Abstract
Green fluorescent protein (GFP) and GFP-like fluorescent proteins owe their photophysical properties to an autocatalytically formed intrinsic chromophore. According to quantum mechanical calculations, the excited state of chromophore model systems has significant dihedral freedom, which may lead to fluorescence quenching intersystem crossing. Molecular dynamics simulations with freely rotating chromophoric dihedrals were performed on green, yellow, and blue fluorescent proteins in order to model the dihedral freedom available to the chromophore in the excited state. Most current theories suggest that a restriction in the rotational freedom of the fluorescent protein chromophore will lead to an increase in fluorescence brightness and/or quantum yield. According to our calculations, the dihedral freedom of the systems studied (BFP > A5 > YFP > GFP) increases in the inverse order to the quantum yield. In all simulations, the chromophore undergoes a negatively correlated hula twist (also known as a bottom hula twist mechanism).
Introduction
In the past 15 years, green fluorescent protein (GFP) has changed from a nearly unknown protein to a commonly used molecular imaging tool in biology, chemistry, genetics, and medicine.1−7 In 2006, more than 10 000 papers were published that used GFP. GFPs and GFP-like proteins (i.e., chromoproteins and fluorescent proteins) are particularly useful due to their stability and the fact that the chromophore (see Figure 1) is formed in an autocatalytic cyclization of the 65SYG67 sequence that does not require a cofactor.8−15 This means that unlike most other bioluminescent reporters, GFP fluoresces in the absence of any other proteins, substrates, or cofactors. Furthermore, it appears that fusion of GFP to a protein does not alter the function or location of the protein. By changing residue 66 and/or the amino acid residues around the chromophore one can change the color and intensity of GFP’s fluorescence.16−18
Figure 1.
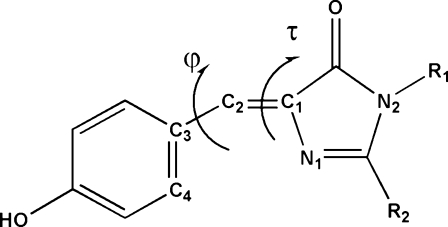
τ(N1−C1−C2−C3) and φ(C1−C2−C3−C4) dihedral angles of the GFP chromophore. In the protein, R1 is Gly67 and R2 is Ser65, and in HBDI, an often-used model compound, R1 = R2 = CH3. In τ one-bond flips (τ-OBF) the dihedral rotation occurs around the τ torsional angle, in a φ-OBF it is around the φ dihedral angle, and in a positively correlated hula-twist (+HT) the φ and τ dihedral angles concertedly rotate in the same direction (as shown above), while in a negatively correlated hula twist (−HT) they concertedly rotate in opposite directions. A plot of the φ and τ dihedrals for a perfectly correlated negative HT will have a slope of −1. If the chromophore cavity is complementary with a planar chromophore, then the φ and τ best fit line will pass through the origin and all the φ and τ angles will be centered around the origin. A nonzero intercept along the φ or τ axis (see, for example, Figure 4) or φ and τ dihedrals centered in quadrant II (φ > 0; τ < 0) or quadrant IV (φ < 0; τ > 0; see, for example, Figure 8) are indications of a cavity that is not complementary with a planar chromophore.
The fluorescent emission of the chromophore within GFP occurs with high efficiency (quantum yield Φfl = 0.8) and a respectable fluorescence lifetime (τ ≈ 3 ns).(19) When the protein is denatured the fluorescence yield decreases by at least 3 orders of magnitude.(20) Model compounds of the chromophore do not fluoresce in solution (quantum yield Φfl < 10−3), unless the rotation of the aryl−alkene bond is restrained.(21) Fluorescence can, however, be obtained by lowering the temperature to 77 K; this freezes the solution. It has been suggested that twisting between the phenolate and imadazolidinone groups of the chromophore is the mechanism for the ultrafast fluorescence quenching internal conversion process.22−26 The photophysical behavior of GFP is summarized in Figure 2.4,9,27,28
Figure 2.
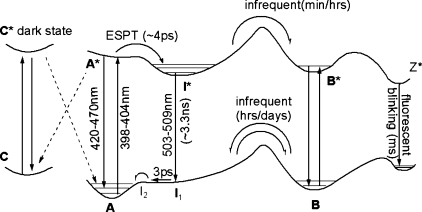
Neutral form (A) of the chromophore, with the phenolic oxygen protonated, can convert to the anionic species (B) by going through an intermediate state (I). The change from forms A to I is solely a protonation change, while the change from I to B is a conformational change with most changes occurring at Thr203. Upon excitation of the A state, an excited-state proton transfer (ESPT) occurs in which the proton is transferred from the chromophore to Glu222 in a time scale of the order of picoseconds. Following radiative relaxation from the excited-state intermediate (I*), the systems returns to the ground state A through the ground-state intermediates I1 and I2.(29) Excitation of the anionic B state results in direct emission from the excited state (B*) at 482 nm. Recently, a nonfluorescent dark state, state C, has been observed that is distinct from states A and B and absorbs at higher energies.(30) The C state, perhaps the neutral trans form of the chromophore, may be populated by nonradiative decay from A* and it may be depopulated by excitation to the excited C* state with trans−cis isomerization to repopulate state A. Fluorescent blinking has been ascribed to nonadiabatic crossing and conversions between the neutral and anionic states,(31) or a possible dark (Z) zwitterionic state.32,33
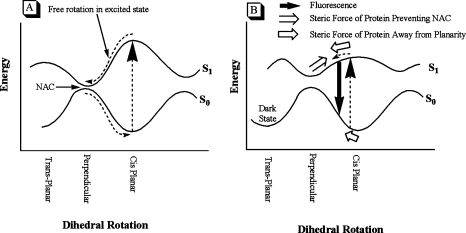
While the ground-state minimum of the GFP chromophore is close to planar, this is not necessarily so for the excited state; in fact, in some cases the excited state has an energy minimum with a twisted chromophore in which both rings are 90° to each other.(32) According to quantum mechanical calculations, the ground and excited states for the τ one-bond flip (OBF) and hula twist (HT) in the neutral form (A) and the φ OBF in the zwitterionic form come very close to each other. It has been proposed that in the absence of the protein matrix, which surrounds the chromophore and prevents twisting, this process can lead to fluorescence quenching internal crossing;(32) see Figure 3. Recent calculations on the GFP chromophore model compound HBDI suggest that the anionic form of HBDI may also undergo a τ-OBF that leads to a favored radiationless decay channel, which is particularly efficient in solvent.(34)
Figure 3.
