Abstract
We report the X-ray crystal structure of human potassium channel tetramerization domain-containing protein 5 (KCTD5), the first member of the family to be so characterized. Four findings were unexpected. First, the structure reveals assemblies of five subunits while tetramers were anticipated; pentameric stoichiometry is observed also in solution by scanning transmission electron microscopy mass analysis and analytical ultracentrifugation. Second, the same Bric-a-brac, Tramtrack, Broad Complex (BTB) domain surface mediates assembly of five KCTD5 and four voltage-gated potassium (Kv) channel subunits; four amino acid differences appear crucial. Third, KCTD5 complexes have well-defined N- and C-terminal modules separated by a flexible linker that swivels ~30°; the C-module shows a new fold and is required to bind Golgi re-assembling stacking protein 55 with ~1 μM affinity as judged by surface plasmon resonance and ultracentrifugation. Fourth, despite the homology reflected in its name, KCTD5 does not impact operation of Kv4.2, Kv3.4, Kv2.1 or Kv1.2 channels.
Keywords: KCTD5, BTB, POZ, Golgi, GRASP55, T1, potassium channel, tetramerization domain
INTRODUCTION
The human genome has 21 KCTD genes that encode K+ channel tetramerization domain-containing (KCTD) proteins. KCTD genes are so-named because the N-termini of KCTD proteins and some voltage-gated K+ (Kv) channels are homologous 1,2. Four Kv channel subunits aggregate to create a transmembrane ion conduction pathway; subunit assembly is expedited by N-terminal tetramerization (T1) domains that contain a BTB fold 3,4. KCTD proteins are soluble with N-terminal BTB domains and variable C-termini 5. Because Kv channels operate in tissue specific fashion due to interaction with accessory subunits that modify subcellular location, level of expression, activity and pharmacology 6, KCTDs had been anticipated to bind to and regulate Kv channels.
Originally identified in Drosophila melanogaster zinc finger proteins, BTB (also called Poxvirus zinc finger, POZ) motifs are protein-protein interaction modules that mediate both self-association and interaction with non-BTB partners. Structures of proteins with BTB domains show significant conservation of the core fold; the domains are known to mediate homodimeric and homotetrameric assembly of transcription factors and Kv channels, respectively, using different surfaces of the fold 7.
The biological function of KCTD proteins remains unclear. Robust expression of transcripts in fetal tissues and low levels in adults suggests they play a role during development 8. Thus, KCTD12 (pfetin/Ron) is implicated in maturation of inner ear neurons by linkage to a progressive, dominant, auditory neuropathy 9 and KCTD11 proposed to suppress Hedgehog signaling and promote tumor growth in medulloblastoma 10,11. A point mutation in KCTD7 has been associated with neurodegeneration and progressive myoclonic epilepsy 12 and KCTD5 reported to interact with human Golgi reassembly stacking protein 55 (GRASP55) 13.
We studied human KCTD5 because it had both the highest predicted solubility and strong homology to Kv channels among KCTD proteins. X-ray single-wavelength anomalous dispersion (SAD) technique was used to determine the structure at 3.1 and 3.3 Å resolution after crystallization in two salt conditions; the N-terminal BTB-containing module was solved on its own at 1.9 Å. It is rare but known for a domain to produce several oligomerization states using the same protein-protein interface 14. Unexpectedly, KCTD5 reveals how four substitutions in a BTB domain permit the same interface to bring together five KCTD5 subunits rather than four Kv channel T1 domains.
RESULTS
KCTD5 crystallizes in pentameric assemblies
Human KCTD5 from residue 34 to the C-terminus (201 residues) was expressed in bacteria. Native and seleno-methionine labeled proteins were purified to homogeneity and crystallized. Two orthorhombic P212121 crystal forms (Table 1) were obtained, one in high-salt and one in low-salt conditions (Methods). The structure was solved by SAD technique at 3.1 Å resolution with seleno-methionine labeled protein crystals grown in high-salt. The low-salt structure was solved by molecular replacement at 3.3 Å using native crystals. Surprisingly, the structure revealed KCTD5 to be pentameric, Fig. 1a–d, rather than tetrameric like Kv channels 1,2.
Table 1.
Data collection and refinement statistics for KCTD5 crystals
| High-salt KCTD5 Se-Met PDBid 3DRX | Low-salt KCTD5 Native PDBid 3DRY | N-terminal domain PDBid 3DRZ | |
|---|---|---|---|
| Data collection | |||
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Cell dimensions | |||
| a, b, c (Å) | 72.9, 128.6, 152.5 | 103.0, 106.8, 110.1 | 59.03, 90.72, 110.16 |
| α,β,γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolutionb) (Å) | 20–3.1 (3.2–3.1) | 20–3.3 (3.4–3.3) | 20–1.9 (1.95–1.90) |
| Rmerge* | 0.072 (0.46) | 0.089(0.56) | 0.065 (0.45) |
| I/σ* | 12.4 (2.3) | 11.3 (2.0) | 15.4 (2.5) |
| Completeness (%)* | 98.9 (93.8) | 99.7 (98.5) | 99.7 (98.4) |
| Redundancy* | 4.4 (4.5) | 4.3 (4.4) | 4.5 (4.5) |
| Refinement | |||
| Resolution (Å) | 20–3.1 | 20–3.3 | 20–1.9 |
| No. reflections | 25,986 | 18,687 | 46,924 |
| Rwork/Rfree | 0.227/0.275 | 0.252/0.308 | 0.193/0.232 |
| No. atoms | |||
| Protein | 6915 | 6815 | 4095 |
| Water | - | - | 359 |
| B-factors | |||
| Protein | 67 | 73 | 35.1 |
| Water | - | - | 42.0 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.008 | 0.012 | 0.009 |
| Bond angles (°) | 1.05 | 1.28 | 1.196 |
Legend: One crystal was used for each data collection. Asterisks (*) indicate where data in parentheses correspond to last resolution shell.
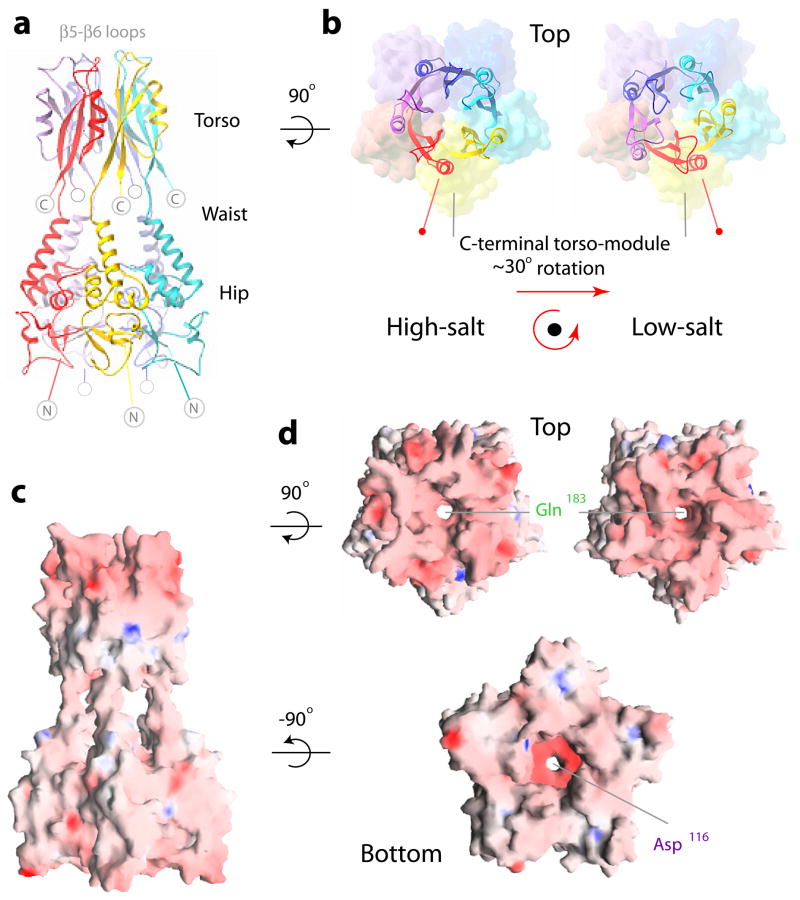
Figure 1. Architecture of KCTD5 by X-ray crystallography.
a. Side-view ribbon representation of KCTD5 complexes (residues 44–211, high-salt crystal form); the five subunits are in red, yellow, cyan, blue and violet. The N-module (hip), inter-domain linker (waist), C-module (torso), and β5-β6 loops are labeled. N-terminal amino acids 1–33 and C-terminal residues 212–234 are not in the crystal structure and indicated by gray circles.
b. Orientation of the C-module (residues 155–211, ribbons) relative to the N-module (residues 44–149, molecular surface). Top views for two different crystal forms, high-salt (left) and low-salt (right). Color scheme as in a. To highlight the change in orientation, N-modules in high- and low-salt crystals are aligned and α6 marked. The β5-β6 loops are modeled as in the well-defined electron density maps of the blue (high-salt) or cyan (low-salt) subunits; more detail on these loops is in Fig. S1.
c. Electrostatic surface representation side view high-salt crystal form. Blue is electropositive and red is electronegative.
d. Electrostatic surface representation for top views (upper) for high-salt (left) and low-salt (right) crystal forms aligned via their N-modules; bottom view (lower) high-salt crystal form. Color scheme as in c.
