Abstract
The ‘balance hypothesis' predicts that non-stoichiometric variations in concentrations of proteins participating in complexes should be deleterious. As a corollary, heterozygous deletions and overexpression of protein complex members should have measurable fitness effects. However, genome-wide studies of heterozygous deletions in Saccharomyces cerevisiae and overexpression have been unable to unambiguously relate complex membership to dosage sensitivity. We test the hypothesis that it is not complex membership alone but rather the topology of interactions within a complex that is a predictor of dosage sensitivity. We develop a model that uses the law of mass action to consider how complex formation might be affected by varying protein concentrations given a protein's topological positioning within the complex. Although we find little evidence for combinatorial inhibition of complex formation playing a major role in overexpression phenotypes, consistent with previous results, we show significant correlations between predicted sensitivity of complex formation to protein concentrations and both heterozygous deletion fitness and protein abundance noise levels. Our model suggests a mechanism for dosage sensitivity and provides testable predictions for the effect of alterations in protein abundance noise.
Keywords: balance hypothesis, dosage sensitivity, heterozygous deletion, protein abundance noise, protein interaction networks
Introduction
Essentially all biological processes involve proteins frequently acting as multi-component complexes (Eisenberg et al, 2000; Vidal, 2005; Gavin et al, 2006; Krogan et al, 2006). However, it remains a challenge to characterize how quantitative interaction parameters, such as rates, affinities and protein concentrations, affect function at the cellular and organismal levels (Kuriyan and Eisenberg, 2007). The balance hypothesis posits that an imbalance in the relative concentrations of proteins involved in a protein complex can disrupt complex formation and should thus be deleterious. As a corollary, it has been suggested that proteins involved in complexes should be more likely to be dosage sensitive than other proteins (Papp et al, 2003).
Several means exist by which stoichiometric imbalances could disrupt complex formation and lead to adverse phenotypic effects: first, reducing the abundance of a component of a protein complex, as might occur through a heterozygous deletion mutation, would be predicted to have a measurable effect on fitness. Accordingly, it has been shown that a twofold reduction in the amount of a component protein can result in a many fold reduction in complex formation, and thus have an amplified effect on cell phenotype (Veitia, 2002, 2003). A second, somewhat less intuitive mechanism is referred to as the pro-zone effect or combinatorial inhibition (CI) (Bray and Lay, 1997; Burack and Shaw, 2000; Ferrell, 2000; Levchenko et al, 2000). CI can occur when a stoichiometric excess of one component of a protein complex is added to a solution containing only moderate amounts of the other components. If the component in excess satisfies certain topological conditions in its interaction with other components from the complex, and particularly if it acts as a bridge between two separate parts of the complex, then this excess will typically inhibit the formation of the full complex, by instead favoring the formation of many incomplete subspecies (Bray and Lay 1997). Third, if overabundance or insufficient concentrations of dosage-sensitive proteins significantly affect cell function, it may be beneficial to reduce protein abundance noise for these proteins. Hence, it may be possible to detect evolutionary selection for reduced protein noise.
Several studies have investigated the balance hypothesis and its corollaries. Papp et al (2003) have argued in favor of the balance hypothesis based in part on the finding of enrichment for complex membership among the products of haploinsufficient genes. However, Deutschbauer et al (2005) argued that the mechanism of haploinsufficiency is not due to stoichiometric imbalances, but instead reflects insufficient protein production for a given rate of growth based on the fact that for 136 out of 184 genes in Saccharomyces cerevisiae, haploinsufficiency is relieved under the slow-growth conditions produced by growing in a minimal medium. Moreover, through a large-scale gene overexpression study in S. cerevisiae, Sopko et al (2006) concluded that there is no significant enrichment for overexpression phenotypes among genes products participating in protein complexes and no correlation between genes with overexpression and haploinsufficiency phenotypes. Consistent with the idea that reduced or increased levels of protein complex members could cause deleterious stoichiometric imbalances, Fraser et al (2004) found that proteins with predicted lower expression noise are enriched for complex membership. In contrast, a large-scale study that measured protein abundance noise in single cells did not find a significant association between protein level variations and participation in protein–protein interactions (PPIs) (Newman et al, 2006).
To reconcile these contradictory findings, we reasoned that complex membership alone might be an insufficient condition to give rise to significant dosage sensitivity. Are there additional topological requirements for the complex or the protein's positioning in that complex that might be needed to generate significant sensitivity to increased and decreased protein levels? The work of Bray and Lay (1997) and Veitia (2003) suggests that this could be the case: for example, while overexpression of bridge proteins could lead to the formation of incomplete non-functional subcomplexes and thus CI, proteins at the periphery of a complex and linked by a single complex subunit interaction would appear to have relatively little effect on complex formation if overexpressed; in addition, closed, multiply bonded topologies typically show only a weak tendency toward CI (Bray and Lay, 1997) (Figure 1A and B). Hence, if a substantial fraction of proteins within complexes were peripherally located, then one might not expect to see a strong correlation between complex membership and overexpression phenotypes. Similarly, topology may be related to effects of decreases in protein abundance: dependent on a protein's topological position in a complex, a reduction in the protein's abundance could result in a higher than proportional decrease in complex formation (Supplementary Figure 1A). For these proteins, there may be a significant fitness effect on heterozygous deletions, whereas for others the consequences could be less severe (Supplementary Figure 1B).