(A) Model compounds of the GFP chromophore in the ground state (S0) can be excited to the first singlet state (S1) in which a HT or OBF can freely occur. Upon reaching the perpendicularly twisted conformation, fluorescence quenching NAC (nonadiabatic crossing) occurs. (B) In the ground state (S0), the residues surrounding the GFP chromophore exert a twisting force on the chromophore (⇐). Upon excitation, the conjugation across the ethylenic bridge of the chromophore is reduced and it will twist; however, the protein matrix prevents the chromophore from reaching the perpendicularly twisted conformation (⇒) and fluorescence quenching internal crossing is prevented.
Recently, the results from a number of interesting experiments that provide evidence for chromophore twisting and/or fluorescence quenching internal crossing (IC) have been published.35−45 For example, a molecular dynamics simulation of the chromophore of cyan fluorescent protein found that the average φ dihedral angle was about 0°; however, when the surrounding protein was considered in the simulation the average shifted by approximately 5°, showing that the protein matrix of CFP twists the chromophore. On the basis of their calculations, the authors concluded that “the driving force for this twist comes from the strong short-range repulsion by four residues (Ile167, Val150, Phe165, and Thr203) surrounding the average position of the chromophore.”(46)
Photoswitching fluorescent proteins can be switched back and forth between the naturally occurring green state and a dark state by 405 nm irradiation (e.g., Dronpa, mTFP0.7, KFP1). A cis−trans isomerization of the chromophore has been proposed as the structural basis for the photoswitching observed in Dronpa.(47) This is supported by the fact that mutating either Val157 or Met159 with smaller residues accelerates photoswitching, presumably by decreasing steric hindrance to cis−trans isomerization.(48) The M159T and V157G mutations also decrease the quantum yield of Dronpa from 0.85 to quantum yields of 0.23 and 0.77, respectively.(48) Recently, it has been suggested that adoption of a trans configuration cannot solely be responsible for the nonfluorescent form.(49) On the basis of NMR analyses, Miyawaki et al. propose that “the fluorescence of the protein is regulated by the degree of flexibility of the chromophore but is not necessarily accompanied by cis−trans isomerization.”(49)
Interestingly, GFP is not unique—in photoactive yellow protein (PYP) the protein matrix also prevents the chromophore from adopting a completely planar structure. In PYP the asymmetric protein−chromophore interaction probably serves as the initial accelerant for the light induced photocycle,(50) which ultimately leads to a cis−trans isomerization.(51)
The chromophore in wild-type GFP is planar due its extended π system (see Figure 1); however, the energy barrier to deformation is low and the protein matrix in wild-type GFP exerts some strain away from planarity.52−55 When the chromophore is computationally permitted to freely rotate, it will adopt a conformation that complements the protein matrix. Recently, we have used computational methods to show that wild-type GFP is not an anomaly and that all GFP and GFP-like proteins in the protein databank have a protein matrix that is not complementary with a planar chromophore.(54) In most cases the freely rotating chromophore undergoes φ rotations of at least 20°. In some cases these rotations are accompanied by an equal but opposite rotation of the τ dihedral angle (a negatively correlated HT). None of the proteins examined have a cavity that causes a rotation solely around the τ dihedral angle.(54) These calculations were done by minimizing, with freely rotating τ and φ dihedral angles, the crystal structure of 38 GFP analogues and mutants found in the PDB. They found the energy minimum conformation of a freely rotating chromophore in the protein matrix of the GFP mutant or GFP-like protein examined; however, they did not provide any information about the range of low-energy conformations available to a freely rotating chromophore. To get this information, we ran molecular dynamics simulations of some of the interesting GFP mutants in the protein databank.(56) By running molecular dynamics simulations, with freely rotating τ and φ dihedrals, we have been able to determine the range of conformations available to chromophores with complete rotational freedom.
Experimental Section
The coordinates of three crystal structures (1GFL,(57) 1MYW,(58) 2EMD(59)) were obtained from the Protein Data Bank (PDB),(56) and hydrogen atoms were added to protein and solvent atoms as required. The OPLS_2005 force field of MacroModel v9.0016(60) was used. Starting structures for mutants for which no crystal structure has been determined were calculated by graphically mutating a known structure and undertaking a conformational search. Conformational searches were conducted using the Monte Carlo torsional and molecular position variation method.61,62 The flexible dihedral angles of all the side chains of residues that are within 8.00 Å of the chromophore were randomly rotated by between 0 and 180°, and all solvent molecules in that sphere were randomly rotated and translated by between 0 and 1.00 Å in each Monte Carlo (MC) step.(63) 15 000 MC steps were taken in each search. Structures within 50 kJ/mol of the lowest energy minimum were kept and a usage-directed method(62) was used to select structures for subsequent MC steps. Structures found in the conformational search were considered unique if the least-squared superimposition of equivalent non-hydrogen atoms found one or more pairs separated by 0.25 Å or more. The lowest energy structure obtained in the Monte Carlo torsional and molecular position variation search was further subjected to a 5000-step large-scale low-mode conformational search.64,65
The final structures obtained from the MC search (Monte Carlo torsional and molecular position variation and large-scale low mode) or fully minimized PDB structures were used to initiate MD simulations with freely rotating τ and φ dihedral angles (V1 = V2 = V3 = 0.000). MD simulations were carried out at 300 K with 1.5 fs steps. The predynamics simulation was set for 100 ps and the full MD simulation for 5000 ps. All molecular dynamics calculations used the OPLS_2005 force field and SHAKE constrained hydrogens. Two thousand structures were sampled in each MD simulation.
