Abstract
Targeted deletion of Tssk1 & 2 resulted in male chimeras which produced sperm/spermatogenic cells bearing the mutant allele, however this allele was never transmitted to offspring, indicating infertility due to haploinsufficiency. Morphological defects in chimeras included failure to form elongated spermatids, apoptosis of spermatocytes and spermatids, and the appearance of numerous round cells in the epididymal lumen. Characterization of TSSK2 and its interactions with the substrate, TSKS, were further investigated in human and mouse. The presence of both kinase and substrate in testis was confirmed, while persistence of both proteins in spermatozoa was revealed for the first time. In vivo binding interactions between TSSK2 and TSKS were established through co-immunoprecipitation of TSSK2/TSKS complexes from both human sperm and mouse testis extracts. A role for the human TSKS N-terminus in enzyme binding was defined by deletion mapping. TSKS immunoprecipitated from both mouse testis and human sperm extracts was actively phosphorylated. Ser285 was identified as a phosphorylation site in mouse TSKS. These results confirm both TSSK 2 and TSKS persist in sperm, define the critical role of TSKS′ N-terminus in enzyme interaction, identify Ser 285 as a TSKS phosphorylation site and indicate an indispensable role for TSSK 1 & 2 in spermiogenesis.
Keywords: Haploinsufficiency, infertility, gene targeting deletion, TSSK1, TSSK2, TSKS, spermatozoa, protein interaction, protein phosphorylation
Introduction
The testis specific ser/thr kinase family consists of at least 5 members (Spiridonov et al. 2005, Hao et al. 2004, Chen et al. 2005), several of which have generated interest as candidate male contraceptive drug targets because of their pattern of testicular expression and the testis specific expression of the substrate, TSKS. The nomenclature of the TSSK family was standardized by Manning et al. (2002). Mouse Tssk1 was cloned by a PCR based strategy searching for previously unknown kinases using degenerate oligonucleotide primers for conserved motifs within the kinase catalytic domain (Bielke et al. 1994). Subsequently, low stringency colony hybridization with the Tssk1 gene as the probe identified mouse Tssk2 (Kueng et al. 1997). A yeast two-hybrid screen led to identification of mouse TSKS, the testis specific kinase substrate, that formed complexes with TSSK1 and TSSK2 (Kueng et al. 1997). Human homologues of TSSK1 & 2 and TSKS were first cloned by Hao et al. (2004).
Murine and human TSSK3 were cloned about the same time by two research groups (Zuercher et al. 2000, Visconti et al. 2001) using PCR based methods. TSSK4 and SSTK (small ser/thr kinase) were discovered by Blast searches of human and mouse genomes (Spiridonov et al. 2005, Hao et al. 2004, Chen et al. 2005). TSSK3 can be activated by PDK1 (phosphoinositide-dependent protein kinase1) through phosphorylation of the TSSK3 T-loop domain (Bucko-Justyna et al. 2005). A central transcription factor, cAMP responsive element binding protein (CREB), was identified as a TSSK4-interacting protein via a yeast two-hybrid analysis (Chen et al. 2005). Recently, targeted deletion of Sstk gene was achieved in mice, resulting in male sterility. A defect in DNA condensation in Sstk null mutants indicated that SSTK was required for proper post-meiotic chromatin remodeling and male fertility (Spiridonov et al. 2005).
Mouse Tssk1 & 2 share high sequence similarity and the same kinase substrate, TSKS, and thus they may be functionally redundant. These genes are closely linked on mouse chromosome 16 by an intergenic region of only 3 kb. This small intergenic region makes individual targeted deletions of these genes and the creation of a double knockout by hybridization extremely difficult if not impossible. Thus it was necessary to simultaneously knockout Tssk1 & 2 to study their functions. Current cDNA array data, which are available online at http://www.mrg.genetics.washington.edu, show that mouse Tssk1 & 2 mRNA expression is specific to spermatids, with the expression of Tssk1 & 2 in spermatogonia, pachytene spematocytes, sertoli cells and myoid cells being lower than the threshold value and the Tssk2 and Tssk1 mRNA levels being 24 and 16 times higher in round spermatids than in pachytene spematocytes, respectively. Therefore, our investigation focused on the role of Tssk1 & 2 in post-meiotic spermatids and spermatozoa.
In this study, we disrupted one allele of Tssk1 & 2 in mouse embryonic stem cells derived from the 129Svj strain, and injected the cells into blastocysts derived from the C57BL/6 strain. Extensive breeding of 21 male chimeras resulted in either infertility or only wild type offspring, due apparently to haploinsufficiency. Testes of chimeras exhibited arrested spermatogenesis, failure to form elongated spermatids, increased apoptosis and increased round spermatogenic cells in the epididymides. In order to better understand the mechanisms underlying this infertility, TSSK2 was characterized in detail and the interactions between TSSK2 and TSKS in testis and sperm of humans and mice were studied.
Materials and Methods
Construction of targeting plasmids
A targeting vector was designed to delete both Tssk1 & 2 genes (Fig. 1A). This construct was made based on knockout vector R2-Neo-TK, in which a 6 kb genomic region including the Tssk1 and Tssk2 coding region and a 3 kb intergenic region was replaced by a neomycin cassette. The neo gene in this vector served as a positive selection marker and thymidine kinase served as a negative selection marker. Two FRT sites were also added at either end of the neo gene. The 5′ and 3′ homology arms of the targeting vector, 3.5 kb and 5 kb respectively, which were PCR amplified from mouse genomic DNA (strain 129Svj), were carefully verified by restriction mapping and DNA sequencing to ensure precise sequence identity to optimize targeting efficiency.
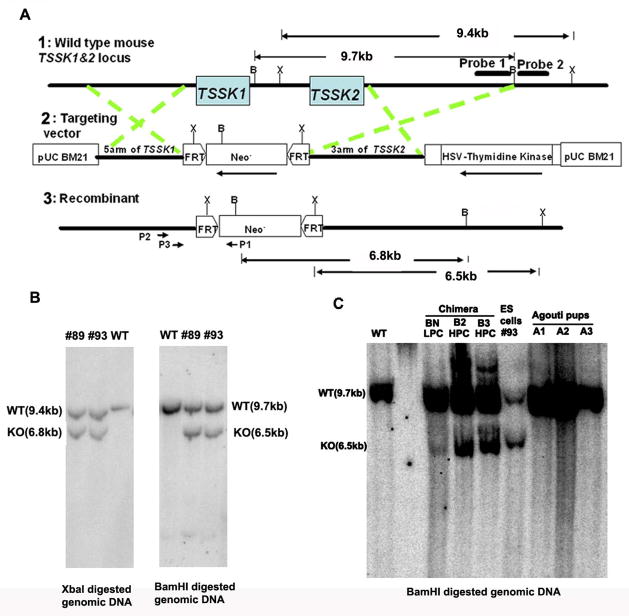
Figure 1. Targeting deletion of mouse Tssk1 & 2.
(A) Schematic representation of the strategy used to disrupt Tssk1 & 2. Line 1 indicates the genomic structure of the Tssk1 and Tssk2 genes. Mouse Tssk1 & 2, both intronless genes, are closely linked with one another by an intergenic region of only 3 kb. Lines 2 and 3 indicate the structure of the targeting vector and the mutant allele, respectively. X: XbaI, B: BamHI. (B) Southern analysis of the targeted ES cell clones. The external probe (probe 2, left panel) gave rise to a single 9.4 kb band in wild type (WT) genomic DNA digested with XbaI and an additional 6.8 kb knockout band in the mutant allele. The internal probe (probe 1, right panel) gave rise to a single 9.7 kb band in WT genomic DNA digested with BamHI and an additional 6.5 kb band in the mutant allele. (C) Southern analysis of the tail DNA from three chimeras and three agouti offspring sired by chimera BN. The probe 1 detected a single 9.7 kb band in WT genomic DNA digested with BamHI and an additional 6.5 kb band in the chimeras BN, B2 and B3 with different densities. However, in agouti mice sired by the chimera BN only the 9.7 kb wild type band was detected.
Transfection of ES cells
The targeting constructs were linearized and purified, and the linearized plasmids were electroporated into 129Svj embryonic stem (ES) cells (of proven germline transmission and bearing XY chromosomes for male germline transmission) cultured using mouse embryo fibroblast (MEF) feeder cells. Three hundred and fifty colonies resistant to both neomycin and gancyclovir were selected and expanded.
Genomic DNA extracted from the ES colonies established above was digested by XbaI, and blotted to hybridization membrane. A 380 bp probe (probe 2, Fig. 1A) targeting the flanking region of one recombinant arm was utilized to conduct Southern analysis. To confirm the screening result, genomic DNA extracted from the ES colonies, which passed the first round of screening, was digested by BamHI, and another 1300 bp probe (Probe1, Fig. 1A) specially targeting the end of the recombinant arm was employed to conduct second round Southern analysis. The dually positive ES clones were subcloned to insure pure lineages.
Generation of mutant mice
Mice were handled according to approved protocols following guidelines of the Instituitional Animal Care and Use Committee (IACUC) of the University of Virginia. Two ES cell lines bearing deletions of Tssk1 & 2 passed karyotyping analysis, then 10–20 cells at a time were injected into C57BL/6 blastocysts. Blastocysts were transferred to the uterine horns of 3 day pseudopregnant females for implantation. Chimeric pups derived from the targeted ES cell lines were identified by agouti coat color, which differs from the black background of the C57BL/6 foster mothers. Chimeric male mice then were mated to C57BL/6 wild type females. The chimera and offspring agouti mice were genotyped by Southern analysis with tail DNA.