Figure 1.
Complex formation model and response curve parameters. (A) A hypothetical protein complex represented as a graph. Each node is colored to correspond to the response curve it generates. Component 1 (orange) acts as a bridge in the graph. Component 2 is peripherally located. (B) Response curves showing complex formation as a function of the amount of each component protein in (A) assuming all interactions are of micromolar strength. (C) Log-width-at-half-max of a response curve is computed to quantify tendency toward CI. (D) An effective Hill coefficient is computed to quantify response curve steepness.
Here, we hence ask the question whether dosage sensitivity characterized by overexpression phenotypes, measurements of fitness effects of heterozygous deletions and quantification of protein abundance noise may be related to the topologies of interactions within complexes rather than just complex membership. Recently, Maslov and Ispolatov (2007) have used a model based on the law of mass action to study the propagation of concentration changes across the S. cerevisiae PPI network. We have developed a similar approach that instead focuses on the local effects of protein concentrations on complex formation by generating complex formation response curves for each protein (Figure 1). A response curve is defined as the dependence of the total amount of full complex (i.e. all proteins in a complex interacting simultaneously) on variation of the concentration of one of its protein components. We evaluate two parameters describing the dosage sensitivity based on each protein's response curves: (i) the tendency toward CI (Figure 1C) or (ii) the steepness (high Hill coefficient (HC)) of the response curve (Figure 1D). To compare these computed measures of dosage sensitivity to experimental characterization of heterozygous gene deletion, gene overexpression and protein abundance noise, we apply our model to manually curated complexes from the Munich Information Center for Protein Sequences (MIPS) database (Mewes et al, 2004) combined with high-confidence PPI data. Affinity purification mass spectrometry experiments, a major source of experimental data on protein complexes (Gavin et al, 2006; Krogan et al, 2006), do not contain explicit information on protein complex topologies. To address this problem, we derive topologies in our analysis in three different ways, using two major interaction sets integrating data from multiple sources (Batada et al, 2006; Collins et al, 2007) as well as a separate set weighted to be enriched for direct physical interactions (Kiemer et al, 2007) (see Materials and methods). Contrary to our initial expectation, we find no significant correlation between response curves that indicated CI and overexpression phenotypes. However, we do find a significant correlation for all three topology sets between steeply sloped response curves and both haploinsufficiency and low noise. Our results therefore suggest that not complex membership alone, but features of complex topologies reflected in our simple model (despite the fact that there are undoubtedly errors in our topology assignments) can be linked to global experimental observables.
Results
Representation of protein complexes
The model for protein complexes implemented here was inspired by that previously described by Bray and Lay (1997). Proteins are represented as nodes in a graph. These nodes are linked by edges to represent binding interactions between proteins; the overall organization of edges and nodes into a graph thus describes a protein complex (Figure 1A). Rather than considering hypothetical complexes as in Bray and Lay (1997), here we aim to generate graphs representing experimentally determined complex topologies. We built graphs representing complexes from the high-confidence manually curated set in the MIPS database (Mewes et al, 2004). A separate graph was defined for each of 123 curated complexes. Edges were drawn between the nodes (proteins) in each graph if there was a binary interaction as indicated by the high-confidence interaction network compiled by Kiemer et al (2007) to be enriched for direct physical interactions. In separate trials, binary interactions were identified from interaction networks compiled by either Batada et al (2006) or Collins et al (2007) (for further details, see Materials and methods and Supplementary information). Complex subspecies were determined by recursively deconstructing the full complex into a set of subgraphs in a manner similar to the algorithm described in Lay and Bray (1997). Our analysis yields similar results using all three topology sets (Supplementary Table I). Unless stated otherwise, we will be referring to our analysis using the Kiemer interaction set (results using the Batada or Collins interaction sets are shown in the Supplementary information).
We make a number of simplifying assumptions about the interactions between proteins and the formation of complexes. We assign simplified association constants for all complexes and their subspecies. To compute association constants, each edge is given a strength: for example, if all edges in a complex were assigned 106 or micromolar interaction strengths, the association constant of the complex would be K=106X, where X is the number of edges in the complex. In the case of a dimer of two interacting proteins, there would be one edge, and hence the association constant of the dimer would be K=106. If both proteins can simultaneously interact with another protein then the trimer formed would have three edges resulting in K=1018 as the association constant for the trimer. In separate trials, we assign different uniform interaction strengths of 108, 106 or 104 to all edges in all complexes. In addition, to test whether our results would hold in the more realistic situation where interactions do not all have the same strength, we also allowed edge strengths to vary between either 108 or 104 and sampled each complex using strengths randomly assigned to each edge. We also apply a simple model of cooperativity and anticooperativity between interactions. We define association constants for cooperativity (the free energy of a pair of interactions is greater than the sum of the free energies of the individual interactions) as K=106X0.85, and for anticooperativity (less than the sum of the free energies of the individual interactions) as K=106X0.85.