The Cambridge Structure Database (CSD) v5.29 was released in January 2008; it comprises 436 384 small molecule crystal structures.
The area of the convex hull of the φ and τ graphs was calculated as was the smallest convex set containing the region.
Results and Discussion
Molecular mechanics and dynamics calculations have been used to examine the steric environment of the chromophore in GFP in its ground state.46,54,66 They are techniques that are based on classical physics and were designed to model structural and not electronic properties; therefore, molecular mechanics and dynamics simulations of GFP cannot examine the excited state of the chromophore. Quantum calculations are an excellent technique to determine the energy profiles of the ground and excited states of the chromophore; however, they are CPU-intensive and it would be cost prohibitive to do conformational searches on the excited state of a series of GFP and GFP-like proteins. Therefore, in an attempt to supplement the quantum calculations, we have examined the conformational space available to the chromophore within GFP using a freely rotating chromophore that is an approximation (based on QM calculations32,34) of the conformational space available to the chromophore in its excited state.
Green Fluorescent Protein (GFP)
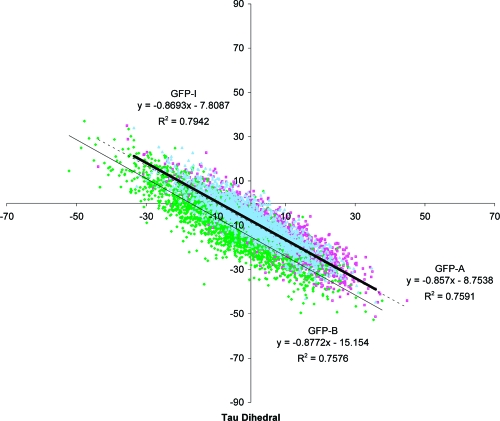
As shown in Figure 2, GFP adopts two states, the neutral state (A) of the chromophore with the phenolic oxygen protonated, and the anionic species (B). They can interconvert by going through an intermediate state (I). The change from forms A to I is solely a protonation change, while the change from I to B is a conformational change with most changes occurring at Thr203.(67) In order to see whether the GFP chromophore has different dihedral freedom in forms A, B, and I, we conducted molecular dynamical simulations with a freely rotating chromophore, as described in the . Not surprisingly, Table 1 and Figure 4 show that forms A, B, and I of GFP have similar dihedral freedom and that all undergo similar negatively correlated (same slopes of best fit lines in Figure 4) hula twists.
Table 1. Parameters Describing the Dihedral Freedom (deg) of GFP (Forms A, B, and I), BFP, A5, and YFP (Phenolic and Phenolate Forms of Tyr66).
| τmax | τmin | τrange | τav | τmedian | φmin | φmax | φrange | φav | φmedian | |
|---|---|---|---|---|---|---|---|---|---|---|
| GFP-A | 44.90 | −43.71 | 88.62 | 5.62 | 6.11 | −51.28 | 34.93 | 86.21 | −13.57 | −13.87 |
| GFP-B | 37.71 | −52.10 | 89.81 | −6.95 | −7.14 | −52.67 | 37.02 | 89.69 | −9.06 | −8.87 |
| GFP-I | 35.85 | −33.50 | 69.35 | 0.76 | 0.73 | −44.76 | 34.02 | 78.78 | −8.47 | −8.73 |
| 2EMD | 68.57 | −24.38 | 92.95 | 26.38 | 27.59 | −54.51 | 43.34 | 97.85 | −9.62 | −10.18 |
| A5 | 50.35 | −35.20 | 85.55 | 2.98 | 2.92 | −48.30 | 46.31 | 94.61 | 2.08 | 2.22 |
| YFP- RO | 49.06 | −54.33 | 103.40 | −12.97 | −13.68 | −50.44 | 43.36 | 93.80 | −3.05 | −2.37 |
| YFP- ROH | 22.05 | −55.46 | 77.51 | −16.22 | −16.86 | −27.45 | 54.04 | 81.49 | 15.53 | 16.14 |
| slope | intercept | area of convex hull | 80% area | Φfl | brightness(18) (E*QY × 1000) | |
|---|---|---|---|---|---|---|
| GFP-A | −0.857 | −8.75 | 2575 | 627 | 0.77(18) | 16.1 |
| GFP-B | −0.877 | −15.15 | 3592 | 786 | − | − |
| GFP-I | −0.869 | −7.81 | 2070 | 445 | − | − |
| 2EMD | −0.906 | 14.27 | 4792 | 1661 | 0.20(70)−0.34(71) | 5.0 |
| A5 | −0.851 | 4.62 | 3725 | 957 | 0.48(71) | − |
| YFP- RO | −0.823 | −13.74 | 3894 | 1136 | − | − |
| YFP- ROH | −0.878 | 1.28 | 3020 | 869 | 0.59(18) | 41.9 |
Figure 4.
Plot of the τ vs φ dihedral angles (see Figure 1 for nomenclature) for the 2000 GFP-A (pink), GFP-B (green), and GFP-I (light-blue) structures obtained in the freely rotating molecular dynamics simulation.
However, there are subtle differences; forms A and I are closer to the planar ground-state conformation, while the B form adopts conformations further from planarity when given rotational freedom around the τ and φ dihedrals (intercept further from τ = 0°); and the anionic B form has the most freedom (largest area, and τ and φ range), while the neutral A form has the least freedom. The A form has robust hydrogen bonds between both Arg96 and Gln94 and the imidazolidinone carbonyl group that remain intact throughout the simulations. In the intermediate (I) form these hydrogen bonds are supplemented by an additional hydrogen bond between the phenolic oxygen of the chromophore and His148 (see Table 2). The main difference between the anionic B form and the intermediate form is that Thr203 has to rotate in order to hydrogen bond to the phenolic oxygen in form I, otherwise they have the same hydrogen-bonding interactions throughout the simulation.