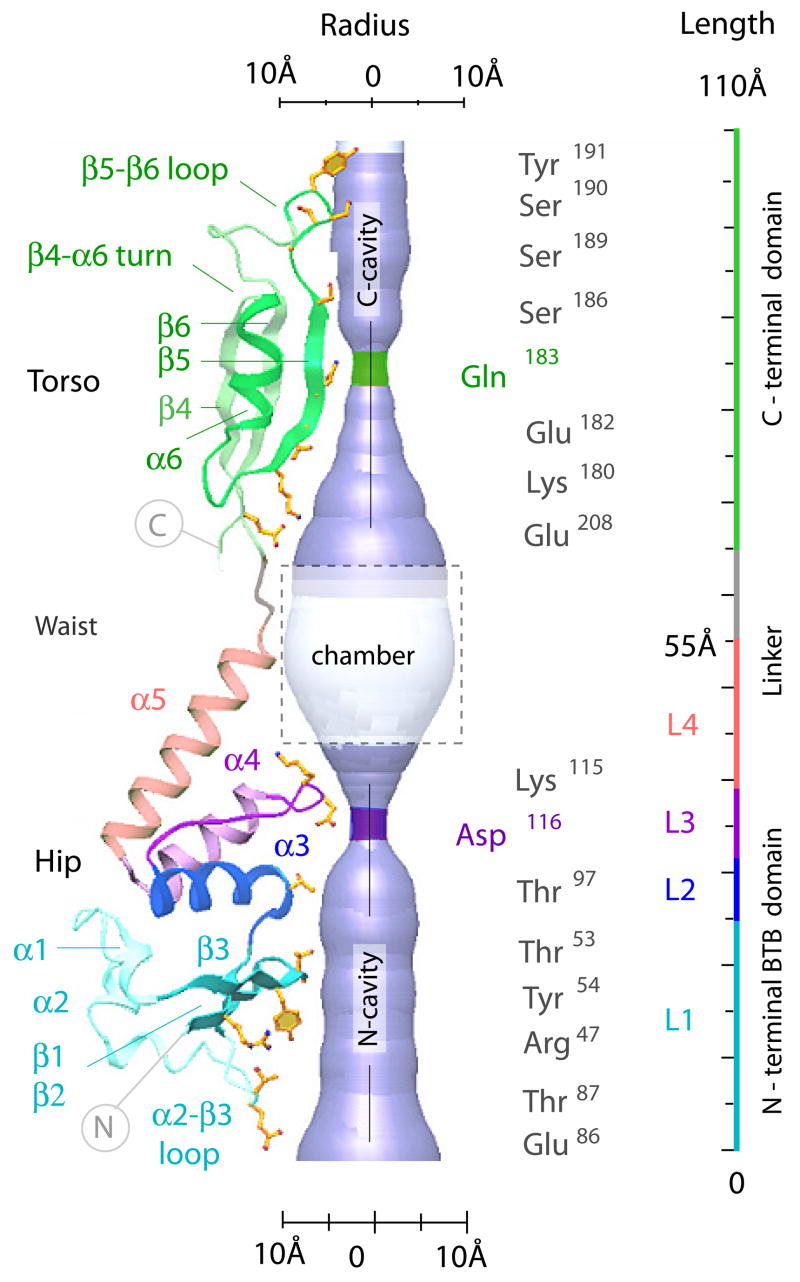
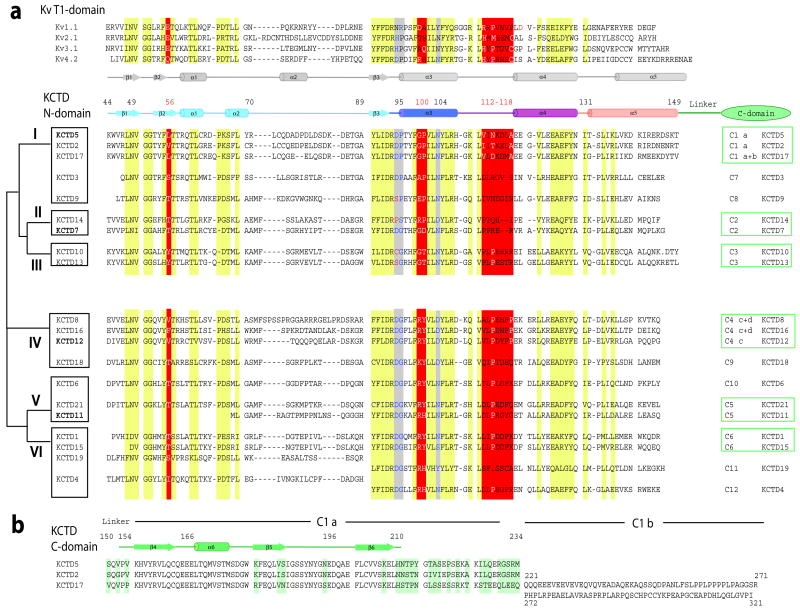
To assess the physical basis for KCTD5 pentamerization, the C-terminal domain was deleted and the N-terminal BTB domain crystallized alone (Methods). This structure was determined from orthorhombic P212121 crystals at 1.9 Å resolution by molecular replacement; again, a pentameric organization was revealed (Table 1). The structure of an individual KCTD5 subunit is shown in Fig. 2. Alignment of KCTD5 sequence and secondary structure are shown in Fig. 3. The KCTDs can be sorted into six groups and KCTD20 based on homology of their N-termini (Fig. 3). Residues 44–211 form the core of the assembly forming six α-helices (α1-α6) and six β-strands (β1-β6). The N-terminal residues 44–149 adopt a familiar BTB fold with a three-stranded β-sheet (β1-β3) and five α-helices (α1-α5). Superposition of the KCTD5 BTB domain and Kv4.2 channel T1 domain (PDBid 1NN7) shows significant structural conservation with an average root mean square deviation (RMSD) value of ~1.7 Å over 92 Cα atoms. Four layers (L1–L4) in the BTB fold are defined as for the Kv channel T1 domain 15. C-terminal residues 155–211 adopt a novel three-dimensional arrangement with a three-stranded β-sheet (β4-β6) and one α-helix (α6) (Fig. 2 and Fig. S1) without a Dali search match 16.
Figure 2. Structural determinants of the KCTD5 central cavity.
A KCTD5 subunit in ribbon presentation showing the four segments (L1–L4) of the BTB fold and the central cavity of the pentamer assembly in cut-away. The residues that shape the central cavity are labeled in ball-and-stick mode and also shown are the solvent accessible surfaces and cavity dimensions.
Figure 3. Sequence and secondary structure alignments.
KCTD BTB domains share 33% to 43% sequence homology with T1 domains of Kv channels 1,2 with most differences in the middle and C-terminus of the fold. Zinc binding motifs in T1 domains of Kv2, Kv3 and Kv4 channels are not found in KCTDs.
a. Alignment of T1-domains of select Kv channels and KCTD N-terminal BTB domains based on X-ray structures: KCTD5 (this work), Kv1.1 (PDBid 1T1D), Kv3.1 (PDBid 3KVT), Kv4.2 (PDBid 1NN7). Secondary structure elements are those determined for Kv4.2 and KCTD5. Conserved patches are highlighted in yellow. Key differences are highlighted in red. The alignment of KCTD proteins was by ClustalW. KCTD20 is omitted. The schematic on left is a KCTD group classification. Residues before the BTB domain are omitted. The KCTD5 N-terminal BTB fold is 106 residues long (residues 44–149). The consensus sequence (including insertions) is 110 amino acids with 26 residues in the diverse α2-β3 loop (corresponds to KCTD5 residues 70–89) and 14 residues in α5 (KCTD5 residues 132–149). Sequences of the α2-β3 loop and α5-helix display homology only between proteins of the same group.
The C-termini of 14 KCTDs show significant identity with at least one other variant while seven are distinct. The % similarity of groups C1, C2, C3, C4, C5, and C6 are 88, 50, 93, 69, 48, and 83, respectively.
b. Alignment of KCTD5, KCTD2 and KCTD17 C-termini; differences are highlighted in green. Residues 212–234 are disordered in the KCTD5 crystal structure.
While neither is present in the crystal structures, the KCTD5 termini are predicted to be exposed in the cytoplasm. Residues 1–33 were not encoded in the protein because they were predicted to be unstructured by the algorithm of Li and coworkers 17. C-terminal residues 212–234 were not visible in electron density maps. Residues 34–43 adopt different conformations in high- and low-salt crystals. Other flexible regions include residues 77–82 near the α2-helix and residues 188–200 near theβ5-β6 loops.
Gown-shaped assemblies with two modules
The architecture of KCTD5 complexes (residues 44–211) revealed by X-ray crystallography is shown from the side (Fig. 1a, c) and bottom (Fig. 1d) in high-salt and from the top (Fig. 1b, d) in both high- and low-salt conditions. The five subunits assemble in a circular manner with N- and C-terminal residues forming distinct N- and C-modules. Views from top and bottom show each module to be pentameric with each subunit related by ~72° rotation. The overall assembly is gown-like and ~120 Å tall with diameters of ~60 Å at the torso, ~40 Å at the waist linker and ~75 Å at the hips (Figs. 1a, c and 2). The hips are formed by N-terminal residues 44–149 (β–sheets 1–3 and α–helices 1–5 with the N-termini of α3 and α4 facing the center of the assembly). The torso is formed by C-terminal residues 155–211 (β–sheets 4–6 and α–helix 6 with β5 facing the center of the assembly). The inter-domain waist (residues 150–154) is flexible and allows rotation of ~30° between the N- and C-modules; this is demonstrated by comparison of the crystal forms obtained in high- and low-salt (Fig. 1b, d).
Central cavity
The KCTD5 assembly displays a central cavity of variable diameter that is shown from top and bottom (Fig. 1d) and in cut-away (Fig. 2). The cavity appears to be continuous through the 110 Å extent of the central portion of the assembly, 10–15 Å in diameter at its ends and 20 Å at a windowed chamber with access to bulk solvent formed by the inter-domain linker and α-helices atop the N-module (Fig. 1c). The cavity narrows twice to ~4.5 Å at five Gln183 residues in the C-module and five Asp116 residues in the N-module. The cavity is lined predominantly by the hydrophilic residues (Fig. 2). The C-terminal portion of the cavity is formed by β5-strand residues Glu182, Gln183, Ser186 and Gly188. The N-terminal portion is formed mainly by residues Thr53, Tyr54, Thr97, Lys115, and Asp116 that are scattered across the β2-strand, α3-helix and tip of the α3-α4 loop. Around ninety well-ordered water molecules are apparent within the N-module portion of the cavity in the 1.9 Å structure. A cation could fit in the assembly at the five Asp116 residues but the electron density difference map (above 2σ) was not revealing.