Computing response curves
Given a complex, a list of its subspecies, an idealized association constant for each species and an assumed total concentration of each protein, we can compute the equilibrium concentration of the fully formed complex (Storer and Cornish-Bowden, 1976), which is both stable and unique. We start by assuming micromolar stoichiometric quantities of each protein, which are in the range of the association constants we assign. By varying the concentration of one protein while holding the concentrations of the other proteins constant, we compute a response curve for the formation of complex as a function of varying amounts of one of its protein components. The shape of this curve can then be used as a description of the sensitivity of the system to changing amounts of the protein. Examples of such curves are shown in Figure 1B. We use two parameters to describe the response curves: their width and their steepness, as illustrated in Figure 1C and D and described in the next two sections.
Measuring CI
To quantify a protein's tendency toward CI, we measure the log width-at-half-max of the response curve (Figure 1C). Narrow widths correspond to strong CI (Figure 1B, orange) and broad widths correspond to mild CI potentially only occurring at non-physiologically high protein concentrations (Figure 1B, green). In cases where the amount of complex increases monotonically with increasing amounts of protein (Figure 1B, pink), such as is characteristic of peripherally located proteins, we label those proteins as being incapable of CI (or width=∞).
Measuring steepness
We compare the steepness of response curves by computing the log width between the protein concentration at 10% of the curve's maximum and 90% of the curve's maximum (on the left-hand, increasing side of the curve, Figure 1D). Dividing Log(81) by this number corresponds to an effective HC. Although our computed HC values should not be quantitatively correct, it is a useful measure to distinguish qualitatively between steep and shallow response curves within the context of our model.
Relationship between topologies and computed response curve characteristics
CI has a simple relationship to topology where bridge nodes correspond to cases of strong CI, whereas multiply connected non-bridging nodes show weaker CI and peripherally connected nodes are incapable of CI (Figure 1A and B). Although the relationship between topology and response steepness is less intuitive than with CI, a correspondence is discernible. Aside from the simple case of shallow response curves represented by dimers, nodes of arbitrary degree can also have shallow response curves when their adjacent nodes are less densely connected, and nodes of equal degree can have different steepnesses (Figure 2A). This effect may be thought of as being related to the CI effect, but in this case complex formation increases less steeply with increasing protein concentration because a small portion of the protein ends up forming incomplete complexes rather than the full complex despite a relative overabundance of the other complex components. The measures of clustering coefficient (the number of links between adjacent nodes divided by how many could exist) and betweenness (the number of shortest paths that pass through a node relative to how many shortest paths exist) thus relate to response curve steepness, as illustrated in Figure 2B. High betweenness correspond to proteins with shallow response curves and high clustering coefficients correspond to proteins with steep response curves (Figure 2B).
Figure 2.
Relationship between complex topology and response curve steepness. (A) Three complexes with the same size where protein 1 has the same number of interactions, but is associated with different average HCs. (B) Plot of clustering coefficient and betweenness of proteins with steep and shallow response curves. Proteins in dimer complexes represent a trivial case of nodes with shallow response curves and are omitted.
Sensitivity of width and steepness to varying interaction strength
Because response curves depend not only on the topology of interactions but also on the interaction strengths themselves, we sampled interaction strengths to test whether our conclusions hold when interaction strengths vary from strong to weak. For each complex, we separately assigned a random strength to an edge of either 100 μM or 10 nM and for each protein we computed a response curve and measured its width and HC. We repeated this process 16 times and recorded the average width and HC value for each protein. (If there are 16 or fewer possible assignments, then the assignments are enumerated instead of being sampled.) Our general findings comparing the predictions of our model to experimental measurements of overexpression phenotypes (Sopko et al, 2006), fitness effects of heterozygous deletions (Deutschbauer et al, 2005) and quantification of protein abundance noise (Newman et al, 2006) (see sections below) remain unchanged whether we consider average steepnesses or widths derived from sampling or those derived assuming uniform interaction strengths for all edges. Unless we state otherwise, we will be referring to average HCs or widths.
Haploinsufficiency is linked to response curve steepness
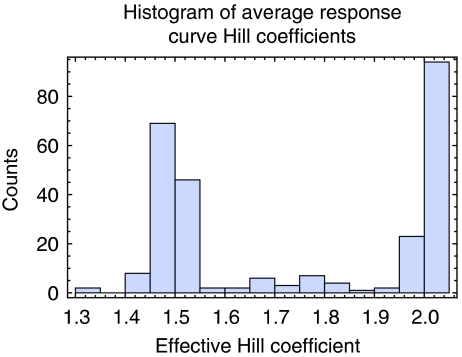
An imbalance in subunit amounts can be created by a reduction in the amount of a component protein. In this situation, the steepness of the left portion of the response curve may indicate how severely complex formation may be affected for a specified reduction in the amount of a component protein. Steeper curves should suggest a higher sensitivity to reduction in protein concentration (Veitia, 2002). We first describe the behavior of our model under the assumptions of different binding strengths and then present a comparison of the model's predictions with experimental data. The average HCs we computed (Figure 3), tend to group into two extremes. We observe a negative correlation between the average HC and the variance of HCs derived from sampling varying interaction strengths of each protein, suggesting that proteins associated with shallower response curves (low HC) might be made steeper by varying interaction strength, whereas proteins associated with very steep response curves (HC>2) are not affected very much by changes in interaction strengths (Supplementary Figure 2). The response curve steepness within our model is related to the number or strength of interactions with more or stronger interactions often leading to steeper curves, but reaching a maximum steepness, as measured by the HC, of ∼2 (Supplementary Figure 3). If interaction strengths are not sampled, but are instead fixed uniformly at micromolar interactions, the HCs group tightly into two populations centered around ∼1.1 and ∼2 (Supplementary Figure 4A). However, these populations broaden out under the anticooperative assumption as the formation of incomplete subcomplexes becomes more favorable (Supplementary Figure 4B).