Table 2. Distances between Residues and Waters around the Chromophore in the A, B and I Forms of GFP That Can Form Hydrogen Bondsa.
|
bond distances to chromophore (Å) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| GFP-B |
GFP-A |
GFP-I |
|||||||
| amino acid | max | min | avg | max | min | avg | max | min | avg |
| Gln94 | 3.52 | 1.69 | 2.36 | 2.59 | 1.76 | 2.1 | 2.42 | 1.69 | 2.06 |
| Arg96 | 2.1 | 1.49 | 1.72 | 2.03 | 1.51 | 1.71 | 1.99 | 1.5 | 1.7 |
| His148 | 2.54 | 1.8 | 2.15 | 5.31 | 3.65 | 4.67 | 3.07 | 1.86 | 2.25 |
| Thr203 | 2.46 | 1.61 | 1.96 | 3.85 | 2.71 | 3.3 | 5.95 | 3.8 | 5.26 |
Bolded values = hydrogen bonds.
Blue Fluorescent Protein (BFP)
The Y66H mutant of GFP exhibits blue fluorescence and has therefore been termed blue fluorescent protein. The crystal structure of BFP (not to be confused with blue fluorescent protein from aequorin BFP-aq(68)) has been solved at pH 4.5(27) and pH 8.5;(59) the overall fold of the protein is identical to wild-type GFP and consists of a chromophore surrounded by an 11-stranded β-barrel. While GFP has absorption maxima at 395 and 475 nm and emits at 508 nm, BFP absorbs at 382 nm and emits at 448 nm.(9) Unfolding of BFP results in an absorption red shift of 15 nm,(27) and quantum mechanical calculations suggest that the 15 nm shift might be due to the protein-induced nonplanarity of the chromophore.(69) Blue fluorescent protein (BFP) has a much lower fluorescence quantum yield than GFP (Φfl = 0.20 vs 0.80). It has been suggested that this is due to the fact that His66 (BFP-chromophore) forms fewer hydrogen bonds with the surrounding protein than Tyr66 (GFP-chromophore) does, and that the smaller imadazole ring (His66) in BFP may have more conformational freedom than the larger phenol (Tyr66), which leads to more intersystem crossing.(27)
In a very elegant series of experiments, Boxer et al. have shown that the fluorescence quantum yield of blue fluorescent protein increases from Φfl = 0.20 to 0.35 when the pressure is increased from atmospheric pressure to 570 MPa.(70) “Analysis of the fluorescence lifetimes in the picosecond and nanosecond regimes reveals that the enhancement of the fluorescence quantum yield is due to the inhibition of fast quenching processes. Temperature-dependent fluorescence measurements reveal two barriers (19 and 3 kJ/mol, respectively) for the transition into nonfluorescing states. These steps are probably linked with dissociation of the hydrogen bond between the chromophore and His148 or an intervening water molecule and to the barrier for chromophore twisting in the excited state, respectively.”(70)
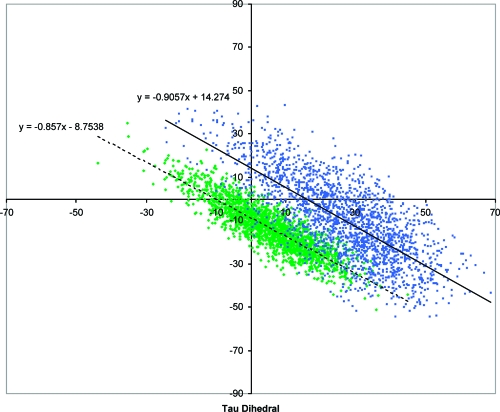
In order to establish the consequences of the Y66H mutation on the flexibility of the chromophore, molecular dynamics simulations of the pH 8.5 BFP crystal structure (PDB code 2EMD with neutral imidazole rings for His66 and His148) with a freely rotating chromophore were run. [We thank one of the referees for suggesting the pH 8.5 (2EMD) over the pH 4.5 (1BFP) structure.] Figure 5 and Table 1 show that the chromophore in BFP has significantly more rotational freedom than that available to the chromophore in GFP, and that the BFP cavity is less complementary to a planar chromophore than the GFP cavity is; that is, the BFP sampled structures are further from a planar chromophore (τ = φ = 0°) and the intercept for the best fit line through the BFP structures is further from the origin than that of GFP.
Figure 5.
Plot of the τ vs φ dihedral angles for the 2000 BFP (blue) and GFP-A (green) structures obtained in the freely rotating molecular dynamics simulation.
Analysis of the 2000 BFP structures revealed a hydrogen bond between the NτH of the His66 imidazole ring and Glu222, which is retained through-out the entire simulation (see Figure 7).
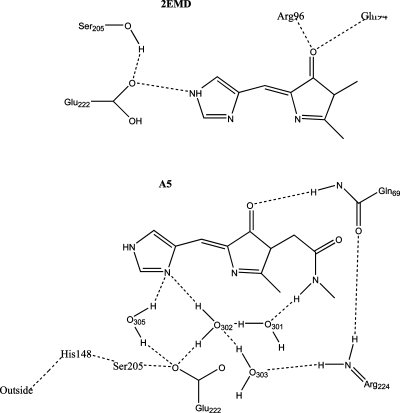
Figure 7.
Hydrogen-bonding interactions (---) for 2EMD (top) and A5 (bottom) chromophore. See text for description.
The smaller imadazole ring of the BFP chromophore results in more dihedral freedom for the chromophore in BFP than phenol in GFP; however, there seems to be no large difference in the number and stability of hydrogen bonds formed by the chromophore in the two FPs.
