Abstract
The S100 proteins are a family of dimeric calcium binding proteins that function in response to changing calcium levels. Several S100 binding proteins have been identified however, the exact biological functions of the S100 proteins are largely unknown as there are several factors which modulate their functions. To address these issues, the specificity of binding of representative members of the human S100 proteins to short N-terminal peptides of annexin I (AnI) and annexin II (AnII) was investigated under controlled experimental conditions. AnI and AnII have been shown previously to interact with S100A11 and S100A10, respectively. This provided a unique opportunity to determine their binding specificity with the other members of the human S100 protein family. It was found that the AnI binds S100A6 or S100A11 while the AnII binds S100A10 or S100A11. This is the first report of the interaction between S100A6 and AnI. The fact that AnI and AnII bind to selected members of the S100 protein family shows that these interactions are specific and that the mode of binding is different to that of calmodulin, as it was found not to bind AnI or AnII. From the analysis of the thermodynamics of interactions the binding seems to be entropically driven. It was found that both AnI and AnII undergo a coil-to-helix transition upon binding to their respective binding partners. The observation that there is an overlap in functionality is not surprising due to considerable sequence homology between S100 protein family members. In fact, the functional overlap can explain previous failures of S100 knockout constructs to show any detectable changes in phenotype despite numerous implications of these proteins in important cellular processes.
The S100 protein family has attracted significant interest due to their role in responding to changing calcium levels in the cell (for review see (1–5)). These proteins have been implicated to play roles in the regulation of several key cellular processes ranging from cytoskeleton dynamics to apoptosis (6, 7). However, the understanding of the biological role of S100 proteins is far from comprehensive. In part, this is a result of the possible overlap in their function, which makes traditional approaches such as gene knockout ineffective.
The proteins of the S100 family generally exist as homodimers, with two EF-hand calcium binding motifs per monomer (1). Upon calcium binding, S100 proteins undergo a conformational change which exposes a region on the surface of the protein which has been shown to be responsible for binding several target proteins or peptides (1, 8). There have been several reports on identifying proteins which interact with the S100 proteins providing some insight into their function (for review see (1)). It is generally suggested that the interactions between S100 proteins and their target proteins are specific due to observations that only certain members of the S100 protein family were found to interact with specific target proteins. However, in many cases these observations are based mainly on interactions screened for under cellular environments which are difficult to define and control. This makes it difficult to identify additional S100 members which might also interact with the target protein, as only the interacting partners with the highest affinity will be identified. In addition, there are several other factors that can modulate the function of the S100 proteins. These include changing calcium levels in the cells, the presence of other divalent cations, such as zinc and copper, which have been shown to bind at positions distant from the calcium binding site (for review see (9)), tissue specific expression of certain members of the S100 family (10) and post-translational modification of target proteins (8).
For some of the interactions between the S100 proteins and their target proteins, relatively short peptide regions of the target protein have been found to retain the ability to bind the S100 target protein (for example see (11, 12)). Several crystal structures of the S100-peptide complexes have shown that these peptides all bind in the same region which is generally exposed upon calcium binding (13–15).
The fact that short peptides retain the ability to bind S100 proteins provides a unique opportunity to determine whether the S100 proteins can discriminately bind these peptides, which would allow for further insight into the degree of S100 specificity and thus their functions. The question of binding specificity is highlighted by the interaction of S100A10 with an N-terminal peptide of the annexin II protein (AnII), and the interaction of S100A11 with the N-terminal peptide of the annexin I protein (AnI). It was initially shown that these binding partners do not show cross-reactivity i.e. S100A10 binds AnII but not AnI, and S100A11 binds AnI but not AnII (15). However, in a more recent study (16), it was suggested that S100A11 does show cross reactivity as it was shown to interact with AnII. Even though these interactions might be specific in the case of S100A10 and S100A11, could these short peptides bind to any of the other members of the S100 protein family? To address this question, sixteen members of the human S100 proteins, which represent a pool of biologically relevant variants, and calmodulin were screened for binding with AnI and AnII using several biophysical methods.
Materials and methods
Peptide purification
Annexin I peptide (AnI; amino acid sequence: Ac-AMVSEFLKQAWFIE) and Annexin II peptide (AnII; amino acid sequence: Ac-STVHEILSKLSLEGY), both with their N-termini acetylated, were synthesized using standard Fmoc chemistry at the Penn State College of Medicine Macromolecular Core Facility. The AnI and AnII peptide sequences are the same sequences as those used by Rety et al (14). In addition, a Tyr residue was introduced at the C-terminus of the AnII peptide to allow for quantification using UV absorbance spectroscopy. It is not expected that this residue would alter the binding specificity or affinity as it has been shown that it is the N-terminal region of the peptide which determines binding. In addition, in the crystal structure of the S100A10-AnII complex, the C-terminal region of the AnII peptide does not have well defined electron density which suggests that that region of the peptide is not highly ordered which could also indicate that this region is not important for binding to S100A10 (14). The peptides were purified using C18 reverse phase HPLC using a 0–100% acetonitrile gradient in the presence of 30 mM ammonium acetate pH 5.5 for AnI or in the presence of 0.065–0.05% TFA for AnII. The fractions containing peptide were subjected to three cycles of lyophilization/resuspension in Milli-Q water to remove residual TFA, or ammonium acetate. The molecular mass of the peptides were confirmed using MALDI-TOF and were found to be 1,741.2 Da and 1,717.7 Da for AnI and AnII, respectively. The molecular masses are in excellent agreement with the expected molecular weights of 1,741.0 Da and 1,717.9 Da for AnI and AnII, respectively based on their amino acid compositions and N-terminal acetylation. Peptide concentrations were determined using the molar extinction coefficients at 280 nm (ε280nm) of 5,500 M−1 cm−1 and 1,490 M−1 cm−1 for AnI and AnII, respectively.
Analytical equilibrium ultracentrifugation
Analytical equilibrium ultracentrifugation experiments were performed on AnI and AnII to determine their oligomeric states using a Beckman XLA centrifuge at 4°C at 35,000 rpm in 20 mM Tris, 1 mM Tris (2-Carboxyethyl) phosphine hydrochloride (TCEP), 0.2 mM EDTA, pH 7.5 in the absence or presence of 5 mM calcium. The samples were considered to be at equilibrium when there was no difference between three consecutive radial distribution scans, with 6 hours between each scan. The data was fitted using non-linear regression software (NLREG) to the following equation, which describes the radial distribution of a single species at equilibrium:
| (1.) |
where Cr is the peptide concentration at radius r, Cro is the concentration of monomeric peptide at r0, ω is the angular velocity, R is the gas constant equal to 8.134 × 107 erg/(mol·K), T is the temperature in Kelvin, M is the monomer molecular mass, v̄ is the partial specific volume of the peptide (which is 0.748 cm3/g and 0.745 cm3/g for AnI and AnII, respectively (17)), and ρ is the density of the solvent. From the fitted data, the molecular mass of AnI and AnII was found to be 1.9 ± 0.07 kDa and 1.9 ± 0.16 kDa, respectively. These molecular masses are consistent with the ones expected from amino acid composition of monomeric peptides.
Cloning of S100A6, S100A10 and S100A11
S100A6, S100A10 and S100A11 were cloned from Matched cDNA Pairs library (Clontech; Palo Alto, CA, USA). The oligonucleotides used to PCR amplify the cDNA fragments for both proteins were based on their cDNA sequences complementary to the 5′ and 3′ ends. The PCR product for S100A10 was inserted into the expression vector pGIA, using SphI and PstI restriction endonucleases, and the S100A6 and S100A11 cDNA was inserted into the pVEX expression vector, using the NdeI and PstI restriction endonucleases. Both pVEX and pGIA expression vectors are under the control of the T7 promoter and confer ampicillin resistance. Mutations on the S100A6 protein that introduce Y73F, Y84S and Y73F/Y84S substitutions were made using the QuikChange approach. The DNA sequences were confirmed using an ABI 3130XL Capillary sequencer.