KCTD5 is also pentameric in solution
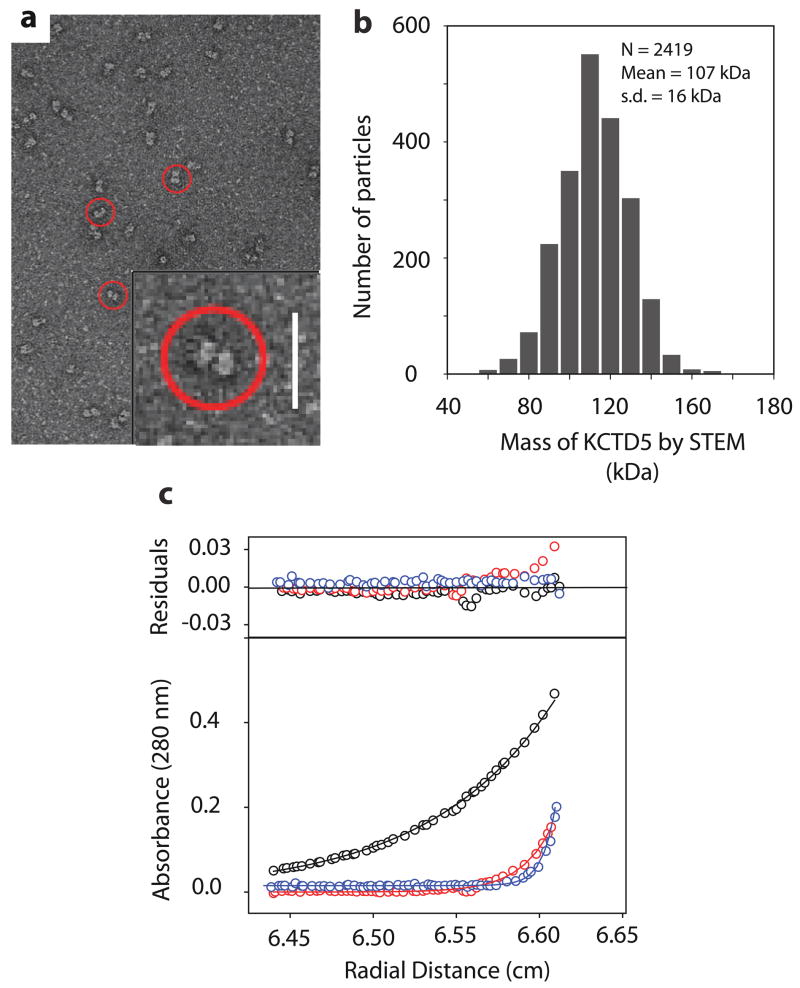
The two modules and five subunits observed in crystallized KCTD5 complexes were observed as well for assemblies studied in solution (Fig. 4). Electron microscopy (EM) of individual particles stained with uranyl acetate showed two domains linked by a thin stalk with a long dimension of ~110 Å and width of ~70 Å (Fig. 4a). Quantitative mass analysis using scanning transmission electron microscopy (STEM) revealed a unimodal distribution with a mean value of 107 +/− 16 kDa, in good agreement with the expected mass of KCTD5 (34–234) pentamers (~115 kDa) (Fig. 4b). Sedimentation velocity (SV) studies also gave the expected masses for KCTD5 and N-module pentamers (~115 kDa and ~ 62 kDa, respectively, not shown). While analytical gel-filtration chromatography suggested a mass about twice the expected (~ 230 kDa, not shown) this appeared to be due to confounding effects of an extended conformation. Sedimentation equilibrium (SE) analyses estimate mass independent of shape and the concentration profile by this method was best fit by a mass of ~114.7 kDa, consistent with pentameric assembly (Fig. 4c).
Figure 4. Electron microscopic and analytical ultracentrifugation studies.
a. Electron micrograph of KCTD5 negatively stained with 2% (w/v) uranyl acetate shows particles with a two domain architecture (circled) at 72,000X. Inset scale bar, 20 nm.
b. Histogram of quantitative STEM mass analysis of KCTD5 (34–234) particles reveals a mass distribution with a mean of 107 kDa. Scatter from individual particles (N = 2419) and conversion to absolute mass as per Methods. s.d. = standard deviation.
c. Sedimentation equilibrium data for KCTD5 (34–234) fit using SEDPHAT. KCTD5 absorbance (280 nm) is shown at 12,000 rpm (black), 24,000 rpm (red) and 36,000 rpm (blue) for analyses with 0.35 mg/ml. The upper panel shows residual difference between the calculated fit and experimental data.
Electrostatic surfaces
The electrostatic properties of the N- and C-modules differ significantly. The N-module exhibits uneven charge distribution with acidic (red) and basic (blue) patches (Fig. 1c, d). In contrast, acidic and basic residues in the C-module are secluded at the upper and lower portions, respectively. The most C-terminal acidic patches are within the α4-α6 turns (Glu165–167) and β5-β6 loops (Glu196, Asp197) at the top of the assembly. The β5-β6 loops crowning the C-terminal KCTD5 module are able to adopt well-defined conformations only when they interact with the neighboring KCTD5 assemblies in the crystal lattice and re-fold in crystals formed in different ionic conditions (Fig. S1). In the absence of close packing contacts, electron density maps were poorly defined for two of the five β5-β6 loops in high-salt (and four of the five in low-salt) indicative of flexibility.
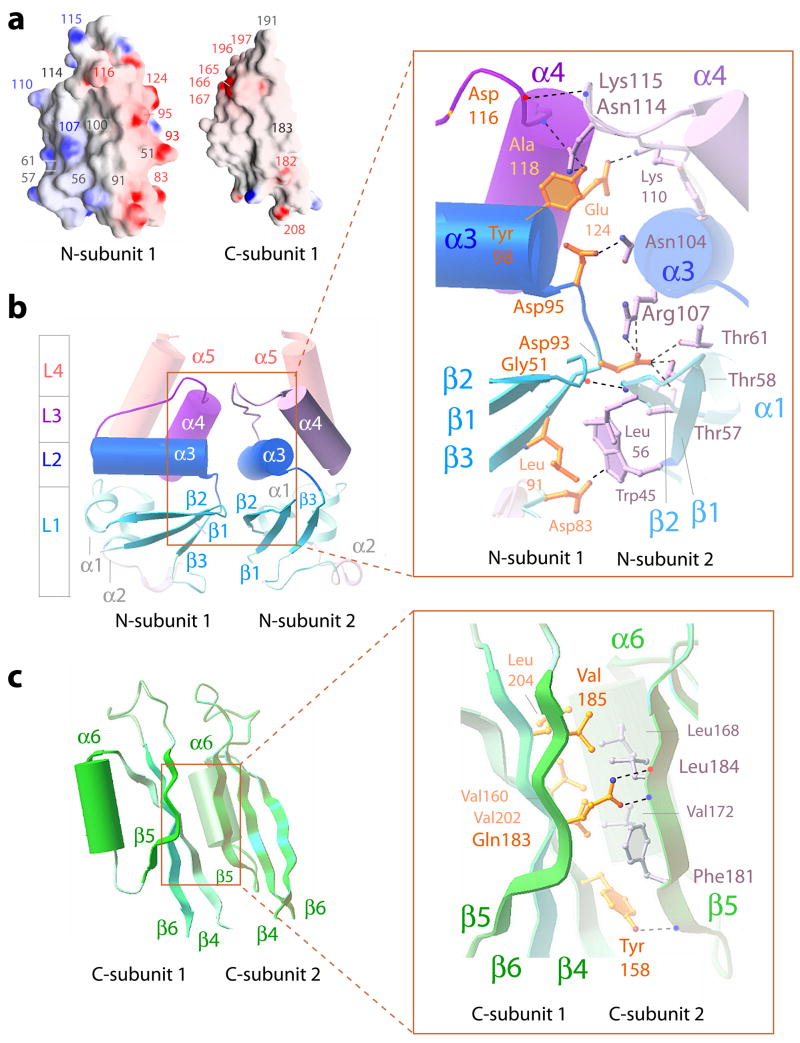
Electrostatic surfaces of residues that face the central cavity and at the interfaces between neighboring subunits are shown in Fig. 5a. Interface interactions in the N-module are mediated by polar residues while those in the C-module are mostly hydrophobic. Similar to Kv channel T1 domains 1,2, acidic and basic residues are isolated on opposite sides of the BTB domains in KCTD5 promoting circular side-to-side arrangement of subunits. In contrast, BTB folds in transcription factors lack polarized distribution of acidic and basic amino acids and their dimeric assembly is via an additional, N-terminal α-helix 18. Pentamer assemblies of KCTD5 and the N-module alone are nearly equivalent, showing an average RMSD of ~0.8 Å. This suggests the N-module has a lead role in homomeric assembly. Overexpression of the C-module (residues 154–234) led to accumulation of insoluble material in inclusion bodies consistent with inability to co-assemble, a finding noted by others 19.
Figure 5. Intermolecular interfaces in KCTD5 assemblies.
a. Potential surfaces of individual KCTD subunit N- and C-modules. Indicated in red are acidic Asp83, Asp93, Asp95, Asp116, Glu124, Glu165–167, Glu196, Asp197, Glu182, and Glu208; basic residues Arg107, Lys110 and Lys115 are indicated in blue. Gly51, Leu56, Thr57, Thr61, Leu91, Gly100, Asn114 and Gln183 are noted in gray.
b. Secondary structural elements at the interface of adjacent subunits in the N-module viewed from the center of the assembly. α-helices and β-strands are depicted as cylinders and arrows, respectively. The L1–L4 segments of the BTB fold are shown in colors per Fig. 2. Inset is expanded view of boxed area. Residues that form interfaces are shown in ball-and-stick in yellow (subunit 1) and gray (subunit 2). H-bonds (2.6–3.4 Å) are indicated with dotted lines. Asp116 (subunit 1) and Lys115 (subunit 2) interact via main chain atoms and their side chains are omitted for clarity. H-bond pairs and distances are noted in further detail in Fig. S2a
c. Secondary structural elements at the interface of adjacent subunits in the C-module viewed from the center of the assembly as in b. Inset is expanded view of boxed area as in b. H-bond pairs and distances are noted in further detail in Fig. S2b
Intermolecular interfaces
Two neighboring subunits in the N-terminal module are shown from the center of the assembly in Fig. 5b. Adjacent subunits assemble via polar edges of all three layers of the BTB fold (L1, L2 and L3) with β1-β2, β3-α3 and α4 residues of subunit 1 facing the β2, α3 and α3-α4 loop residues on subunit 2, respectively. Subunit 1 employs acidic Asp93 and Asp95 in the β3-α3 turn and Glu124 in α4 to interact via H-bonds with the subunit 2 Arg107 in α3 and Lys110 in the α3-α4 loop. The interface surface is ~37 Å in length and ~22 Å in breadth with a buried area for each subunit of ~710 Å2. A network of H-bonds between the subunits spans all three layers, (Fig. 5b, inset and S2a).