Blue Fluorescent Protein with Enhanced Brightness (A5)
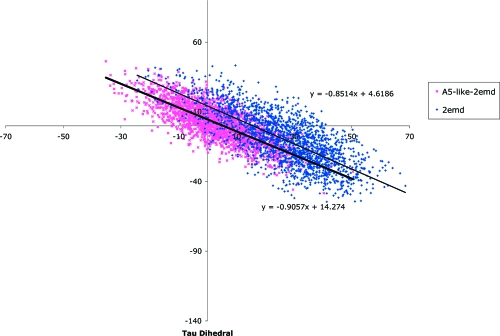
Recently, two new blue fluorescent proteins (Azurite and A5) with enhanced brightness and photostability were created.71,72 The methodology applied to find the brighter BFP mutants was based on the concept that replacing the residues surrounding the chromophore with bulkier amino acids would constrain the chromophore’s motion and thereby increase the proteins brightness. We therefore compared the rotational freedom of A5 and BFP. The crystal structure of A5 has yet to be solved. In order to find a starting conformation for the MD simulation, the 2EMD crystal structure was graphically mutated to A5 and a thorough conformational search was undertaken (see ). Given the high structural similarity between all the solid state structures of GFP mutants in the protein databank, a thorough conformational search should find the lowest energy conformations of A5 by making the relevant mutations to the 2EMD structure.
Figure 6 and Table 1 show that the conformational space available to the chromophore in BFP is larger than that in A5 and that the BFP conformations tend to be further from planarity; i.e., for A5 τmedian = 2.92°, φmedian = 2.22° and intercept of best-fit line is 4.62 vs for BFP τmedian = 27.59°, φmedian = −10.18° and intercept of best-fit line 14.27. Similar results were found for MD simulations carried out from all of the five lowest energy conformational families sampled in the conformational search of A5 and in a MD simulation initiated from an A5 structure obtained by taking the 2EMD structure, graphically mutating it to A5, and thoroughly minimizing the structure without undertaking a conformational search.
Figure 6.
Plot of the τ vs φ dihedral angles for the 2000 BFP (dark blue) and A5 (pink) structures obtained in the freely rotating molecular dynamics simulation.
The Nπ of the His66 imidazole ring hydrogen bonds with water305 (2EMD numbering) in all the sampled structures. This water connects His66 to the surface of the protein via a robust hydrogen-bonding network to His148 and is also hydrogen bonded to Glu222. In half the structures sampled, the Nπ of the His66 imidazole ring hydrogen bonds to water302 which is in an extensive hydrogen-bonding network with Val68, Arg224, and Gln69. In contrast to 2EMD there are no hydrogen-bonding interactions with the Nτ hydrogen; see Figure 7.
These results seem to indicate that the enhanced brightness of A5 is at least in part due to the fact that its chromophore movement is restricted relative to BFP. This restriction can be due to both steric factors and the increased hydrogen-bonding networks in A5.
Yellow Fluorescent Protein
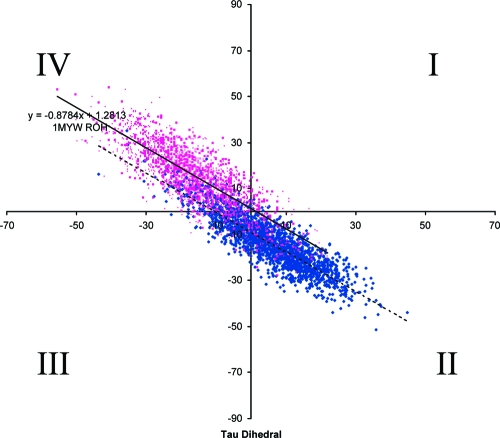
In the first rationally designed mutant based on the crystal structure of GFP-S65T, it was decided to mutate T203 into a tyrosine so that it could π stack with the phenolic group in the chromophore. (73) The resultant mutant, YFP, is red-shifted by 16 nm relative to GFP-S65T and does indeed have a π stacking interaction between the chromophore and Tyr203.(74) On the basis of the crystal structure of YFP, Remington et al. proposed that the red shift was due to the additional polarizability of the π-stacked Tyr203, the hydrogen-bond pattern around the chromophore, or an out-of-plane distortion of the chromophore (in analogy with out-of-plane distortions in porphyrin systems).(74) Comparison with the T203V mutant revealed that the T203Y substitution leads to a significant I-form population and only about 10 nm of the shift can be ascribed to the π−π interaction.(75) A new varient of YFP, Venus, with improved brightness and maturation properties as well as a reduced environmental sensitivity was developed and crystallized.(58) Since some researchers have argued that the presence of Tyr203 leads to a decrease in chromophore flexibility (van der Waals volume for Tyr = 141 Å3 vs 93 Å3 for Thr),74,76 and others argue that this is not necessarily so,(32) we have examined the conformational space available to a freely rotating chromophore in YFP (PDB code 1MYW) with the phenol of Tyr66 in both the protonated and unprotonated forms. Both Table 1 and Figure 8 show that the chromophore (in the phenolic form) in GFP and YFP has differing rotational freedom. In YFP the chromophore has approximately 1.2 times more dihedral freedom than in GFP and although the dihedrals undergo negatively correlated hula-twisting in both proteins, in YFP the average conformation sampled is further from planarity than that found in GFP; i.e., the majority of the sampled conformations are located in quadrant IV.
Figure 8.
Plot of the τ vs φ dihedral angles (see Figure 1 for nomenclature) for the 2000 structures obtained in the freely rotating molecular dynamics simulation of GFP (dark blue) and YFP (pink) in their phenolic forms. The majority of the YFP conformations sampled are nonplanar (i.e., in quadrant IV φ < 0 and τ > 0).
During the entire simulation the hydrogen bonds between Tyr66 and Ser205, Arg96 and the imidazolone carbonyl, as well as between Tyr203 and water354 remain stable, while those between Tyr203 and Gln69 were less stable.
We also examined the distance between the centroid of Tyr66 (the choromophore) and Tyr203. The distance is fairly short and does not change much during the simulation (minimum = 3.43 Å, maximum = 4.18 Å, average = 3.75 Å, SD = 0.103 Å) while the angle between the planes of the phenols varies between 0.0° and 22.5° (average = 6.9°, SD = 4.11°). The short distance between the phenol rings is indicative of π stacking, which may be responsible for the red shift observed in YFP; however, the variability in the angle between the planes of the phenols shows that Tyr203 does not significantly restrict the rotational freedom of the excited chromophore.
