89-94 [DOI] [PubMed] [Google Scholar]
- 70.Vogel, R. O., Dieteren, C. E., van den Heuvel, L. P., Willems, P. H., Smeitink, J. A., Koopman, W. J., and Nijtmans, L. G. (2007) J. Biol. Chem. 282, 7582-7590 [DOI] [PubMed] [Google Scholar]
- 71.Crawford, M. J., Thomsen-Zieger, N., Ray, M., Schachtner, J., Roos, D. S., and Seeber, F. (2006) EMBO J. 25, 3214-3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guler, J. L., Kriegova, E., Smith, T. K., Lukes, J., and Englund, P. T. (2008) Mol. Microbiol. 67, 1125-1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pagliarini, D. J., Calvo, S. E., Chang, B., Sheth, S. A., Vafai, S. B., Ong, S. E., Walford, G. A., Sugiana, C., Boneh, A., Chen, W. K., Hill, D. E., Vidal, M., Evans, J. G., Thorburn, D. R., Carr, S. A., and Mootha, V. K. (2008) Cell 134, 112-123 [DOI] [PMC free article] [PubMed] [Google Scholar]