The interface between neighboring subunits in the C-module are formed mainly by hydrophobic residues and shown from the center of the assembly in Fig. 5c. The interface surface is ~40 Å in length and ~16 Å in breadth with a buried area of ~580 Å2. The β–sheets 4–6 of one subunit lie against the α6-β5 junction of the adjacent subunit employing subunit 1 residues Tyr158, Val160, Val185, Val202, and Leu204 and subunit 2 residues Leu168, Val172, Phe181, and Ile184 (Fig. 5c, inset). The side chains of five Gln183 residues on β5 face the center of the assembly to shield hydrophobic residues and shape the central cavity. This arrangement is stabilized by three H-bonds between the side chains of Tyr158 and Gln183 in subunit 1 and the main chain atoms of Phe181 and Ile184 in subunit 2 (Fig. 5c, inset and S2b).
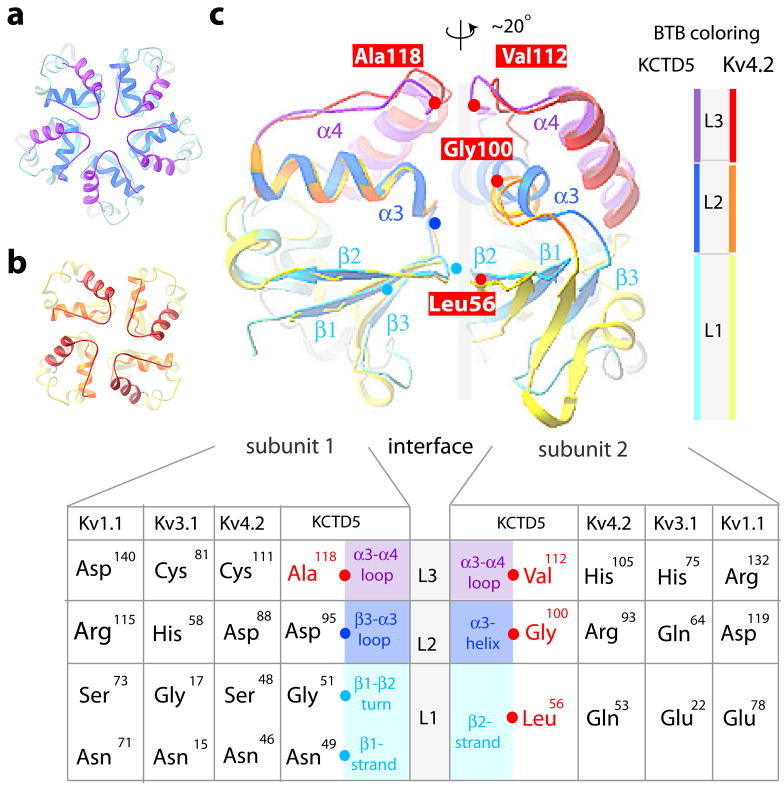
BTB folds of KCTD5 and Kv channel T1 domains
To assess how a fifth subunit is stably incorporated, KCTD5 was compared to the crystal structures of T1 tetramers in Kv1.1 and Kv3.1 15 and Kv4.2 20 (Figs. 6 and 7). While secondary structural elements are similar, KCTD5 and T1 differ in relative orientation by ~20° to accommodate five (Fig. 6a) or four subunits (Fig. 6b). Superimposition of the interfacial regions of KCTD5 and the T1 in Kv4.2 revealed salient differences (Figs. 6c, 7 and S2c). The BTB fold in KCTD5 has four aliphatic substitutions (Leu56 in β2, Gly100 in α3, and Val112 and Ala118 in the α3-α4 loop) at sites that are hydrophilic in T1 (Fig. 6c) and known to be important to formation of tetrameric assemblies 15,20. KCTD5 employs other charge-charge interactions found in T1 domains, notably, Asp93 (β3-α3 turn) and Glu124 (α4-helix) with Arg107 (α3-helix) and Lys110 (α3-α4 loop) at the interface between adjacent subunits (Fig. 5b).
Figure 6. BTB protein-protein interfaces in KCTD5 and T1 domain assemblies.
BTB layers are: L1 in cyan (KCTD5) and yellow (Kv4.2); L2 in blue (KCTD5) and gold (Kv4.2); L3 in violet (KCTD5) and red (Kv4.2). L4 α5 helix is omitted for clarity. X-ray structures used: KCTD5 (high-salt), Kv1.1 (PDBid 1T1D), Kv3.1 (PDBid 3KVT), and Kv4.2 (PDBid 1NN7).
a. Top view of a pentameric assembly of KCTD5 N-modules.
b. Top view of a tetrameric assembly of Kv4.2 T1 domains.
c. Comparison of pairs of adjacent subunits for the KCTD5 N-module and the Kv4.2 T1 domain. To highlight the ~20° difference in orientation, subunit pairs were aligned on subunit 1 (left). Cα atoms of four key aliphatic residues in KCTD5 are noted (red spheres) and labeled; Cα atoms of their partners are in cyan (L1) or blue (L2). Inset. Correspondence of KCTD5 and T1 residues by layer with KCTD5 secondary structure elements noted. Interface H-bond pairs and distances are in Fig. S2c.
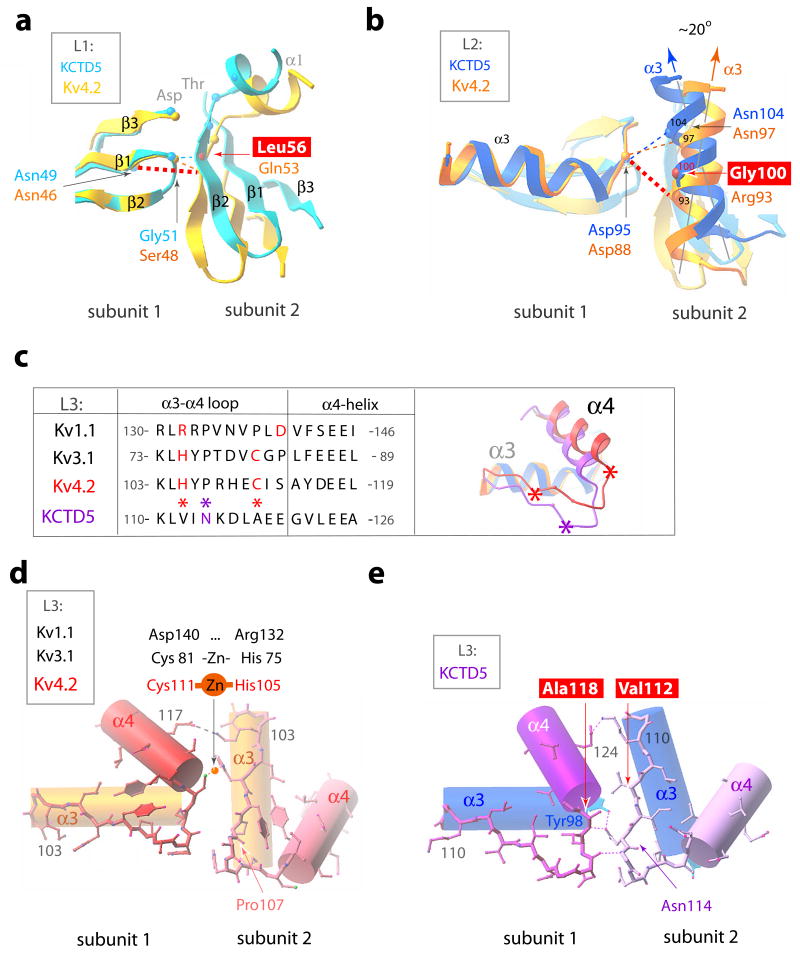
Figure 7. BTB intra-molecular interactions in KCTD5 and T1 domains.
KCTD5 (high-salt) and Kv4.2 (PDBid 1NN7) structures are shown in the color scheme of Fig. 6. H-bonds indicated as dashed lines. Four key residue differences between the KCTD5 and T1 interfaces are boxed.
a. BTB L1. Interacting residues shown as small spheres at the Cα-atoms. The view is perpendicular to the image in Fig. 6c.
b. BTB L2. Presentation as in panel a. The ~20° difference in orientation of α3-helix axes of KCTD5 and Kv4.2 is indicated.
c. BTB L3 sequence alignment and ribbon presentation for KCTD5 and Kv4.2 viewed from top. Val112, Ala118 and Asn114 in KCTD5 are marked with asterisks.
d. BTB L3 in Kv4.2 α3- and α4 helices are shown as cylinders and the α3-α4 loop in ball-and-stick mode. Arg108 side chain omitted for clarity.
e. BTB L3 in KCTD5. Representation as in d. Lys115 side chain omitted for clarity.
At level L1 (Figs. 6c and 7a), KCTD5 Leu56 in the β2-strand (subunit 2) interacts with β1-β2 turn residue Gly51 (subunit 1) via the main chain N-amino group whereas in T1 domains hydrophilic Gln or Glu in the β2-strand interacts via an H-bond network with a β1-β2 turn Ser or Gly and β1-strand Asn via main and side chain atoms, respectively. All KCTDs have the β1-strand Asn (Asn49 in KCTD5) found in T1 domains but none have Gln (or Glu) in the β2-strand (Fig. 3).
At level L2 (Figs. 6c and 7b), KCTD5 bears Gly100 in the middle of α3 (subunit 2) whereas T1 domains have Arg, Gln or Asp that form an H-bond network with β3-α3 turn residues Asp, His or Arg (subunit 1), respectively. KCTD5 does bear Asp95 at the end of the β3-α3 turn (analogous to Asp88 in Kv4.2) and this interacts with Asn104 (analogous to Asn97 in Kv4.2) but Gly100 in the next helical turn (rather than Arg93 in Kv4.2) lacks the capacity to contribute to the network (Fig. 7b). As a result, the KCTD5α3 helix tilts by ~20° compared to T1.