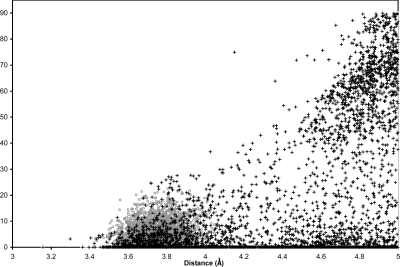
The Cambridge Structural Database (CSD)(77) v5.29 comprises 436 384 small molecules crystal structures. It has 5551 structures with two phenol rings in separate molecules that have centroids within 5.00 Å of each other. The closest ones are 3.16 Å from each other, and only 832 structures are within 3.75 Å; i.e., the average distance between the phenol centroids in the freely rotating YFP. A plot of the distance vs the angle between the phenol planes in our freely rotating simulation and the CSD structures shows that the protein matrix of YFP restricts the distance between Tyr66 and Tyr203 from being larger than 4.18 Å, but that the angles between the two phenols have as much freedom in YFP as in the small molecule structures with intermolecular distances of less than 4.18 Å (see Figure 9). It does not seem as if the increased fluorescence lifetime of YFP is due to a decrease in the dihedral freedom of the chromophore. It is more likely due to electronic effects such as those proposed by Jung et al.(78) They suggested that the anionic form of the chromophore can be described by two mesomeric forms, the benzoidal and the quinoidal structure. The benzoidal form with the majority of its negative charge on the phenol is stabilized by hydrogen bonding to Thr203, which is not possible in YFP and therefore the T203Y mutation favors the quinoidal form, which may have a longer fluorescence lifetime.(78)
Figure 9.
Distance vs angle between the phenol planes in YFP (○) and all structures in the CSD that have two unconnected phenols separated by less than 5.00 Å (+).
Conclusion
There are many factors responsible for the fluorescence intensity of fluorescent proteins. Some have suggested that the crystal structures of strongly fluorescent GFP and GFP-like proteins have their chromophores in a cis configuration, while those of the nonfluorescent proteins are in a trans configuration.66,79−81 However, the trans-planar form of eqFP611 is an important exception: it is fluorescent.(82) Remington et al. have suggested that coplanarity might be more important than isomer configuration in determining the efficiency of fluorescence.53,83 Most current theories (see ) suggest that a restriction in the rotational freedom of the fluorescent protein chromophore will lead to an increase in fluorescence brightness and/or quantum yield. Our calculations show that this is the case for the systems examined. For BFP, A5, YFP, and GFP, there is an inverse correlation between the dihedral freedom of the chromophore and the quantum yield; see Table 1.
In all simulations, the protein matrix is not complementary with a planar chromophore, and in all cases the freely rotating chromophore undergoes a negatively correlated hula twist (also known as a bottom hula twist mechanism).
Acknowledgments
M.Z. is a Henry Dreyfus Teacher-Scholar and the Barbara Zaccheo Kohn ‘72 Professor of Chemistry. The research was funded by the NIH (Area Grant # R15 GM59108-02).
Funding Statement
National Institutes of Health, United States
References
- Zimmer M.Glowing Genes: A Revolution in Biotechnology; Prometheus Books: Amherst, NY, 2005. [Google Scholar]
- Tsien R. Y. FEBS Lett. 2005, 579, 927. [DOI] [PubMed] [Google Scholar]
- Green Fluorescent Protein: Properties, Applications and Protocols, 2nd ed.; Chalfie M., Kain S. R., Eds.; John Wiley & Sons: Hoboken, NJ, 2006. [Google Scholar]
- Tozzini V.; Pellegrini V.; Beltram F.. CRC Handbook of Organic Photochemistry and Photobiology, 2nd ed.; CRC Press: Boca Raton, FL, 2004. [Google Scholar]
- Chudakov D. M.; Lukyanov S.; Lukyanov K. A. Trends Biotechnol. 2005, 23, 605. [DOI] [PubMed] [Google Scholar]
- Shaner N. C.; Steinbach P. A.; Tsien R. Y. Nature Methods 2005, 2, 905. [DOI] [PubMed] [Google Scholar]
- Shaner N. C.; Patterson G. H.; Davidson M. W. J. Cell Sci. 2007, 120, 4247. [DOI] [PubMed] [Google Scholar]
- Cody C. W.; Prasher D. C.; Westler W. M.; Pendergast F. G.; Ward W. W. Biochemistry 1993, 32, 1212. [DOI] [PubMed] [Google Scholar]
- Heim R.; Prasher D. C.; Tsien R. Y. Proc. Natl. Acad. Sci. U.S.A. 1994, 91, 12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchini B. R.; Nemser A. R.; Zimmer M. J. Am. Chem. Soc. 1998, 120, 1. [Google Scholar]
- Rosenow M. A.; Huffman H. A.; Phail M. E.; Wachter R. M. Biochemistry 2004, 43, 4464. [DOI] [PubMed] [Google Scholar]
- Barondeau D. P.; Putnam C. D.; Kassmann C. J.; Tainer J. A.; Getzoff E. D. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barondeau D. P.; Kassmann C. J.; Tainer J. A.; Getzoff E. D. Biochemistry 2005, 44, 1960. [DOI] [PubMed] [Google Scholar]