Figure 3.
Histogram of the computed average HC for each protein. HCs tend to group into two extremes.
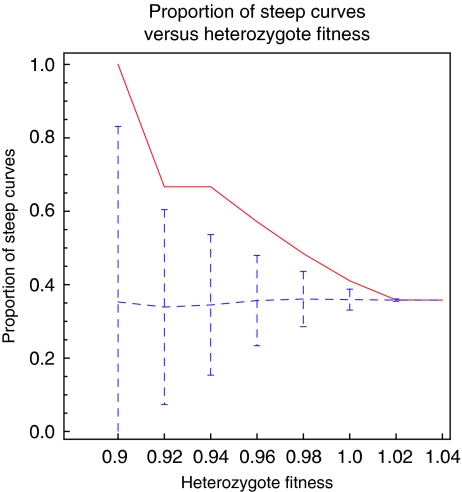
Although approximately 3% of S. cerevisiae genes have been identified as haploinsufficient under rich medium conditions (Deutschbauer et al, 2005), we noted that protein complex members within our set show some enrichment for haploinsufficiency (Papp et al, 2003) with approximately 6% identified as haploinsufficient (N=436, P∼0.001). Proteins in our set identified as having steep response curves (HC>2) show further enrichment beyond 6% to near 10% haploinsufficient (N=94, P∼0.04). Additionally, Figure 4 and Supplementary Figure 5 show a clear increasing trend in the proportion of steep response curves (HC>2) as the fitness of haploinsufficient mutants is decreased. This trend remains qualitatively similar under our different assumptions about interaction strength or cooperativity, different choices for the HC threshold (HC=1.7, 1.8 and 1.9) separating steep and shallow responses, and additionally, on removal of dimer complexes that represent a significant portion of the low HC response curves (Supplementary Figure 6). Thus, proteins with steep response curves according to our model appear to be associated with haploinsufficiency.
Figure 4.
The proportion of steep response curves as a function of degree of haploinsufficiency (red) shows a steady increasing trend as the fitness of heterozygous deletion mutants decreases. This is compared to the expected mean and standard deviation for the proportion of steep response curves if fitness and steepness are randomly assigned (blue dotted lines and error bars).
Proteins with steep response curves tend to have lower noise
It has been suggested that proteins that are members of complexes may be expected to have lower abundance noise (Fraser et al, 2004). We wondered whether we could detect a reduction in noise not just for all members of protein complexes but also specifically for proteins where our model predicts complex formation to be most sensitive to protein concentration (steep response curves). This analysis of noise in the context of concentration sensitivity is complicated by the fact that there is a strong global correlation between mean protein abundance and abundance noise defined by the coefficient of variation (CV), as shown by Newman et al (2006) in a large-scale study of protein abundance noise in S. cerevisiae. The authors, however, find a finer structure in noise levels by defining DM values, which represent the distance from a running median of CV values around a given abundance, effectively normalizing measured noise in CV against protein abundance. Consistent with our hypothesis of a relation between complex topology and dosage sensitivity, we find that proteins with steep response curves (HC>2) tend to have lower noise as defined by their DM values than other proteins with shallow response curves (HC<2) (Figure 5; Supplementary Figure 7). To test the significance of this difference, we compared the median DM values of sets of proteins with steep (median=−1.01, N=50) and shallow (median=0.114, N=82) response curves, using a randomization test (see Materials and methods). The medians appear to be drawn from different distributions with a P∼0.002 level of significance (alternatively, the Wilcoxon rank sum test gives P∼0.003). The difference in noise levels remains significant (P<0.01) under the different assumptions about interaction strength or cooperativity, different choices for the HC threshold separating steep and shallow responses, and on limiting the set to either essential or non-essential proteins, or excluding haploinsufficient proteins (data not shown). We did not find a significant correlation between noise levels and response curve width.
Figure 5.
Histograms of DM values for proteins with either steep (red) or shallow (blue) response curves. The distributions of DM values for proteins with steep and shallow response curves are significantly different (P∼0.002). One shallow response with DM=61 was omitted from the histogram for clarity.
