Abstract
Two recent genome-wide association studies have independently identified a prostate cancer susceptibility locus on chromosome 10q11.2. The most significant single-nucleotide polymorphism (SNP) marker reported, rs10993994, is 57 bp centromeric of the first exon of the MSMB gene, which encodes β-microseminoprotein (prostatic secretory protein 94). In this study, a fine-mapping analysis using HapMap SNPs was conducted across a ≈65-kb region (chr10: 51168330–51234020) flanking rs10993994 with 13 tag SNPs in 6,118 prostate cancer cases and 6,105 controls of European origin from the Cancer Genetic Markers of Susceptibility (CGEMS) project. rs10993994 remained the most strongly associated marker with prostate cancer risk [P = 8.8 × 10−18; heterozygous odds ratio (OR) = 1.20, 95% confidence interval (CI): 1.11–1.30; homozygous OR = 1.64, 95% CI: 1.47–1.86 for the adjusted genotype test with 2 df]. In follow-up functional analyses, the T variant of rs10993994 significantly affected expression of in vitro luciferase reporter constructs. In electrophoretic mobility shift assays, the C allele of rs10993994 preferentially binds to the CREB transcription factor. Analysis of tumor cell lines with a CC or CT genotype revealed a high level of MSMB gene expression compared with cell lines with a TT genotype. These findings were specific to the alleles of rs10993994 and were not observed for other SNPs determined by sequence analysis of the proximal promoter. Together, our mapping study and functional analyses implicate regulation of expression of MSMB as a plausible mechanism accounting for the association identified at this locus. Further investigation is warranted to determine whether rs10993994 alone or in combination with additional variants contributes to prostate cancer susceptibility.
Keywords: genome-wide association studies, prostate cancer genetics, CREB transcription factor
Prostate cancer is the most common noncutaneous malignancy and the second leading cause of cancer-related deaths of men in the developed world, with an incidence of ≈170 per 100,000 in the United States (1). Well-established risk factors include age, ethnicity, and family history (2), and although it is believed that genetic factors contribute to disease etiology, until recently, there have been few validated genetic candidates associated with prostate cancer risk. To date, many hypothesis-based candidate gene studies have been performed, but none have been convincingly replicated; most were underpowered or had design problems (3).
Recent advances in human genomics, specifically the development of dense genotyping technologies, have provided the opportunity to scan the genomes of large numbers of individuals in genome-wide association studies (GWASs) rapidly. In the studies reported to date, single-nucleotide polymorphisms (SNPs) are scanned across the genome with a fixed panel of thousands of SNPs, chosen on the basis of regular intervals or SNPs chosen to represent independent variation (tag SNPs) (4). A key feature of well-designed GWAS is replication of the most promising findings (5). So far, GWAS have discovered over 400 genomic regions in over 75 diseases or human traits (6).
Several robust GWAS of prostate cancer have provided strong evidence for at least 14 independent loci that reach the statistical level of genome-wide significance (7–15). Interestingly, 2 independent groups have reported an association with a SNP (rs10993994) on chromosome 10q11.2, in close proximity to the MSMB gene (7, 14), which encodes β-microseminoprotein [also known as prostatic secretory protein 94 (PSP94)]. The gene product of MSMB is a member of the Ig binding factor family and is synthesized by epithelial cells in the prostate gland before secretion into the seminal plasma (16). Both PSP94, the gene product of MSMB, and its binding protein, PSPBP, have been reported to be serum markers for early detection of high-grade prostate cancer (17, 18), and it has been suggested that the gene product of MSMB could function as a tumor suppressor (19). During development of prostate cancer from early to late stages, the expression of MSMB progressively decreases (20–22). Loss of expression of MSMB is also associated with disease recurrence after radical prostatectomy (18). Recently, it was shown that MSMB is a target of a putative oncogene, EZH2, a member of the Polycomb group proteins, which acts as an inhibitor of the expression of MSMB.
To localize the reported association of SNP marker rs10993994 with prostate cancer risk further, we performed a first-generation fine mapping of the region and genotyped an additional 12 tag SNPs across a ≈65-kb region of chromosome 10 that contains the MSMB gene in 6,118 prostate cancer cases and 6,105 controls. The rs10993994 SNP remained the most significant of the 13 tested markers. Consequently, the analysis focused on characterizing the functional consequences of rs10993994 and the other common SNPs in the proximal promoter region. Thus, our study unites the functional and genetic mapping observations and provides the basis for further investigation of this common variant as a susceptibility allele for prostate cancer.
