Abstract
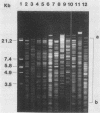
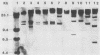
Eleven strains comprising representatives of different subspecies, biotypes, and serotypes of Campylobacter jejuni and reference strains of C. coli, C. fetus subsp. fetus, C. hyointestinalis, and C. sputorum subsp. sputorum were studied to assess the utility of different DNA profiles for measuring fine differences between allied bacteria. Strains were compared by analyses of HaeIII and XhoI digest patterns of chromosomal DNA and Southern blot hybridization patterns of XhoI digests obtained with an Escherichia coli 16S + 23S rRNA gene probe. Visual comparisons and numerical analyses of the HaeIII and XhoI digest patterns both revealed clear differences between the five Campylobacter species and between representatives of C. jejuni subspecies and biotypes. Only strains with the same Penner serotype gave identical total digest polymorphisms. The advantages of XhoI total digests and Southern blot hybridization patterns were that they were less complex than the HaeIII patterns and easier to compare visually. However, numerical analysis of XhoI data resulted in reduced discrimination. We conclude that DNA fingerprinting using either HaeIII or XhoI fragment polymorphisms has considerable potential as a generally applicable method for identification of Campylobacter isolates, especially at the infrasubspecific level.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ades A. E., Garrett A. J., Cowell J., Chin K. S., Peckham C. S. Computer matching of oligonucleotide patterns on electrophoretic gels: an application to the epidemiology of cytomegalovirus. Epidemiol Infect. 1988 Jun;100(3):467–479. doi: 10.1017/s0950268800067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D., Razin S., Glaser G. Ribosomal RNA genes in Mycoplasma. Nucleic Acids Res. 1982 Jul 24;10(14):4215–4222. doi: 10.1093/nar/10.14.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovier H., Kafri O., Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986 May 14;136(3):1136–1141. doi: 10.1016/0006-291x(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Bradbury W. C., Pearson A. D., Marko M. A., Congi R. V., Penner J. L. Investigation of a Campylobacter jejuni outbreak by serotyping and chromosomal restriction endonuclease analysis. J Clin Microbiol. 1984 Mar;19(3):342–346. doi: 10.1128/jcm.19.3.342-346.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D. M., Ross D. E. Restriction endonuclease analysis of Campylobacter strains with particular reference to Campylobacter fetus ss. fetus. J Med Microbiol. 1984 Aug;18(1):117–124. doi: 10.1099/00222615-18-1-117. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Hartmann R. K., Toschka H. Y., Ulbrich N., Erdmann V. A. Genomic organization of rDNA in Pseudomonas aeruginosa. FEBS Lett. 1986 Jan 20;195(1-2):187–193. doi: 10.1016/0014-5793(86)80158-2. [DOI] [PubMed] [Google Scholar]
- Kakoyiannis C. K., Winter P. J., Marshall R. B. Identification of Campylobacter coli isolates from animals and humans by bacterial restriction endonuclease DNA analysis. Appl Environ Microbiol. 1984 Sep;48(3):545–549. doi: 10.1128/aem.48.3.545-549.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakoyiannis C. K., Winter P. J., Marshall R. B. The relationship between intestinal Campylobacter species isolated from animals and humans as determined by BRENDA. Epidemiol Infect. 1988 Jun;100(3):379–387. doi: 10.1017/s0950268800067133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenberg W., Rauws E. A., Widjojokusumo A., Tytgat G. N., Zanen H. C. Identification of Campylobacter pyloridis isolates by restriction endonuclease DNA analysis. J Clin Microbiol. 1986 Sep;24(3):414–417. doi: 10.1128/jcm.24.3.414-417.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lior H. New, extended biotyping scheme for Campylobacter jejuni, Campylobacter coli, and "Campylobacter laridis". J Clin Microbiol. 1984 Oct;20(4):636–640. doi: 10.1128/jcm.20.4.636-640.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lior H., Woodward D. L., Edgar J. A., Laroche L. J., Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982 May;15(5):761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski S. I., Goodwin C. S. Restriction endonuclease analysis of the genome of Campylobacter pylori with a rapid extraction method: evidence for considerable genomic variation. J Infect Dis. 1988 Mar;157(3):465–471. doi: 10.1093/infdis/157.3.465. [DOI] [PubMed] [Google Scholar]
- Mégraud F., Gavinet A. M., Camou-Junca C. Serogroups and biotypes of human strains of Campylobacter jejuni and Campylobacter coli isolated in France. Eur J Clin Microbiol. 1987 Dec;6(6):641–645. doi: 10.1007/BF02013060. [DOI] [PubMed] [Google Scholar]
- Owen R. J., Beck A., Borman P. Restriction endonuclease digest patterns of chromosomal DNA from nitrate-negative Campylobacter jejuni-like organisms. Eur J Epidemiol. 1985 Dec;1(4):281–287. doi: 10.1007/BF00237103. [DOI] [PubMed] [Google Scholar]
- Owen R. J., Costas M., Sloss L., Bolton F. J. Numerical analysis of electrophoretic protein patterns of Campylobacter laridis and allied thermophilic campylobacters from the natural environment. J Appl Bacteriol. 1988 Jul;65(1):69–78. doi: 10.1111/j.1365-2672.1988.tb04319.x. [DOI] [PubMed] [Google Scholar]
- Owen R. J. Nucleic acids in the classification of Campylobacters. Eur J Clin Microbiol. 1983 Aug;2(4):367–377. doi: 10.1007/BF02019473. [DOI] [PubMed] [Google Scholar]
- Penner J. L., Hennessy J. N., Mills S. D., Bradbury W. C. Application of serotyping and chromosomal restriction endonuclease digest analysis in investigating a laboratory-acquired case of Campylobacter jejuni enteritis. J Clin Microbiol. 1983 Dec;18(6):1427–1428. doi: 10.1128/jcm.18.6.1427-1428.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner J. L., Hennessy J. N. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol. 1980 Dec;12(6):732–737. doi: 10.1128/jcm.12.6.732-737.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yoshinaga K., Ono Y., Nagata A., Yamada T. Organization of rRNA genes in Mycobacterium bovis BCG. J Bacteriol. 1987 Feb;169(2):839–843. doi: 10.1128/jb.169.2.839-843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen B., Falk E. S., Wisløff-Nilsen E., Bjorvatn B., Kristiansen B. E. Multivariate analysis of Neisseria DNA restriction endonuclease patterns. J Gen Microbiol. 1985 Nov;131(11):3099–3104. doi: 10.1099/00221287-131-11-3099. [DOI] [PubMed] [Google Scholar]