Abstract
Despite the importance of subcellular localization for cellular activities, the lack of high-throughput, high-resolution imaging and quantitation methodologies has limited genomic localization analysis to a small number of archival studies focused on C-terminal fluorescent protein fusions. Here, we develop a high-throughput pipeline for generating, imaging, and quantitating fluorescent protein fusions that we use for the quantitative genomic assessment of the distributions of both N- and C-terminal fluorescent protein fusions. We identify nearly 300 localized Caulobacter crescentus proteins, up to 10-fold more than were previously characterized. The localized proteins tend to be involved in spatially or temporally dynamic processes and proteins that function together and often localize together as well. The distributions of the localized proteins were quantitated by using our recently described projected system of internal coordinates from interpolated contours (PSICIC) image analysis toolkit, leading to the identification of cellular regions that are over- or under-enriched in localized proteins and of potential differences in the mechanisms that target proteins to different subcellular destinations. The Caulobacter localizome data thus represent a resource for studying both global properties of protein localization and specific protein functions, whereas the localization analysis pipeline is a methodological resource that can be readily applied to other systems.
Keywords: bacteria, Caulobacter, genomics, quantitative image analysis, high-throughput imaging
The emergence of high-throughput methods to assay the levels of cellular components such as mRNAs and proteins has enabled the study of biological processes at a systems level, enhancing the understanding of both individual proteins and functional modules. Meanwhile, the subcellular localizations of proteins that mediate such cellular processes as growth, division, and motility indicate that many important aspects of cell biology require precise spatial organization. The significance of this localization holds true in both eukaryotes and prokaryotes. Large-scale efforts have cataloged the localizations of C-terminal GFP fusions to most of the proteins of Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Escherichia coli (1–3). However, the utility of these libraries has been limited by the laborious nature of generating, imaging, and scoring fluorescent fusions, thereby restricting their reanalysis under different conditions and reconstruction as N-terminal fusions or fusions to other fluorescent proteins. Furthermore, protein localization has classically been analyzed via qualitative descriptors that limit their utility for quantitative systems-level characterizations.
Despite their small size and lack of internal compartmentalization, bacterial cells are highly organized (4). Relatively small genomes and experimental tractability thus make bacteria powerful systems for studying universal cell biological processes such as cell division, cell cycle progression, and macromolecular trafficking. However, the extent to which bacterial cells employ protein localization has remained unclear from gene-specific studies. We have focused on the Gram-negative aquatic bacterium, Caulobacter crescentus. Caulobacter is a particularly useful model for studying protein localization because of its asymmetric morphology, which readily allows the 2 cell poles to be distinguished by the presence of a stalk that protrudes from only 1 pole. In addition, a number of important Caulobacter proteins have been shown to assume specific subcellular localizations (5). These proteins serve as positive controls for genomic studies and establish proof-of-principle examples that protein localization plays an important role in the regulation of this organism's biological activities. A recent transposon-mediated forward-genetic screen identified 11 additional localized proteins (6), but Caulobacter protein localization has yet to be systematically studied at a genomic scale.
Here, we have begun to address the classical limitations of genomic localization analysis by developing a pipeline of high-throughput, high-resolution methods for generating, imaging, and analyzing fluorescent protein fusions. This approach enables the rapid, efficient, and repeated study of spatial processes on the scale of an entire genome and allowed us to reimage the localization of both N- and C- terminal mCherry fusions. The identification of 289 localized proteins represents a nearly 10-fold increase in the number of localized proteins in Caulobacter. By using a projected system of internal coordinates from interpolated contours (PSICIC), a recently developed software suite for automated image analysis (7), we quantitatively analyzed the accuracy and distributions of these localizations, leading to the appreciation of new aspects of Caulobacter proteome localization. These data thus enable the cell biological analysis of both individual proteins of interest and the general properties of the Caulobacter proteome.
Results
Rapid, Efficient, and Scalable Generation of Fluorescent Protein Fusions.
To study protein localization at the genomic scale, we developed rapid and efficient methods for generating, imaging, and analyzing fluorescent Caulobacter fusion proteins. To enable future modularity, we adopted the Gateway (Invitrogen) system of recombinational subcloning (8), which we modified for improved ease and scalability. In the Gateway system, each ORF of interest is first cloned into a “donor” vector to create an “entry” vector. This library of entry vectors, also known as an ORFeome (9), is then mobilized into any “destination” vector of choice to create an “expression” vector for specific applications. Each of the cloning steps is mediated by highly efficient site-specific recombinases, and efficiency is further enhanced by a negative selection strategy wherein the ORF is designed to replace a toxic ccdB-containing cassette (8). We have modified the generation of expression vectors such that it can be performed in living cells without the need for purified reagents.