Genotyping the sperm and spermatogenic cells from chimeric mice
Sperm and spermatogenic cells were collected from the uteri of C57BL/6 females following mating with chimeric males. The pellets were treated with proteinase K (1mg/ml) with 0.1M DTT and 2% SDS overnight, and DNA was isolated by standard phenol/chloroform extraction.
Sperm/spermatogenic cells genotyping was carried out by PCR analysis using the primers that amplify a microsatellite in the D2Mit94 locus of mouse chromosome 2. The PCR products were 194 bp for 129Svj and 160 bp for C57BL/6 mice strain (Cho et al. 2001).
Primers sets were also designed to detect the mutant allele. One primer was specific for neo: P1(5′ ggtgagaacagagtacctac 3′); two other primers were specific to one homology arm: P2(5′-GCACTGAGTACAAGATCCAGG 3′) and P3(5′ AGAGCCGCCGTTAAGAGTCC 3′) as shown in Fig. 1A.
Light microscopy of testis and epididymis
Testes and epididymides were collected from chimeric mice after fixation by cardiac perfusion with 4% paraformaldehyde. Testes and epididimides were then further fixed in 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1M phosphate buffer pH 7.0 at 4°C overnight. Following paraffin embedding and sectioning, the tissue slides were counterstained with hematoxylin and eosin.
Germ cell apoptosis was examined immunohistochemically with the monoclonal antibody F7-26 (Apostain; Alexis Corporation, San Diego, CA) directed against single-stranded DNA (ssDNA). Testes were immersed in Bouins fixative for 6 h, and paraffin-embedded. The Apostain technique was performed according to the protocol described by Lysiak et al. 2001.
Extraction of sperm proteins and Western analysis
Human semen samples were collected from healthy volunteers. All donors provided informed consent using University of Virginia Human Investigation Committee approved forms. After liquefaction of the semen, the mature sperm were separated from the seminal plasma by washing twice in Ham’s F-10 medium. Prior to the last centrifugation, the cells were counted. The sperm obtained from 8–12 individuals were pooled and frozen immediately until further use. Mouse spermatozoa were collected from cauda epididymides by allowing sperm to swim out of minced caudae in a 1 ml drop of PBS solution for 15 minutes. Epididymal sperm were washed and concentrated by centrifugation, and the sperm yield was determined by hemocytometer counting.
Human and mouse sperm pellets were solubilized in Celis lysis buffer (Celis et al.1992) containing 2% (v/v) NP-40, 100 mM dithiothreitol, 9.8 M urea, and a 10 μl/ml cocktail of protease inhibitors (Sigma Chemical Company, St. Louis, MO). Sperm cells were solubilized by constant shaking at 4°C for 45 min. Insoluble material was removed by centrifugation at 10,000 × g for 5 min. Protein concentrations were measured with the protein assay reagent (Bio-Rad, Inc.). Laemmli sample buffer (Bio-Rad, Inc.) was added to the protein lysate extracted by Celis buffer. After 5 minutes of boiling, the samples were loaded at 80 μg/lane and resolved by 1-D electrophoresis. For 2-D electrophoresis, sperm protein was extracted by Celis lysis buffer with 0.3% Ampholines (PH 3.5–10) and 0.2% Ampholines (PH 2.5–5), and ~200 μg of sperm protein were loaded onto immobilized pH gradient (IPG) strips. First dimension isoelectric focusing and second dimension SDS-PAGE were performed following the manufacturer’s instructions (Bio-Rad, Inc.).
Sperm proteins separated in 1-D or 2-D gels were eletrophoretically transferred to PVDF membranes. After blocking, the blots were incubated with the polyclonal rat anti-TSKS serum (Hao et al. 2004) or rabbit anti-TSSK2 serum respectively, at 4°C overnight, and then were incubated with enzyme-conjugated secondary antibodies. Horseradish peroxidase conjugates were visualized by enhanced chemiluminescence (ECL) (Amersham, Buckinghamshire, UK) and subsequently visualized with the TMB substrate (Kirkergaard & Perry Laboratories).
Antibody purification
Anti-TSKS polyclonal antibodies were purified using the method described by Rao et al. (Rao et al. 2003) with modifications. Briefly, purified human TSKS recombinant protein was separated on 1-D gels, and electrophoretically transferred to nitrocellulose membrane. The membrane strip with transferred TSKS was cut out, washed, and blocked with 5% dry milk in PBS for 1 hr. After blocking, the membrane was incubated overnight at 4°C with anti-TSKS antiserum. Then, the membrane strip was washed, and was eluted with elution buffer (Pierce) for 5 min at 22°C. The eluted sample was immediately neutralized by adding 1 M Tris-HCl (pH 8.0), and was dialyzed against PBS at 4°C. The resulting affinity-purified antibodies were stored at 4°C.
Characterization of interactions between human TSSK2 and human TSKS in a yeast two-hybrid system
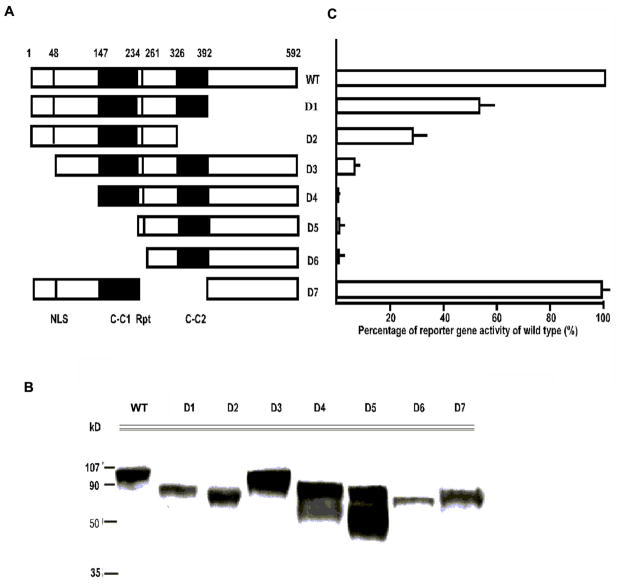
The entire open-reading frame (ORF) of human TSSK2 was fused with the GAL4 DNA binding domain (DBD) and a myc tag in frame in the pGBKT7 two-hybrid vector (Match-Maker yeast two-hybrid reaction kit, Clonetech). The entire ORF of human TSKS and a series of deletion mutants of various TSKS domains (Fig. 6) were fused with the GAL4 DNA activation domain (AD) with a HA tag in frame in pGADT7. After confirmation of correct sequence alignment by DNA sequencing, plasmid pairs containing TSSK2 and TSKS were introduced into the yeast two-hybrid host strain AH109 by co-transformation and selected for both nutritional markers carried on the two plasmids. Single colonies containing all plasmid pairs were tested for their ability to grow on medium selective for the histidine reporter gene as well as for color formation (using alpha galactosidase). Single colonies were inoculated in liquid medium, and the expression levels of alpha-galactosidase were quantified according to the method described previously (Hao et al. 2004). Binding strengths have been shown to be linearly related to expression levels of alpha-galactosidase (Clonetech). The seven deletion mutants were: D1 Δ 392–592; D2 Δ 326–592; D3 Δ 1–48; D4 Δ 1–147; D5 Δ 1–234; D6 Δ 1–261; and D7 Δ 234–392.
Figure 6. Analysis of the essential domains of human TSKS required for interaction with TSSK2.
A: Schematic drawing of TSKS ORF highlighting known motifs and deletion mutants. Two putative coiled-coil domains [C-C1, C-C2], the six amino-acid repeats (Rpt) and the amino terminal nuclear localization signal (NLS) are marked. A total of 7 deletion mutants of TSKS were created and fused with the Gal4 activation domain in the pGAD two hybrid vectors. Fusion proteins of each mutant and the Gal-AD were co-transformed with Gal-DBDTSSK2 into the yeast host strain AH109. B: The expression of the fusion proteins of each mutant was analyzed by Western analysis. C: Culture supernatants of each pair were assayed for alpha-galactosidase activity. The activity shown by each deletion mutant is expressed as the percentage of wild type (WT) TSKS. The corresponding deletion mutant is shown in A. The N-terminus of TSKS is crucial to its interaction with TSSK2. Error bars indicate variation among 3 experiments.
Immunoprecipitation
Immunoprecipitation was performed by protein G following the manufacturer’s instructions (Immunoprecipitation Kit, Roche Molecular Biochemicals). The starting biological samples were: 1.5 × 109 washed human sperm, or 4 decapsulated mouse testes. These samples were homogenized in pre-chilled lysis buffer. The lysate was then cleared by centrifugation at 12,000 × g for 20 min at 4 °C. Protein concentrations were measured with the Bio-Rad protein assay reagent, and adjusted to 15 μg/μl. To reduce background, 50 μl of protein-G agarose suspension was added to each milliliter of protein extract and incubated for 3 hrs at 4 °C on a rocking platform. The resultant pre-cleared protein extracts were then incubated with an appropriate amount of anti-TSKS rat serum or anti-TSSK2 rabbit serum for 2 hrs. Subsequently, 50 μl of protein-G agarose suspension was added to each milliliter of protein extract and rocked overnight at 4 °C. The agarose pellets were collected and washed in lysis buffer twice, and in 10 mM Tris buffer, pH7.5, twice. For protein gel electrophoresis, Laemmli loading buffer was added to the agarose pellets. After boiling for 5 min, the samples were loaded on SDS-PAGE gels. Normal rat and rabbit sera were used as immunoprecipitation controls.
In vitro kinase assay
TSKS/TSSK complexes immunoprecipitated from human sperm protein extracts or mouse testis protein extracts were subjected to in vitro kinase assay by incubating complexes bound to protein-G agarose beads in a kinase assay buffer described by Hao et al. (2004) and 5 μCi [32P]γATP in a volume of 20 μl at 37°C for 20 min. The reaction was stopped by adding 20 μl of 2X Laemmli sample buffer, and reaction products were subjected to SDS-PAGE. After staining with Coomassie dye to confirm equal loading, gels were dried and autoradiographed to detect phosphorylation.