Protein purification
Sixteen representative members of the human S100 protein family (for S100 nomenclature used see (18)) - S100A1, S100A2, S100A3, S100A4, S100A5, S100A7, S100A8, S100A9, S100A12, S100A13, S100B, S100P, S100Z – as well as calmodulin were expressed and purified as described (19–21). Protein expression was performed using E. coli BL21 (DE3) cells as the protein expression is under the control of a T7 promoter. Cells were grown at 37 °C in 2xYT media in the presence of 100 μg/ml ampicillin to an optical density at 600 nm of approximately 1 unit when Isopropyl β-D-1-thiogalactopyranoside was added to a final concentration of 1 mM to induce protein expression. Protein expression was allowed to continue for 4 hours at 37 °C and 225 rpm, after which the cells were harvested by centrifugation at 5000 × g at 4 °C for 40 minutes. The cells were resuspended in 50 mM Tris, 0.2 mM EDTA, pH 7.5 and stored at −20 °C. After thawing, the cells were lysed using a French pressure cell, after which 3 mM dithiothreitol (DTT) and 5 mM calcium was added, and the cellular debris removed by centrifugation at 16 000 × g at 4° C for 45 minutes. The supernatant was loaded onto a Phenyl-Sepharose column equilibrated with 20 mM Tris, 0.2 mM EDTA, 5 mM calcium, 3 mM DTT, pH 7.5 and washed with 15 column volumes of the same buffer to remove unbound proteins. Proteins were eluted using 20 mM Tris, 3 mM DTT, 5 mM EGTA, pH 7.5 buffer and fractions containing the protein of interest were passed through a G-75 Sephadex column equilibrated with 20 mM Tris, 0.2 mM EDTA, 5 mM calcium, 3 mM DTT, pH 7.5. For S100A10, 8 M urea was used for elution instead of EGTA and the fractions containing the protein of interest were refolded by dialyzing the samples against 20 mM Tris, 3 mM DTT, 0.2 mM EDTA, pH 7.5 buffer, after which the protein was passed through a Sephadex G-75 column. The purity of the fractions containing the protein of interest was assessed using SDS-PAGE. In the cases where the protein of interest was not sufficiently pure, the samples were further purified using C18 reverse phase HPLC with a 0–100% acetonitrile gradient in the presence of 0.065 – 0.05 % TFA and the purity determined using SDS-PAGE. After the proteins were found to be pure (greater than 95 %), the proteins were dialyzed against double-deionized water with 0.001 % ammonium hydroxide, lyophilized and stored at −20°C.
Intrinsic Trp fluorescence experiments
Steady state fluorescence experiments were performed using a FluoroMax spectrofluorimeter. The AnI peptide contains a Trp residue which, based on the crystal structure of the AnI-S100A11 complex, becomes buried in a hydrophobic region upon binding to S100A11 (15). Thus changes in Trp-fluorescence emission intensity and/or emission maximum can be used to monitor interactions between the AnI and the S100 proteins. The experiments were performed at a constant temperature, 25°C, which was maintained using a thermostated cell holder and a circulating water bath. The buffer used was 20 mM Tris, 0.2 mM EDTA, 3 mM DTT pH 7.5, with 5 mM calcium (CaCl2) or, as a control, in the absence of calcium. The Trp residue in AnI was selectively excited at 295 nm and the emission monitored from 305–400 nm. The initial concentration of AnI in the fluorescence cell was 2 μM. Small aliquots of concentrated protein solution, in the assay buffer, were added stepwise. The final concentration of protein was 10 μM resulting in a final protein to peptide ratio of 5:1 at the end of titration. For the S100 proteins which contain Trp residues in their sequence (S100A1, S100A3, S100A8, S100A9 and S100A13) concentrated volumes of AnI was added to 2 μM protein until the ratio of peptide to protein was 5:1. The titration data was corrected for by subtracting the emission spectra for protein added to buffer, in the absence of peptide.
The molar extinction coefficients at 280 nm (ε280nm) used to determine the protein concentrations are as follows: S100A1 - 8,480 M−1 cm−1; S100A2 - 2,980 M−1 cm−1; S100A3 - 14,440 M−1 cm−1; S100A4 - 2,980 M−1 cm−1; S100A5 - 4,470 M−1 cm−1; S100A6 - 4,470 M−1 cm−1; S100A7 - 4,470 M−1 cm−1; S100A8 - 11,460 M−1 cm−1; S100A9 - 6,990 M−1 cm−1; S100A10 - 2,980 M−1 cm−1; S100A11 - 4,470 M−1 cm−1; S100A12 - 2,980 M−1 cm−1; S100A13 - 6,990 M−1 cm−1; S100B - 1,490 M−1 cm−1; S100P - 2,980 M−1 cm−1; S100Z - 2,980 M−1 cm−1 and calmodulin - 2,980 M−1 cm−1. These values were calculated as described in (22).
Far-UV Circular Dichroism
The far-UV CD spectra (260-200 nm) of free AnI and AnII was measured on a Jasco-715 spectropolarimeter in a 1 mm light path length cuvette. Measurements were done under three different conditions: buffer A was 20 mM Tris, 3 mM DTT, 0.2 mM EDTA pH 7.5; buffer B was same as buffer A with additional 5 mM calcium, buffer C same as buffer B in the presence of 30% TFE. Peptide concentrations were 0.017 mg/ml, which corresponds to approximately 30 μM for each peptide. The ellipticity values (Θ) for the peptides were corrected by subtracting the corresponding values for buffer only and converted to mean residue ellipticity, [Θ] using the following expression:
| (2.) |
where MR is the mean molecular mass of the amino acids in each peptide (124 Da for AnI and 114 Da for AnII), l is the light path length in centimeters and c is the peptide concentration in mg/ml. The fraction helicity, fH, was determined as:
| (3.) |
where [Θ]222 is the experimentally determined ellipticity at 222 nm, [Θ]C is the ellipticity of the fully coiled state, and [Θ]H is the ellipticity of the fully helical state. The ellipticity of the fully coiled state [Θ]C has a temperature dependence described by (23):
| (4.) |
where T is the temperature in degrees Celsius. The ellipticity of the fully helical state, [Θ]H, for a protein or peptide consisting of Nr residues, has a temperature dependence described by (23):
| (5.) |
To determine whether AnI and AnII form helical structures in the presence of their binding partners (i.e. S100A11, S100A10 and S100A6), far-UV CD spectra were measured from 250 nm to 200 nm for 10 μM protein in the presence of 50 μM AnI or 50 μM AnII, in binding buffer containing 5 mM calcium. The difference spectra, Θdiff, were determined as follows:
| (6.) |
where Θcomp, Θpep and Θprot are the ellipticities measured for the protein-peptide complex, free peptide and free protein, respectively. Using equation (2), Θdiff was converted to mean residue ellipticity assuming that the protein binding sites are saturated with bound peptide.