Dosage sensitivity is not a simple function of complex size
Because proteins participating in dimer complexes universally tend toward shallow response curves within our model, it is possible that our observed proclivity for lower noise among proteins with steep response curves might also reflect a tendency for lower noise among proteins participating in higher order complexes versus dimers. This would be consistent with the earlier hypothesis that dosage sensitivity should be related to complex size: if disruption of complex formation by a dosage imbalance of one subunit has a fitness cost proportional to the wasted production of other subunits in the complex (Fraser et al, 2004), one might expect that, as the number of subunits in a complex increased, there would be a greater waste, and consequently higher fitness costs for the disruption of complex formation. To determine whether our observed correlation might be due to complex size rather than curve steepness, we looked for a correlation between the number of subunits in a complex and the amount of abundance noise observed for component proteins while excluding dimer complexes (as dimer complexes are all associated with shallow response curves). Such a correlation did not exist (Spearman's rank correlation r=−0.052, P∼0.49, N=199) and there were no significant differences between the median noise levels on any partitioning of the set with respect to complex size. The most significant partitioning compared members of trimer complexes (median=0.05, N=30) to members of pentamer or larger complexes (median=−0.14, N=134) giving P∼0.7. In contrast and in accordance with our model, when excluding dimer complexes, proteins with steep response curves (median DM value=−1.01, N=50) and those with shallow response curves (median DM value=0.12, N=35) still show distinct median noise levels (P∼0.01, or by Wilcoxon rank sum test P∼0.01). Thus, our observations support the idea that lower abundance noise is correlated with steeper response curves and not more simply complex size.
Degree alone may not explain reduced noise or haploinsufficiency
Although there is a correlation between the number of interactions (degree) and the HC in our model, the correlation between HC and noise may not solely be explained as being due only to the previously observed correlation between degree and noise (Batada et al, 2006). Proteins with the same number of interactions and in the same size complex may show distinct HCs based on differing position within the overall complex topology (Figure 2A provides an illustration). To test the influence of degree and modeled steepness (HC), we investigated whether steep response curve proteins were likely to be less noisy than shallow response curve proteins with equal degree. Out of 213 pairs of steep and shallow HC proteins, where the steep HC protein had a degree equal to the shallow protein, in ∼61% of pairs the steep protein has the lower DM value (P∼0.0005). The same analysis considering heterozygous deletion fitness data also finds a significant difference between steep and shallow HC proteins with the same degree: out of 968 pairs, in ∼53% of pairs the steep protein has a lower fitness (P∼0.02). As an additional control designed to reassign topologies while preserving degree to test for the influence of incorrect topology assignment, we randomized the computed HC values within groups of proteins having the same degree. We found that this assignment of steep or shallow response curves based only on degree did not show a significant difference in median DM values (P∼0.1). These results suggest that the relation between computed steepness and observed reduced noise or haploinsufficiency may not be explained only by degree.
CI may play a limited role in overexpression phenotypes
CI could be one of the causes of dosage sensitivity under the balance hypothesis in cases of substantial excess of one of the components. Although a previous study has linked the lethality of overexpression to complex membership (Papp et al, 2003), a subsequent large-scale study (Sopko et al, 2006) was unable to find a significant increase in complex membership among proteins displaying overexpression phenotypes. Because complex membership alone is not sufficient to result in CI, we asked whether a stronger CI signal might be observed using our model to distinguish between complex member proteins that are capable of CI and those that are not.
Using our width-at-half-max measurement for CI (Figure 1C), we computed response curve widths for all proteins in our set of complexes. A histogram of the average widths measured for each protein in our set is shown in Supplementary Figure 8. The histogram shows widths grouped into populations centered around ∼1.2, ∼5, ∼10, and infinity (CI incapable). When using either the cooperative or anti-cooperative assumption, most widths shift to become significantly narrower or wider, respectively (Supplementary Figure 9B and C). This makes intuitive sense within the simple model because the cooperative assumption shifts the association constants to favor the formation of the full complex over smaller subcomplexes, whereas the anticooperative assumption significantly reduces the association constant for the full complex, but has less effect on the association constants of smaller subcomplexes.
For our analysis of the relationship between complex topology and overexpression phenotype, we first used widths computed using the assumption of micromolar interaction strengths without cooperativity. We considered essential proteins from our set and compared the mean overexpression lethality scores (OLSs) (Sopko et al, 2006, ranging from 1: lethal to 5: no effect) of two groups: those predicted to be capable of CI (width≠∞, N=105, mean OLS=4.69) and those not capable (width=∞, N=18, mean OLS=4.52). Using a randomization test (see Materials and methods), we were unable to find a significant difference between the two sets (P>0.3). At large response curve widths, CI of complex formation may occur only at non-physiological protein concentrations (Figure 1B, green). Therefore, we tested whether this lack of significance persisted when we compared proteins with narrow widths (where inhibitory effects might take place at lower physiologically achievable protein concentrations) against all other proteins. This was the case under all binding strength assumptions (100, 1 μM, 10 nM, cooperative and anticooperative). Thus, based on available data we are unable to identify a role for CI in overexpression phenotypes.