- Wachter R. M. Acc. Chem. Res. 2007, 40, 120. [DOI] [PubMed] [Google Scholar]
- Lemay N. P.; Morgan A. L.; Archer E. J.; Dickson L. A.; Megley C. M.; Zimmer M. Chem. Phys. 2008, 348, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G. H. Nat. Biotechnol. 2004, 22, 1524. [DOI] [PubMed] [Google Scholar]
- Shaner N. C.; Campbell R. E.; Steinbach P. A.; Giepmans B. N. G.; Palmer A. E.; Tsien R. Y. Nat. Biotechnol. 2004, 22, 1567. [DOI] [PubMed] [Google Scholar]
- McNamara G.; Boswell C. A.. A Thousand Proteins of Light: 15 Years of Advances in Fluorescent Proteins. In Modern Research and Educational Topics in Microscopy; Méndez-Vilas A., Diaz J., Eds.; Formatex: Badajoz, Spain, 2008; p 287. [Google Scholar]
- Striker G.; Subramaniam V.; Seidel C. A. M.; Volkmer A. J. Phys. Chem. B 1999, 103, 8612. [Google Scholar]
- Niwa H.; Inouye S.; Hirano T.; Matsuno T.; Kojima S.; Kubota M.; Ohashi M.; Tsuji F. I. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. X.; Burgess K. J. Am. Chem. Soc. 2008, 130, 4089. [DOI] [PubMed] [Google Scholar]
- Webber N. M.; Litvinenko K. L.; Meech S. R. J. Phys. Chem. B 2001, 105, 8036. [Google Scholar]
- Litvinenko K. L.; Webber N. M.; Meech S. R. Chem. Phys. Lett. 2001, 346, 47. [Google Scholar]
- Litvinenko K. L.; Webber N. M.; Meech S. R. Bull. Chem. Soc. Jpn. 2002, 75, 1065. [Google Scholar]
- Litvinenko K. L.; Webber N. M.; Meech S. R. J. Phys. Chem. A 2003, 107, 2616. [Google Scholar]
- Litvinenko K. L.; Meech S. R. Phys. Chem. Chem. Phys. 2004, 6, 2012. [Google Scholar]
- Wachter R. M.; King B. A.; Heim R.; Kallio K.; Tsien R. Y.; Boxer S. G.; Remington S. J. Biochemistry 1997, 36, 9759. [DOI] [PubMed] [Google Scholar]
- Chattoraj M.; King B. A.; Bublitz G. U.; Boxer S. G. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennis J. T. M.; Larsen D. S.; van Stokkum I. H. M.; Vengris M.; van Thor J. J.; van Grondelle R. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nifosi R.; Ferrari A.; Arcangeli C.; Tozzini V.; Pellegrini V.; Beltram F. J. Phys. Chem. B 2003, 107, 1679. [Google Scholar]
- Fron E.; Flors C.; Schweitzer G.; Habuchi S.; Mizuno H.; Ando R.; De Schryver F. C.; Miyawaki A.; Hofkens J. J. Am. Chem. Soc. 2007, 129, 4870. [DOI] [PubMed] [Google Scholar]
- Weber W.; Helms V.; McCammon J.; Langhoff P. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico G.; Diaspro A.; Cannone F.; Collini M.; Bologna S.; Pellegrini V.; Beltram F. Chemphyschem 2005, 6, 328. [DOI] [PubMed] [Google Scholar]
- Altoe P.; Bernardi F.; Garavelli M.; Orlandi G.; Negri F. J. Am. Chem. Soc. 2005, 127, 3952. [DOI] [PubMed] [Google Scholar]
- Mandal D.; Tahara T.; Webber N. M.; Meech S. R. Chem. Phys. Lett. 2002, 358, 495. [Google Scholar]
- Mandal D.; Tahara T.; Meech S. R. J. Phys. Chem. B 2004, 108, 1102. [Google Scholar]
- Toniolo A.; Granucci G.; Martinez T. J. J. Phys. Chem. A 2003, 107, 3822. [Google Scholar]
- Toniolo A.; Olsen S.; Manohar L.; Martinez T. J. Faraday Discuss. 2004, 127, 149. [DOI] [PubMed] [Google Scholar]
- Toniolo A.; Ben-Nun M.; Martinez T. J. J. Phys. Chem. A 2002, 106, 4679. [Google Scholar]
- Loos D. C.; Habuchi S.; Flors C.; Hotta J. I.; Wiedenmann J. R.; Nienhaus G. U.; Hofkens J. J. Am. Chem. Soc. 2006, 128, 6270. [DOI] [PubMed] [Google Scholar]
- Usman A.; Mohammed O. F.; Nibbering E. T. J.; Dong J.; Solntsev K. M.; Tolbert L. M. J. Am. Chem. Soc. 2005, 127, 11214. [DOI] [PubMed] [Google Scholar]
- Follenius-Wund A.; Bourotte M.; Schmitt M.; Iyice F.; Lami H.; Bourguignon J. J.; Haiech J.; Pigault C. Biophys. J. 2003, 85, 1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strongin D. E.; Bevis B.; Khuong N.; Downing M. E.; Strack R. L.; Sundaram K.; Glick B. S.; Keenan R. J. Protein Eng., Des. Sel. 2007, 20, 525. [DOI] [PubMed] [Google Scholar]
- Abbyad P.; Childs W.; Shi X. H.; Boxer S. G. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 20189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solntsev K. M.; Poizat O.; Dong J.; Rehault J.; Lou Y. B.; Burda C.; Tolbert L. M. J. Phys. Chem. B 2008, 112, 2700. [DOI] [PubMed] [Google Scholar]
- Demachy I.; Ridard J.; Laguitton-Pasquier H.; Durnerin E.; Vallverdu G.; Archirel P.; Levy B. J. Phys. Chem. B 2005, 109, 24121. [DOI] [PubMed] [Google Scholar]