Isothermal titration calorimetry experiments
Isothermal titration calorimetry (ITC) experiments were performed on a VP-ITC instrument (Microcal Inc. Northampton, MA) as described in (24) and (25). The ITC experiments were performed in 20 mM Tris, 0.2 mM EDTA, 1 mM TCEP, pH 7.5 without calcium or with 5 mM calcium. To minimize differences in buffer composition, protein and peptide samples were dialyzed simultaneously with three buffer changes at 4°C. For the screening of binding between AnII and the S100 family members, as well as calmodulin, 2 μL of approximately 0.75 mM AnII was injected into the sample cell containing approximately 20–25 μM protein. Since the heat of binding strongly depends on temperature, the experiments were performed at 10°C and 25°C to ensure that the heat of binding can be observed. Heats of peptide dilution were determined by performing 2 μL peptide injections into buffer and were found to be insignificant. In cases when large heat effects, indicative of binding events, were observed, the titration was allowed to continue until saturation was achieved. In cases when no heat effect was observed, the titration was terminated after 5 injections.
ITC experiments were performed for the protein-peptide binding partners at 5 different temperatures. Due to limitations of AnI solubility, all titrations performed with AnI were performed by having the peptide in the cell and the protein in the syringe. Heats of dilution were less than 1% of the total heat when compared with a protein-peptide titration.
The heat of the reaction, Q, was obtained by integrating the peak after each injection of either peptide or protein into the cell, using scripts provided by the VP-ITC manufacturer (26). The heat of the reaction, after each injection, is related to the calorimetric enthalpy of binding ΔHcal, and the other thermodynamic parameters in a model dependent way (26). For a homodimeric system, the two simplest models that were considered are as follows:
Two identical (per dimer) binding sites model
| (7.) |
where
| (8.) |
n=2 is the stoichiometry of the peptide-protein complex, Ka is the association constant, [cell]t is the concentration of solute in the ITC cell with a volume Vo, and [syringe]t is the total concentration of injectant.
Two sequential binding sites model (per dimer)
| (9.) |
where K1 and K2 are the association constants for binding to sites one and two, and ΔHcal1 and ΔHcal2 are the calorimetric enthalpies for binding to sites one and two, respectively.
Structure based calculation of ΔCp
Structure-based calculation of thermodynamic parameters frequently allows validation of structural models (27). These calculations are usually based on changes in accessible surface area, ASA. To determine the total change in accessible surface upon binding (ΔASAtot), the ASA of protein in the absence of peptide (ASAdim − pep), and the ASA of unfolded peptide (ASAunf pep) were subtracted from the ASA of the protein in complex with the peptide (ASAdim + pep):
| (10.) |
The changes in ΔASA were subdivided into aliphatic surface area, aromatic surface area, peptide backbone surface area and polar surface area (28). The ΔASAtot was then converted into ΔCp using the following relation (28):
| (11.) |
where ΔASAalp, ΔASAarm, ΔASApol and ΔASAbb are the changes in ASA for aliphatic amino acids, aromatic amino acids, polar amino acids and the polypeptide backbone, respectively.
Results and discussion
Specificity of AnI or AnII binding to S100 family members and calmodulin
The AnI peptide, corresponding to the N-terminal sequence of annexin I, has been shown to bind with relatively high affinity to S100A11 (16, 29). Similarly, the AnII peptide, which corresponds to the N-terminal sequence of annexin II, was shown to bind S100A10 (15). The same group has also suggested that the interactions of AnI with S100A11 and AnII with S100A10 are specific (14). This conclusion was based on the observations that in their experiments no cross-reactivity (i.e. of AnI with S100A10 and AnII with S100A11) was observed. However, a more recent report suggests that S100A11 does indeed interact with AnII (16). In this work, the binding specificity of the AnI and AnII peptides to sixteen members of the S100 protein family (S100A1-S100A13, S100B, S100P, S100Z) and calmodulin was investigated using intrinsic Trp fluorescence and/or isothermal titration calorimetry (ITC). Calmodulin has been included as a control for several reasons. It is a member of a member of the EF-hand family and has four EF-hand motifs (30). It interacts with a rather diverse set of peptide sequences in a Ca2+-dependent manner (27, 31). Upon binding to calmodulin, peptides adopt a helical conformation (2, 27, 31). It is known that at least one peptide, melittin, has the ability to bind both S100P (25) and calmodulin (27) however, these interactions seem to have little biological relevance. All these properties make calmodulin a relevant control in the context of the present study addressing the issues of binding specificity of AnI and AnII peptides.
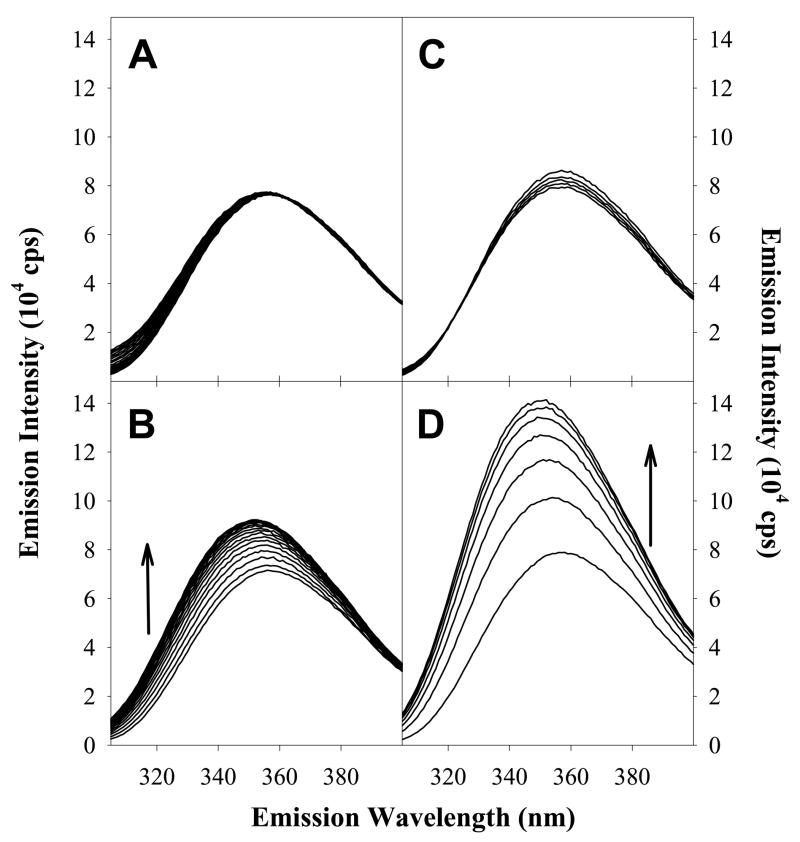
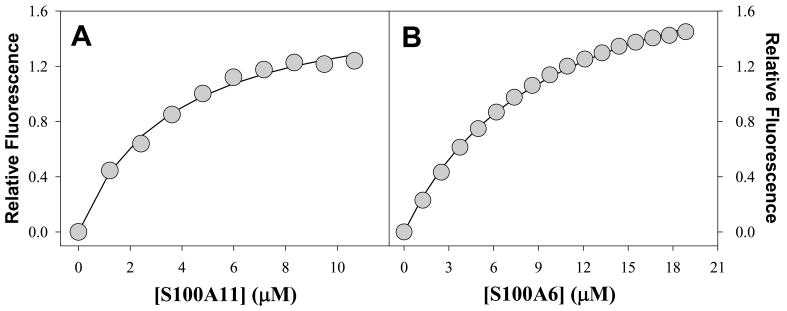
The fluorescence emission properties of tryptophan residues are a sensitive probe because they change depending to the polarity of the local environment. Trp residues which are highly exposed to polar environments (e.g. aqueous solution) have emission maxima in the range of 350 to 360 nm, whereas Trp residues in hydrophobic environments have emission maxima ranging between 330 to 345 nm (32). Figure 1A shows the fluorescence properties of the free AnI peptide in solution and of the AnI peptide in the presence of Ca2+-S100A11. It can be seen that the fluorescence emission maximum wavelength shifts from approximately 359 nm for free AnI to 350 nm in the presence of Ca2+-S100A11. The shift in emission maximum is also accompanied by a large increase in emission intensity suggesting that the environment of Trp residue on AnI becomes more hydrophobic. This observation is consistent with the crystal structure of the AnI-Ca2+-S100A11 complex which shows that the Trp residue in AnI becomes buried in a hydrophobic environment upon binding (15). In the absence of calcium, there is very little change in the emission spectra, which is expected, as the S100A11-AnI interaction has been shown to be calcium dependent (29). The dissociation constant, Kd, for the binding of AnI to Ca2+-S100A11, determined using Trp fluorescence, was found to be 5 μM (see Figure 2A) which is consistent with previous estimates (16). In addition, the Kd for the interaction of Ca2+-S100A11 and AnI measured using Trp fluorescence is in excellent agreement with the Kd measured using ITC (see below), which validates the dual-method approach for detecting binding and determining the binding affinity.