We do not see this as inconsistent with the importance of topology in affecting dosage sensitivity. Instead, the lack of correspondence may be due to a principle difference between response curve steepness, which may result from having stronger interactions or a higher clustering coefficient and smaller betweenness (Figure 2), and CI, which results primarily from the presence of a bridging-type interaction (Figure 1A and B) that may be especially difficult to infer from available data. Therefore, due to the substantial sensitivity of the CI effect to complex topology (i.e. small errors in assigning topologies may mask the detection of small true dependency), current data may be insufficient to reveal correlations. In addition, cooperativity effects could reduce the potential for CI even for proteins with bridge-like interactions. For example, a scenario of sequential complex assembly, where one protein needs to bind a bridging protein in a trimeric complex before the third protein can bind would prevent CI (Veitia, 2002). For other, non-bridging proteins, a limited role for CI in overexpression phenotypes could be explained by the requirement for non-physiologically high concentrations to cause the effect (Figure 1B). A final possible explanation for the absence of a clear CI phenotype follows from the hypothesis that nonspecific interactions not modeled here could be deleterious at elevated concentrations for a wide range of ‘sticky' proteins, independent of their topologies within their functional complexes (Zhang et al, 2008).
Discussion
Relying on the basic principles of the law of mass action, we make qualitative predictions about the amount of complex formation as a function of the changing concentrations of individual subunits within protein complexes. Through this simple model, we observe correlations between sharper responses in complex formation and both reduction in protein abundance noise and greater likelihood of haploinsufficiency. Our key assumptions going into this analysis are the interaction strengths and types of (non-)cooperative behavior of the PPIs that form complexes and perhaps most importantly, our ability to infer complex topologies from available data (see below). Within our study, we consider proteins from data sets that are limited to relatively abundant proteins. Thus, the equilibrium model we use to compute complex formation seems applicable. To account for the assumptions we have made about association constants, we have tested our results under varying interaction strengths and different assumptions about the cooperative nature of interactions. We observe similar correlations using most of these different assumptions about interaction strengths (the only exception is the extreme case when all interactions are assumed to be very weak, 100 μM).
The interaction sets we have used as indicators of direct physical interactions between proteins are based at least in part on affinity purification data that by their nature identify interactions between proteins that exist in the same complex, but may not interact directly. Ideally, the interactions that we model would be based solely on experimental evidence that represented direct physical interactions less ambiguously, such as crystal structures or yeast two-hybrid assays. However, given that state-of-the-art yeast two-hybrid interactomes cover only ∼20% interactions (Yu et al, 2008), the analysis was not possible with such a limited interaction set. Although there are sure to be cases where incorrectly assigned complexes, topologies or association constants lead us to the wrong conclusion about response curve characteristics, these misclassifications may tend to underestimate the significance of our observations relating interaction topology with protein abundance noise and haploinsufficiency. This is consistent with the observation that when we used the original set of interactions obtained by Krogan et al (2006), the identified relations were weak, whereas using the higher confidence interactions from combined data sets by Batada et al (2006) or Collins et al (2007) and overlaying them with manually curated complexes, we were able to observe significant correlations. Because neither the Batada et al or Collins et al interactomes were specifically designed to identify direct physical interactions between proteins and thus might be prone to higher false-positive rates when used to define direct physical interactions represented as edges in our graphs, we chose to perform our analysis using an interaction set created by Kiemer et al (2007) that was enriched for direct physical interactions (although we cannot exclude the possibility that this set still contains indirect interactions). Using the Kiemer interaction set reduced the total number of interactions by ∼40% compared to the Batada set, while reducing the total number of connected graphs required for our analysis by only ∼10% (Supplementary Table I). Despite this significant change in interactome size favoring the removal of indirect interactions, the correlations observed in our model remained significant.
It might be argued that HCs of 1 and 2 do not seem sufficiently different to cause the observed effects. However, simple estimates show a substantial reduction in the complex formation under noisy expression of a protein with a steep response curve versus a shallow response curve (see Supplementary information and Supplementary Figure 10). Additionally, the range of HCs produced in our model is likely to be more confined than the actual range. Specific cooperativity effects not modeled here might extend this range further. The upper and lower bounds of our computed HCs could also be broadened if the abundance of one protein component was closely correlated with the abundance of another component such that a change in the concentration of one protein component implied a similar change in the concentration of another and their abundances varied simultaneously. Hence, the effects of observed coexpression of complex subunits (Stuart et al, 2003) could be significant. The range of HCs would additionally be extended if a complex contained multiple copies of the same protein. Information describing complex stoichiometry is an element missing from our model. In some situations, such as when a protein interacts with a larger complex as a tight homodimer, it may be possible to treat the homodimer as a single entity. In this case, one would obtain results that are qualitatively similar to those presented here. However, in general complex stoichiometry may have a significant impact on complex formation. We also set aside modeling situations where complexes share or compete for subunits as well as potential non-functional interactions (Zhang et al, 2008).