To generate an entry-vector ORFeome library for Caulobacter, we PCR-amplified each predicted gene and recombined it into a Gateway-compatible donor vector. We initially succeeded in isolating entry vectors for 3,184 of the 3,763 predicted Caulobacter genes (85%), of which 2,786 (74%) were sequence-verified as full-length ORFs of the correct identity (Table 1). These 2,786 entry vectors constitute version 1.0 of the Caulobacter ORFeome.
Table 1.
Results of the high-throughput pipeline for generating, imaging, and analyzing Caulobacter N- and C-terminal mCherry fusions
| Pipeline step | Number (%) |
|---|---|
| Predicted Caulobacter proteins | 3763 |
| Successful PCR reactions | 3744 (99.5% of all proteins) |
| Entry vectors recovered | 3184 (85.0% of PCRs and 84.6% of all proteins) |
| Sequence-verified entry vectors | 2786 (87.5% of BPs and 74.0% of all proteins) |
| C-terminal Pxyl mCherry fusions generated and imaged | 2786 (100% of entry vectors and 74.0% of all proteins) |
| Localized and sequence-verified C-terminal fusions | 187 (6.7% of C-terminal fusions and 5.0% of all proteins) |
| N-terminal Pxyl mCherry fusions generated and imaged | 2786 (100% of entry vectors and 74.0% of all proteins) |
| Localized and sequence-verified N-terminal fusions | 165 (5.9% of N-terminal fusions and 4.4% of all proteins) |
| Proteins localized as both N- and C-terminal fusions | 63 (33.6% of localized C-terminal fusions, 38.4% of localized N-terminal fusions, and 1.7% of all proteins) |
| Unique localized proteins | 289 (10.4% of proteins examined and 7.7% of all proteins) |
To assess the subcellular localization of each protein in this ORFeome, we generated fusions to the mCherry red fluorescent protein (10) because mCherry, unlike GFP, can fold and fluoresce in all cellular compartments (11). To reduce complications associated with variable or low expression or protein toxicity, we constructed a destination vector, gXRC, that can be used to express C-terminal mCherry fusions as single-copy chromosomal integrants at the xylose-inducible promoter (Pxyl) locus (12).
Traditionally, the gateway left–right (LR) recombination reaction that mobilizes ORFs from an entry vector into a destination vector is performed in vitro. As a more labor and cost-efficient alternative, we implemented a high-throughput “in vivo LR” recombination strategy in which the recombination is performed in E. coli that express the proper LR recombinases (detailed in SI Experimental Procedures). This strategy eliminates the need for purified components although maintains >99% efficiency. We recombined all 2,786 entry vector ORFs into gXRC through in vivo LR reactions, and subsequent transfer of these fusions into Caulobacter generated a C-terminal mCherry fusion library (Table 1).
Pedestal Slides Enable High-Throughput High-Resolution Imaging of Protein Localization.
Having generated 2,786 Caulobacter strains, each bearing a different mCherry fusion protein, our next goal was to rapidly image these strains. Commercially available high-throughput imaging systems operate with air-immersion objectives that provide data whose resolution is too low to be useful for detailed studies of protein localization. Meanwhile, traditional high-resolution oil-immersion imaging is labor-intensive. We addressed this problem by designing a “pedestal slide” system for imaging 48 separate strains at high-resolution on 1 slide. Each pedestal slide contains a 6 × 8 array of agarose pedestals whose spacing prevents cross-contamination of neighboring strains. Combined with a robotic microscope stage and a semiautomated image acquisition script, this system increases the throughput of the traditional process of imaging Caulobacter at maximal wide-field epifluorescence resolution (≈200 nm) nearly 50-fold, without any sacrifice in image quality.
To image the subcellular distribution of the C-terminal mCherry fusion library, each of the 2,786 C-terminal mCherry fusions was induced with xylose for a period that is long enough for robust expression but brief enough to minimize toxicity effects. We imaged these fusions in asynchronous cell populations, enabling us to study proteins that only localize during specific stages of the cell cycle. Each strain was subsequently imaged on pedestal slides twice and scored independently by 2 people using a script that automated file handling and scoring nomenclature. The proteins with nonuniform, localized patterns were independently reimaged and rescored, yielding a final set of 187 confirmed C-terminal localized fusions (Table 1 and Table S1). Additionally, we identified multiple proteins that localized to the cell periphery in a uniform manner, but these proteins were not defined as localized for the purposes of this study to focus on proteins with distinct heterogeneous localizations.