Figure 1.
Changes in Trp fluorescence emission spectra at 20°C of AnI for: 1.2 μM incremental additions of S100A11 in the absence of calcium (Panel A) and in the presence of 5 mM calcium (Panel B), 1.2 μM incremental additions of S100A6 in the absence of calcium (Panel C) and in the presence of 5 mM calcium (Panel D). The black arrows indicate the direction of the enhancement in Trp fluorescence emission for AnI in the presence of increasing concentrations of S100A11 or S100A6, which were taken to represent binding.
Figure 2.
Changes in intrinsic Trp fluorescence of AnI as a function of concentrations of Ca2+-S100A11 (Panel A) and Ca2+-S100A6 (Panel B). The symbols represent the experimental values. The solid lines represent the fits of the experimental data to equation (7). The dissociation constants for the binding of AnI to S100A11 and S100A6 were found to be 5±1 μM and 17±4 μM, respectively.
Trp fluorescence was used to screen for binding of AnI to all sixteen S100 protein family members and calmodulin (see Table S1 in Supplementary Materials for a summary of the results). Addition of any other S100 proteins or calmodulin to AnI did not show changes in Trp fluorescence (i.e. emission maximum or emission intensity). Surprisingly, the only exception was found to be Ca2+-S100A6. Figure 1B shows that AnI undergoes a large blue shift in its emission maximum wavelength, from approximately 359 nm to 352 nm, upon addition of Ca2+-S100A6. The shift in emission maximum is also accompanied by a large increase in emission intensity. No changes in Trp-fluorescence of the AnI peptide, in the presence of S100A6, are observed if calcium is omitted from the binding buffer. Thus, addition of Ca2+-S100A6 shows similar changes of Trp fluorescence of AnI as the addition of Ca2+-S100A11, suggesting that AnI also binds to the Ca2+-S100A6 protein in a calcium-dependent manner. Interactions of AnI with the S100A6 protein have not been observed to date. Analysis of the titration profile (Figure 2B) yields a dissociation constant, Kd, for this interaction to be on the order of 17±4 μM which is three-fold lower than the Kd for AnI binding to S100A11, 5±1 μM.
The AnII peptide sequence does not contain any Trp-residues, but only one Tyr residue. The fluorescence properties of Tyr are not as sensitive to its environment as is Trp. In addition, S100 proteins contain Tyr residues in their sequence, which also prevents the use of Tyr fluorescence spectroscopy for binding assays. As a result, the binding specificity of AnII to all the members of the S100 protein family was determined using ITC, a method of choice for characterizing the thermodynamics of macromolecular interactions. A summary of the binding interactions of sixteen S100 proteins and calmodulin with AnII as determined by ITC are shown in Table S1. It can be seen that AnII binds S100A10, which has been observed previously (14, 33). Moreover, the binding is independent of calcium, which is also consistent with previous findings that S100A10 does not have the ability to bind calcium and is locked in the ligand binding conformation (14).
Interestingly, in addition to binding S100A10, AnII is also found to bind Ca2+-S100A11 and this binding to S100A11 is also modulated by calcium. The interaction between Ca2+-S100A11 and AnII was initially shown not to occur (15). Recently, however, Rintala-Dempsey et al (16) using NMR spectroscopy have shown that AnII can interact with Ca2+-S100A11. A possible reason for the conflicting reports pertaining to the interaction between S100A11 and AnII is that Réty et al (15) labeled AnII with Prodan, a fluorescent tag, which could have sterically hinder binding. In our work, and in the report by Rintala-Dempsey et al (16), the AnII peptide was not modified by the addition of any extrinsic fluorescent probe.
In summary, AnI and AnII peptides were screened for interactions with sixteen members of S100 protein family and calmodulin. Neither peptide was found to bind to calmodulin, suggesting differences in specificity between the calmodulin and the S100 protein family. AnI has been found to bind Ca2+-S100A6 and Ca2+-S100A11, and AnII has been found to bind S100A10 and Ca2+-S100A11. The observation that AnI and AnII each have a unique binding partner, i.e. S100A6 binding to AnI, or S100A10 binding to AnII, and share a common binding partner i.e. S100A11 binds both AnI and AnII, is quite remarkable. To gain a better understanding of these interactions, the binding between S100A11, S100A10, S100A6 and the AnI and AnII were further characterized using far-UV CD and isothermal titration calorimetry.
AnI and AnII undergo coil-to-helix transition upon binding
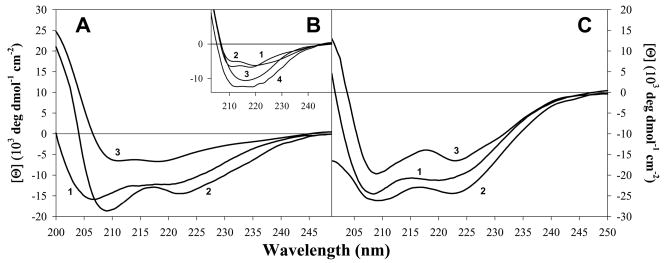
Far-UV circular dichroism (CD; 250 nm to 190 nm) has been widely used to characterize the secondary structural content of proteins and peptides. A protein, or peptide, with high content of helical structure would have a CD spectrum with characteristic minima at 222 nm and 208 nm, whereas a spectrum corresponding to a β-sheet, has a minimum at approximately 215 nm (34). Unstructured polypeptides tend to have far-UV CD spectra lacking the aforementioned features. Far-UV CD spectra of free AnI and AnII were measured in the absence and presence of calcium to determine whether they are structured and whether the presence of calcium has an effect on their secondary structure (data not shown). The helical propensities of the AnI and AnII peptides according to AGADIR (35) are expected to be relatively low, less than 2%. Indeed, the experimentally measured far-UV CD spectra of AnI and AnII do not show the characteristic minima for either α-helices or β-sheets. Thus it can be concluded that AnI and AnII are both devoid of regular secondary structure in solution. To determine the potential for helix formation by AnI and AnII, trifluoroethanol (TFE) was added to each peptide up to a final concentration of 60 %, in the presence and absence of calcium. It is well known that TFE promotes helix formation for peptides which are devoid of regular secondary structure (36). In the presence of TFE both AnI and AnII have far-UV CD spectra characteristic of helices, which are similar irrespective of whether calcium is present or not (see Figure 4). Taken together, these results show that even though the intrinsic helical propensities of AnI and AnII in aqueous solution are low, they can form helices in the presence of TFE.
Figure 4.