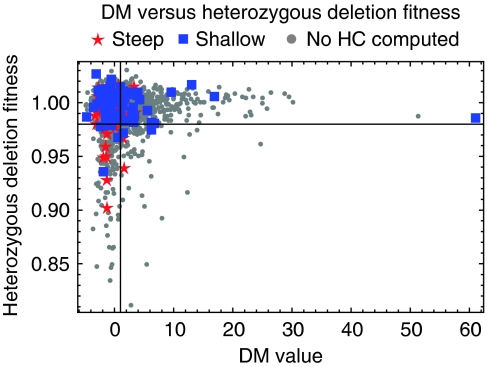
The role of noise in biological systems has recently gained attention. Although noise may serve useful purposes in certain situations (Samoilov et al, 2006), we find that noise tends to be reduced when there is a sharp relationship between a protein's concentration and the formation of a complex. We hence speculate that dosage sensitivity, which may occur, in part, due to complex topology, leads to selection against the noisy expression of a given protein. Consistent with this idea we note that there are fewer proteins with high noise (DM>1) and low heterozygous deletion fitness (<0.98) than might be expected if DM values and heterozygous deletion fitness were paired randomly (N=1917, P<0.00001) (Batada and Hurst, 2007). Proteins with low noise (DM<1) and low heterozygous deletion fitness (<0.98) correspond to dosage-sensitive proteins whose noise levels are near the minimum. On the other hand, proteins with high noise and high heterozygous deletion fitness correspond to dosage-insensitive proteins that may have larger levels of abundance noise without detrimental effects. The former group tends to be populated by proteins with steep response curves (11 proteins with steep versus 4 proteins with a shallow response curve), whereas the latter group tends to be populated by proteins whose response curves are shallow (9 proteins with steep versus 29 proteins with shallow response curves, P=0.001, Fisher's test; Figure 6).
Figure 6.
A scatter plot of DM versus heterozygous deletion fitness is divided into quadrants representing low fitness–low noise, low fitness–high noise, high fitness–low noise and high fitness–high noise. Red stars represent proteins classified as being likely to have steep response curves. Blue squares represent proteins classified as having shallow response curves. Gray dots represent proteins with known heterozygous deletion fitness values and DM values that were not analyzed with our model because they were not members of MIPS complexes or because the complexes did not generate connected graphs of nine or less nodes when combined with the interaction data set.
Finally, we note that correlations with DM values typically yielded higher levels of significance than correlations with heterozygous deletion fitness (in rich media) and appeared slightly more robust to varying model parameters or interaction data sets. We believe that this is because the effect of protein dosage on growth rate is dependent on the growth environment and, thus, significant growth defects due to reductions in specific complexes may only appear under a subset of environmental conditions. For example, there is only a weak correlation between heterozygous deletion fitnesses in minimal and rich media (Kendall's τ=0.25). On the other hand, one might expect protein expression noise to be less variable between different environments (Kendall's τ=0.56 for minimal and rich media CVs) and optimized to satisfy constraints imposed by many possible environments. Thus, the degree to which expression noise is tuned higher or lower may reflect, in part, a dosage sensitivity that is generalized to the variety of environments that a cell might find itself in.
Our simple model for the response of complex formation to varying concentrations of component proteins provides a possible mechanistic explanation for the observed significant correlation between dosage sensitivity and the steepness of the formation response as classified by the model. Dosage sensitivity in our model is dependent in large part on the topological arrangement of the protein within the complex (Figure 2) and it is the topological element of this effect that our conclusions are based on. Hence, the model, although based on significant assumptions about complex topology, makes predictions that are testable. For example, increasing the noise levels of proteins with predicted steep response curves should have measurable fitness effects, dependent on the position of the protein's mean abundance relative to that optimal for complex formation. Moreover, altering the abundance noise levels for proteins within the same complex (and hence functionally related) but with different predicted steepnesses in their response curves could have differential effects on fitness. The analyses we describe may be relevant to understanding copy number variation or predicting interactions that would be more sensitive to removal. We find it remarkable that under relatively broad assumption, models such as this and that earlier produced by Maslov and Ispolatov (2007) are sufficient to extract important trends and correlations from large-scale genetic and proteomic data. As proteomic data become more detailed and more accurate, we look forward to seeing how more complex models can highlight local network properties leading to hypotheses for specific complexes (Supplementary Figure 11) that can be characterized in mechanistic detail.
Materials and methods
Complex and interaction data
To keep response curves readily computable, we limit our set of complexes to those composed of nine proteins or less and further omit any complexes from the MIPS database that did not result in connected graphs when overlaid with edges from interaction sets. Supplementary Table I lists details for the three interaction sets used in our analysis. If a protein appeared in multiple complexes, HC and CI values were averaged over both complexes. There was general agreement between steepness classifications using the different datasets (Supplementary Figure 12).
Fitness and viability data
For our analysis related to dosage sensitivity, we used deletion viability data from the MIPS database (Mewes et al, 2004), heterozygous deletion data from Deutschbauer et al (2005), overexpression data from Sopko et al (2006) and abundance noise data from Newman et al (2006). For all cases, except deletion viability data, we consider results from growth in rich medium.
Response curve algorithm
To compute response curves for concentrations of complexes, complex subspecies and free concentrations of proteins, we used an algorithm described by Storer and Cornish-Bowden (1976). This algorithm has most recently been explained by Maslov and Ispolatov (2007). Given all total concentrations, Ai, of n proteins (we fix all proteins at 1 μM concentrations except for the protein of varying concentration in the response curve), all association constants, Kj, for m complexes, and matrix elements αij describing the number of occurrences of protein i in the jth complex, the free concentration of each of the proteins is obtained by iteratively solving equation (1) starting with the initial condition of ai=Ai inserted into the right-hand side of the equation. We allow iterations to continue until the change in free concentration is less than 0.1% for each protein during a single iteration. We have implemented this algorithm in the C programming language and in Mathematica. We find convergence to be generally very rapid matching results published by Bray and Lay (1997) and Lay and Bray (1997) who used a different algorithm.