At level L3 (Figs. 6c and 7c–e), the V-shaped α3-α4 loop in KCTD5 differs significantly from T1 domains. Here, Kv4.2 and Kv3.1 deploy Cys and His, respectively, to bind zinc and Kv1.1 bears Asp and Arg to form a salt-bridge (Fig. 7c, d). In contrast, KCTD5 bears the small aliphatic residues Ala118 and Val112 at these sites (Fig. 7e). In KCTD5, the main chain amino group in Ala118 (subunit 1) forms an H-bond with the carbonyl group of Asn114 (subunit 2). In addition, the Asn114 amide interacts with the hydroxyl of α3 residue Tyr98 (subunit 1). The H-bond network propagates to the tip of the α3-α4 loop via the main chain carboxyl and N-amino groups of Asp116 (subunit 1) and Lys115 (subunit 2), respectively. As a result, the L3 segment rotates in KCTD5 relative to T1 (Fig. 7c), a difference that appears to be favorable due to the absence of bulky aliphatic side chains present in T1 (Tyr versus Ile113 and Phe versus Val122) and the presence of Gly121 in α4 that stacks on top of the side chain of Tyr98 and interacts with Asn114. The KCTD5 α3-α4 loop sequence is homologous only to KCTD2 and KCTD17 (Fig. 3).
Thus, incorporation of a fifth subunit in the KCTD5 assembly is facilitated by four aliphatic substitutions compared to T1 domains: Leu56 and Gly100, that lead to loss of two H-bonds and relaxation of the interface at L1 and L2 (Figs. 7a, b) and Val112 and Ala118 (Figs. 7c–e), that allow adjacent L3 segments to come closer (~1 Å) in KCTD5 (Fig. 6c) and eliminate zinc-binding or salt-bridging T1 residues. It is interesting to note how KCTD maintains stability despite losing these interactions. KCTD5 employs Asn114 to bridge L3 to L2 on adjacent subunits via Ala118 and Tyr98 (Figs. 5b and 7e), Asp93 is held within a pocket formed by Thr57, Thr58, Thr61 and Arg107 in L1 of the neighboring subunit (Figs. 5b, 7b and Fig. S2a), the L1–L2 interface is stabilized by interaction of Trp45 with Asp83 and Leu91 on the next subunit (Fig. 5b) and, as in T1, Glu124 and Lys110 at the periphery of L3 interact on flanking subunits (Fig. 7e).
The KCTD5 C-terminus is important for interaction with GRASP55
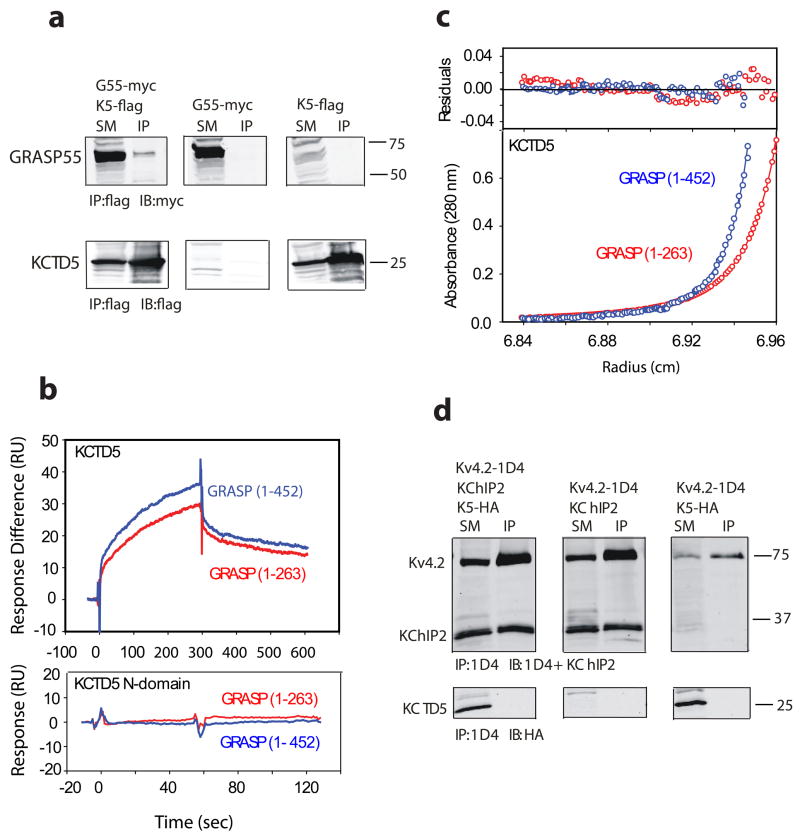
GRASP proteins contribute to the stacked organization of Golgi cisternae and are thought to participate in protein trafficking 21,22. As reported by others 13, we found that GRASP55 and KCTD5 expressed in the Human embryonic kidney cell line 293T (HEK293) co-immunoprecipitate (Fig. 8a). Surface plasmon resonance (SPR) and SE analyses were therefore used to study interaction of the purified proteins.
Figure 8. Binding of KCTD5 and GRASP55 in solution.
a. KCTD5 (K5) co-purifies with GRASP55 (G55) when co-expressed in HEK293 cells. Starting material (SM) was detergent-soluble lysate that was incubated with antibody to flag to yield immunoprecipitate (IP); SM and IP were resolved by SDS-PAGE and visualized by Western blot (IB) with antibody to myc or flag. Upper panels show that GRASP55 is isolated specifically with KCTD5 and not seen either in the absence of KCTD5 or GRASP55. Lower panels show KCTD5 is isolated in complexes with GRASP55. Numbers indicate apparent mass in kDa.
b. KCTD5 and GRASP55 studied by SPR. Upper panel depicts binding of full length GRASP55 (blue line) or truncated GRASP55 (1–263, red line) to KCTD5 (residues 34–234) immobilized on the surface. GRASP proteins injected at 5 μM. Measured biophysical parameters listed in Table 2. Lower panel shows that GRASP55 does not bind to the KCTD5 N-terminal domain.
c. Equilibrium sedimentation shows association of KCTD5 and GRASP55. Here, KCTD5 residues 34–234 (9.2 μM) and full length GRASP55 (blue line, 2.6 μM) or truncated GRASP55 (1–263, red line, 2.5 μM) are shown at 24,000 rpm in the lower panel. The upper panel shows residual difference between the calculated fit and experimental data. Nine data sets (three speeds, six concentrations) were analyzed globally using SEDPHAT 4.3 software (Methods) yields an equilibrium binding affinity of 1.41 ± 0.26 μM and 8.26 ± 1.6 μM for full-length and truncated GRASP55, respectively. Estimated partial specific volumes and solvent densities of KCTD5 with full length and truncated GRASP55 were 0.733 cm3 g−1 and 0.99823 g ml−1, respectively.
d. Kv4.2 and KChIP2 form complexes but not KCTD5 on co-expression in COS cells. Lysates were incubated with antibody to 1D4, resolved by SDS-PAGE and the interaction visualized by Western blot analysis with antibodies to KChIP2 and HA. Upper panels show that KChIP2 is purified with Kv4.2. Lower panels show that KCTD5 does not co-immunoprecipitate with Kv4.2 in the presence or absence of KChIP2 despite synthesis of the protein. Numbers indicate apparent mass in kDa.
KCTD5 was observed by SPR to bind with micromolar affinity to both full-length GRASP55 and the GRASP55 N-terminal domain (residues 1–263) in accord with a simple 1:1 binding model (Fig. 8b, upper panel and Table 2). This argued for assembly of one KCTD5 pentamer and one GRASP55 monomer with an apparent equilibrium affinity of ~1 μM. Indicating that binding requires an intact KCTD5 C-module, the pentameric KCTD5 N-module (residues 40–145) bound neither full-length nor truncated GRASP55 (Fig. 8b, lower panel and Table 2).
Table 2.
SPR (Biacore) kinetic data for KCTD5 and GRASP55 binding
| Immobilized protein | Injected protein | ka, M−1 s−1 | kd, s−1 | KD, M−1 |
|---|---|---|---|---|
| KCTD34–234 | GRASP551–452 | 0.87 ± 0.1 ×103 | 1.03 ± 0.2 × 10−3 | 1.18 ± 0.2 × 10−6 |
| KCTD34–234 | GRASP551–263 | 2.59 ± 0.4 ×103 | 2.23 ± 0.4 ×10−3 | 8.6 ± 1.2 × 10−7 |
Legend. KCTD40–145 (the N-module assembly) bound neither GRASP1–452 nor GRASP551263. ka – association rate constant; kd – dissociation rate constant; KD –equilibrium binding constant.
Analytical ultracentrifugation at three speeds and nine protein concentrations analyzed using SEDPHAT 4.3 indicated masses of ~172 kDa and ~147 kDa for complexes of KCTD5 with full-length and truncated GRASP55 protein, respectively (Fig. 8c). These are the expected values for KCTD pentamers and a GRASP55 monomer, respectively, based on masses of ~115, 48 and 28 kDa for KCTD5, full-length GRASP55 and truncated GRASP55. The concentration profile gave a good fit with a single site model to yield an equilibrium binding affinity of 1.41 ± 0.26 μM and 8.26 ± 1.6 μM for full-length and truncated GRASP55, respectively, in-keeping with the affinity estimation by SPR.
KCTD5 does not alter the function of four Kv channel subtypes
Kv channels require a multitude of regulatory accessory subunits to operate in vivo 6 and KCTD proteins were expected to serve this purpose. To our surprise, electrophysiological and biochemical methods failed to support expectations. KCTD5 shares sequence homology with the BTB-bearing T1 domains of Kv channels, most significantly, those in the Kv4 subfamily (Fig. 3). Whereas, KCTD5 was without effect on Kv4.2, the known regulator Kv channel interacting protein 2 (KChIP2) that interacts via the channel N-terminus 23 in Xenopus laevis oocytes increased current density, slowed inactivation and speeded recovery from inactivation (Table 3). Further, the effects of KChIP2 were unaltered by co-expression with KCTD5. Similarly, KCTD5 neither impacted directly nor interfered with KChIP2 regulation of Kv4.2 channels studied in Chinese Hamster Ovary (CHO) cells using whole-cell patch clamp (not shown). Moreover, KCTD5 had no effect on the biophysical operation of three other Kv channels from additional subfamilies with BTB motifs: Kv1.2, Kv2.1 and Kv3.4.