Phosphorylation sites mapping by IMAC-LC-MS/MS
Immune-complexes immunoprecipitated with anti-TSKS (IP-TSKS) were submitted for phosphopeptide analysis by IMAC-LC-MS/MS. Fe3+-immobilized metal affinity chromatography (IMAC) was employed to enrich the digest for peptides containing phospho-amino acids. To decrease nonspecific binding to the IMAC column, acidic residues were converted to esters before binding. Eluent from the IMAC column was directly analyzed in an LC-MS/MS system consisting of a Thermo Electron LTQFT mass spectrometer interfaced with a Protana nanospray ion source and a C18 reversed-phase capillary column. The TSKS IP digest was analyzed using the double play capability of the instrument acquiring a full scan mass spectrum to determine peptide molecular weights followed by sequential product ion spectra (ten) to determine amino acid sequences. This mode of analysis produces many MS/MS spectra of ions ranging in abundance over several orders of magnitude. The MS/MS data were then analyzed by database searching using the Sequest search algorithm against TSKS. Putative phosphopeptides were manually verified.
Results
In addition to Tssk1–4 and Sstk reported previously in the mouse genome (Chen et al. 2005, Spiridonov et al. 2005, Zuercher et al. 2000, Visconti et al. 2001, Kueng et al. 1997), a putative Tssk5 (NP_898922) located on chromosome 15 is now listed in the NCBI database. All six Tssk genes are located on somatic chromosomes. A maximal parsimony tree (supplemental Figure 1) which depicts phylogenetic relationships between protein members of the mouse TSSK family was generated. It was noted that TSSK1 & 2 form a closely related clade (100% certainty score). Both TSSK1 and TSSK2 have a conserved kinase domain while also having unique carboxyl terminal extensions. Tssk1 and Tssk2 are intronless genes linked to each other with an intergenic region of only 3 kb, implying one is a retrogene duplicated from an ancestral Tssk by a retroviral mechanism during evolution, while the other may have resulted from duplication of the retrogene. A recent study of primate genomes reported that the majority of retrogenes are specifically expressed in testis, whereas their parental genes show broad expression patterns, suggesting these retrogenes may involve enhancing male germline function (Marques et al. 2005).
Generation of Tssk1 & 2 knockout allele
Because of the proximity of Tssk1 and Tssk2 on mouse chromosome 16 (only 3 kb separate Tssk1 from Tssk2), it is nearly impossible to approach targeting these genes with the conventional strategy of creating separate knockouts of Tssk1 and Tssk2 and then establishing the double knockout by crossing. Also, because Tssk1 & 2 share high sequence similarity as well as the same kinase substrate, arising likely from gene duplication, it is highly likely they are functionally redundant. Therefore, a simultaneous double knockout of Tssk1 & 2 was performed (Fig. 1A). The knockout construct was electroporated into mouse ES cells (129Svj strain), and double selection was used to identify targeted clones.
Correct targeting was verified by Southern blot analyses using external and internal probes. An external 380 bp probe (probe 2, Fig. 1A) for the flanking region of one homology arm was utilized to conduct Southern hybridization with XbaI digested genomic DNA. The presence of a 6.8 kb knockout band in addition to a 9.4 kb wild type band indicated that homologous recombination had interrupted one copy of the Tssk1 & 2 genes, resulting in heterozygous ES cell lines (Fig. 1B left panel). This result was confirmed with a second Southern using BamHI digested genomic DNA from the ES colonies and an internal 1300 bp probe (Probe 1, Fig. 1A) located at the end of one homology arm. The presence of a 6.5 kb knockout band in addition to a 9.7 kb wild type band further confirmed homologous recombination had interrupted one copy of the Tssk1 & 2 genes (Fig. 1B right panel). The positive ES clones, #89, #93, carrying verified targeting deletions were subcloned to ensure pure lineages and injected into blastocysts derived from the C57BL/6 strain.
A total 34 chimeric mice were obtained; 21 of them were male, including 10 high-percentage chimeric (HPC) males (estimated by fur color mosaicism) and 11 low-percentage chimeric (LPC) males. Southern analysis of tail DNA showed the presence of 6.5 kb knockout bands in addition to 9.7 kb wild type bands, indicating that the correct mutant allele integrated into the chimeras (Fig. 1C chimeras BN, B2 and B3). The intensity of the knockout bands vs. wild type bands varied in tail DNA retrieved from chimeras, and was consistent with their variable percentage of agouti fur color.
Twenty one chimeric males were mated with C57BL/6 wild type female mice. During 6–10 months of intensive breeding during which those mice that were fertile had more than 10 litters, the following outcome was observed: 4 HPC and 1 LPC were completely infertile; 6 HPC were fertile but only sired wild type black offspring; and ten LPC were also fertile but only sired wild type black offspring.
In only one instance, were agouti pups produced: 8 agouti pups resulted from a total 99 offspring from one LPC. Southern techniques were used to identify if heterozygous (+/−) mice were among the agouti offspring. Surprisingly, none of these agouti mice were heterozygous. Although the genotype of the chimeric father (BN) clearly had integrated the targeted mutant allele (Fig. 1C, BN), all offspring were wild type (Fig. 1C, A1, A2, A3 shows three representative genotypes of these agouti mice). In contrast, it was predicted that 50% of these agouti mice would be heterozygous while there is only a 1 in 256 chance of having no heterozygotes within 8 agouti offspring.
Genotyping the testis, epididymis and sperm from chimeric males confirmed germline transmission
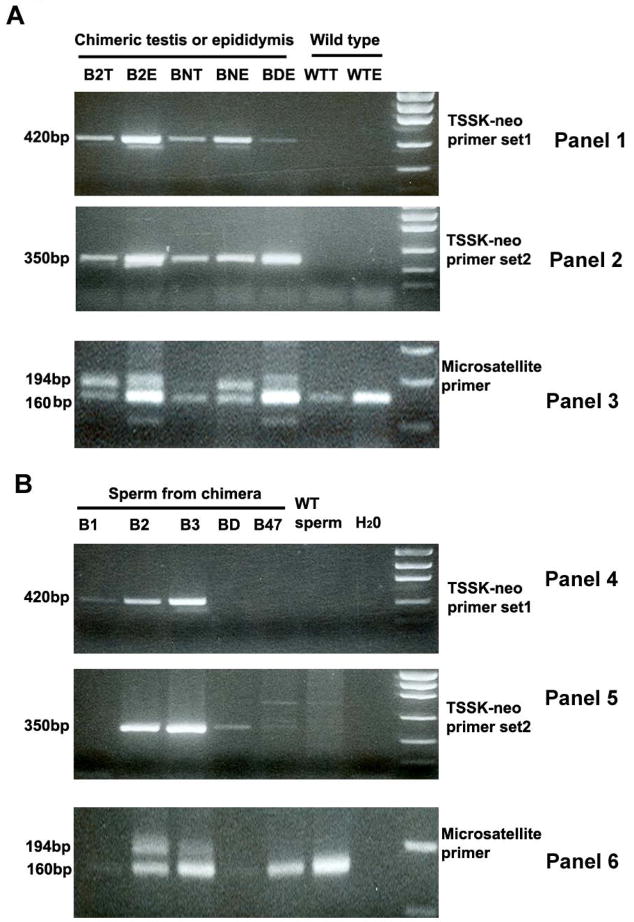
The testes and epididymides were collected from wild type (WT) as well as the chimeric males, DNA was extracted, and PCR genotyping was conducted using three sets of primers: 1) a neo-Tssk primer set 1 (shown in Fig. 1A, P1 with P2) produced a 420 bp DNA fragment representing the mutant allele; 2) a neo-Tssk primer set 2 (shown in Fig. 1A, P1 with P3) amplified a 350 bp band also representing the mutant allele; and 3) microsatellite primers targeting the D2Mit94 locus of mouse chromosome 2 were designed to PCR amplify 194 bp and 160 bp bands representing the microsatellite polymorphism of D2Mit94 loci in 129Svj and C57BL/6 strains, respectively. As shown in Fig. 2A, 420 and 350 bp bands representing the mutant allele were detected in the testes and epididymides of chimeric mice (eg. B2, BN and BD in panel 1 & 2 in Fig. 2A). This result was further supported by the detection of both C57BL/6 and 129Svj microsatellite polymorphisms at the D2Mit94 loci (same three chimeras B2, BN and BD in panel 3 in Fig. 2A). Neither the mutant allele nor the 129Svj D2Mit94 locus was detected in wild type mice.
Figure 2. PCR genotyping of chimeric testes and epididymides (A) and sperm/spermatogenic cells collected from uteri of females mated with male chimeras (B).
PCR for mutant alleles is shown in panels 1 & 2 and 4 & 5. PCR to detect the 129Svj (194 bp) and C57BL/6 (160 bp) microsatellite DNA at D2Mit94 locus is shown in Panels 3 & 6. A: The mutant alleles were not detected in wild type testis and epididymides (WTT and WTE), but were detected in DNA from the chimeric testes (B2T, BNT) and epididymides (B2E, BNE and BDE), consistent with the detection of both 129Svj and C57BL/6 genomes in the samples from chimeric mice. B: The mutant alleles were also detected in several DNA samples (B2, B3) from sperm/spermatogenic cells collected after mating chimeric mice (panel 4 & 5), consistent with detection of both 129Svj and C57BL/6 microsatellite DNA at the D2Mit94 locus (panel 6).