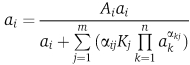 |
Statistics and randomization testing
Randomization tests were used to determine whether the observed difference between the medians of two sets was large enough to reject the null hypothesis that the values in the two sets are drawn from the same probability distribution. These tests were performed by first computing the observed difference of the medians of the two sets. The values of the two sets were then pooled and sampled without replacement to generate a sample of two new sets where medians are computed and the differences are recorded. This was repeated to generate 10 000 samples of the difference of medians and a P-value was determined from this distribution.
To test randomization while preserving degree, HC assignments were shuffled within groups of proteins of the same degree 10 000 times to produce 10 000 shuffled assignments that preserved any overall relationship between degree and HC. A P-value was then determined by comparison of the difference in steep and shallow median DMs of the 10 000 degree preserving assignments to 10 000 reshuffled assignments not forced to preserve degree.
Computing clustering coefficient and betweenness
The clustering coefficient is defined as the number of links between adjacent nodes divided by the number of links that could possibly exist between them. Betweenness is defined as the fraction of shortest paths that pass through a node and was normalized by the number of pairs of nodes. Both clustering coefficient and betweenness were computing using the NetworkX python package, available at http://networkx.lanl.gov/.
Supplementary Material
Supplementary Discussion, Supplementary Table 1, Supplementary Figures 1-12 and Supplementary References
Acknowledgments
We thank Hana El-Samad and Matt Eames for many stimulating discussions and a critical reading of the manuscript, and Ala Trusina and the Kortemme laboratory for valuable comments. RO was supported by a graduate fellowship in Complex Biological Systems funded by HHMI and NIBIB. This study was additionally supported the Sandler Program in Basic Sciences. TK is a fellow of the Alfred P Sloan Foundation in Molecular Biology.
References
- Batada NN, Hurst LD (2007) Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet 39: 945–949 [DOI] [PubMed] [Google Scholar]
- Batada NN, Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hurst LD, Tyers M (2006) Stratus not altocumulus: a new view of the yeast protein interaction network. PLoS Biol 4: e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D, Lay S (1997) Computer-based analysis of the binding steps in protein complex formation. Proc Natl Acad Sci USA 94: 13493–13498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack W, Shaw A (2000) Signal transduction: hanging on a scaffold. Curr Opin Cell Biol 12: 211–216 [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ (2007) Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics 6: 439–450 [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G (2005) Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169: 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Marcotte EM, Xenarios I, Yeates TO (2000) Protein function in the post-genomic era. Nature 405: 823–826 [DOI] [PubMed] [Google Scholar]
- Ferrell J (2000) What do scaffold proteins really do? Sci STKE 2000: pe1. [DOI] [PubMed] [Google Scholar]
- Fraser H, Hirsh A, Giaever G, Kumm J, Eisen M (2004) Noise minimization in eukaryotic gene expression. PLoS Biol 2: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Kiemer L, Costa S, Ueffing M, Cesareni G (2007) WI-PHI: a weighted yeast interactome enriched for direct physical interactions. Proteomics 7: 932–943 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrín-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Kuriyan J, Eisenberg D (2007) The origin of protein interactions and allostery in colocalization. Nature 450: 983–990 [DOI] [PubMed] [Google Scholar]
- Lay S, Bray D (1997) A computer program for the analysis of protein complex formation. Comput Appl Biosci 13: 439–444 [DOI] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, Sternberg P (2000) Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci USA 97: 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S, Ispolatov I (2007) Propagation of large concentration changes in reversible protein-binding networks. Proc Natl Acad Sci USA 104: 13655–13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Amid C, Arnold R, Frishman D, Güldener U, Mannhaupt G, Münsterkötter M, Pagel P, Strack N, Stümpflen V, Warfsmann J, Ruepp A (2004) MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res 32: D41–D44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow D, Noble, DeRisi J, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst L (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Samoilov M, Price G, Arkin A (2006) From fluctuations to phenotypes: the physiology of noise. Sci STKE 2006: re17. [DOI] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver S, Cyert M, Hughes TR, Boone C, Andrews B (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21: 319–330 [DOI] [PubMed] [Google Scholar]
- Storer A, Cornish-Bowden A (1976) Concentration of MgATP2- and other ions in solution. Calculation of the true concentrations of species present in mixtures of associating ions. Biochem J 159: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Veitia R (2002) Exploring the etiology of haploinsufficiency. Bioessays 24: 175–184 [DOI] [PubMed] [Google Scholar]
- Veitia RA (2003) Nonlinear effects in macromolecular assembly and dosage sensitivity. J Theor Biol 220: 19–25 [DOI] [PubMed] [Google Scholar]
- Vidal M (2005) Interactome modeling. FEBS Lett 579: 1834–1838 [DOI] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Maslov S, Shakhnovich EI (2008) Constraints imposed by non-functional protein–protein interactions on gene expression and proteome size. Mol Syst Biol 4: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Discussion, Supplementary Table 1, Supplementary Figures 1-12 and Supplementary References