Table 3.
Biophysical properties of Kv channels expressed with KCTD5
| Kv4.2 | Kv4.2 + KCTD5 | Kv4.2 + KChIP | Kv4.2 + KCTD5 + KChIP | Kv3.4 | Kv3.4 + KCTD5 | |
|---|---|---|---|---|---|---|
| Peak Current (μA at 40 mV) | 4.7 ± 1.0 | 3.9 ± 1.0 | 5.2 ± 1.2 | 4.8 ± 0.9 | 1.9 ± 0.8 | 2.3 ± 0.6 |
| Time to peak (ms at 40 mV) | 8.2 ± 1.1 | 7.9 ± 0.8 | 9.2 ± 1.1 | 9.4 ± 1.0 | 9.3 ± 0.3 | 9.7 ± 0.6 |
| Activation V0.5 (mV) | −28 ± 2 | −25 ± 2 | −29 ± 5 | −28 ± 3 | 7.8 ± 2.3 | 8.2 ± 1.1 |
| Inactivation (t0.5decay, ms at 40 mV) | 25 ± 4 | 24 ± 5 | 48 ± 6* | 49 ± 2* | 8 ± 1 | 8 ± 1 |
| Inactivation V0.5 (mV) | −64 ± 1 | −62 ± 3 | −56 ± 2* | −56 ± 1* | −51 ± 4 | −50 ± 4 |
| Recover inactivation (τ, ms at 40 mV) | 109 ± 6 | 111 ± 9 | 22 ± 3* | 23 ± 2* | _ | _ |
| Kv2.1 | Kv2.1 + KCTD5 | Kv1.2 | Kv1.2 + KCTD5 | |||
|
|
||||||
| Peak Current (μA at 40 mV) | 7.9 ± 1.0 | 7.1 ± 0.7 | 2.3 ± 0.9 | 1.8 ± 0.6 | ||
| Activation (τ, ms at 40 mV) | 2.7 ± 0.3 | 2.4 ± 0.1 | 4.3 ± 0.6 | 4.0 ± 0.5 | ||
| Activation V0.5 (mV) | 2 ± 3 | 2 ± 2 | −20 ± 3 | −20 ± 4 | ||
| Deactivation (τfast, ms at 40 mV) | 3.1 ± 0.3 | 3.6 ± 0.3 | 4.5 ± 0.8 | 4.9 ± 0.5 | ||
| Deactivation (τslow, ms at 40 mV) | 63 ± 7 | 63 ± 5 | 49 ± 9 | 50 ± 7 | ||
| Deactivation af/(af + as) | 0.046 ±0.003 | 0.055 ±0.007 | 0.087 ±0.014 | 0.091 ±0.005 | ||
Legend: Data obtained as mean ± s.d. for six to nine Xenopus oocytes injected with cRNA for the indicated proteins and using two-electrode voltage-clamp after 24h expression at room temperature. A ratio of 1:5 for channel and KCTD cRNA was used except for Kv3.4 where a ratio of 1:1 was used or in cells with KChIP2 where the channel cRNA was decreased 20-fold to compensate for KChIP2-induced increase in surface expression. KChIP2 slows inactivation and speeds recovery from inactivation of Kv4.2 channels in the presence or absence of KCTD5 (*), P<0.001 by t-test. Voltage protocols and curve fits as described in Methods.
When KCTD5, KChIP2 and Kv4.2 were expressed in African Green Monkey COS-7 cells using methods we previously employed 24, KChIP2 was co-purified with Kv4.2 while KCTD5 was not isolated with Kv4.2 whether KChIP2 was present or absent from the cells (Fig. 8d). The same observations were made with CHO cells and for studies with Kv4.3 channels (not shown).
DISCUSSION
We report the X-ray crystal structure of human KCTD5, the first member of the family to be so characterized. The structure reveals a gown-shaped pentameric assembly with N-and C-modules and a central, complex-spanning cavity (Figs. 1 and 2). Comparing two crystal forms revealed the modules can swivel by ~30° around the inter-module linker (Fig. 1b, d). Coiled-coils show unusual versatility allowing assembly of subunits with more than one stoichiometry using the same portion of the fold 14. While BTB motifs had previously been demonstrated to mediate aggregation of two and four subunits via separate portions of the domain 7, we demonstrate here that the same BTB surfaces mediate pentamer and tetramer formation in KCTD5 and Kv channels, respectively (Figs. 6 and 7).
While the BTB in KCTD5 preserves many interactions found in T1 domains to array polar edges at subunit interfaces (Fig. 5b), it has four aliphatic substitutions that allow incorporation of a fifth subunit (Figs. 6 and 7): Leu56 in L1, Gly100 in L2, and Val112 and Ala118 in L3; these sites in T1 domains are hydrophilic and important for tetrameric assembly 15,20. As a result, KCTD5 lacks two H-bonds and one ionic interaction, relaxing the inter-subunit interface. Moreover, Val112 and Ala118 permit adjacent subunits to come closer than in T1 domains and eliminate zinc-binding or salt-bridging residues at the corresponding positions.
High sequence identity (>70%) of KCTD5 with N-termini of KCTD2 and KCTD17 suggests the latter will also form pentameric assemblies. Although, KCTD3 and KCTD9 bear only ~60% homology in the BTB domain with KCTD5 and have unique C-termini, we suspect they also will form pentamers because they are more similar to KCTD5 than T1 domains (Fig. 3). The number of subunits in other KCTD complexes is less apparent because, for example, groups IV-VI have Arg where KCTD5 has Gly100 in α3 (Fig. 3).
Testing was far from exhaustive; however, we gathered no evidence that KCTD5 assembles with or influences the function of Kv channels. Sequence similarities suggest that like KCTD5, KCTD2, KCTD3 and KCTD17 will be without effect on Kv4.2, Kv3.4, Kv2.1 and Kv1.2. The 21 KCTD genes predict proteins that can be sorted into seven groups based on alignment of their N-terminal sequences (Fig. 3). Some KCTD genes are single exon loci (4, 11, 12 and 14) while others appear to be subject to splice variation that would increase diversity. Group I contains KCTD 2, 3, 5, 9, and 17. KCTD5 is most closely related to KCTD2 and KCTD17 with ~70% and ~80% identity to their N- and C-modules, respectively, while KCTD3 and KCTD9 display ~60% similarity to the KCTD5 N-module and have dissimilar C-termini. The KCTD5 N-module displays ~20% identity (~40% similarity) to the T1 domains of Kv1, Kv2, Kv3 and Kv4 channels.
KCTD5 does assemble with GRASP55 13. Purified Human KCTD5 and GRASP55 showed an affinity of ~1 μM by SPR and SE (Fig. 8b, c). The KCTD5 N-module forms pentameric complexes on its own but does not bind GRASP55 supporting a role for the C-terminus.
KCTD5 reveals a novel fold in the C-module composed of aligned β4/α6 and β5/β6 hairpins that expose loop residues to solvent (Fig. 1 and Fig. S1). This domain may be involved in assembly with GRASP55 via an electronegative patch atop the module (Glu196 and Asp197 in the β5-β6 loop and Glu165–167 in the β4-α6 turns, Fig. 1c, d and Fig. S1) and Tyr and Ser in the β5 and β6 loops (residues that are important in other flexible loops for protein-protein recognition 25,26). KCTD5 crystal structures in high and low salt demonstrate flexibility of the β5 and β6 loops (Fig. S1). The KCTD5 C-module also appears to have a role in binding the adeno-associated virus type 2 proteins Rep78 and Rep68 because removing the last 31 residues of KCTD5 blocks Rep protein binding and nuclear accumulation 27. The rotational flexibility of KCTD5 inter-module linker and refolding of the loops crowning the C-module suggests that these changes may be required to bind and respond to ligands.
METHODS
Cloning, expression and purification
DNA for human KCTD5 (NP_061865) was amplified by PCR from adult brain cDNA (Clontech, Palo Alto, CA), inserted into TOPO 2.1 (Invitrogen Inc., Carsbad, CA) and subcloned into pMax, an in-house dual-purpose expression vector containing a CMV promoter for mammalian expression and a T7 promotor for cRNA synthesis. For biochemistry and electrophysiology studies, KCTD5 (residues 1–234) constructs with and without an N-terminal epitope tag for Hemagglutinin A (HA) were used, respectively. For structural and binding analysis, truncated versions of KCTD5 were produced carrying residues 34–234 (“full length”) or 40–145 (“N-module”); these were subcloned into a modified pET28a vector (Invitrogen) with six His residues and a TEV protease cleavage site replacing the thrombin site and then expressed in E. coli BL21 (DE3) (Novagen, Madison, WI). Seleno-methionine labeled proteins were produced as described 28. Native and seleno-methionine proteins were purified on Ni2+-NTA affinity columns (Qiagen, Valencia, CA), subjected to TEV protease cleavage and further purified through a second Ni2+-NTA column and gel filtration on a Superdex-200 column (GE Healthcare, Piscataway, NJ) equilibrated with 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2 mM DTT. The KCTD5 C-terminal residues 154–234 were expressed in the same E. coli strain and found exclusively in inclusion bodies. Human GRASP55 (NP_056345) was produced with residues 1–452 and 1–263 in the same manner.
Crystallization and data collection
Crystals were produced by the hanging-drop diffusion technique at 19º C using protein at 20 mg/ml. Preliminary crystallization screening was performed with the INDEX screen (Hampton Research, Aliso Viejo, CA). After several rounds of optimization, two crystal forms of KCTD5 were obtained: high-salt (3.2 M NaCl, and 100 mM Tris-HCl, pH 8.5) and low-salt (0.2 M proline, 100 mM HEPES, pH 7.5, and 7% (w/v) PEG 3350). The N-terminal KCTD5 domain was crystallized in 24% (w/v) PEG 3350, 100 mM MgCl2, and 100 M HEPES, pH 7.5. Crystals were cryoprotected in the presence of glycerol (22–25% (v/v)) or mineral oil and flash-frozen in liquid nitrogen. X-ray diffraction experiments were performed at 100K using the GM/CA CAT 23IDD beam line of the Advanced Photon Source (Argonne National Laboratory, IL). X-ray data sets were processed and scaled with HKL2000 29.