Results
Genotyping Completion and Concordance.
Genotyping completion rates were determined for each sample and assay for each study separately, the details of which are included in supporting information (SI) Table S1. Overall sample completion rates for informative loci were 93.08%, 95.67%, 98.93%, 96.85%, 96.84%, and 99.39% for the CPS-II, ATBC study, CeRePP study, HPFS, PHS, and PLCO trial, respectively. Concordance rates for known duplicates were consistently higher than 99%. Five SNPs (rs11006207, rs10826223, rs10993994, rs7904463, and rs10994675) were successfully genotyped for the CeRePP study. Estimates of MAFs are included in Table S2. Tests of deviations from Hardy-Weinberg Proportions (HWP) were performed for each SNP in the control individuals on a study-by-study basis (Table S1). No significant deviations (P < 0.001) from HWP were observed for SNPs in the CPS-II, ATBC study, CeRePP study, HPFS, and PLCO trial. However, 3 SNPs in the PHS deviated from HWP with a probability value less than 0.001 (rs10826223, rs17178655, and rs10994675). These departures could be attributable to chance or to bias introduced by whole-genome amplification of DNA before genotyping the PHS.
Association Results.
For each study and for all studies combined, tests of association were performed. The results for the combined analysis are shown in Table 1; individual results per study are included in Table S3. No evidence was observed to suggest that rs10993994 is associated with the type of prostate cancer (aggressive vs. nonaggressive disease). Overall, rs10993994 was most significantly associated with prostate cancer risk (P = 9.7 × 10−19). Four other SNP markers (rs11004422, rs7071471, rs4630240, and rs11006207) also exhibited a highly significant (P < 10−8) association (i.e., lower than the threshold for genome-wide significance) with prostate cancer risk, although it is unlikely that these represent associations that are independent of rs10993994 because of the strong LD within this region. The correlations between each of these 4 SNPs and rs10993994 are as follows: r2 = 0.68, 0.75, 0.34, and 0.75, respectively.
Table 1.
SNP association results for 6,118 prostate cancer cases and 6,105 controls from 5 studies in the CGEMS prostate cancer study
| Locus | Alleles | MAF | χ2, 2 df | P | Het OR | 95% CI | Hom OR | 95% CI |
|---|---|---|---|---|---|---|---|---|
| rs11004422 | T, C | 0.489 | 34.17 | 3.8E-08 | 1.08 | 0.97–1.20 | 1.42 | 1.25–1.60 |
| rs7071471 | C, T | 0.457 | 40.31 | 1.8E-09 | 1.11 | 1.00–1.23 | 1.49 | 1.31–1.69 |
| rs11593319 | G, T | 0.071 | 9.36 | 0.009 | 0.81 | 0.71–0.93 | 0.90 | 0.52–1.53 |
| rs10826075 | C, G | 0.248 | 9.28 | 0.010 | 1.08 | 0.98–1.19 | 1.30 | 1.09–1.56 |
| rs4630240 | C, T | 0.381 | 32.44 | 9.0E-08 | 0.79 | 0.72–0.87 | 0.71 | 0.62–0.82 |
| rs11006207 | C, T | 0.464 | 58.39 | 2.1E-13 | 1.14 | 1.05–1.24 | 1.49 | 1.34–1.65 |
| rs10826223 | G, A | 0.096 | 6.64 | 0.036 | 0.88 | 0.80–0.97 | 0.92 | 0.67–1.25 |
| rs10993994 | C, T | 0.407 | 82.95 | 9.7E-19 | 1.20 | 1.11–1.30 | 1.64 | 1.47–1.82 |
| rs7076948 | T, C | 0.376 | 8.09 | 0.012 | 1.03 | 0.94–1.13 | 1.21 | 1.06–1.39 |
| rs10994470 | G, A | 0.036 | 1.04 | 0.596 | 0.92 | 0.78–1.10 | 0.74 | 0.23–2.33 |
| rs7904463 | C, T | 0.327 | 5.77 | 0.056 | 1.06 | 0.98–1.14 | 1.15 | 1.02–1.30 |
| rs17178655 | G, A | 0.211 | 1.42 | 0.491 | 0.99 | 0.90–1.09 | 0.88 | 0.72–1.08 |
| rs10994675 | G, A | 0.416 | 9.12 | 0.011 | 1.07 | 0.99–1.16 | 1.17 | 1.06–1.30 |
Het, heterozygous; Hom, homozygous.
MSMB Promoter Polymorphism and Promoter Activity.
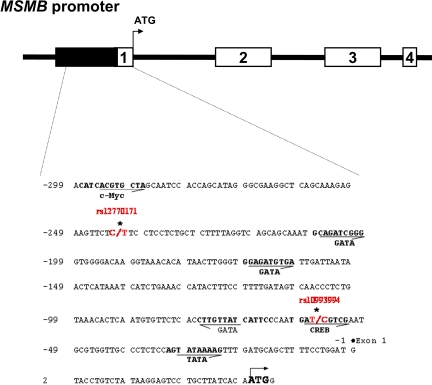
Sixty-seven cancer cell lines were chosen for a detailed analysis of sequence variation in the MSMB promoter region. Promoter polymorphisms were identified by bidirectional sequencing of PCR-generated clones of the promoter region from each cell line. Only the 2 previously identified common SNPs in the proximal promoter region were observed: rs12770171 at −242 and rs10993994 at −57, respectively (Fig. 1). The rs12770171 variant was not genotyped as a part of HapMap Phase 2. Otherwise, no previously unreported mutations were observed.
Fig. 1.
A schematic of the MSMB gene. Four exons are indicated by the numbered white rectangles. Nucleotide sequence of 336-bp MSMB gene, including 5′-region and Exon 1, is shown with the putative transcription factor binding sites in bold. Numbering is relative to the transcriptional start site at exon 1. Transcription orientations are indicated by arrows.
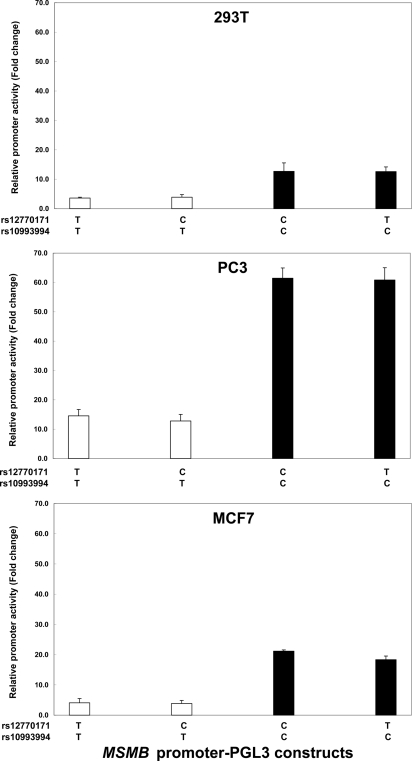
To elucidate whether sequence variations observed in the MSMB promoter region might influence promoter activity, 4 DNA fragments containing SNP rs12770171 and SNP rs10993994 (−299 to +36) were cloned into the pGL3 vector and the promoter activities were determined in 293T, PC3, and MCF7 cells. As shown in Fig. 2, the transcriptional activities of the MSMB promoter fragments with C at rs10993994 are higher than those of fragments with T at rs10993994. There was no effect of variation at SNP rs12770171 on promoter activity. The presence of a C residue at SNP rs10993994 is associated with a putative CREB binding site; therefore, the increased promoter activity of the MSMB promoter is likely attributable to the generation of a CREB site by the SNP. Conversely, the SNP rs12770171 is not associated with any predicted transcription factor binding sites, suggesting that it should not affect promoter activity.
Fig. 2.
The SNP rs10993994 contributes to MSMB promoter activity. Effect of MSMB promoter polymorphisms on promoter activity in 293T, PC3, and MCF7 cells is shown. Values represent fold increase of luciferase activity relative to empty pGL3 vector. The mean and SD of at least 3 independent experiments are shown.
CREB Binding Has a Strong Effect on MSMB Promoter Activity.
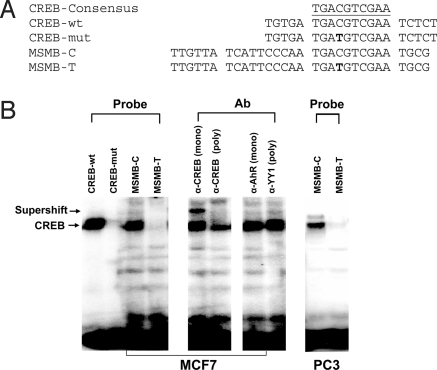
Modulation of MSMB promoter activity could be partially explained by variants in a putative CREB site (Fig. 1). The CREB transcription factor binding site is downstream of a GATA site and close to the TATA box. In vitro promoter assays have demonstrated that disruption of the CREB site is associated with decreased promoter activity. To investigate the effect of SNP rs10993994 within the proximal MSMB promoter region on CREB binding to the MSMB promoter, we performed electrophoretic mobility shift assay analysis with oligonucleotide probes containing the polymorphisms observed in the MSMB promoter region and nuclear protein extracts of MCF7 and PC3 cells (Fig. 3). Similar data was obtained for 293T cells (Fig. S1). As shown in Fig. 3B, the allele C of rs10993994 has increased promoter activity (MSMB-C), and thus stronger CREB binding, whereas the allele already shown to have weak promoter activity (MSMB-T) had undetectable CREB binding. The addition of anti-CREB antibody consistently reduced the intensity of the oligonucleotide-protein complex (Fig. 3B, Right). These results are consistent with the proposed modulation of the MSMB promoter activity by CREB binding.
Fig. 3.
Binding of CREB transcription factor to the promoter of the MSMB gene. Electrophoretic mobility shift assay (EMSA) analysis of the CREB binding site corresponding to the SNP rs10993994 observed in MSMB promoter region. (A) Oligonucleotides used for EMSA analysis. The sense strand of oligonucleotide probes corresponding to the predicted CREB binding site of the MSMB promoter is shown. (B) EMSA analysis performed on MCF7 nuclear extracts with probes indicated in A. The right panels show supershift analysis of the MSMB-C probe from MCF7 extracts in the presence of specific Abs.
Quantitation of MSMB mRNA Expression Levels.
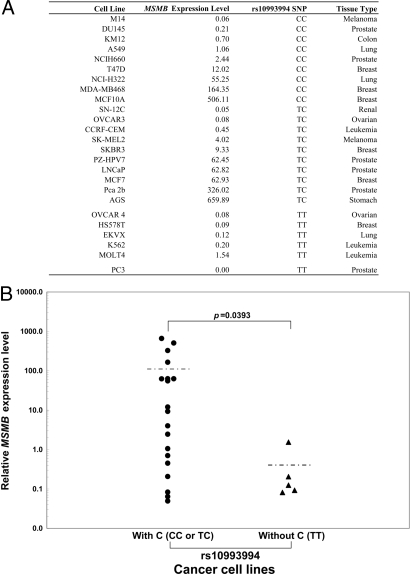
To confirm the predicted effect of the observed changes in promoter activity associated with SNP rs10993994 in the MSMB promoter, the mRNA expression levels of the MSMB gene were measured by real-time quantitative RT-PCR. Fig. 4A, Table 2, and Table S4 show the result of quantitation of MSMB mRNA expression in 65 cancer cell lines with MSMB genotypes that have either strong (with C for rs10993994) or weak (without C for rs10993994) promoter activity based on the transfection data shown in Fig. 2. Nineteen cancer cell lines had detectable MSMB mRNA expression, whereas the mean of MSMB mRNA expression level with rs10993994-C was significantly higher than that with rs10993994-T (Fig. 4 and Table 2).
Fig. 4.
Quantitative analysis of MSMB mRNA expression by Taqman real-time RT-PCR. (A) List of cell lines with detectable MSMB mRNA expression. The relative mRNA expression level of MSMB gene was normalized by the following formula: (copy number of target gene)/(copy number of 18S rRNA) × 107. (B) Distribution of MSMB mRNA expression relative to with or without C at SNP rs10993994.
Table 2.
Detection of MSMB expression in cancer cell lines
| rs10993994 SNP | Number of cell lines |
MSMB+ rate, % | Mean MSMB expression level in MSMB+ cell lines, 95% CI | ||
|---|---|---|---|---|---|
| MSMB+ | MSMB− | Total | |||
| With C (CC or TC) | 19 | 31 | 50 | 38.00 | 101.59 (10.43–192.75) |
| Without C (TT) | 5 | 10 | 15 | 33.33 | 0.41 (−0.38–1.20) |
Discussion
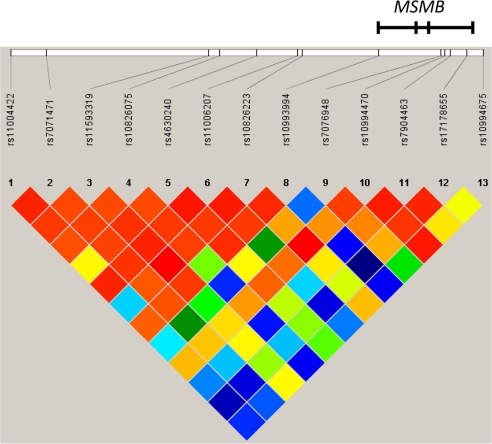
To explore the contribution of genetic variation in a locus on 10q11.2 identified by 2 independent GWAS for prostate cancer further, we have performed mapping of the region, including the MSMB gene (Fig. 5), using a highly correlated tag SNP approach across the region (6, 7). After genotyping 13 tag SNPs from a ≈65-kb region of LD flanking the initial SNP marker (rs10993994), the T allele of rs10993994 remained the most highly significant SNP in the association testing of 6,118 prostate cancer cases and 6,105 controls.
Fig. 5.
Location of and LD (D') among 13 SNPs genotyped for association with prostate cancer. Data shown are from 6,105 prostate cancer control individuals. The region shown is chr10:51168330-51234020.
Although it is possible that the functional variant is in strong LD with one of the established markers, we present evidence that rs10993994 probably contributes to the association signal. Promoter constructs with the C allele of rs10993994 display strong promoter activity, whereas the T allele destroys the binding site of the CREB transcription factors. Our work extends the findings of 2 studies, both of which suggest that the T allele of rs10993994 is associated with lower expression levels of MSMB than the C allele in vitro using luciferase assays; the promoter SNP was singled out in a rapid screen of a large number of candidate promoters by Buckland et al. (23), whereas Chang et al. (24) recently reported this finding in an exploration of the region. We have further investigated the allelic differences between the T and C variants of rs10993994 in several important ways. The proximal promoter region was investigated by (i) sequence analysis of exons, which did not identify common SNPs not previously recognized (14), and (ii) deep sequence analysis of the proximal promoter region in more than 60 cancer cell lines reported herein, which identified 1 SNP that was not reported in HapMap or well tagged by HapMap Phase 2. In functional studies of the previously unreported SNP, the results were unremarkable, thus making it highly unlikely that this variant directly contributes to prostate cancer susceptibility.
It is notable that in tumor cell lines of diverse origin, the C allele appears to be required (although not sufficient) for expression. Because the expression of MSMB is principally found primarily in normal prostate and salivary gland tissue, we speculate that TT individuals have lower expression, especially under conditions in which CREB is induced. We detected high expression of MSMB in a sample of normal prostate tissue (an average value of 8,168) and a 10-fold lower level in prostate tumor tissue (Table S4). This preliminary observation suggests that it will be important to investigate the relation between germline genotype and tumor expression of MSMB.
There are likely to be other proximal regulatory elements for the MSMB gene; in fact, a ≈273-bp GC-rich region 2.8 kb upstream of the ATG is methylated in some nonexpressing cell lines, and azacytidine treatment leads to partial derepression of expression (data not shown). Three lines of evidence suggest that rs10993994 is functionally important and could partially account for the observed association with prostate cancer susceptibility: (i) MSMB expression is reported to be reduced during the progression of prostate cancer, (ii) loss of MSMB is associated with cancer recurrence after radical prostatectomy, and (iii) lower expression of MSMB is associated with the risk allele. To characterize the promoter region of MSMB fully, however, all common genetic variations will need to be cataloged by deep resequencing across a larger region (25). This approach will determine whether additional variants, namely SNPs or insertions/deletions that lie on a haplotype with rs10993994, could also influence expression of MSMB.
Materials and Methods
Definition of the Region and SNP Selection for Fine Mapping.
The pattern of linkage disequilibrium (LD) was assessed in a ≈65-kb region (chr10: 51168330–51234020; see Fig. 5) flanking the strongest signal, rs10993994, using genotype data from HapMap Phase 2 unrelated CEPH Utah CEU subjects (n = 120 chromosomes). Although the MSMB gene itself appears to lie outside the block of LD that contains rs10993994, we included this region for the selection of tag SNPs because of its proximity to MSMB and the fact that it resides in the proximal promoter.
A conservative tagging strategy was used to maximize coverage of the region of LD (chr10: 51168330–51234020); tag SNPs were selected from HapMap Phase 2 CEU unrelated subjects based on an r2 ≥0.975, minimum minor allele frequency (MAF) ≥0.05. This strategy was chosen to maximize coverage across this region, because only very highly correlated SNPs are not chosen as tags. The 2 most promising SNPs associated with prostate cancer risk previously cited were obligate inclusions (rs10993994 and rs11006207; ref. 14), treated as in the binning process. A total of 15 SNPs met these criteria: for 13 SNPs (rs10826075, rs10826223, rs10993994, rs10994470, rs10994675, rs11004422, rs11006207, rs11593319, rs17178655, rs4630240, rs7076948, rs7904463, and rs7071471), TaqMan (Applied Biosystems) assays were successfully designed and validated; for 2 SNPs (rs2072701 and rs7081532), assays were not successfully designed and/or manufactured. However, rs7081532 is monitored by rs11006027 at an r2 at 0.95, whereas rs2072701 is not adequately (r2 >0.8) monitored by any other SNP in the panel. Overall, these 13 SNPs monitor a total of 49 HapMap SNPs in Phase 2.
Subjects: Genotyping.
Genotype analysis included a total of 6,118 prostate cancer cases and 6,105 controls from the 5 studies in the Cancer Genetic Markers of Susceptibility (CGEMS) initial GWAS and 4 follow-up studies (15). The initial study consisted of 1,175 cases and 1,100 controls from the Prostate, Lung, Colorectal, and Ovarian (PLCO) trial. The follow-up studies included 1,784 cases and 1,786 controls from the American Cancer Society's Cancer Prevention Study-II (CPS-II), 946 cases and 935 controls from the Alpha-Tocopherol Beta Carotene (ATBC) study, 606 cases and 621 controls from the Health Professionals Follow-up Study (HPFS), and 656 cases and 656 controls from the French Prostate Case-Control (CeRePP) study. The CPS-II, ATBC study, HPFS, PLCO trial, and CeRePP study designs have been described elsewhere (15). For the present study, an additional 938 cases and 983 controls were included from the Physician's Health Study (PHS).
The PHS was a randomized trial of aspirin and β-carotene for cardiovascular disease and cancer among 22,071 US male physicians aged 40–84 years at randomization; none had a cancer diagnosis at baseline. From 1982 to 1984, blood samples were collected from 14,916 physicians before randomization. Participants are sent yearly questionnaires to ascertain end points. Whenever a physician reports cancer, permission is requested to obtain the medical records, and cancers are confirmed by pathology report. Death certificates and pertinent medical records are obtained for all deaths. Follow-up for nonfatal outcomes in the PHS is over 97% complete, and follow-up for mortality is over 99% complete (26).
Genotyping.
Thirteen SNPs were genotyped for 5 studies, whereas only 5 SNPs were genotyped in the CeRePP study because of DNA depletion; these included rs11006207, rs10826223, rs10993994, rs7904463, and 10994675. TaqMan probes, primers, and assay conditions are available on the SNP500Cancer database (http://snp500cancer.nci.nih.gov) (27).
Completion, concordance, MAF estimations, deviations from Hardy-Weinberg proportions (HWPs), pair-wise LD, and tag SNP selection were all computed using the Genotype Library and Utilities (GLU) software package (http://code.google.com/p/glu-genetics).
Association Analyses.
Association testing was performed with the GLU software package using unconditional logistic regression, adjusted for age in 5-year categories, study, and center for the 2 studies in which the center for recruitment was available (i.e., PLCO trial and CeRePP study). Three significant principal components were derived for stage 1, whereas 4 significant principal components were used for stage 2. We analyzed each study separately as well as the combined studies for both the dichotomous (case vs. control) and the trichotomous (aggressive case vs. nonaggressive case vs. control) phenotypes. The odds ratios for each SNP were modeled multiplicatively. Probability values were computed based on a genotype score test with a maximum of 2 df for all cases compared with controls.
Generation of Luciferase Reporter Plasmids.
Promoter fragments were generated by PCR using unique oligonucleotide primers: forward primer MSMBpro-For 5′-ACATCACGTGCTAGCAATCCAC-3′ and reverse primer MSMBpro-Rev 5′-CCATTGTGATAAGCAGGACTCC-3′, corresponding to the promoter region from −299 to +36 relative to the start codon of the MSMB gene. PCR reactions were performed in a volume of 25 μL containing 100 ng of genomic DNA and 20 pmol of each primer using Platinum PCR SuperMix (Invitrogen). Thermal cycling conditions included 35 cycles of 94 °C for 20 s, 59 °C for 30 s, and 73 °C for 2 s. PCR products were cloned into the TOPO-TA vector (Invitrogen) according to the manufacturer's directions. Subclones were sequenced to identify the promoter allele of MSMB gene using either the M13 reverse or T7 primer. Putative transcription factor binding sites were identified in silico using the TFSEARCH web site (http://www.cbrc.jp/research/db/TFSEARCH.html) and using a Genomatix package.
Inserts were excised with the restriction endonucleases, SacI/XhoI, directionally cloned into a transfection vector, pGL3 (Promega), and were verified by bidirectional sequencing. Sequence analysis was performed with the SeqWeb package at the National Cancer Institute (NCI)-Frederick supercomputing center and Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0.
Cell Transfection and Luciferase Assays.
Cell lines 293T (human embryo kidney), PC3 (prostate cancer), and MCF7 (breast cancer) were used for transfection experiments. The cells were plated at 1 × 105 cells per well in a 6-well plate the day before transfection and incubated overnight at 37 °C in 5% CO2. For each well, 5 μL of FuGENE 6 transfection reagent (Roche) was diluted in 95 μL of growth medium without serum and incubated at room temperature for 5 min. The DNA mixture containing 1 μg of the specific reporter construct plus 1 ng of the Renilla luciferase pRL-SV40 control DNA was then added to each well and incubated at room temperature for 20 min.
Luciferase activity was assayed at 48 h using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Measurement of the firefly luciferase activity of the MSMB promoter constructs was normalized relative to the activity of the Renilla luciferase produced by the pRLSV40 control vector, and each construct was tested in triplicate in at least 3 independent experiments.
Electrophoretic Mobility Shift Assay of CREB Binding to the MSMB Promoter Element.
Nuclear extracts were prepared from the MCF7, 293T, and PC3 cell lines using the CellLytic NuCLEAR extraction kit (Sigma-Aldrich). Protein concentration was measured with a Bio-Rad protein assay, and samples were stored at −70 °C until use. Four double-stranded DNA oligonucleotide probes corresponding to the predicted CREB binding sequence of the MSMB promoter element were synthesized (Fig. 3A, sense strand shown). Sense and antisense oligonucleotides were annealed to generate double-stranded oligonucleotides and labeled with [α-32P]dCTP (3,000 Ci/mmol; Perkin-Elmer) using the Klenow fragment of DNA polymerase I (Invitrogen). 32P-labeled double-stranded oligonucleotides were purified using mini Quick Spin Oligo Columns (Roche GmbH). DNA protein-binding reactions were performed in a 20-μL mixture containing 5 μg of nuclear protein and 1 μg of poly(dI-dC)poly(dI-dC) (Sigma-Aldrich) in 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, and 10 mM Tris-HCl (pH 7.5). After a 10-min incubation on ice, samples were incubated with 1 μL of 32P-labeled oligonucleotide probe (20,000 cpm) at room temperature for 20 min and then loaded on a 5% polyacrylamide gel (37:5:1). Electrophoresis was performed in 0.5 × Tris/Borate/EDTA (TBE) buffer for 2 h at 130 V, and the gel was visualized by autoradiography after 1 day of exposure at −70 °C. For antibody supershift experiments, nuclear extracts were incubated with 2 μL of antibody for 1 h on ice before the addition of 32P- labeled DNA probe. After the addition of labeled DNA probe, the binding reaction was incubated for an additional 20 min at room temperature. The antibodies used were anti-CREB-1 (24H4B, mouse monoclonal; C-21, rabbit polyclonal) from Santa Cruz Biotechnology Company. The mouse monoclonal to Ah Receptor (C-4) and rabbit polyclonal to Yin Yang-1 (YY1) (C-20) (Santa Cruz Biotechnology Company) were used as antibody controls.
Real-Time Quantitative RT-PCR.
Total cellular RNA extracted from NCI 60 cancer cell lines was received from the resource of the Developmental Therapeutics Program (DTP), Information Technology Branch, NCI. The cancer cell lines AGS, MCF10A, NCIH660, MDA-PCa-2b, SKBR3, PZ-HPV7, and LNCaP were obtained from American Type Culture Collection (ATCC). Total RNA was further purified using the RNeasy Clean Up Kit (QIAGEN) according to the manufacturer's instructions. cDNA synthesis was carried out using Random Hexamer primer, Taqman Reverse Transcription Reagents kit (Applied Biosystems).
The Taqman Gene Expression Assay primer and probe (FAM-labeled) set (Applied Biosystems) was used for real-time quantitative PCR analysis of MSMB (assay ID: Hs00159303_ml). A TaqMan Gene Expression Assay mix of primer and probe (VIC-labeled) of 18S rRNA was used as an internal control. The PCR reactions were performed in a volume of 10 μL containing 8 ng of cDNA, 1 × Master Mix (TaqMan Universal PCR Master Mix; ABI), 900 nM each primer, and 200 nM each probe, respectively. All assays were performed in triplicate, were repeated 3 or more times, and each plate contained a positive quality control sample from human prostate normal tissue (Clontech Laboratories).
The standard curves were generated using a dilution series of plasmids containing full-length cDNA of MSMB (GenBank accession no. BC005257.1; ATCC). The copy number of plasmid cDNA was calculated by optimal density according to the exact molar mass derived from the sequences. Serial dilutions were made to obtain 101 to 107 copies. The observed efficiency of the standard curve for MSMB and 18S rRNA in this study is greater than 99%, and r2 is greater than 0.99. The relative mRNA expression level of MSMB was normalized by the following formula: (copy number of target gene)/(copy number of 18S rRNA) × 107.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902104106/DCSupplemental.
References
- 1.Crawford ED. Epidemiology of prostate cancer. Urology. 2003;62:3–12. doi: 10.1016/j.urology.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Johns LE, Houlston RS. A systematic review and meta-analysis of familial prostate cancer risk. BJU Int. 2003;91:789–794. doi: 10.1046/j.1464-410x.2003.04232.x. [DOI] [PubMed] [Google Scholar]
- 3.Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13:R103–R121. doi: 10.1093/hmg/ddh072. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 4.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 6.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 8.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 9.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 11.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 12.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 15.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 16.Mbikay M, et al. Molecular cloning and sequence of the cDNA for a 94-amino-acid seminal plasma protein secreted by the human prostate. DNA Cell Biol. 1987;6:23–29. doi: 10.1089/dna.1987.6.23. [DOI] [PubMed] [Google Scholar]
- 17.Bjartell AS, et al. Association of cysteine-rich secretory protein 3 and beta-microseminoprotein with outcome after radical prostatectomy. Clin Cancer Res. 2007;13:4130–4138. doi: 10.1158/1078-0432.CCR-06-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves JR, Dulude H, Panchal C, Daigneault L, Ramnani DM. Prognostic value of prostate secretory protein of 94 amino acids and its binding protein after radical prostatectomy. Clin Cancer Res. 2006;12:6018–6022. doi: 10.1158/1078-0432.CCR-06-0625. [DOI] [PubMed] [Google Scholar]
- 19.Beke L, Nuytten M, Van Eynde A, Beullens M, Bollen M. The gene encoding the prostatic tumor suppressor PSP94 is a target for repression by the Polycomb group protein EZH2. Oncogene. 2007;26:4590–4595. doi: 10.1038/sj.onc.1210248. [DOI] [PubMed] [Google Scholar]
- 20.LaTulippe E, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 21.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 22.Stanbrough M, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 23.Buckland PR, et al. Strong bias in the location of functional promoter polymorphisms. Hum Mutat. 2005;26:214–223. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- 24.Chang BL, et al . Fine mapping association study and functional analysis implicate a SNP in MSMB at 10q11 as a causal variant for prostate cancer risk. Hum Mol Genet. 2009;18:1368–1375. doi: 10.1093/hmg/ddp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeager M, et al. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Hum Genet. 2008;124:161–170. doi: 10.1007/s00439-008-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J, et al. Prediagnostic body-mass index, plasma C-peptide concentration, and prostate cancer-specific mortality in men with prostate cancer: A long-term survival analysis. Lancet Oncol. 2008;9:1039–1047. doi: 10.1016/S1470-2045(08)70235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Packer BR, et al. SNP500Cancer: A public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.