In order to further characterize this phenotype, sperm/spermatogenic cells from the chimeric and wild type males were collected from uteri after mating with C57BL/6 females (e.g. chimeras B1, B2, B3, B47, BD and WT in Fig. 2B) and were genotyped with the set of primers described in the preceding paragraph. The 420 and 350 bp bands representing the mutant allele were clearly detected in sperm/spermatogenic cells produced by chimeras B2 and B3 (panel 4 & 5 in Fig. 2B) and both C57BL/6 and 129Svj type genomes were present. Neither the mutant allele nor the 129Svj of D2Mit94 locus was detected from the wild type mouse and chimera B47.
A faint band representing the mutant allele was detected in the B1 (panel 4 in Fig. 2B) and BD (panel 5 in Fig. 2B) chimeras, however PCR amplification of microsatellite DNA of D2Mit94 locus failed due to limited amount of DNA extracted from the uterine flushing. In fact, the chimera B1 was an infertile mouse, it only formed a plug in one female mouse in 5 mating experiments, and only a few sperm were observed in the uterine flushing. Biopsy of testis and epididymis of B1 indicated that very few sperm reached maturation, and only a few round spermatogenic cells occupied the lumen of the epididymis (Fig. 3). Thus very few sperm/spermatogenic cells could be collected from this chimera.
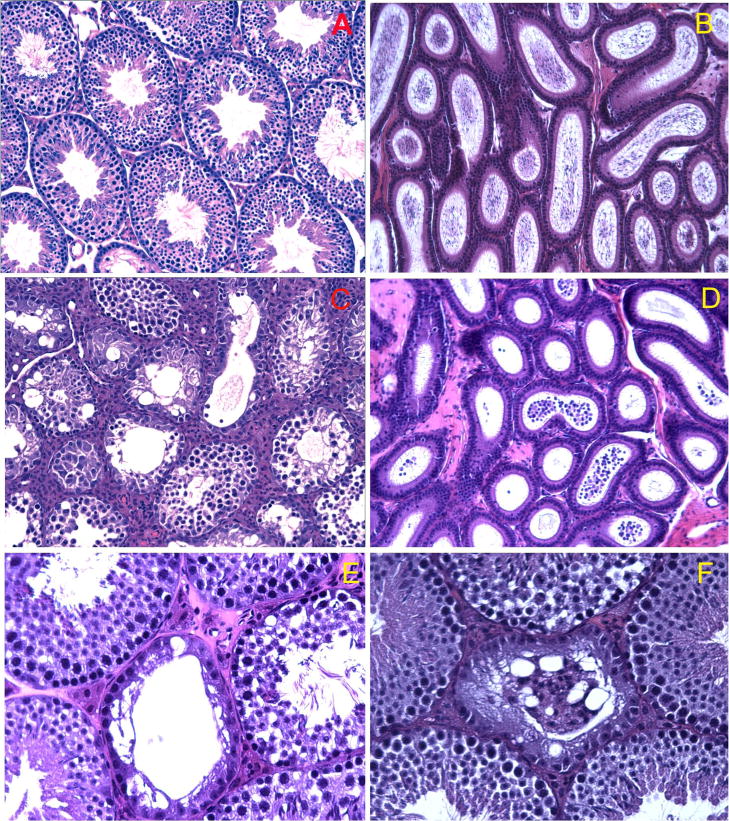
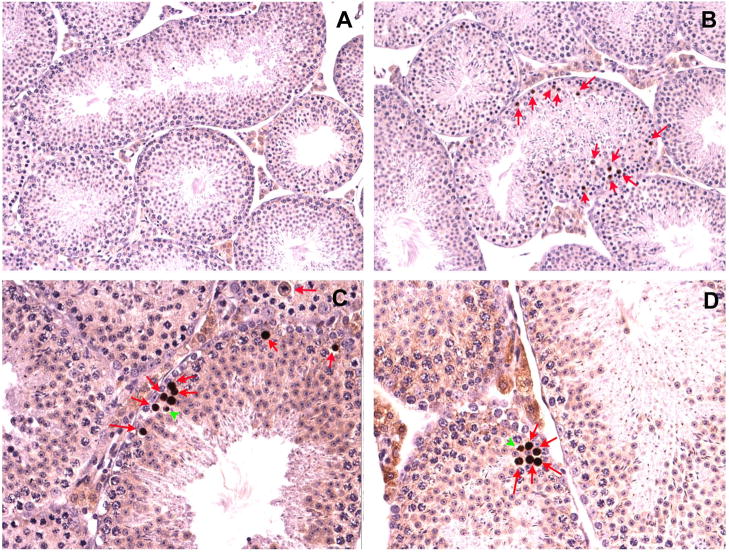
Figure 3. Histological analysis of testes and epididymides from wild type and chimeric mice.
Testes and epididymides from wild type C57BL/6 mice show normal stages of spermatogenesis (A) and spermatozoa fill the epididymal lumen (B). (C to F) Light micrographs of testes (C, E and F) and epididymides (D) from three chimeric mice. Abnormal morphology in testes from chimera B1 (C) is characterized by absence of elongated spermatids from most seminiferous tubules. Abnormal morphology in the epididymis of chimera B1 is characterized by absence of sperm and the presence of numerous round cells in the epididymal lumen (D). (E) Abnormal morphology of testes from chimera BN. A tubule (at the central) with no evident germ cells is accompanied by a few tubules showing apparently normal spermatogenesis. (F) Abnormal testis morphology of chimera BD. Vacuoles and degenerating symplast-like cells were found in one seminiferous tubule surrounded by tubules with normal morphology.
In addition to the chimeras shown in Fig. 2B, a total of 10 chimeric males from 21 tested chimeric males produced sperm/spermatogenic cells bearing the mutant allele and 129Svj type of D2Mit94 locus. These findings confirmed that germline transmission of the mutant allele did occur and that chimeric mice were producing 129Svj type sperm/spermatogenic cells. Among these 10 chimeras, 6 of them were high percentage chimeras including infertile B1 (judging from the fur color, B1 is a more than 95% chimera); 4 of them were low percentage chimeras including BN which produced 8 agouti offspring.
Thus, the targeted allele was transmitted into the germline and into sperm/spermatogenic cells of these chimeric males but only wild type offspring resulted. Identical results were obtained from chimeric mice generated from two independent ES clones. These results indicate that targeted deletion of one copy of Tssk1 & 2 genes causes haploinsufficiency which leads to male infertility.
To rule out the possibility that the deletion of Tssk1 & 2 caused the arrest of early embryonic development instead of male infertility, the litter sizes produced by 10 chimeric mice with evidence of germline transmission were compared with those of 7 fertile chimeric mice without detectable germline transmission. “In the experimental group, except for infertile mouse B1, the average litter size for individual males range from 5.65 to 8.25, the mean of average litter size for this group was 6.54, which was not significantly different than the control group, where the average litter size for each male ranged from 5.93 to 8.23, and the mean of average litter size for the control group was 6.67. Therefore, it is unlikely that the deletion of Tssk1 & 2 led to early embryonic lethality.”
Phenotyping the testes and epididymides from chimeric males
Histological analyses focused on the testes and epididymides from 10 male chimeras in which both integration of ES cells into the testis and production of sperm/spermatogenic cells bearing the mutant allele had been confirmed by PCR. These testes showed a range of altered testicular morphologies. For example, in a HPC (>95%) mouse, which was entirely infertile, both testes were small (<50% of wild type in weight and size), and 98% of the seminiferous tubules displayed severely altered spermatogenesis (Fig. 3C) with an absence of elongating and elongated spermatids, lack of testicular sperm in the tubule lumen and presence of round cells rather than spermatozoa in the epididymal lumen (Fig. 3D). 2 HPC and 2 LPC had increased numbers of abnormal seminiferous tubules, 19.2%, 8.9%, 11.5%, 9.2% tubules being disrupted in this group, compared with age-matched wild type mice in which the mean incidence of abnormal tubules was 2.8%. Along with this increased incidence of abnormal seminiferous tubules in chimeric mice, increased numbers of round cells were observed in the epididymal lumen compared to age-matched wild type controls. In the abnormal seminiferous tubules from these 5 animals, spermatogenesis was arrested before step 7. Sixty three percent of these abnormal seminiferous tubules had early spermatids up to step 7, but lacked elongating and elongated spermatids. Twenty eight percent of the abnormal seminiferous tubules had spermatogonia and spermatocytes and lacked spermatids, while 9% showed the “Sertoli-only” phenotype with very few spermatogonia. The severely disrupted tubules often had multiple vacuoles and aggregated germ cells (the central tubules in Fig. 3E and F). Thus, a full complement of TSSK1 & 2 proteins may be essential for spermiogenesis after step 7, when the critical processes of flagellar formation and nuclear condensation occur. Since cDNA array data (http://www.mrg.genetics.washington.edu) indicate both Tssk1 and Tssk2 are expressed only in spermatids, a finding confirmed by the immunolocalization of Tssk2 in our companion paper, it is posited that more severe and earlier arrest in some of the tubules may be caused by the arrest of spermatid development which disrupt the cellular communication between germ cells.
Apoptosis in chimeric testes
Another 3 HPC and 2 LPC which produced sperm/spermatogenic cells bearing the mutant allele had only a few obvious abnormal seminiferous tubules (<5%), which was not statistically different from the incidence of abnormal tubules observed in the wild type mice (2.8%), and very few round cells were noted in the epididymal lumen of these chimeras. Despite the lack of obvious morphological disruption of the stages of the seminiferous cycle, differences were noted, when the testes from these animals were subjected to apoptosis analysis. As shown in Fig. 4, testis sections from the wild-type mice revealed very few apoptotic nuclei (Fig. 4A); spermatogonia were the major cell type among those undergoing apoptosis and the apoptotic cells were interspersed with normal cells (data not shown). Sections from chimeric mice, on the other hand, showed a 6 to 8.5 fold increase in the number of apoptotic nuclei (Fig. 4B). Spermatocyte (Fig. 4C and D, arrow) and early round spermatid nuclei (Fig. 4C and D, arrowhead) were the predominant cell types undergoing apoptosis in chimeric mice, and the apoptotic cells often appeared in clusters (Fig. 4C and D), consistent with the interpretation that the apoptotic cells represented clonally derived daughter cells. Thus, although the stages of spermatogenesis appeared morphologically intact in this group of chimeras, cohorts of spermatocytes and early spermatids underwent markedly increased apoptosis compared to the wild type controls. These findings indicate that increased apoptosis occurs as a result of the lower the dose of TSSK1 & 2, which appears to be critical for spermatogenesis.