Structure determination and validation
The structure was solved by SAD on the high-salt crystal form of the seleno-methionine labeled full-length KCTD5. A heavy atom search was performed in SHELXD 30 using a 3.5 Å X-ray resolution cut-off. SAD phasing, electron density modification and non-crystallographic (five-fold) averaging were done in the SOLVE/RESOLVE suite 31,32. The initial model building allowed ~25% of the main chain atoms with the RESOLVE basic script. The electron density map and auto-built model were examined in TURBO-FRODO 33. Model building of the C-terminal domain that bears two seleno-methionine residues (M171 and M175) was completed first. SAD and calculated phases from the C-terminal domain allowed improvement of the electron density and completion of the model for the N-terminal region that has no Met residues. Simulated annealing and preliminary positional refinement were performed in CNS1.1 34. The N-module was used to solve the crystal structure of the truncated N-terminal domain by molecular replacement using MOLREP 35. The structure of the N-terminal domain was then refined at 1.9 Å X-ray resolution in REFMAC5 36 using the protocol implemented in CCP4 37 that included the search for water molecules, the restrained positional and individual B-factor refinement and the TLS (translation, librations and screw-axis rotation) and bulk-solvent parameter refinement and electron density map calculations. After refinement, the structure of the N-terminal domain was used to verify the model of the N-terminal domain in the high-salt crystal form of the seleno-methionine labeled full-length KCTD5. The corrected model of the full-length KCTD5 was then refined in REFMAC5 using non-crystallographic symmetry and a protocol that included the restrained positional and B-factor refinement followed by the TLS and bulk-solvent parameter refinement. The low-salt crystal structure of the native full-length KCTD5 was solved by molecular replacement using the refined N- and C-terminal pentameric modules from the high-salt crystals as the trial models in the dyad-search implemented in MOLREP. The refinement was carried out in REFMAC5 using a protocol similar to that used to refine the high-salt crystal form. Crystal data, X-ray data collection and refinement statistics are listed in Table 1. During refinement, R-free was monitored by setting aside 5% of the reflection as a test set. A Ramachandran plot calculated in CCP4 indicated that 100% of non-Gly and non-Pro residues in the N-terminal KCTD5 structure lie in the most favored or additional allowed regions. The full-length KCTD5 structures have 99.1% (high-salt KCTD5) and 97.7% (low-salt KCTD5) of non-Gly and non-Pro residues in the most favored or additional allowed regions with the others in the generously allowed regions of the Ramachandran plot and only partially ordered in the crystal structures. These residues belong to the loop adjoining the α2-helix and β5 and β6 loops. Residues 188–199 constituting β5 and β6 loops adopt well-defined conformations only when they interact with the neighboring protein molecules in the crystal lattice. The packing arrangements are different in the two crystal forms for KCTD5. More pronounced involvement of the β5 and β6 loops in the crystal packing was found in the high-salt form where three loops were defined in the electron density maps and modeled. The Sigma A-weighted 2Fobs-Fcalc electron density map is shown in Figure S1e. In contrast, only one subunit exhibits partially ordered β5-β6 conformation in low-salt crystal and is unlike the high-salt structure.
The surface area calculations were performed using the Protein-Protein Interaction Server (http://www.biochem.ucl.ac.uk/bsm/PP/server). The central cavity was calculated using the HOLE program 38. The three-dimensional figures using RIBBONS 39. The potential surface was prepared using the program GRASP 40.
Sequence Analysis
Classification of KCTD proteins into groups I-VI was by multiple sequence alignment in ClustalW2 (www.clustal.org) and then confirmed by sequence identity/similarity calculations in pairwise alignment with EMBOSS 41. Sequence alignment between human KCTD proteins and Kv T1-domains was performed with ClustalW2 and confirmed based on X-ray crystal structures (Fig. 3): KCTD5 (this work), Kv1.1 (PDBid 1T1D), Kv3.1 (PDBid 3KVT), Kv4.2 (PDBid 1NN7).
Electron microscopy
For negatively stained imaging, purified KCTD5 was diluted to a concentration of 0.17 mg/ml in 200 mM NaCl, 20 mM Tris-HCl, pH 7.5 with 2 mM DTT and stained with 2% (w/v) uranyl acetate over a glow discharge thin carbon film. Images were recorded on CCD with a FEI Morgagni microscope at an acceleration voltage of 80 keV, magnification of 72,000 × and 1 μm defocus, giving a pixel size at the specimen level of 3.33 Å.
Quantitative mass analysis by STEM was performed at the Brookhaven National Laboratory (BNL, Upton, NY) with a custom-built electron microscope. Purified KCTD5 was diluted to 0.05 mg/ml in 150 mM NaCl, 10 mM HEPES, pH 7.4, with 1 mM DTT. Grid preparation and image acquisition per http://www.biology.bnl.gov/stem/stem.html. Data were analyzed using PCMass29 with a customized model for KCTD5. A total 2149 particles were automatically selected from 50 images and processed. The KCTD5 measurements were calculated using Tobacco mosaic virus (TMV) calibration for each image. Histogram generation and statistics were by SigmaPlot9.0.
Analytical ultracentrifugation
SV and SE experiments were carried out using an Optima XL-A analytical ultracentrifuge (Beckman Coulter, Fullerton, CA) equipped with a Ti60 rotor. SV data for KCTD5 (residues 34–234 or 40–145) and GRASP55 (full length or residues 1–263) were obtained at 40,000 rpm using an Epon two-channel centerpiece. Absorbance of the samples at 280 nm was monitored in a continuous mode time interval of 360–480 s and a step size of 0.003 cm. Multiple scans at different time points were fitted to the continuous size distribution (c(s)) model using SEDFIT 11.3. The partial specific volume of the proteins and buffer density were calculated using SEDNTERP.
SE experiments were conducted at rotor speeds of 12,000, 24,000 and 36,000 rpm at 25º C. Data were acquired as an average of 10 absorbance measurements at a wavelength of 230 and 280 nm, depending on the concentration of the proteins, and a radial spacing of 0.001 cm. Molecular mass determination of KCTD5 (34–234) was performed at 0.1 mg/ml, 0.35 mg/ml and 0.5 mg/ml in 150 mM NaCl, 10 mM HEPES, pH 7.4. For binding experiments, KCTD5 and GRASP55 were in 150 mM NaCl, 10 mM HEPES, pH 7.4. Ultrascan9.1 software was used to determine the time necessary to reach equilibrium at all three speeds: 38, 12, and 6 hr, respectively. Nine data sets (three speeds, six concentrations) were analyzed globally using SEDPHAT 4.3 software. Protein concentrations were determined by absorbance at 280 nm using extinction coefficients of 2.62 × 104 M−1 cm−1 for KCTD5 and 2.71 × 104 M−1 cm−1 for both full length and truncated GRASP55.
Co-immunoprecipitation
Mammalian expression constructs for GRASP55 and KCTD5 expressing epitope tags for myc and flag, respectively, were generously shared by the A. Pandey and co-immmunoprecipitations performed according to Gandhi et al. 13 Briefly, cDNAs were co-transfected into HEK293 cells at a ratio of 1:1 μg using Lipofectamine 2000 (Invitrogen Inc., Carsbad, CA) and lysed after 48 hrs. Proteins were solubilized in 1% NP40 and clarified by centrifugation at 16,000×g for 10 min. The supernatant was subjected to immunoprecipitation overnight with mouse anti-flag M2 affinity gel (Sigma) and SDS-PAGE electrophoresis. Western Blots with rabbit anti-c-myc antibody (1:200 dilution, Sigma) and goat anti-rabbit Alexa Fluor 680nm fluorescent secondary antibody (1:5000 dilution, Invitrogen) were imaged using the LI-COR Odyssey (LI-COR Biosciences, Lincoln, NE). As previously 23,42, hKv4.2 (AH009258) expressing an epitope tag for 1D4 and untagged hKChIP2 (AAF33683) were transfected with HA tagged hKCTD5 into CHO or COS-7 cells at a ratio of 1:2:2 μg, Kv4.2:KChIP:KCTD5 and lysed after 48 hrs. Protein was solubilized in 1% Triton for 1 hr and clarified by centrifugation at 50,000×g for 30 min. The supernatant was subjected to immunoprecipitation overnight anti-mouse monoclonal 1D4 and SDS-PAGE. Western Blots with rat anti-HA (1:1000 dilution, Roche) and mouse-anti KChiP2 (J. Trimmer, University of California, Davis, CA) and Alexa Fluor 680nm and 800nm fluorescent secondary antibodies (Invitrogen) and the LI-COR Odyssey were used.
SPR binding
Interaction of KCTD5 and GRASP55 was analyzed by SPR using optical biosensor Biacore 3000 (GE Healthcare, Piscataway, NJ) at 25º C. KCTD5 (34–234 or 40–145) was covalently coupled to the dextran surface of CM5 biosensor chips to a density of 4000–5000 response units (RU) using amine chemistry (per protocol). Purified GRASP55 full length or residues 1–263 in running buffer (150 mM NaCl, 20 mM HEPES, pH 7.4, 0.005% (v/v) surfactant P20) was applied over immobilized KCTD5 by injection in triplicate of various concentrations (0.1 to 5 μM) using a flow rate of 20 μl min−1. Data were prepared by the method of “double referencing” where parallel injections of analyte over a control dextran surface were performed as well as running buffer injections over both the immobilized KCTD and control dextran surfaces. In the derived sensogram, association and dissociation were fit locally and globally using BIAevaluation 4.1 software (GE Healthcare). Affinity constants were estimated by fitting the data to simple 1:1 binding model 43 and analyzed using: dR/dt = kaC(Rmax-R) − kdR, where R is the SPR signal in RU; ka – the association rate constant in M−1 s−1; kd – the dissociation rate constant s−1; C – the concentration of GRASP55 in M; Rmax – the maximum GRASP55 binding capacity in RU; and, dR/dt the rate of change of the SPR signal. The equilibrium binding constants (KD) of the complexes were determined as the ratio of ka/kd. The concentrations of purified proteins were determined by absorbance at 280 nm using calculated extinction coefficients noted above. To confirm the specificity of KCTD5/GRASP55 binding, the following purified proteins were used: KChiP2 (residues 73–252), rat MiRP2 (NP_071571, residues 1–61), human SUMO1 (NP_001005781, residues 1–97), and BSA (Pierce, Rockford, IL). There was no binding observed between these proteins and immobilized full length KCTD5.