- Andresen M.; Stiel A. C.; Trowitzsch S.; Weber G.; Eggeling C.; Wahl M. C.; Hell S. W.; Jakobs S. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiel A. C.; Trowitzsch S.; Weber G.; Andresen M.; Eggeling C.; Hell S. W.; Jakobs S.; Wahl M. C. Biochem. J. 2007, 402, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H.; Kumar Mal T.; Waelchli M.; Kikuchi A.; Fukano T.; Ando R.; Jeyakanthan J.; Taka J.; Shiro Y.; Ikura M.; Miyawaki A.. Proc. Natl. Acad. Sci. U.S.A., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A.; Ishikura T.; Yamato T. Proteins—Struct. Funct. Bioinform. 2004, 55, 1063. [DOI] [PubMed] [Google Scholar]
- Hellingwerf K. J.; Hendriks J.; Gensch T. J. Phys. Chem. A 2003, 107, 1082. [Google Scholar]
- Chen M. C.; Lambert C. R.; Urgitis J. D.; Zimmer M. Chem. Phys. 2001, 270, 157. [Google Scholar]
- Henderson J. N.; Remington S. J. Physiology 2006, 21, 162. [DOI] [PubMed] [Google Scholar]
- Maddalo S. L.; Zimmer M. Photochem. Photobiol. 2006, 82, 367. [DOI] [PubMed] [Google Scholar]
- Nifosi R.; Tozzini V. Chem. Phys. 2006, 323, 358. [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. Nucleic Acids Res. 2000, 28, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F.; Moss L.; Phillips G. Nat. Biotechnol. 1996, 14, 1246. [DOI] [PubMed] [Google Scholar]
- Rekas A.; Alattia J. R.; Nagai T.; Miyawaki A.; Ikura M. J. Biol. Chem. 2002, 277, 50573. [DOI] [PubMed] [Google Scholar]
- Palm G. J.; Zdanov A.; Gaitanaris G. A.; Stauber R.; Pavlakis G. N.; Wlodawer A. Nat. Struct. Biol. 1997, 4, 361. [DOI] [PubMed] [Google Scholar]
- Mohamadi F.; Richards N. G. J.; Guida W. C.; Liskamp R.; Lipton M.; Caufield C.; Chang G.; Hendrickson T.; Still W. C. J. Comput. Chem. 1990, 11, 440. [Google Scholar]
- Chang G.; Guida W. C.; Still W. C. J. Am. Chem. Soc. 1989, 111, 4379. [Google Scholar]
- Saunders M.; Houk K.; Wu Y.-D.; Still W.; Lipton M.; Chang G.; Guida W. J. Am. Chem. Soc. 1990, 112, 1419. [Google Scholar]
- Bartol J.; Comba P.; Melter M.; Zimmer M. J. Comput. Chem. 1999, 20, 1549. [Google Scholar]
- Kolossvary I.; Keseru G. M. J. Comput. Chem. 2001, 22, 21. [Google Scholar]
- Keseru G. M.; Kolossvary I. J. Am. Chem. Soc. 2001, 123, 12708. [DOI] [PubMed] [Google Scholar]
- Andresen M.; Wahl M. C.; Stiel A. C.; Grater F.; Schafer L. V.; Trowitzsch S.; Weber G.; Eggeling C.; Grubmuller H.; Hell S. W.; Jakobs S. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K.; Sixma T. K.; Kitts P. A.; Kain S. R.; Tsien R. Y.; Ormo M.; Remington S. J. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S.; Sasaki S. FEBS Lett. 2006, 580, 1977. [DOI] [PubMed] [Google Scholar]
- Lopez X.; Marques M. A. L.; Castro A.; Rubio A. J. Am. Chem. Soc. 2005, 127, 12329. [DOI] [PubMed] [Google Scholar]
- Mauring K.; Deich J.; Rosell F. I.; McAnaney T. B.; Moerner W. E.; Boxer S. G. J. Phys. Chem. B 2005, 109, 12976. [DOI] [PubMed] [Google Scholar]
- Mena M. A.; Treynor T. P.; Mayo S. L.; Daugherty P. S. Nat. Biotechnol. 2006, 24, 1569. [DOI] [PubMed] [Google Scholar]
- Treynor T. P.; Vizcarra C. L.; Nedelcu D.; Mayo S. L. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormoe M.; Cubitt A. B.; Kallio K.; Gross L. A.; Tsien R. Y.; Remington S. J. Science 1996, 273, 1392. [DOI] [PubMed] [Google Scholar]
- Wachter R. M.; Elsiger M. A.; Kallio K.; Hanson G. T.; Remington S. J. Structure 1998, 6, 1267. [DOI] [PubMed] [Google Scholar]
- Kummer A.; Wiehler J.; Rehaber H.; Kompa C.; Steipe B.; Michel-Beyerle M. J. Phys. Chem. 2000, 104, 4791. [Google Scholar]
- Pal P. P.; Bae J. H.; Azim M. K.; Hess P.; Friedrich R.; Huber R.; Moroder L.; Budisa N. Biochemistry 2005, 44, 3663. [DOI] [PubMed] [Google Scholar]
- Allen F. H. Acta Crystallogr. Sect. B—Struct. Sci. 2002, 58, 380. [DOI] [PubMed] [Google Scholar]
- Jung G.; Wiehler J.; Zumbusch A. Biophys. J. 2005, 88, 1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott M.; Ling M.; Beddoe T.; Oakley A. J.; Dove S.; Hoegh-Guldberg O.; Devenish R. J.; Rossjohn J. Structure 2003, 11, 275. [DOI] [PubMed] [Google Scholar]
- Wilmann P. G.; Petersen J.; Devenish R. J.; Prescott M.; Rossjohn J. J. Biol. Chem. 2005, 280, 2401. [DOI] [PubMed] [Google Scholar]
- Wilmann P. G.; Petersen J.; Pettikiriarachchi A.; Buckle A. M.; Smith S. C.; Olsen S.; Perugini M. A.; Devenish R. J.; Prescott M.; Rossjohn J. J. Mol. Biol. 2005, 349, 223. [DOI] [PubMed] [Google Scholar]
- Petersen J.; Wilmann P. G.; Beddoe T.; Oakley A. J.; Devenish R. J.; Prescott M.; Rossjohn J. J. Biol. Chem. 2003, 278, 44626. [DOI] [PubMed] [Google Scholar]
- Quillin M. L.; Anstrom D. A.; Shu X. K.; O’Leary S.; Kallio K.; Chudakov D. A.; Remington S. J. Biochemistry 2005, 44, 5774. [DOI] [PubMed] [Google Scholar]