Figure 4. Immunohistochemical examination of germ cell apoptosis in chimeric testes.
(A) Representative testis section from a wild type C57BL/6 mouse showing few apoptotic cells. (B) Numerous apoptotic germ cells (arrows) are present in a chimeric testis. (C and D) Spermatocytes (arrows) and round spermatids (arrowheads) are major cell types undergoing apoptosis in chimeric testes.
Presence of TSSK2 and TSKS proteins in mature mouse and human spermatozoa
The failure of Tssk1 & 2 chimeras to propagate the null allele and the arrest of spermatogenesis at the spermatid stage prompted us to study further the presence and localization of these two kinases. Since TSSK1 and TSSK2 have high similarity and share the same substrate TSKS, the study focused on one of the kinases, TSSK2, and the interaction of TSSK2 with TSKS.
TSSK1 & 2 and TSKS proteins were previously reported (Kueng et al. 1997) to be abundant in mouse testis, but absent in the epididymis where sperm undergo final maturation and storage. However, in this study with 80 μg testis or sperm proteins loaded in each lane, antibody to mouse TSSK2 not only detected 41 kDa TSSK2 in mouse testis extracts but also in mouse epididymal sperm (Fig. 5D), and 42 kDa TSSK2 in human ejaculated sperm extracts (Fig. 5A). Pre-immune sera resulted in only a very faint background. These Western results differed from previous reports which showed no TSSK2 in sperm (Kueng et al. 1997). The present demonstration differed methodologically from the previous report in that Celis buffer was employed in the present extraction protocol, which may have been more efficient in extracting immunoreactive TSSK2 from sperm.
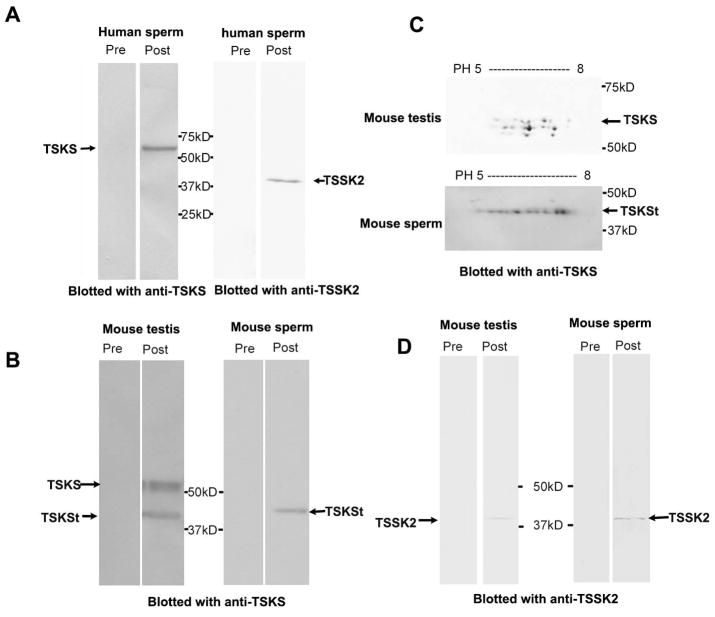
Figure 5. Western blot analyses of TSKS and TSSK2 protein expression in testes and spermatozoa from mouse and human.
A: rat anti-TSKS or rabbit anti-TSSK2 sera detected 65 kDa TSKS and 42 kDa TSSK2, respectively, on 1-D Western blots of human sperm protein extracts. B: Protein extracts from mouse testis and epididymal sperm on 1-D Western blots stained with rabbit anti-TSSK2, rat anti-TSKS or corresponding pre-immune sera. 41 kDa TSSK2 was recognized in both mouse testis and sperm by the immune sera, while pre-immune sera were negative. A broad TSKS band at ~60 kDa and a ~40 kDa truncated TSKS [TSKSt] were noted in testis extracts. The ~40 kDa TSKSt was the major isoform detected in epididymal spermatozoa. Pre-immune control sera were negative. C: Charge and mass variants of TSKS in 2-D Western blots indicated that, in the mouse testis, ~60 kDa TSKS showed protein microheterogeneity with both mass and charge variants (upper panel), while ~40 kDa TSKSt showed charge variants at a single mass (lower panel) in mouse sperm.
To generate a specific immunoreagent, recombinant human TSKS was expressed in E.coli, purified, and employed to generate polyclonal antibodies in rats. The anti-TSKS antibodies immunoreacted with both human and mouse TSKS (Fig. 5), which are 83% identical at the amino acid level. In protein extracts from human sperm, the anti-TSKS antibody recognized a single 65 kDa band, which is the predicted size of full length human TSKS (Fig. 5A, left pair), as previously published for human testis (Hao et al. 2004). Control pre-immune serum did not recognize any proteins in the sperm extracts. This is the first demonstration that TSKS is found in ejaculated human sperm.
In addition to the reported human TSKS sequence (Hao et al. 2004), an alternative splice variant of human TSKS was detected by cDNA cloning from human testis Marathon-ready cDNA (Clontech) [now listed in the NCBI database with accession number AF411384]. Whereas the full length human TSKS has 592 aa, the variant lacks the first 5 exons while retaining exons 6–10 and adds a unique first exon, yielding a predicted 42.3 kDa protein of 386 aa which, interestingly, did not appear in human sperm extracts in Western analyses (Fig. 5A).
In mouse testis extracts (Fig. 5B) anti-TSKS antibody recognized a broad band at ~60 kDa, as well as a band of ~40 kDa, which we refer to truncated TSKS, or TSKSt. The ~60 kDa TSKS was the major isoform detected in mouse testis, while the ~40 kDa TSKSt was the major isoform in mouse epididymal spermatozoa (Fig. 5B) where the ~60 kDa TSKS was undetectable at 80 μg sperm protein/lane. The current NCBI database lists two mouse TSKS cDNAs, NM_001077591 and NM_011651, that encode 57.3 and 63.5 kDa proteins, respectively, the former having a 9 aa deletion in the middle (exon 5) and an additional 51 aa deletion at C-terminus (exon 11). The ~60 kDa TSKS may be encoded by one or both of these variants.
Three lines of evidence argue that the ~60 kDa and ~40 kDa proteins in Fig. 5B are both authentic mouse TSKS isoforms. 1) As shown below, anti-TSKS antibody was employed to immnoprecipitate TSKS from mouse testis extracts, the immunoprecipitated products were submitted for mass spectrometry, and multiple peptides covering 221 aa of mouse TSKS were detected. 2) To establish the immunological identity of the ~60 kDa and ~40 kDa proteins as authentic TSKSs, an affinity purified antibody to TSKS was generated through binding and eluting antibody from recombinant TSKS protein. This affinity purified antibody recognized the ~60 kDa and ~40 kDa TSKS protein from mouse testis and sperm on 1-D gels. 3) This antibody revealed considerable TSKS protein microheterogeneity. High resolution 2-D gel immunoblots of ~60 kDa TSKS isoforms from mouse testis showed charge trains, as well as three slight mass variants, indicating possible phosphorylation at multiple sites as well as other post-translational modifications (Fig. 5C). The ~40 kDa TSKS isoform extracted from mouse sperm exhibited protein microheterogeneity with a prominent charge train at one consistent mass, a feature of phosphorylated proteins (Fig. 5C).
The finding of several mass variants of ~60 kDa TSKS on 2-D blots is consistent with the broad TSKS band observed on 1-D Western blots. The 57.3 and 63.5 kDa mouse TSKS variants mentioned above may be responsible for the mass variants of ~60 kDa TSKS shown in Fig. 5C. The ~40 kDa mouse TSKSt protein may be encoded by an unknown splicing variant of TSKS or could represent a proteolytic product. As noted in our companion paper, a population of TSKS was localized in the residual cytoplasm of late stage spermatids where proteolysis may cause the formation of truncated TSKS.
The essential interacting domain of human TSKS with TSSK2
We previously reported that human TSKS strongly interacts with TSSK2 in yeast two-hybrid analyses (Hao et al. 2004). To dissect the essential domain(s) required for TSKS interaction with TSSK2, bioinformatic analyses were first performed. A number of interesting structural motifs were noted within human TSKS. These included a putative nuclear localization signal (NLS, aa 48–51), two coiled-coil domains (C-C1, aa 147–234; C-C2, aa 324–392), and three 6 amino acid repeats (Rpt), QEPEEK, located at aa 244–261 (Fig. 6A). Based on these analyses, a total of 7 deletion mutants of TSKS were created (Fig. 6A). The 7 deletion mutants (namely D1 to D7) were fused with the Gal4 activation domain in the pGAD, and their expression in the two-hybrid host strain was first tested by Western blot analysis (Fig. 6B). These fusion proteins were then tested for their ability to interact with TSSK2 fused with pGBD. A quantitative analysis of the reporter gene activity (alpha-galactosidase) was performed (Fig. 6C). The activity shown by each mutant is expressed as the percentage of wild type TSKS (Fig. 6C) adjacent to the corresponding deletion mutant (Fig. 6A). In mutant D1 (Δ 392–592), which represented a deletion of a major part of the TSKS C terminus, TSSK2 binding was reduced by 43%. In mutant D2 (Δ 326–592), in which both the C-terminus and the second coiled-coil domain were deleted, TSSK2 binding was reduced by 68%. Thus, some TSSK2 binding persisted in the absence of the second coiled-coil region. In mutant D7 (Δ 234–392) where the second coiled-coil domain and the region between the two coiled–coil domains, including the six amino acid repeat motif, were absent but the C-terminus was intact, binding was close to wild type levels. This indicated that the second coiled-coil domain and the six amino acid repeats play no role in TSSK2 binding, and suggested that the C-terminus may play some role in TSSK2 interaction but is not the critical binding site. Deletion of the first 48 amino acids (D4) reduced TSKS binding to 5% while deletion of the first 147 amino acids (D5, D6, D7) abolished TSKS binding entirely. These results showed that the N-terminus of TSKS is essential for its interaction with TSSK2.