Far-UV CD difference spectra for AnI and AnII binding to their respective S100 protein binding partners, in the presence of 5 mM calcium, determined using equation 6. Panel A shows the far-UV CD spectrum for AnI in 60 % TFE (curve 1; shown for comparison), the difference spectra for AnI bound to S100A11 (curve 2) and for AnI bound to S100A6 wild type (curve 3). Panel B shows the difference spectra for AnI bound to: S100A6 wild type (curve 1; shown for comparison), S100A6-Y84S (curve 2), S100A6-Y73F (curve 3) and S100A6-Y73F-Y84S (curve 4). Panel C shows the far-UV CD spectrum for AnII in the presence of 60 % TFE (curve 1; shown for comparison), the far-UV CD difference spectra for AnII bound to S100A10 (curve 2) and S100A11 (curve 3).
It is well established that AnI and AnII adopt helical conformations when bound to Ca2+-S100A11 and S100A10, respectively (Protein Data Bank codes 1QLS (15) and 1BT6(14)). It is however not known whether AnI and AnII undergo the same conformational change upon binding to Ca2+-S100A6 and Ca2+-S100A11, respectively. For this reason the far-UV CD spectra of peptides in the presence corresponding S100 proteins (i.e. AnI in the presence of Ca2+-S100A6 and Ca2+-S100A11 and AnII in the presence of S100A10 and Ca2+-S100A11) were determined. Figure 4 shows the difference far-UV CD spectra for all the binding partners.
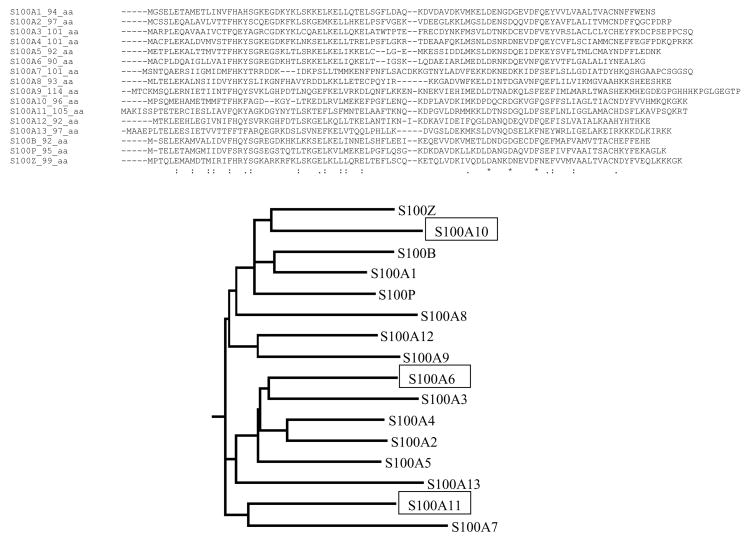
It can be observed that in most cases the peptides form helices upon binding to their respective binding partners. A noted lower elipticity is observed for the far-UV CD spectrum for AnI binding to Ca2+-S100A6. There are two possible explanations for this. First, it is possible that upon binding to S100A6 AnI does not form as extensive a helix as it does when it binds to S100A11, and second, the unusual far-UV CD spectrum could be due to the contribution of aromatic side chains to the measured CD spectrum. Chakrabarty et al (37) have shown that aromatic amino acids, Tyr and Trp, have opposite effects on far-UV CD spectra of peptides. They have shown that the difference spectrum for a peptide containing a Tyr residue has a positive Cotton effect (i.e. leading to an apparent decrease in helical content) whereas a Trp containing peptide has a negative cotton effect (i.e. leading to an overestimate in helical content). The fact that both AnI and AnII have the same difference spectra when binding Ca2+-S100A11, with fractional helicities of 0.59 and 0.57 respectively, supports the argument that it is the environment on the binding surface of Ca2+-S100A6 which could be responsible for the observation made with the AnI and the S100A6 complex. Based on the sequence comparison of S100A11 and S100A6 (Figure 3) there are two additional Tyr residues in the sequence of S100A6 (at positions 73 and 84) that are Phe and Cys respectively, in S100A11. It is possible that upon peptide binding, the environments of the Tyr residues change leading to a positive Cotton effect which could decrease the overall far-UV CD spectrum. Consequently, this could mask the characteristic helical far-UV CD spectrum for the bound peptide. To test this hypothesis, Tyr73 and Tyr84 in S100A6 were substituted with Phe and Ser, respectively. The Tyr84 was substituted with Ser and not Cys, as found in S100A11, so as to avoid possible disulfide bond formation. It appears that Y73F contributes to the apparent lower intensity of far-UV CD. This can be seen from the comparison of far-UV CD spectrum of AnI upon binding to the wild type, Y73F, Y84S or Y73F/Y84S variants of S100A6. Indeed, the difference spectrum for the Y84S variant has a similar spectrum to that of the wild type, while the difference spectra for Y73F and Y73F/Y84S are similar to that of the difference far-UV CD spectrum of AnI upon binding to S100A11 or AnI in 60% TFE. Importantly, the substitutions do not have a significant effect on the binding affinity (data not shown). These results support the notion that both AnI and AnII peptides undergo a coil-to-helix transition upon binding to Ca2+-S100A6 and Ca2+-S100A11 or S100A10 and Ca2+-S100A11, respectively.
Figure 3.
Sequence alignment of human S100 family members generated using ClustalW. Also shown is the phylogenetic tree based on the sequence alignment. For clarity, S100A6, S100A10 and S100A11 are highlighted in the phylogenetic tree.
Thermodynamics of peptide binding
From the Trp fluorescence and ITC screening results, as well as the far-UV CD results, it has been shown that Ca2+-S100A6 and Ca2+-S100A11 bind AnI, whereas S100A10 and Ca2+-S100A11 bind AnII. The thermodynamics of these interactions were further characterized in detail by performing ITC experiments at different temperatures. ITC, in many cases, is the method of choice for measuring the thermodynamics of protein-protein interactions. It simultaneously provides the estimates for binding constant (and thus Gibbs energy of binding), the enthalpy of binding and the stoichiometry of binding. A drawback of ITC, however, is that the analysis of data is model-dependent.
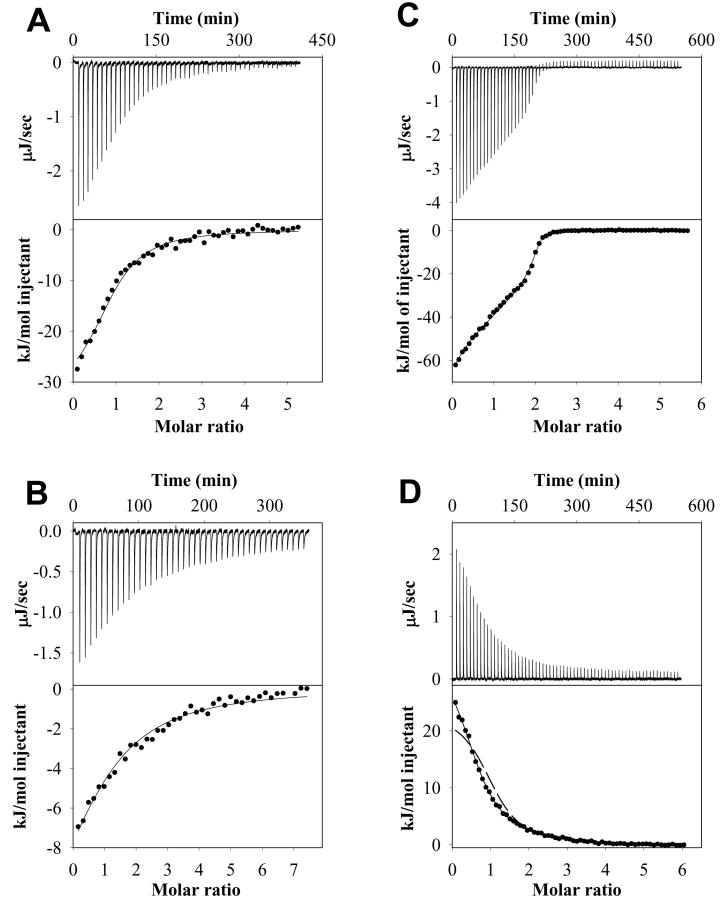
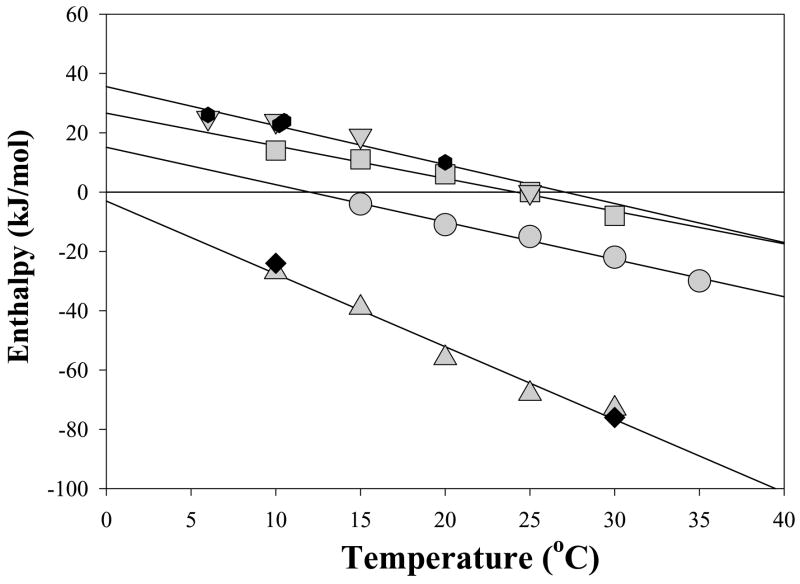
Figure 5A shows an example of an ITC experiment where Ca2+-S100A11 was titrated into AnI. The ITC titration data was best fit to a simplest model, two identical non-interacting binding sites per dimer of Ca2+-S100A11 described by equation 7. This model is essentially equivalent to a model with one peptide binding site per Ca2+-S100A11 monomer (see Table 1 for a summary of the thermodynamic data). The Kd per binding site for the interaction of Ca2+-S100A11 with AnI was found to be 5±2 μM which is in excellent agreement with the 5±1 μM Kd determined using Trp fluorescence. ITC experiments were performed for the temperature range from 15°C to 35°C. Figure 6 shows the dependence of the enthalpy of binding, ΔHcal, on temperature. The enthalpy of binding of AnI to Ca2+-S100A11 is always negative and decreases with an increase in temperature from −4 kJ/mol at 15°C to −30 kJ/mol at 35°C. At the same time the equilibrium dissociation constant does not depend significantly on temperature (see Table S2 in Supplementary Materials), leading to a weak temperature dependence of the free energy of binding, ΔG=−RTln(Kd)≈30 kJ/mol, per binding site. This suggests that the binding of AnI to Ca2+-S100A11 is entropically driven.
Figure 5.
Examples of ITC experiments for AnI or AnII binding to their specific S100 protein binding partners. Shown for each example are the heat effects (μJ/sec) as a function of time, the cumulative heat effects (kJ/mol and represented by symbols) as a function of the molar ratio of peptide to protein, and the fits to the experimental data (solid lines) for: AnI binding to S100A11 at 35 °C (Panel A; data fitted to equation 7), AnI binding to S100A6 at 35 °C (Panel B; data fitted to equation 7), AnII binding to S100A10 at 15 °C (Panel C; data fitted to equation 9) and AnII binding to S100A11 at 6 °C (Panel D; data fitted to equation 9). For comparison, the dashed line in Panel D shows the experimental binding data fitted to a model describing identical binding sites (equation 7).
Table 1.
Summary of the thermodynamic properties of S100A6, S100A10, and S100A11 binding to AnI and AnII determined using Trp fluorescence titrations and ITC experiments as well as the comparison with structure-based calculations
| Protein | AnI Kd (μM) | AnII Kd1 and Kd2 (μM) | AnI ΔCp,exp kJ/(mol·K) | AnI ΔCp,calc kJ/(mol·K) | AnII ΔCp,exp kJ/(mol·K) | AnII ΔCp,calc kJ/(mol·K) |
|---|---|---|---|---|---|---|
| S100A6 | 13±4 (17±4)a | −1.1±0.3 | −1.6±0.5 | |||
| S100A11 | 5±2 (5±1)a | −1.3±0.2 | −1.5±0.3 | |||
| S100A11 | 1.7±1.2 and 9.2±1.9 | −1.1±0.2 | −1.4±0.3 | |||
| S100A10 | 0.5±0.4 and 0.5±0.3 | −2.4±0.3 | −1.7±0.3 |
Values in parentheses were determined using Trp fluorescence
Figure 6.
Temperature dependence of the enthalpies, ΔHcal, of AnI and AnII binding to their specific S100 protein binding partners: AnII binding to S100A10 in the presence of 5 mM calcium (▲) and in the absence of calcium (◆), AnII binding to S100A11 in the absence of NaCl (▼) and in the presence of 100 mM NaCl ( ), AnI binding to S100A11 (■) and AnI binding to S100A6 (●). Solid lines represent linear fits of the ΔHcal temperature dependencies for each S100-peptide interaction. The slopes of these lines represent the change in heat capacity, which are summarized in Table 2.
), AnI binding to S100A11 (■) and AnI binding to S100A6 (●). Solid lines represent linear fits of the ΔHcal temperature dependencies for each S100-peptide interaction. The slopes of these lines represent the change in heat capacity, which are summarized in Table 2.
The binding of AnI to Ca2+-S100A6 is a novel interaction, which was identified by us using Trp fluorescence and was further confirmed using ITC. Figure 5B shows an example of an ITC experiment where Ca2+-S100A6 was titrated into AnI. The data was best fit to a model with two identical non-interacting binding sites per dimer of Ca2+-S100A6 described by equation 7. The Kd for the interaction was found to be 13±4 μM (see Table 1 for a summary of the thermodynamic parameters). This compares well with the 17±4 μM Kd for AnI binding to Ca2+-S100A6 determined using Trp fluorescence to monitor the binding (Figure 2B). It is also comparable to the Kd of 5±1 μM for AnI binding to Ca2+-S100A11 determined from Trp-fluorescence titration experiments (Figure 2A) and ITC (Figure 5). The similarity in the binding thermodynamics of the AnI peptide to Ca2+-S100A11 and Ca2+-S100A6 extends beyond the similarities of Kd. Figure 6 compares the enthalpies of binding of AnI to Ca2+-S100A11 and Ca2+-S100A6 at different temperatures. The enthalpies of binding are either negative or positive but small relative to the free energy of binding ΔG=−RTln(Kd)≈26 or 30 kJ/mol, per binding site, for AnI binding to Ca2+-S100A11 or Ca2+-S100A6, respectively. This suggests that binding of AnI to both these proteins is entropically driven. The other remarkable similarity for thermodynamics of binding for AnI to Ca2+-S100A11 and Ca2+-S100A6 is that both have similar slopes for ΔHcal dependencies on temperature, −1.3±0.2 kJ/(mol·K) and −1.1±0.2 kJ/(mol·K), respectively. The slope of the ΔHcal dependence on temperature represents the heat capacity changes upon binding, ΔCp. The values of ΔCp for protein-protein interactions can be calculated from the structural information on the interacting proteins before and after complex formation (see Materials and Methods for details). Using the structure of AnI in complex with Ca2+-S100A11, solved by x-ray crystallography (15), and the approach described in the Material and Methods (see equations 10–11) the changes in ΔCp for AnI binding to Ca2+-S100A11 is predicted to be −1.5±0.3 kJ/(mol·K) which is in a very good agreement with the experimentally measured values (see Table 1).
There are currently no structures available for the AnI peptide bound to Ca2+-S100A6 as this is the first description of this interaction. There is however ample evidence that this complex is probably structurally similar to the AnI/Ca2+-S100A11 complex (15). Consequently, the structure of the complex was modeled by threading the S100A6 sequence into the structure of S100A11 or threading the sequence of AnI into the structure of Ca2+-S100A6 complex with another unrelated peptide (38). It was found that the experimentally determined ΔCp (−1.1±0.2 kJ/mol K) and the expected ΔCp (−1.6±0.5 kJ/mol K) are in relatively good agreement taking into consideration that the AnI/Ca2+-S100A6 complex is a homology model. This correspondence of the calculated and experimental values of ΔCp for AnI-binding to Ca2+-S100A6, together with the CD data (see Figure 4), provides strong support for the similarity of the binding mode of AnI to these two distinctive proteins, S100A6 and S100A11.
Figure 5C shows representative ITC profile for the titration of AnII into S100A10. The binding isotherm cannot be fit to the simplest model (two identical noninteracting binding sites per dimer of S100A10) represented by the equation 7 because of its clear biphasic shape. Since ITC data analysis is model dependent and should therefore follow the principle of Occam’s Razor, the next simplest model could be one that has two non-identical non-interacting binding sites. However, this model cannot hold true due to the symmetry of the S100A10 homodimer. Thus the binding isotherm was fitted to a model that assumes two sequential binding sites per S100A10 dimer. The fit to this model is very good not only at 15°C (shown in Figure 5B) but at all other temperatures and solvent conditions (data not shown). The dissociation constants from the fit are summarized in Table 1. The implication of this model is that one of the two sites of the S100A10 homodimer must be occupied before the other. Such an order of binding events is possibly due to the fact that there are several interactions made with the peptide from both subunits of the homodimer. The interactions formed upon binding the first peptide are transduced through a slight adjustment of the dimer interface onto the other subunit of the S100A10 homodimer, resulting in enhanced binding at the second binding site.
Figure 6 shows the ΔHcaltot, the total enthalpy from both binding sites, plot versus temperature. It is remarkable that the enthalpy is large and negative for the entire range of temperatures, again suggesting that AnII binding to S100A10 is entropically driven. This entropically driven binding of AnII to S100A10 is similar to the entropically driven binding of AnI to Ca2+-S100A6 and Ca2+-S100A11, however in these latter two cases the binding is modulated by calcium ions. S100A10 is an exception among all other S100 proteins in that it does not bind calcium or any other divalent ions (14). It has been suggested that S100A10 is “locked” in the ligand-binding-competent state. Consequently, it is expected that the binding of AnII to S100A10 should be independent of calcium as S100A10 does not have the ability to bind calcium. Indeed, the ITC experiments for binding of AnII to S100A10 performed in the presence of calcium are very similar to the data obtained in the absence of calcium (see Figure 6), suggesting that the entropically driven peptide binding to these S100 proteins is not related to calcium binding.
The temperature dependence of the enthalpy of AnII binding to S100A10 is very similar to the ΔCp observed previously for the peptide melittin binding to Ca2+-S100P (25). For the latter system, the ΔCp was estimated to be −2.5±0.5 kJ/(mol·K) which compares well with the ΔCp of −2.4±0.3 kJ/(mol·K) for AnII binding to S100A10. Interestingly, in both cases, the values for ΔCp predicted from structure based calculations according to the equations (11) gave similar albeit lower estimates (−1.7 kJ/(mol·K)) than the experimentally measured ΔCp (Figure 6).
Even though it was initially shown that Ca2+-S100A11 and AnII do not interact (14), recent NMR experiments suggest that there is an interaction (16). By screening for interactions of S100 proteins with AnII using ITC, we found independently that S100A11 indeed does bind the AnII. The enthalpy of binding of AnII to Ca2+-S100A11 is endothermic while the enthalpy of AnII binding to S100A10 is exothermic. To further investigate these differences in sign of heat effects, ITC experiments for AnII binding to Ca2+-S100A11 were performed in 100 mM NaCl. The higher ionic strength however did not have any effect on the thermodynamics of binding (see e.g. Figure 6), thus eliminating the possibility that this is non-specific electrostatically driven interaction. ITC titration data cannot be fit to the simplest model, two identical non-interacting binding sites per dimer of Ca2+-S100A11, represented by the equation 7 (see Figure 5D). However, as for the AnII S100A10 ITC titration profile, the ITC data can be fit well to a model that assumes two sequential binding sites for AnII per Ca2+-S100A11 dimer (see Figure 5D). The dissociation constants for this model were found to be 2±1 μM and 9±2 μM. As in the case of AnII-S100A10 interactions, the two sequential binding sites model that describes best the AnII-Ca2+-S100A11 interactions, suggests a thermodynamic linkage between binding sites.
The enthalpy of interaction between AnII and Ca2+-S100A11 is endothermic but small relative to the Gibbs energy of binding, indicating that as in all previous cases binding is entropically driven. This entropically driven binding is observed at all studied temperatures. This is despite the fact that the enthalpy of interactions is strongly temperature dependent with experimentally determined ΔCp of −1.1 kJ/(mol·K), a value very similar to that of −1.4 kJ/(mol·K) calculated from the modeled structure of AnII complex with Ca2+-S100A11 (using equation 11). Such good correspondence between calculated and experimental values of ΔCp implies that there are sufficient degrees of similarities in the structures that can be predicted by homology modeling. Despite this, it is not sufficient to explain, for example, why and how AnII becomes more helical when bound to S100A10 compared to when it is bound to S100A11 or to explain why AnI and AnII peptides bind to only these S100 proteins and not the others.
Conclusions
By screening for binding for AnI and AnII against representatives of the human S100 protein family under defined conditions, it has been shown that very few members of the protein family bind the two peptides. This suggests that the observed binding interactions are specific. In addition to demonstrating this specificity, we also confirmed the recently suggested interaction between AnII and Ca2+-S100A11. Furthermore, we have identified a novel interaction between Ca2+-S100A6 and AnI. These interactions, as were suggested previously, are modulated by calcium. Detailed thermodynamic and spectroscopic characterization of the interactions revealed additional common properties. First, binding in all four studied pairs of interacting partners is entropically driven. Second, both AnI and AnII peptides, unstructured when free in solution, fold into helical conformation upon binding.
The exact cellular functions of the S100 proteins are not known even though there are several reports identifying different target proteins. One of the main limitations of the approaches used to screen for potential S100 interacting partners is that the target protein with the highest binding affinity would be identified. This could lead to only a single target protein being identified to interact with the specific S100 protein family member, which would suggest that the interaction is specific. The overlap in binding specificity observed here could provide a reason for why gene knockout approaches to map function of S100 proteins have not lead to an identifiable phenotypic change (see (3) for examples). The response to changing cellular calcium levels is critical to the cells survival and it is therefore not unexpected that the functionality of the S100 proteins could overlap.
Supplementary Material
Footnotes
Acknowledgments: This work was supported in part by a grant from the National Institutes of Health (GM54537).
SUPPORTING INFORMATION AVAILABLE
Summary of AnI and AnII interactions with S100 protein family members (Table S1) and thermodynamic parameters determined using ITC for AnI and AnII binding their respective S100 protein binding partners (Table S2). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya S, Bunick CG, Chazin WJ. Target selectivity in EF-hand calcium binding proteins. Biochim Biophys Acta. 2004;1742:69–79. doi: 10.1016/j.bbamcr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 4.Zimmer DB, Wright Sadosky P, Weber DJ. Molecular mechanisms of S100-target protein interactions. Microsc Res Tech. 2003;60:552–559. doi: 10.1002/jemt.10297. [DOI] [PubMed] [Google Scholar]
- 5.Rintala-Dempsey AC, Rezvanpour A, Shaw GS. S100-annexin complexes - structural insights. Febs J. 2008 doi: 10.1111/j.1742-4658.2008.06654.x. [DOI] [PubMed] [Google Scholar]
- 6.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60:540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 7.Ghavami S, Kerkhoff C, Chazin WJ, Kadkhoda K, Xiao W, Zuse A, Hashemi M, Eshraghi M, Schulze-Osthoff K, Klonisch T, Los M. S100A8/9 induces cell death via a novel, RAGE-independent pathway that involves selective release of Smac/DIABLO and Omi/HtrA2. Biochim Biophys Acta. 2008;1783:297–311. doi: 10.1016/j.bbamcr.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Wilder PT, Lin J, Bair CL, Charpentier TH, Yang D, Liriano M, Varney KM, Lee A, Oppenheim AB, Adhya S, Carrier F, Weber DJ. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim Biophys Acta. 2006;1763:1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Heizmann CW, Cox JA. New perspectives on S100 proteins: a multi-functional Ca(2+)-, Zn(2+)- and Cu(2+)-binding protein family. Biometals. 1998;11:383–397. doi: 10.1023/a:1009212521172. [DOI] [PubMed] [Google Scholar]
- 10.Ravasi T, Hsu K, Goyette J, Schroder K, Yang Z, Rahimi F, Miranda LP, Alewood PF, Hume DA, Geczy C. Probing the S100 protein family through genomic and functional analysis. Genomics. 2004;84:10–22. doi: 10.1016/j.ygeno.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Ivanenkov VV, Jamieson GA, Jr, Gruenstein E, Dimlich RV. Characterization of S-100b binding epitopes. Identification of a novel target, the actin capping protein, CapZ. J Biol Chem. 1995;270:14651–14658. doi: 10.1074/jbc.270.24.14651. [DOI] [PubMed] [Google Scholar]
- 12.Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca(2+)-dependent interaction of S100B(beta beta) with a peptide derived from p53. Biochemistry. 1998;37:1951–1960. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 14.Rety S, Sopkova J, Renouard M, Osterloh D, Gerke V, Tabaries S, Russo-Marie F, Lewit-Bentley A. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat Struct Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- 15.Rety S, Osterloh D, Arie JP, Tabaries S, Seeman J, Russo-Marie F, Gerke V, Lewit-Bentley A. Structural basis of the Ca(2+)-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure. 2000;8:175–184. doi: 10.1016/s0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- 16.Rintala-Dempsey AC, Santamaria-Kisiel L, Liao Y, Lajoie G, Shaw GS. Insights into S100 target specificity examined by a new interaction between S100A11 and annexin A2. Biochemistry. 2006;45:14695–14705. doi: 10.1021/bi061754e. [DOI] [PubMed] [Google Scholar]
- 17.Makhatadze GI, Medvedkin VN, Privalov PL. Partial molar volumes of polypeptides and their constituent groups in aqueous solution over a broad temperature range. Biopolymers. 1990;30:1001–1010. doi: 10.1002/bip.360301102. [DOI] [PubMed] [Google Scholar]
- 18.Marenholz I, Lovering RC, Heizmann CW. An update of the S100 nomenclature. Biochim Biophys Acta. 2006;1763:1282–1283. doi: 10.1016/j.bbamcr.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Gribenko A, Lopez MM, Richardson JM, 3rd, Makhatadze GI. Cloning, overexpression, purification, and spectroscopic characterization of human S100P. Protein Sci. 1998;7:211–215. doi: 10.1002/pro.5560070123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopalakrishna R, Anderson WB. Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem Biophys Res Commun. 1982;104:830–836. doi: 10.1016/0006-291x(82)90712-4. [DOI] [PubMed] [Google Scholar]
- 21.Gribenko AV, Hopper JE, Makhatadze GI. Molecular characterization and tissue distribution of a novel member of the S100 family of EF-hand proteins. Biochemistry. 2001;40:15538–15548. doi: 10.1021/bi0114731. [DOI] [PubMed] [Google Scholar]
- 22.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 23.Rohl CA, Baldwin RL. Deciphering rules of helix stability in peptides. Methods Enzymol. 1998;295:1–26. doi: 10.1016/s0076-6879(98)95032-7. [DOI] [PubMed] [Google Scholar]
- 24.Lopez MM, Makhatadze GI. Isothermal titration calorimetry. Methods Mol Biol. 2002;173:121–126. doi: 10.1385/1-59259-184-1:121. [DOI] [PubMed] [Google Scholar]
- 25.Gribenko AV, Guzman-Casado M, Lopez MM, Makhatadze GI. Conformational and thermodynamic properties of peptide binding to the human S100P protein. Protein Sci. 2002;11:1367–1375. doi: 10.1110/ps.0202202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 27.Brokx RD, Lopez MM, Vogel HJ, Makhatadze GI. Energetics of target peptide binding by calmodulin reveals different modes of binding. J Biol Chem. 2001;276:14083–14091. doi: 10.1074/jbc.M011026200. [DOI] [PubMed] [Google Scholar]
- 28.Makhatadze GI, Privalov PL. Energetics of protein structure. Adv Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 29.Mailliard WS, Haigler HT, Schlaepfer DD. Calcium-dependent binding of S100C to the N-terminal domain of annexin I. J Biol Chem. 1996;271:719–725. doi: 10.1074/jbc.271.2.719. [DOI] [PubMed] [Google Scholar]
- 30.Babu YS, Bugg CE, Cook WJ. Structure of calmodulin refined at 2.2 A resolution. J Mol Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- 31.Clore GM, Bax A, Ikura M, Gronenborn AM. Structure of Calmodulin Target Peptide Complexes. Current Opinion in Structural Biology. 1993;3:838–845. [Google Scholar]
- 32.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; New York: 2006. [Google Scholar]
- 33.Becker T, Weber K, Johnsson N. Protein-protein recognition via short amphiphilic helices; a mutational analysis of the binding site of annexin II for p11. Embo J. 1990;9:4207–4213. doi: 10.1002/j.1460-2075.1990.tb07868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson WC., Jr Protein secondary structure and circular dichroism: a practical guide. Proteins. 1990;7:205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- 35.Lacroix E, Viguera AR, Serrano L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J Mol Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 36.Nelson JW, Kallenbach NR. Stabilization of the ribonuclease S-peptide alpha-helix by trifluoroethanol. Proteins. 1986;1:211–217. doi: 10.1002/prot.340010303. [DOI] [PubMed] [Google Scholar]
- 37.Chakrabartty A, Kortemme T, Padmanabhan S, Baldwin RL. Aromatic side-chain contribution to far-ultraviolet circular dichroism of helical peptides and its effect on measurement of helix propensities. Biochemistry. 1993;32:5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- 38.Lee YT, Dimitrova YN, Schneider G, Ridenour WB, Bhattacharya S, Soss SE, Caprioli RM, Filipek A, Chazin WJ. Structure of the S100A6 Complex with a Fragment from the C-Terminal Domain of Siah-1 Interacting Protein: A Novel Mode for S100 Protein Target Recognition. Biochemistry. 2008 doi: 10.1021/bi801233z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.