Repeating the High-Throughput Pipeline to Analyze N-Terminal Fusions.
To demonstrate the power of this high-throughput strategy for generating, imaging, and scoring fluorescent protein fusions, we repeated the entire pipeline with a different destination vector, gXRN, which produces N-terminal mCherry fusions. The resulting N-terminal mCherry fusion library identified 165 sequence-verified fusions with localized nonuniform distributions (Table 1 and Table S1). Of these 165 fusions, 63 (38%) were also localized as C-terminal fusions (Table 1). Proteins that only localize as fusions to a single terminus may reflect the disruption of terminal structural or interaction motifs by the mCherry fusion. Consistent with this hypothesis, the percentage of proteins predicted to have N-terminal signal peptides by PSORTb (13) was higher in proteins that localized as C-terminal but not N-terminal fusions (28.2%) than in those that localized only as N-terminal fusions (10.8%) or as both N- and C-terminal fusions (15.9%).
To our knowledge, this dataset represents the first time that both N- and C-terminal protein fusions have been systematically analyzed at a whole-genome scale. Had we limited our analysis to fusions at a single terminus, we would have missed approximately one third of the localized proteins, demonstrating the importance of performing such large-scale studies on multiple types of fusion proteins. The localization screen results (Table S1), including images of the localized fusions are accessible at the Gitai lab website: www.molbio1.princeton.edu/labs/gitai/.
Characterization and Validation of the Caulobacter Localizome.
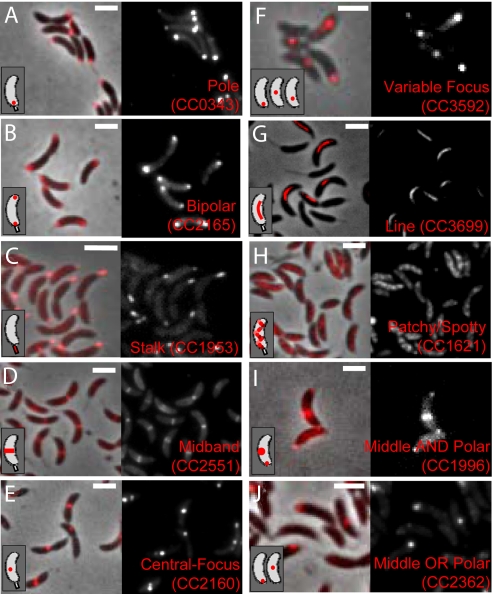
The N- and C-terminal mCherry libraries identified 352 localized fusions that included 289 unique proteins (Table S1). The localization patterns of the fusions could be readily sorted into 10 classes described and enumerated in Fig. 1 and Table 2. Three of these classes consist of proteins that localize to the poles, including unipolar localization, bipolar localization, and localization to the stalk extension (Fig. 1 A–C). The identification of proteins restricted to the Caulobacter stalk is particularly interesting because no proteins with this type of distribution have been previously described. In addition to highlighting the benefits of unbiased screening for localized proteins, the newly appreciated stalk-specific localization suggests that the Caulobacter stalk may function as a type of cellular organelle to which proteins can be specifically targeted and/or restricted.
Fig. 1.
Representative images of the different classes of Caulobacter protein localizations. (A–J) For each class, an overlay of phase contrast and mCherry fluorescence images is shown on the Left, the fluorescence image alone is shown on the Right, and a schematic representation is shown in the Inset. (Scale bars represent 2 μm.)
Table 2.