Co-immunoprecipitation (IP) of TSKS/TSSK2 complexes from human sperm
To test whether in vivo interactions between TSKS and TSSK2 occurred in ejaculated human sperm, co-immunoprecipitation experiments were performed. Precleared protein extracts from human sperm were immunoprecipitated with rabbit anti-TSSK2 serum (IP-TSSK2) or control normal rabbit serum (Rabbit IP-control). The immunoprecipitates were subjected to immunoblotting with rabbit anti-TSSK2 antibody (Fig. 7, panel 1) or rat anti-TSKS antibody (Fig. 7, panel 2). Immunoprecipitates from human sperm brought down with rabbit anti-TSSK2 antibody contained two bands only in the IP-TSSK2 lane at ~65 kDa and ~50 kDa that were recognized by anti-TSKS antibody (Fig. 7, panel 2). No bands were recognized by anti-TSKS antibody in the rabbit IP-control lane. This confirmed that immunoprecipitation of TSSK2 from human sperm brought down its substrate, TSKS. As a positive control, immunoblotting with anti-TSSK2 confirmed that the immunoprecipitates indeed contained TSSK2, which migrated at ~42 kDa only in the IP-TSSK2 lane (Fig. 7, panel 1). In panel 1, background heavy and light chain (arrowheads) immunoglobulin bands were present due to the reactivity of the goat anti-rabbit secondary antibody with the rabbit anti-TSSK2 antibody or normal rabbit serum used in the IP reaction.
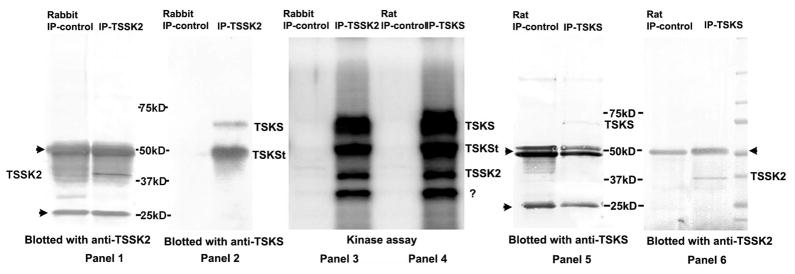
Figure 7. Co-immunoprecipitation and in vitro phosphorylation of TSKS/TSSK2 complexes from human sperm.
Panels 1 and 2: pre-cleared human sperm extracts were immunoprecipitated with either control rabbit serum (Rabbit IP-control) or rabbit anti-TSSK2 serum (IP-TSSK2), and the immunoprecipitates were subjected to immunoblotting with anti-TSSK2 (panel 1) or anti-TSKS (panel 2). 65 kDa and ~50 kDa TSKS isoforms were recognized by anti-TSKS antibody only in the lane immunoprecipitated with anti-TSSK2 antibody while 42 kDa TSSK2 was detected in the immunoprecipitate. Panels 5 and 6: pre-cleared human sperm extracts were immunoprecipitated with either pre-immune rat serum (Rat IP-control), or rat anti-TSKS serum (IP-TSKS), the immunoprecipitates were subjected to immunoblotting with anti-TSKS antibody (panel 5) or anti-TSSK2 antibody (panel 6). 65 kDa TSKS and 42 kDa TSSK2 were detected only in the lanes immunoprecipitated with anti-TSKS. Panels 3 and 4: autoradiograph of TSSK2/TSKS complexes immunoprecipitated with either anti-TSSK2 (IP-TSSK2) or anti-TSKS (IP-TSKS) and incubated with [32P]γATP. Four identical phospho-protein bands were detected in immune complexs precipitated with either anti-TSSK2 (IP-TSSK2) or anti-TSKS (IP-TSKS). Conversely, no proteins were phosphorylated in the controls immunoprecipitated with normal sera (Rabbit IP-control and Rat IP-control). These bands represent phosphorylation of 65 and ~50 kDa forms of TSKS, autophosphorylation of TSSK2 and an unknown 30 kDa phosphoprotein.
The converse experiment gave similar results. Precleared protein extracts from human sperm were immunoprecipitated using rat anti-TSKS serum (IP-TSKS) and normal rat serum (Rat IP-control). The immunoprecipitates were subjected to immunoblotting with anti-TSKS antibody (Fig. 7. panel 5) or anti-TSSK2 antibody (Fig. 7. panel 6), respectively. The immunoblots with anti-TSKS confirmed the immunoprecipitation of ~65 kDa TSKS from human sperm protein extracts only in the lane containing the anti-TSKS immune complexes and not in the lane precipitated with control rat serum (Fig. 7, panel 5). The ~50 kDa TSKS form could not be conclusively identified due to possible masking by the ~56 kDa heavy chain band. The immunoblot with anti-TSSK2 showed a band in the IP-TSKS lane at ~42 kDa (Fig. 7, panel 6), the predicted size of human TSSK2, while this band was not evident in the lane immunoprecipitated with control rat sera. Together these experiments demonstrated that both rabbit anti-TSSK2 and rat anti-TSKS immunoreagents were capable of precipitating complexes containing both TSSK2 and TSKS from human sperm. This confirmed in vivo interactions of this kinase/substrate pair in mature human sperm.
In vitro phosphorylation of TSKS
Immune complexes precipitated from human sperm extracts with either anti-TSKS (IP-TSKS) or anti-TSSK2 (IP-TSSK2) were incubated with [32P]γATP (Fig. 7. panels 3, 4). Four phosphorylated bands were detected in complexes immunoprecipitated with anti-TSSK2 (Fig. 7, panel 3), while the same four bands were detected in complexes immunoprecipitated with anti-TSSK2 (Fig. 7, panel 4). These phosphorylated bands did not appear in lanes immunoprecipitated with either control serum. Based on their alignment with the immunoblots to either side, these bands represent 65 kDa TSKS, the ~50 kDa truncated TSKS (also seen in Fig. 7, panel 2), 42 kDa TSSK2 and an unknown ~30 kDa protein. Since this ~30 kDa protein appeared in immunoprecipitates with both TSKS and TSSK2 antisera but not in controls, it is potentially a new phospho-protein constituent of the TSKS/TSSK2 complex or a proteolytic fragment of either kinase or substrate. This experiment conclusively demonstrated that TSSK2 in human sperm is enzymatically active and capable of phosphorylating TSKS in vivo.
Co-immunoprecipitation (IP) of TSSK2 and TSKS from mouse testis and in vitro phosphorylation of mouse testicular TSKS
TSSK2 and TSKS co-immuoprecipitation experiments were also conducted with protein extracts from mouse testis. Precleared testis extracts were immunoprecipitated using normal rabbit serum (Rabbit IP-control), or rabbit anti-TSSK2 serum (IP-TSSK2), and the immune complexes were subjected to immunoblotting with anti-TSSK2 or anti-TSKS antibodies, respectively (Fig. 8. panels 1 and 2). Anti-TSSK2 antibodies immunoprecipitated 41 kDa TSSK2 as well as ~60 kDa TSKS and ~40 kDa TSKSt from mouse testis protein extracts, whereas control sera did not immunoprecipitate these proteins (Fig. 8. panels 1 and 2). Conversely, anti-TSKS antibodies immunoprecipitated TSKS and TSSK2 proteins from mouse testis, while the control sera did not (Fig. 8. panels 3 and 4). This verified that both anti-TSKS and anti-TSSK2 antibodies are capable of co-immunoprecipitating TSKS and TSSK2 from mouse testis.
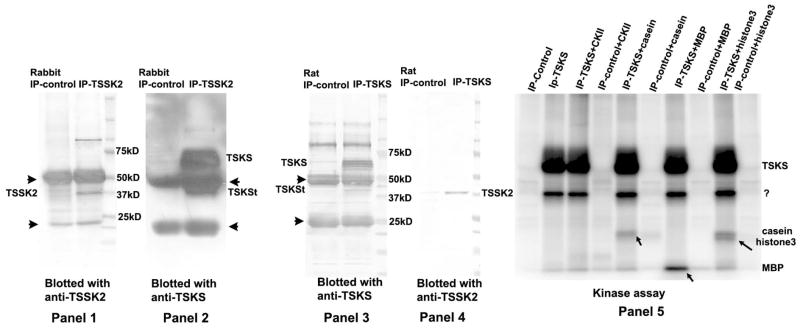
Figure 8. Co-immunoprecipitation and in vitro phosphorylation of TSKS/TSSK2 complexes from mouse testis.
In panels 1 and 2, pre-cleared mouse testis extracts were immunoprecipitated with either rabbit normal serum (Rabbit IP-control), or rabbit anti-TSSK2 serum (IP-TSSK2), and the immunoprecipitates were subjected to immunoblotting with anti-TSSK2 (panel 1) or anti-TSKS (panel 2). TSKS and TSSK2 were both detected in the immune-complex immunoprecipitated with anti-TSSK2 serum. In panels 3 and 4, precleared mouse testis extracts were immunoprecipitated with either normal rat serum (Rat IP-control), or rat anti-TSKS serum (IP-TSKS), and the immunoprecipitated complexes were subjected to immunoblotting with anti-TSKS (panel 3) or anti-TSSK2 antibody (panel 4). Both TSKS and TSSK2 were detected in the immune-complex immunoprecipitated with anti-TSKS serum. Panel 5 shows an autoradiograph of immune-complexes (as described in panel 3 and 4) immunoprecipitated with either anti-TSKS (IP-TSKS) or rat normal serum (Rat IP-control) and subsequently incubated with [32P]γATP in an in vitro kinase assay. A common kinase, casein kinase 2 (CKII) and several common kinase substrates including casein, myelin basic protein (MBP) and histone 3 were added to the reactions as the positive control. In immunoprecipitates with anti-TSKS sera, TSKS was strongly phosphorylated as well as a ~40 kDa protein, while no proteins were phosphorylated in immunoprecipitates with control normal rat sera. The ~40 kDa protein may represent autophosphorylation of TSSK1 and/or TSSK2 or the truncated form of TSKS. Addition of CKII did not result in any additional phosphorylation. Casein, MBP and histone3 were also phosphorylated by the kinases in the precipitated complex, whereas these proteins were not phosphorylated in the controls immunoprecipitated by normal sera.
In vitro phosphorylation of immune-complexes immunoprecipitated with either anti-TSKS (IP-TSKS) or rat normal serum (IP-control) from mouse testis were subsequently incubated with [32P]γATP in an in vitro kinase assay (Fig. 8, panel 5). A common kinase, casein kinase 2 (CKII) and several common kinase substrates including casein, myelin basic protein (MBP) and histone 3 were added to the reactions as positive controls. In the immunoprecipitates with anti-TSKS sera, TSKS was strongly phosphorylated as well as a ~40 kDa protein, while no proteins were phosphorylated by immune complexes precipitated with control normal rat sera. The ~40 kDa protein may represent autophosphorylation of TSSK1 and/or TSSK2 or phosphorylation of the truncated TSKS. Addition of CKII did not result in any additional phosphorylation, indicating either that phosphorylation of TSKS and TSSK is saturated or that they are not substrates of CKII. Casein, MBP and histone3 were also phosphorylated by the kinases in the precipitated complexes (pointed by arrows), whereas these proteins were not phosphorylated in the controls immunoprecipitated by the normal sera. This kinase assay provides a method to screen for inhibitors of TSKS phosphorylation as well as demonstrating that casein, MBP and histone 3 may be used as alternative substrates for TSSK1 & 2.
Identification of the site of TSKS phosphorylation by IMAC-LC-MS/MS
Immune-complexes (as described in Fig. 8) immunoprecipitated with anti-TSKS (IP-TSKS) from mouse testis protein extracts were submitted for phosphopeptide mapping by IMAC-LC-MS/MS. The TSKS phosphopeptide HGLSPATPIQGCSGPPGS*PEEPPR was identified, with serine 285 followed by a proline found to be phosphorylated. Serine 285 represented an in vivo phosphorylation site since it was detected directly from the immunoprecipitate of the testis tissue. Also serine 285 was one of the phosphorylation sites predicted by Netphos 2.0 software with a high confidence score.
Discussion
Analysis of the Tssk1 & 2 targeted deletion
Our mating results with chimeric males are similar to previous observations of haploinsufficiency in the cases of gene deletions affecting testis genes Klhl10, Pf20 (Spag16), Cyp17, and Prm1 or Prm2 (Zhang et al. 2004, Liu et al. 2005, Yan et al. 2005, Cho et al. 2001). In the case of the Klhl10 gene, 47 male chimeras were produced; 16 HPC males were infertile, and 26 LPC only produced black offspring (Yan et al. 2005). In the Prm1 or Prm2 targeted deletions, 51 male chimeras and 71 male chimeras were obtained, respectively, none of which produced agouti offspring (Cho et al. 2001). In the example of the Pf20 (Spag16) targeted deletion, among 23 male chimeras, four males fathered only black pups, six fathered both black and agouti offspring, and the remaining 13 chimeric mice were infertile. Although 204 agouti mice were obtained, none of them carried the mutant allele (Zhang et al. 2004).
In TSSK1 & 2 haploinsufficiency, despite the presence of one functional Tssk1 & 2 allele in the testes, the haploid spermatids originating from targeted ES cells are apparently not producing sufficient TSSK1 & 2 proteins to support normal spermatogenesis and sperm function. It is very interesting that one of the other proteins that shows haploinsufficiency, PF20 (Spag16), is an axonemal protein which has been localized to the flagellum (Zhang et al. 2004, Zhang et al. 2006), and was recently shown by our team to be a TSSK2 substrate (Zhang et al, submitted). Together these studies strongly implicate the TSSK1 & 2 pathway and the TSSK2 substrate PF20 (Spag16) as essential genes for fertility.
Mouse Tssk1 & 2 are located on chromosome 16 in a region that is syntenic to a locus on human chromosome 22q11 which was previously thought to be associated with the DiGeorge syndrome. The human chromosomal region contains TSSK2 but not TSSK1. To create potential mouse models of DiGeorge syndrome, two research groups generated “large” 150 kb and 550 kb targeting deletions within the putative DiGeorge syndrome related region, which included mouse Tssk1 & 2 and several other genes. Homozygous animals bearing deletions of both these areas on chromosome 16 were embryo lethal (Puech et al. 2000, Kimber et al. 1999). It was discovered later that Tbx1, which does not locate in these 150 kb and 550 kb regions, is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome (Merscher et al. 2001). It is noteworthy that the two research groups mentioned above were able to generate chimeric mice and heterozygotes when they mated mice with these “large” deletions containing mouse Tssk1 & 2. This stands in contrast to our findings that deletion of Tssk1 & 2 alone prevents transmission of the Tssk1 & 2 mutant allele from chimeric males to offspring. A similarly puzzling result has been observed in the case of the deletion of the mouse Cyp17 (Cytochrome P450 17 -Hydroxylase/17,20 Lyase) gene. One research group reported that male chimeras of Cyp17 gene consistently failed to generate heterozygous Cyp17 mice, and after five matings chimeric mice stopped mating indicating a change in sexual behavior (Liu et al. 2005). At the same time, another research group reported that they were able to generate chimeric mice and heterozygotes that were reproductively normal, but Cyp17−/− zygotes died by embryonic day 7 (Bair et al. 2004).
Haploinsufficency of the Tssk1 & 2 genes implies that the dose of these germ cell genes is critical to spermatogenesis. A single dose of these genes may not be sufficient to fulfill the full gene function, especially in certain genetic backgrounds. Therefore, it is reasonable to posit that the dose of a certain germ cell gene may be more critical in some mouse strains than in others in which the same gene may effectively be transmitted. The two research groups which conducted the deletion of large fragments of chromosome 16 used WW6 ES cells (Merscher et al. 2001, Puech et al. 2000) and TC1 ES cells respectively (Kimber et al. 1999), while we used the 129Svj ES cells to conduct our targeted deletions. Thus, different mouse strains and different targeting strategies may explain the different effects on fertility. Nevertheless, our extensive mating experiments, and careful analysis on genotype and phenotype of Tssk1 & 2 chimeric mice indicate that the dose of TSSK1 and 2 must be essential for normal male fertility.
Expression, localization, interaction and functional activation of TSSK2 and TSKS
Western blot analyses identified TSSK2 and TSKS in testes and mature sperm of both mice and men. Microscopic studies immunolocalized TSSK2 and TSKS proteins in spermatids and spermatozoa (Xu et al. the companion paper). In situ analysis indicated that mRNAs for these proteins are mainly expressed in post-meiotic spermatids (Xu et al. the companion paper). In addition, high-throughput cDNA array data regarding gene expression in the testis during the progression of spermatogenesis are available now online: http://www.mrg.genetics.washington.edu/. Consistent with our in situ result, the cDNA array analysis showed that Tssk1& 2 and Tsks mRNA expression is specific to spermatids. These expression and localization data are consistent with the phenotype of arrested spermiogenesis noted in testes of chimeric Tssk 1 & 2 mice.
To dissect the function of TSSK2 kinase, it is important to understand the interaction of this enzyme with its major substrate TSKS. Our co-immunoprecipitation experiments demonstrated that both rabbit anti-TSSK2 and rat anti-TSKS were capable of immunoprecipitating complexes from human sperm as well as from mouse testes that contained both the enzyme and its substrate. These findings confirmed that interactions of this kinase/substrate pair occur in both testis and sperm in different species. Our analysis of TSKS deletion mutants revealed that the N-terminus of TSKS is essential for TSSK2 binding and thus contains the critical interaction site. In our companion paper, we discovered TSKS concentrated in spermatid centrioles during flagellogenesis, while TSSK2 partially co-localized at the the same region with addition localization at sperm tail, acrosomal region of mouse sperm and equatorial segment of human sperm. The strong binding activity between TSSK2 and TSKS may be the driving force to recruit TSSK2 to the sites around the centrioles, suggesting that TSSK2/TSKS may have a role in centriolar function during flagellogenesis. This is also consistant with the phenotype of arrested spermiogensis in testes of chimeric Tssk1 & 2. TSSK2 had a wider subcellular localization than TSKS, so it is also possible that TSSK2 has other substrates, and may have functions beyond the centriolar region that affect the phenotype of chimeric Tssk1 & 2 animals.
Most importantly, our experiments indicated that TSSK2 kinase is enzymatically active in both testis and mature sperm. In vitro kinase experiments confirmed that TSSK2 recovered in immunoprecipitates from human sperm and mouse testes was capable of phosphorylating TSKS and itself. The TSKS phosphopeptide HGLSPATPIQGCSGPPGS*PEEPPR was identified, with serine 285 following by a proline found to be phosphorylated in vivo. Serine 285 was one of the phosphorylation sites predicted by Netphos 2.0 software with a high confidence score. This phosphorylation site, located between two coiled-coil domains, is conserved in both mouse and human TSKS. However, it is likely that there are multiple phosphorylation sites on TSKS, because multiple immunoreactive TSKS spots were detected in the charge train in 2-D Western analysis (Fig. 5C), and the Netphos 2.0 predicted multiple high probability phosphorylation sites. In our companion paper, we further discussed several kinase/substrate pairs that play important roles in somatic centrosome function. The infertile phenotype of mice bearing targeted deletion of Tssk1 & 2 suggests an essential role for TSSK2/TSKS in spermatid centriole functions and underscores the contraceptive target candidacy of this first kinase/substrate pair detected in the gamete centriole.
Previous studies of TSSK2 reported the presence of this enzyme in testis (Kueng et al. 1997). The confirmation in the present study of the persistence of this enzyme in sperm in humans and mice opens the possibility of considering TSSK 1 & 2 as targets for both a male contraceptive that acts on the testis as well as for an intra-vaginal spermicidal agent.
Supplementary Material
Acknowledgments
We thank Dr. Robert A. Bloodgood and Dr. Jeffrey J. Lysiak and others at the Center for Research in Contraceptive and Reproductive Health for critique of the manuscript. This work was supported in part by NIH R03 HD055129, R01 HD 045783, R01 HD037416, NIH Fogarty International Center Grant D43 TW/HD 00654, NIH U54 HD29099, the Andrew W. Mellon Foundation and Schering AG.
Contributor Information
Bingfang Xu, Center for Research in Contraceptive and Reproductive Health, Department of Cell Biology, University of Virginia, Charlottesville, VA 22908.
Zhonglin Hao, Center for Research in Contraceptive and Reproductive Health, Department of Cell Biology, University of Virginia, Charlottesville, VA 22908.
Kula N. Jha, Center for Research in Contraceptive and Reproductive Health, Department of Cell Biology, University of Virginia, Charlottesville, VA 22908
Zhibing Zhang, Department of Obstetrics & Gynecology, Virginia Commonwealth University, Richmond, VA, 23298.
Craig Urekar, Center for Research in Contraceptive and Reproductive Health, Department of Cell Biology, University of Virginia, Charlottesville, VA 22908.
Laura Digilio, Center for Research in Contraceptive and Reproductive Health, Department of Cell Biology, University of Virginia, Charlottesville, VA 22908.
Silvia Pulido, Center for Research in Contraceptive and Reproductive Health, Department of Cell Biology, University of Virginia, Charlottesville, VA 22908.
Jerome F Strauss, III, Department of Obstetrics & Gynecology, Virginia Commonwealth University, Richmond, VA, 23298.
Charles J. Flickinger, Center for Research in Contraceptive and Reproductive Health, Department of Cell Biology, University of Virginia, Charlottesville, VA 22908
John C. Herr, Center for Research in Contraceptive and Reproductive Health, Department of Cell Biology, University of Virginia, Charlottesville, VA 22908
References
- Bair SR, Mellon SH. Deletion of the mouse P450c17 gene causes early embryonic lethality. Mol Cell Biol. 2004;24:5383–90. doi: 10.1128/MCB.24.12.5383-5390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielke W, Blaschke RJ, Miescher GC, Zurcher G, Andres AC, Ziemiecki A. Characterization of a novel murine testis-specific serine/threonine kinase. Gene. 1994;139:235–9. doi: 10.1016/0378-1119(94)90762-5. [DOI] [PubMed] [Google Scholar]
- Bucko-Justyna M, Lipinski L, Burgering BM, Trzeciak L. Characterization of testis-specific serine-threonine kinase 3 and its activation by phosphoinositide-dependent kinase-1-dependent signalling. Febs J. 2005;272:6310–23. doi: 10.1111/j.1742-4658.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- Celis JE, Rasmussen HH, Madsen P, Leffers H, Honore B, Dejgaard K, Gesser B, Olsen E, Gromov P, Hoffmann HJ, et al. The human keratinocyte two-dimensional gel protein database (update 1992): towards an integrated approach to the study of cell proliferation, differentiation and skin diseases. Electrophoresis. 1992;13:893–959. doi: 10.1002/elps.11501301198. [DOI] [PubMed] [Google Scholar]
- Chen X, Lin G, Wei Y, Hexige S, Niu Y, Liu L, Yang C, Yu L. TSSK5, a novel member of the testis-specific serine/threonine kinase family, phosphorylates CREB at Ser-133, and stimulates the CRE/CREB responsive pathway. Biochem Biophys Res Commun. 2005;333:742–9. doi: 10.1016/j.bbrc.2005.05.157. [DOI] [PubMed] [Google Scholar]
- Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet. 2001;28:82–6. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- Hao Z, Jha KN, Kim YH, Vemuganti S, Westbrook VA, Chertihin O, Markgraf K, Flickinger CJ, Coppola M, Herr JC, Visconti PE. Expression analysis of the human testis-specific serine/threonine kinase (TSSK) homologues. A TSSK member is present in the equatorial segment of human sperm. Mol Hum Reprod. 2004;10:433–44. doi: 10.1093/molehr/gah052. [DOI] [PubMed] [Google Scholar]
- Kimber WL, Hsieh P, Hirotsune S, Yuva-Paylor L, Sutherland HF, Chen A, Ruiz-Lozano P, Hoogstraten-Miller SL, Chien KR, Paylor R, Scambler PJ, Wynshaw-Boris A. Deletion of 150 kb in the minimal DiGeorge/velocardiofacial syndrome critical region in mouse. Hum Mol Genet. 1999;8:2229–37. doi: 10.1093/hmg/8.12.2229. [DOI] [PubMed] [Google Scholar]
- Kueng P, Nikolova Z, Djonov V, Hemphill A, Rohrbach V, Boehlen D, Zuercher G, Andres AC, Ziemiecki A. A novel family of serine/threonine kinases participating in spermiogenesis. J Cell Biol. 1997;139:1851–9. doi: 10.1083/jcb.139.7.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yao ZX, Bendavid C, Borgmeyer C, Han Z, Cavalli LR, Chan WY, Folmer J, Zirkin BR, Haddad BR, Gallicano GI, Papadopoulos V. Haploinsufficiency of cytochrome P450 17alpha-hydroxylase/17,20 lyase (CYP17) causes infertility in male mice. Mol Endocrinol. 2005;19:2380–9. doi: 10.1210/me.2004-0418. [DOI] [PubMed] [Google Scholar]
- Lysiak JJ, Turner SD, Nguyen QA, Singbartl K, Ley K, Turner TT. Essential role of neutrophils in germ cell-specific apoptosis following ischemia/reperfusion injury of the mouse testis. Biol Reprod. 2001;65:718–25. doi: 10.1095/biolreprod65.3.718. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:1970–79. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–29. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Puech A, Saint-Jore B, Merscher S, Russell RG, Cherif D, Sirotkin H, Xu H, Factor S, Kucherlapati R, Skoultchi AI. Normal cardiovascular development in mice deficient for 16 genes in 550 kb of the velocardiofacial/DiGeorge syndrome region. Proc Natl Acad Sci U S A. 2000;97:10090–5. doi: 10.1073/pnas.97.18.10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J, Herr JC, Reddi PP, Wolkowicz MJ, Bush LA, Sherman NE, Black M, Flickinger CJ. Cloning and characterization of a novel sperm-associated isoantigen (E-3) with defensin- and lectin-like motifs expressed in rat epididymis. Biol Reprod. 2003;68:290–301. doi: 10.1095/biolreprod.102.005983. [DOI] [PubMed] [Google Scholar]
- Spiridonov NA, Wong L, Zerfas PM, Starost MF, Pack SD, Paweletz CP, Johnson GR. Identification and characterization of SSTK, a serine/threonine protein kinase essential for male fertility. Mol Cell Biol. 2005;25:4250–61. doi: 10.1128/MCB.25.10.4250-4261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti PE, Hao Z, Purdon MA, Stein P, Balsara BR, Testa JR, Herr JC, Moss SB, Kopf GS. Cloning and chromosomal localization of a gene encoding a novel serine/threonine kinase belonging to the subfamily of testis-specific kinases. Genomics. 2001;77:163–70. doi: 10.1006/geno.2001.6628. [DOI] [PubMed] [Google Scholar]
- Yan W, Ma L, Burns KH, Matzuk MM. Haploinsufficiency of kelch-like protein homolog 10 causes infertility in male mice. Proc Natl Acad Sci U S A. 2004;101:7793–8. doi: 10.1073/pnas.0308025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kostetskii I, Moss SB, Jones BH, Ho C, Wang H, Kishida T, Gerton GL, Radice GL, Strauss JF., 3rd Haploinsufficiency for the murine orthologue of Chlamydomonas PF20 disrupts spermatogenesis. Proc Natl Acad Sci U S A. 2004;101:12946–51. doi: 10.1073/pnas.0404280101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kostetskii I, Tang W, Haig-Ladewig L, Sapiro R, Wei Z, Patel AM, Bennett J, Gerton GL, Moss SB, Radice GL, Strauss JF., 3rd Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol Reprod. 2006;74:751–9. doi: 10.1095/biolreprod.105.049254. [DOI] [PubMed] [Google Scholar]
- Zuercher G, Rohrbach V, Andres AC, Ziemiecki A. A novel member of the testis specific serine kinase family, tssk-3, expressed in the Leydig cells of sexually mature mice. Mech Dev. 2000;93:175–7. doi: 10.1016/s0925-4773(00)00255-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.