Electrophysiology
As previously 42, cRNA for Kv channels and KCTD5 were made using the T7 cDNA synthesis Kit (Ambion) and injected into Xenopus laevis oocytes. When hKChIP2 was included, channel cRNA was diluted 20-fold. Recording solution comprised in mM: 96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2, and 5 HEPES-NaOH, pH 7.5, for Kv4.2 recordings and 96 NaCl, 4 KCl, 1 MgCl2, 0.3 CaCl2, and 10 HEPES-NaOH, pH 7.5, for all other channels. Electrodes were filled with 3 M KCl and had resistances of 0.2–0.5 MΩ. All experiments were performed at room temperature, 20–38 hours post-injection. Voltage families were obtained with test pulses of −100 to 60 mV for 0.5 or 1 s with 10 mV increments every 10 s, using p/4 protocols. Time to peak current was used as a measure of activation and time to half-maximal decay, as a measure of inactivation for Kv3.4 and Kv4.2 channels. For Kv1.2 and 2.1 channels, activation was fitted with a single and deactivation with a double exponential equation. Steady state inactivation was examined from a holding potential of −100 mV with test pulses from −120 mV to 10 mV held for 2.5 s with a second pulse to −40 mV to measure currents that were not inactivated. V0.5 activation and V0.5 inactivation were calculated by fitting normalized conductance-voltage relationships with a Boltzman function 44. Recovery from inactivation was measured by driving channels to an inactivated state at 40 mV, hyperpolarizing to −110mV and then applying a second pulse to 40 mV for 10 ms incremental intervals. Recovery from inactivation was fit with a single exponential equation. All parameters were analyzed and averaged with 6–9 cells per group.
Accession codes
Coordinates and structure factors have been deposited in the Protein Data Bank with PDB codes 3DRX, 3DRY, 3DRZ.
Supplementary Material
Acknowledgments
This work was supported by funding from the NIH to S.A.N.G. and to A. Kossiakoff for support of V.T. We thank R. Goldstein, C. R. King, A. Sullivan, and M. Toups for technical support and A. Pandey for the hGRASP55-cMyc construct. Use of the APS was supported by the U.S. Department of Energy, Office of Basic Energy Sciences (Contract No. DE-AC02-06CH11357). Use of the GM/CA CAT Sector 23-IDD was funded by the National Cancer Institute (Y1-CO-1020) and General Medical Science (Y1-GM-1104).
Footnotes
Acc No: human KCTD5 (NP_061865); human GRASP55 (NP_056345);human Kv4.2 (AH009258); human KChIP2 (AAF33683);rat MiRP2 (NP_071571), human SUMO1 (NP_001005781)
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
AUTHORS CONTRIBUTIONS
I.S.D., V.T., Z.A.M., E.S. and N.G. designed the experiments. Z.A.M. and V.T. performed sequence and homology analyses. Z.A.M. cloned KCTD5 and performed biochemistry and electrophysiological studies. I.S.D. purified KCTD5 and GRASP55 and conducted crystallization trials, centrifugation studies and SPR with the support of D.A. and E.S. V.T. performed X-ray diffraction experiments, solved and refined structures. C.X. performed TEM analysis. I.S.D., V.T., and S.A.N.G. analyzed crystal structures and wrote the manuscript. S.A.N.G. conceived and directed the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shen NV, Chen X, Boyer MM, Pfaffinger PJ. Deletion analysis of K+ channel assembly. Neuron. 1993;11:67–76. doi: 10.1016/0896-6273(93)90271-r. [DOI] [PubMed] [Google Scholar]
- 2.Lee TE, Philipson LH, Kuznetsov A, Nelson DJ. Structural determinant for assembly of mammalian K+ channels. Biophys J. 1994;66:667–673. doi: 10.1016/s0006-3495(94)80840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 4.Zollman S, Godt D, Prive GG, Couderc J, Laski FA. The BTB Domain, Found Primarily in Zinc Finger Proteins, Defines an Evolutionarily Conserved Family that Includes Several Developmentally Regulated Genes in Drosophila. Proc Natl Acad Sci. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a conserved domain database for interactive domain family analysis. Nucl Acids Res. 2007;35:D237–240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbott GW, Goldstein SAN. A superfamily of small potassium channel subunits: form and function of the MinK-related peptides (MiRPs) Quarterly Reviews Biophysics. 1998;31:357–398. doi: 10.1017/s0033583599003467. [DOI] [PubMed] [Google Scholar]
- 7.Stogios P, Downs G, Jauhal J, Nandra S, Prive G. Sequence and structural analysis of BTB domain proteins. Genome Biology. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamse JT, Kuan YS, Macurak M, Brosamle C, Thisse B, Thisse C, et al. Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development. 2005;132:4869–4881. doi: 10.1242/dev.02046. [DOI] [PubMed] [Google Scholar]
- 9.Resendes BL, Kuo SF, Robertson NG, Giersch ABS, Honrubia D, Ohara O, et al. Isolation from Cochlea of a Novel Human Intronless Gene with Predominant Fetal Expression. J Assoc Res Otolaryngology. 2004;5:185–202. doi: 10.1007/s10162-003-4042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Marcotullio L, Ferretti E, De Smaele E, Argenti B, Mincione C, Zazzeroni F, et al. REN/KCTD11 is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zawlik I, Zakrzewska M, Witusik M, Golanska E, Kulczycka-Wojdala D, Szybka M, et al. KCTD11 expression in medulloblastoma is lower than in adult cerebellum and higher than in neural stem cells. Cancer Genetics and Cytogenetics. 2006;170:24–28. doi: 10.1016/j.cancergencyto.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Van Bogaert P, Azizieh R, Désir J, Aeby A, De Meirleir L, Florence JF, et al. Mutation of a potassium channel-related gene in progressive myoclonic epilepsy. Annals of Neurology. 2007;61:579–586. doi: 10.1002/ana.21121. [DOI] [PubMed] [Google Scholar]
- 13.Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 14.Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Current Opinion in Structural Biology. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bixby KA, Nanao MH, Shen NV, Kreusch A, Bellamy H, Pfaffinger PJ, et al. Zn2+-binding and molecular determinants of tetramerization in voltage-gated K+ channels. Nat Struct Biol. 1999;6:38–43. doi: 10.1038/4911. [DOI] [PubMed] [Google Scholar]
- 16.Holm L, Sander C. The FSSP database: fold classification based on structure-structure alignment of proteins. Nucl Acids Res. 1996;24:206–209. doi: 10.1093/nar/24.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Informatics. 1999;10:30–40. [PubMed] [Google Scholar]
- 18.Li X, Peng H, Schultz DC, Lopez-Guisa JM, Rauscher FJ, III, Marmorstein R. Structure-Function Studies of the BTB/POZ Transcriptional Repression Domain from the Promyelocytic Leukemia Zinc Finger Oncoprotein. Cancer Res. 1999;59:5275–5282. [PubMed] [Google Scholar]
- 19.Bayón Y, Trinidad AG, de la Puerta ML, Del Carmen Rodríguez M, Bogetz J, Rojas A, et al. KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 2008;15:3900–10. doi: 10.1111/j.1742-4658.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- 20.Nanao MH, Zhou W, Pfaffinger PJ, Choe S. Determining the basis of channel-tetramerization specificity by x-ray crystallography and a sequence-comparison algorithm: Family values (FamVal) Proc Natl Acad Sci. 2003;100:8670–8675. doi: 10.1073/pnas.1432840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a Protein Involved in the Stacking of Golgi Cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 22.Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim LA, Furst J, Butler MH, Xu S, Grigorieff N, Goldstein SA. Ito channels are octomeric complexes with four subunits of each Kv4.2 and K+ channel-interacting protein 2. J Biol Chem. 2004;279:5549–5554. doi: 10.1074/jbc.M311332200. [DOI] [PubMed] [Google Scholar]
- 24.Kim LA, Furst J, Gutierrez D, Butler MH, Xu S, Goldstein SA, et al. Three-dimensional structure of I(to); Kv4.2-KChIP2 ion channels by electron microscopy at 21 Angstrom resolution. Neuron. 2004;41:513–519. doi: 10.1016/s0896-6273(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 25.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidhu SS, Kossiakoff AA. Exploring and designing protein function with restricted diversity. Current Opinion in Chemical Biology. 2007;11:347–354. doi: 10.1016/j.cbpa.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Weger S, Hammer E, Götz A, Heilbronn R. Identification of a cytoplasmic interaction partner of the large regulatory proteins Rep78/Rep68 of adeno-associated virus type 2 (AAV-2) Virology. 2007;362:192–206. doi: 10.1016/j.virol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic Structures of the Human Immunophilin FKBP-12 Complexes with FK506 and Rapamycin. Journal of Molecular Biology. 1993;229:105–204. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC. Maximum likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cambillau C, Roussel A. Turbo Frodo, version OpenGL1. University Aix-Marseille II; Marseille: 1997. [Google Scholar]
- 34.Adams PD, Pannu NS, Read RJ, Brunger AT. Cross-validated maximum likelihood enhances crystallographic simulated annealing and refinement. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagin AA, Teplyakov A. An automated program for molecular replacement: MOLREP. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson E. Refinement of macromolecular structures by maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–245. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project, N. The CCP4 suit: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 39.Carson M. Ribbons. Methods in Enzymology. 1996;277:493–505. [PubMed] [Google Scholar]
- 40.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 41.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 42.Soh H, Goldstein SAN. ISA Channel Complexes Include Four Subunits Each of DPP6 and Kv4.2. J Biol Chem. 2008;283:15072–15077. doi: 10.1074/jbc.M706964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myszka DG. Improving biosensor analysis. Journal of Molecular Recognition. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 44.Clay JR. Determining K+ channel activation curves from K+ channel currents. European Biophysical Journal. 2000;29:555–557. doi: 10.1007/s002490000091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.