Summary of the localization patterns observed for mCherry fusions localized only as C-terminal fusions, only as N-terminal fusions, or both N- and C-terminal fusion
| Localization | Only C-terminal | Only N-terminal | Both N- and C-terminal | Total |
|---|---|---|---|---|
| Pole | 25 | 22 | 20 | 67 |
| Stalk | 0 | 1 | 3 | 4 |
| Bipolar | 4 | 3 | 6 | 13 |
| Central focus | 8 | 4 | 11 | 23 |
| Midband | 1 | 3 | 6 | 10 |
| Variable focus | 3 | 2 | 7 | 12 |
| Line | 1 | 0 | 4 | 5 |
| Patchy/spotty | 61 | 43 | 49 | 153 |
| Middle (focus) and pole | 3 | 1 | 2 | 6 |
| Middle (focus) or pole | 18 | 23 | 18 | 59 |
| Total | 124 | 102 | 126 | 352 |
Three other localization classes include proteins that localize to a single structure in the middle portion of the cell. These classes consisted of proteins localized to a cross-sectional band at midcell (midband proteins), proteins localized to a single focus that is reproducibly in the central portion of the cell (central focus proteins), and proteins localized to a single focus whose position in the cell was variable (variable focus proteins) (Fig. 1 D–F). These proteins may function in cell division or other activities that can occur in the cell's middle such as DNA replication (14). Two additional localization classes consist of proteins that form a line down the vertical axis of the cell, and proteins with patchy or spotty distributions that resemble the localizations of helical proteins such as the actin homolog, MreB (15, 16) (Fig. 1 G and H). A recent electron cryotomography study demonstrated that in addition to the known bacterial cytoskeletal proteins, Caulobacter appears to possess as-yet-uncharacterized filament systems (17). These uncharacterized filament-forming proteins may be represented in the set of line or patchy/spotty localization classes that we have identified here.
Two final localization classes displayed combinatorial manifestations of localization to the middle and/or polar regions (Fig. 1 I and J). Strains with individual cells that simultaneously exhibited localization in more than 1 pattern were described as “and” combinations, whereas strains in which different cells displayed 1 of 2 distinct individual patterns were described as “or” combinations. The “or” combinations differed from the “variable focus” proteins in that they were found in only 2 positions, as opposed to a large range of locations. These bimodal localized proteins may reflect cell-cycle-dependent regulation; future protein-specific studies will be needed to confirm such dynamics. Caulobacter cells thus appear to localize their proteins in a discrete number of patterns that can be mixed and matched.
The entry vector library included 29 Caulobacter proteins reported to be localized as full-length fluorescent protein fusions in previous studies (Table S2). The localizations of 24 of these 29 control proteins (83%) were recapitulated in the library, indicating a low rate of false-negative localizations (Table S2). Determining the rate of false positives in our localized set is significantly more difficult. The overwhelming majority of the localized proteins have no known loss-of-function phenotype, and it is not feasible to raise specific antibodies to so many proteins, such that we cannot use phenotypic complementation or immunofluorescence to verify localizations. However, the probability of the 187 C-terminal and 165 N-terminal localized fusions identifying the same 63 overlapping proteins by chance is miniscule (P < E-20). Moreover, these overlapping N- and C-terminal fusions corroborated each other's localization patterns in 58 of 63 cases (92%) (Table S1). The 5 proteins whose N- and C-terminal fusions localize in different patterns may reflect proteins with multiple interactions partners, such that mCherry-mediated occlusion of each terminus results in differential protein targeting. With respect to the 58 proteins with identical N- and C-terminal localizations, the corroborating patterns indicate that these localizations can be viewed with high confidence.
To date there have been no reports of false-positive localizations in Caulobacter, but inclusion body formation can cause false-positive localization in E. coli. Inclusion bodies can be visualized in bacterial cells by the localization of the small heat shock protein, IbpA (18). The Caulobacter homolog of IbpA, CC3592, was localized in our library as a “variable focus” protein. Only 8 other proteins in our library were similarly localized (Table 2), suggesting that the remaining proteins are unlikely to represent inclusion bodies. We consequently expect that most of the localized proteins identified here are true positives, but future studies will be needed to definitively confirm these localizations.
Localized Proteins Function in Spatio-Temporally Regulated Pathways.
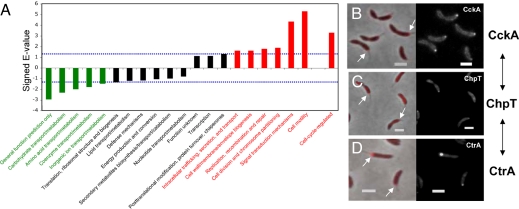
To generate insight into the cellular functions carried out by the Caulobacter proteins with nonuniform distributions, we performed an analysis of their Gene Ontology (GO) functional classifications (19). Genes involved in metabolic processes such as inorganic ion, amino acid, coenzyme, and carbohydrate transport and metabolism were significantly under-represented among localized proteins (Fig. 2A). The under-representation of proteins involved in small molecule metabolism suggests that the ability of small molecules to rapidly diffuse throughout the cell could obviate the need to localize their biosynthetic machinery. The over-represented GO term subcategories included cell motility, signal transduction mechanisms, cell division and chromosome partitioning, DNA replication/recombination/repair, cell wall/membrane/envelope biogenesis, and intracellular trafficking and secretion (Fig. 2A). