Abstract
The disulfide bond between Cys-110 and Cys-187 in the intradiscal domain is required for correct folding in vivo and function of mammalian rhodopsin. Misfolding in rhodopsin, characterized by the loss of ability to bind 11-cis-retinal, has been shown to be caused by an intradiscal disulfide bond different from the above native disulfide bond. Further, naturally occurring single mutations of the intradiscal cysteines (C110F, C110Y, and C187Y) are associated with retinitis pigmentosa (RP). To elucidate further the role of every one of the three intradiscal cysteines, mutants containing single-cysteine replacements by alanine residues and the above three RP mutants have been studied. We find that C110A, C110F, and C110Y all form a disulfide bond between C185 and C187 and cause loss of retinal binding. C185A allows the formation of a C110–C187 disulfide bond, with wild-type-like rhodopsin phenotype. C187A forms a disulfide bond between C110 and C185 and binds retinal, and the pigment formed has markedly altered bleaching behavior. However, the opsin from the RP mutant C187Y forms no rhodopsin chromophore.
Keywords: G-protein-coupled receptors, signal transduction, 11-cis-retinal, misfolding, retinitis pigmentosa
Early work indicated the important role of the intradiscal domain in folding in vivo and function of rhodopsin (1). The presence of a disulfide bond between Cys-110 and Cys-187 was established (2) and it was shown later to be required for optimal signal transduction (3). A disulfide bond between two cysteines in the extracellular domain at positions equivalent to the above two cysteines in rhodopsin is known now to be conserved in most of the known G-protein-coupled receptors (4, 5). The presence of a disulfide bond indicated a tertiary structure in the intradiscal domain of rhodopsin. Strong support for this conclusion was forthcoming from extensive designed mutagenic studies as well as from studies of a large number of naturally occurring point mutations in rhodopsin that are associated with retinitis pigmentosa (RP) (6–11). Mutations in the intradiscal domain were shown to cause total or partial misfolding in vivo in rhodopsin, misfolding being operationally defined as the loss of ability to bind 11-cis retinal (6). Further, evidence was presented that misfolding results from the formation of a disulfide bond in the intradiscal domain different from the normal Cys-110—Cys-187 disulfide bond (12). Recently, Oprian and colleagues demonstrated the formation of a disulfide bond under certain conditions between Cys-185 and Cys-187 in a rhodopsin mutant reconstituted from two appropriate rhodopsin fragments (13).
Further studies showed that although replacements of Cys-110 and Cys-187 by serine residues prevented folding of rhodopsin to a functional structure, corresponding replacements by alanine residues allowed the formation of the correctly folded dark-state structure. This result paralleled the earlier findings on the small-protein bovine pancreatic trypsin inhibitor, and, by analogy, suggested a globular structure for the intradiscal domain in rhodopsin (3). Recently, point mutations at the intradiscal cysteines (C110F, C110Y, and C187Y) in rhodopsin associated with RP have been discovered (14–17) and, in addition, mutation of a conserved cysteine equivalent to Cys-187 in cone opsins (C203R) has been shown to cause color vision deficiencies (18, 19).
With the aim of clarifying the effects of the above mutations on folding in the intradiscal domain, as well as to elucidate the nature of the disulfide bond involved in misfolding of rhodopsin (12), the work herein reported was undertaken. Single cysteine-to-alanine replacements have now been carried out at the three intradiscal positions and consequences of these replacements as well as those of the RP mutations C110F, C110Y, and C187Y have been studied (Fig. 1). The mutants C110A, C110F, and C110Y all form a C185–C187 disulfide bond and result in lack of retinal binding. The mutant C185A allows correct folding of rhodopsin by the formation of the normal C110–C187 disulfide bond. Finally, the mutant C187A forms a disulfide bond between C110 and C185 and binds retinal to form a pigment that has a markedly altered bleaching behavior. The RP mutant C187Y forms no rhodopsin chromophore.‡
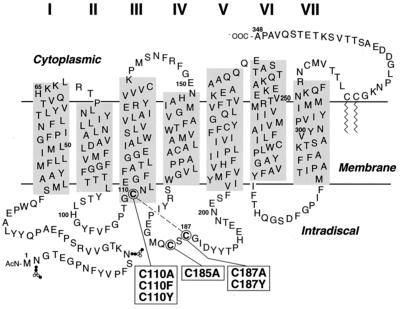
Figure 1.
A secondary-structure model of bovine rhodopsin showing the transmembrane domain with lengths of individual helices determined from electron microscopy by Unger et al. (23) and Baldwin et al. (24). The horizontal line depicting the membrane aqueous boundary on the cytoplasmic face is derived from EPR data (ref. 25 and unpublished work). The three intradiscal cysteines are circled. The mutants studied in this work are shown in boxes connected to the intradiscal cysteines. The dashed line between C110 and C187 indicates the disulfide bond in WT rhodopsin.
MATERIALS AND METHODS
Materials.
The reagents as well as the buffers used were the same as those described in the accompanying paper (20) with the following additions: Buffer I, 2 mM NaH2PO4 (pH 6.0) containing 0.05% dodecyl maltoside (DM); Buffer J, 2 mM NaH2PO4 (pH 6.0)/0.05% DM/100 μM rhodopsin C-terminal nonapeptide; Buffer K, Buffer J containing 150 mM NaCl. 4,4′-dithiodipyridine [pyridine disulfide (PDS)] was from Aldrich.
UV–visible (UV–vis) absorption spectroscopy and the rates of metarhodopsin II decay were measured as described previously (11, 21).
Construction of the Vectors for the Mutant Opsins Genes and Their Expression.
The mutations were introduced by restriction fragment replacement in the pMT4 vector containing the synthetic rhodopsin gene vector by using synthetic DNA duplexes containing the changed codons. For the C110 mutants, the native cysteine codon, TGC, was replaced by TTC (Phe), TAC (Tyr), or GCC (Ala). The duplexes containing these codons were inserted between RsrII–XhoI sites (nucleotides 322 to 340 in the opsin gene). The C185 (TGC) codon was replaced by GCC (Ala), and the duplex was inserted between XbaI–ClaI sites (nucleotides 532 to 555). Similarly the cysteine codon at C187 position was replaced by TAC (Tyr) or GCC (Ala). The procedures for expression in COS-1 cells have been described previously (22).
Retinal Binding in Vivo.
The procedure for treatment of COS-1 cells expressing the mutants with 11-cis-retinal was as described in the accompanying paper (20), except that the incubation with retinal was for 16 hr. The 1D4-Sepharose column was washed with 100 bed volumes of buffer C, and subsequent differential salt elutions were performed by using buffers I, J, and K. The column was washed with 100 bed volumes of buffer I and then eight fractions (300 μl) were eluted with buffer J and seven (300 μl) with buffer K.
Retinal Binding in Vitro and Purification of the Constituted C187A Rhodopsin.
These were as described in the accompanying paper (20).
Sulfhydryl Group Titrations with PDS.
The procedure has been described previously (22). A solution of the protein (2 μg) in buffer I was treated with PDS at a final concentration of 25 μM in SDS (final concentration, 0.5%). The final volume of the reaction mixtures was 250 μl. The reference cuvette contained an identical solution without rhodopsin. The reactions were monitored by UV–vis spectroscopy (250–650 nm) until a plateau was reached at A323. Quantitation of cysteines labeled per rhodopsin was by methods previously described (22). Results shown are the mean ± standard error from at least two separate experiments by using proteins from two separate transfections.
RESULTS AND DISCUSSION
The Mutants C110A, C110F, and C110Y Are Misfolded.
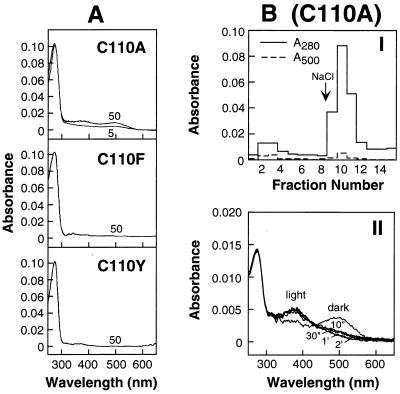
Fig. 2A shows the results of retinal treatment in vivo of the COS-1 cells expressing the mutants C110A, C110F, and C110Y. The mutant C110A at 50 μM shows mostly lack of retinal binding, although a very small amount of rhodopsin-like absorption at 500 nm is observed. The mutant opsins from C110F and C110Y show no retinal binding and are completely misfolded. Further characterization of the protein obtained from the mutant C110A after treatment with retinal is documented in Fig. 2B. In I is shown the elution pattern from 1D4-Sepharose column. The first step, elution at pH 6 with no salt, elutes a small amount of pigment, a behavior expected for the constituted rhodopsin. Most of the 280-nm-absorbing nonchromophore-forming material elutes when salt is present. II shows the UV–vis spectra of the small amount of constituted rhodopsin in the dark and after illumination. These spectra confirm the formation of the rhodopsin-like pigment that undergoes bleaching quite normally to form the 380-nm-absorbing metarhodopsin II-like material.
Figure 2.
Characterization of the expressed mutants C110A, C110Y, and C110F. (A) UV–vis absorption spectra of the mutants isolated after treatment of COS-1 cells with 11-cis-retinal. As indicated, COS-1 cells expressing the opsin from C110A were treated with 5 and 50 μM retinal; the opsins from C110F and C110Y were treated with 50 μM retinal (Methods). Elution of the proteins from 1D4-Sepharose was with Buffer A containing 0.05% DM and 100 μM peptide. (B) I, C110A: Elution profile from 1D4-Sepharose of the in vivo retinal-treated (50 μM) C110A mutant opsin. Elution of the first eight fractions (300 μl each) was with Buffer J; the following seven fractions were eluted with Buffer K, which contains salt. II: Bleaching behavior of the fraction (no. 2) eluted with Buffer J (no salt) in B. The UV–vis spectrum showed an A280/A500 ratio of 3.9, indicating there was contamination from unconstituted opsin.
The conclusion that the opsins from C110A, C110F, and C110Y form C185–C187 disulfide bonds is supported by the previous findings of Oprian and coworkers (13), who demonstrated the formation of the C185–C187 disulfide bond in the C-terminal fragment, amino acids 143–348, in their study of rhodopsin reconstitution from fragments. Titration of sulfhydryl groups in the separated fractions from C110A (Fig. 2BI) under denaturing conditions confirmed the absence of a disulfide bond in the retinal-binding fraction and the presence of one disulfide bond in the nonretinal-binding fraction (Table 1). Previously, RP mutants in the intradiscal domain were found to cause partial (e.g., P23H, D190A) or total (e.g., G188R) misfolding (9). The RP mutations, C110F and C110Y, are now added to the group that cause complete misfolding.
Table 1.
Titration of sulfhydryl groups in WT and mutant opsins under denaturing conditions by using PDS
| Mutant | Fraction* | mol of PDS per mol of opsin
|
|
|---|---|---|---|
| Found | Expected number for (1) or (0) disulfide bond | ||
| WT | F1 | 6.1 ± 0.2 | 6.0 (1) |
| C110A | F1 | 6.0 ± 0.2 | 7.0 (0) |
| F2 | 4.5 ± 0.1 | 5.0 (1) | |
| C185A | F1 | 4.9 ± 0.3 | 5.0 (1) |
| C187A | F1 | 4.9 ± 0.4 | 5.0 (1) |
| F2 | 5.1 ± 0.3 | 5.0 (1) | |
F1 eluted with buffer J (no salt); F2 eluted with buffer K (containing salt).
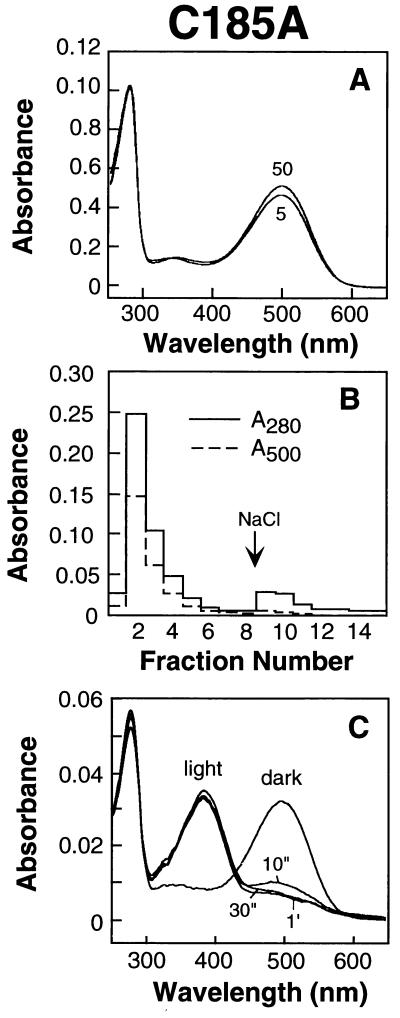
The Mutant C185A Allows Correct Folding and the Formation of the Native C110–C187 Disulfide Bond.
Fig. 3A shows the UV–vis absorption spectra of pigments formed after treatment of COS-1 cells expressing the mutant C185A with 5 and 50 μM retinal. The extent of binding of retinal at the two concentrations is similar and compares with that observed for WT opsin [accompanying paper (20)]. C shows the UV–vis spectra of the purified rhodopsin mutant in the dark and after illumination. The dark spectrum as characterized by A280/A500 ratio and the spectrum after illumination to form the metarhodopsin II intermediate are identical to those established for WT rhodopsin. The elution pattern shown in B from 1D4-Sepharose is also typical for rhodopsin. Most of the pigment as measured at A280, and A500 is eluted at pH 6.0 in the absence of salt. Finally the formation of a disulfide between C110 and C187 is confirmed by the titration of sulfhydryl groups with PDS (Table 1). It is noted that C185 in WT rhodopsin is normally silent to sulfhydryl reagents but is titratable under denaturing conditions. Evidently, alanine at position 185 can substitute for cysteine in the formation of the intradiscal structure.
Figure 3.
Characterization of the mutant rhodopsin C185A. (A) UV–vis spectra of the rhodopsin isolated after treatment of COS-1 cells expressing the opsin mutant in vivo with 5 and 50 μM retinal. Elution from the 1D4-Sepharose column was with Buffer A containing 0.05% DM plus 100 μM peptide. (B) Elution profile from 1D4-Sepharose of the mutant rhodopsin obtained as in A, first with Buffer J (no salt) (first eight fractions) followed by elution with Buffer K (containing salt). Shown are the A280 and A500 profiles of the eluted fractions. (C) Bleaching behavior of the second fraction (B). The dark UV–vis spectrum shown had an A280/A500 ratio of 1.7.
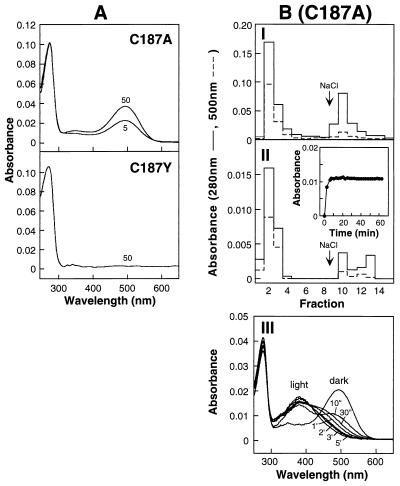
The Mutation C187A but Not C187Y Allows the Formation of a Rhodopsin-Like Pigment.
Fig. 4A shows the behavior of the opsins formed from the mutants C187A and C187Y on treatment in vivo with 11-cis-retinal. The mutant C187A shows concentration-dependent binding of retinal to form a rhodopsin-like pigment, although the mutant C187Y shows no binding of retinal. Fig. 4B shows experiments on characterization of the pigment formed from C187A. Thus, I confirms the elution of A280/A500 material from 1D4-Sepharose at pH6 without salt. More material is eluted with salt. However, pigment formation from the C187A opsin can be made to go to completion by in vitro constitution (II Inset and chromatographic separation on 1D4-Sepharose) by using the method developed in the accompanying paper (20). Thus, the results (Fig. 4A and II of Fig. 4B) show that the binding of retinal to the C187A opsin occurs with reduced affinity but can be driven to completion. III in Fig. 4B shows that the pigment formed from the mutant opsin C187A has abnormal bleaching behavior. Titration of the sulfhydryl groups in the mutant C187A confirmed the presence of a disulfide bond (Table 1). A corresponding experiment to test the presence of a disulfide bond in the mutant C187Y has so far not been possible because of the very low expression level of the mutant opsin in COS-1 cells.
Figure 4.
Characterization of the mutant rhodopsin, formed from expressed opsin C187A, and opsin C187Y. (A) UV–vis absorption spectra of products after treatment of COS-1 cells with 11-cis-retinal at concentrations of 5 μM and 50 μM (C187A) and 50 μM (C187Y). Elution from 1D4-Sepharose column was with Buffer A containing 0.05%DM plus 100 μM peptide. (BI) Elution profile from 1D4-Sepharose of the in vivo-treated (50 μM) C187A. Elution of the first eight fractions (300 μl) was with Buffer J; the following seven fractions were with Buffer K containing salt. II. Elution profile from 1D4-Sepharose of the in vitro-treated (33.3 μM) C187A opsin. As described in I, the first eight fractions (300 μl) were eluted with Buffer J; the following seven fractions were with Buffer K containing salt. The procedure for retinal binding in vitro of the C187A opsin (Inset) was as described (20). III. Bleaching behavior for the second fraction eluted with Buffer J (no salt) in I. The dark UV–vis spectrum shown had an A280/A500 ratio of 1.7.
CONCLUSIONS
Work with the mutants C110A, C110F, and C110Y showed that these result in the formation of a C185-C187 disulfide bond, which causes loss of ability to bind retinal. C185A allowed the formation of WT-like tertiary structure in the intradiscal domain and formed the C110–C187 disulfide bond. The mutant C187A formed a disulfide bond between C110 and C185 but allowed retinal binding to form a pigment with abnormal bleaching behavior. Further confirmation of the identity of all the disulfide bonds encountered in the intradiscal domain and in particular that between C110 and C185 is being sought by mass spectrometric analysis.
Acknowledgments
We thank Professor Daniel Oprian for reading the manuscript. We have benefited greatly from discussions with Professor U. L. Raj Bhandary (Biology Department, Massachusetts Institute of Technology) and our laboratory colleagues. We thank Ms. Judy Carlin for her assistance in the preparation of the manuscript. This work was supported by the National Eye Institute, grant R01-EY11716, and in part by a grant from the National Institute of General Medical Sciences (NIGMS) R01-GM28289, from the National Institutes of Health. J.K.-S. is the recipient of a Howard Hughes Predoctoral Fellowship. J.H. is the recipient of a Howard Hughes Medical Institute Physician Postdoctoral Fellowship.
ABBREVIATIONS
- RP
retinitis pigmentosa
- PDS
4,4′-dithiodipyridine
- DM
dodecyl maltoside
- WT
wild type
- UV–vis
UV–visible. Mutant rhodopsin with amino acid substitutions are designated by the one-letter abbreviations for amino acids. The amino acid to the left of the residue number is the original, whereas that to the right is the substituted amino acid
Footnotes
This is paper no. 35 in the series “Structure and Function in Rhodopsin.” Paper no. 34 is ref. 20.
References
- 1.Karnik S S, Sakmar T P, Chen H-B, Khorana H G. Proc Natl Acad Sci USA. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karnik S S, Khorana H G. J Biol Chem. 1990;265:17520–17524. [PubMed] [Google Scholar]
- 3.Davidson F F, Loewen P C, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4029–4033. doi: 10.1073/pnas.91.9.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strader C D, Fong T M, Tota M R, Underwood D, Dixon R A F. Annu Rev Biochem. 1994;63:101–132. doi: 10.1146/annurev.bi.63.070194.000533. [DOI] [PubMed] [Google Scholar]
- 5.Dohlman H G, Thorner J, Caron M G, Lefkowitz R J. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- 6.Doi T, Molday R S, Khorana H G. Proc Natl Acad Sci USA. 1990;87:4991–4995. doi: 10.1073/pnas.87.13.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushal S, Ridge K D, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anukanth A, Khorana H G. J Biol Chem. 1994;269:19738–19744. [PubMed] [Google Scholar]
- 9.Liu X, Garriga P, Khorana H G. Proc Natl Acad Sci USA. 1996;93:4554–4559. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garriga P, Liu X, Khorana H G. Proc Natl Acad Sci USA. 1996;93:4560–4564. doi: 10.1073/pnas.93.10.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwa J, Garriga P, Liu X, Khorana H G. Proc Natl Acad Sci USA. 1997;94:10571–10576. doi: 10.1073/pnas.94.20.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ridge K D, Lu Z, Liu X, Khorana H G. Biochemistry. 1995;34:3261–3267. doi: 10.1021/bi00010a016. [DOI] [PubMed] [Google Scholar]
- 13.Kono M, Yu H, Oprian D D. Biochemistry. 1998;37:1302–1305. doi: 10.1021/bi9721445. [DOI] [PubMed] [Google Scholar]
- 14.Richards J E, Scott K M, Sieving P A. Ophthalmology. 1995;102:669–677. doi: 10.1016/s0161-6420(95)30972-4. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs S, Kranich H, Denton M J, Zrenner E, Bhattacharya S S, Humphries P, Gal A. Hum Mol Genet. 1994;3:1203. doi: 10.1093/hmg/3.7.1203. [DOI] [PubMed] [Google Scholar]
- 16.Dryja T P. Eye. 1992;6:1–10. doi: 10.1038/eye.1992.2. [DOI] [PubMed] [Google Scholar]
- 17.Nathans J, Maumenee I H, Zrenner E, Sadowski B, Sharpe L T, Lewis R A, Hansen E, Rosenberg T, Schwartz M, Heckenlively J R, et al. Am J Hum Genet. 1993;53:987–1000. [PMC free article] [PubMed] [Google Scholar]
- 18.Nathans J, Davenport C M, Maumenee I H, Lewis R A, Hejtmancik J F, Litt M, Lovrien E, Weleber R, Bachynski B, Zwas F, et al. Science. 1989;245:831–838. doi: 10.1126/science.2788922. [DOI] [PubMed] [Google Scholar]
- 19.Kazmi M A, Sakmar T P, Ostrer H. Invest Ophthalmol Visual Sci. 1997;38:1074–1081. [PubMed] [Google Scholar]
- 20.Reeves P J, Hwa J, Khorana H G. Proc Natl Acad Sci USA. 1999;96:1927–1931. doi: 10.1073/pnas.96.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrens D L, Khorana H G. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 22.Cai K, Langen R, Hubbell W L, Khorana H G. Proc Natl Acad Sci USA. 1997;94:14267–14272. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unger V M, Hargrave P A, Baldwin J M, Schertler G F X. Nature (London) 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 24.Baldwin J M, Shertler G F X, Unger V M. J Mol Biol. 1997;272:144–164. doi: 10.1006/jmbi.1997.1240. [DOI] [PubMed] [Google Scholar]
- 25.Yang K, Farrens D L, Hubbell W L, Khorana H G. Biochemistry. 1996;35:12464–12469. doi: 10.1021/bi960848t. [DOI] [PubMed] [Google Scholar]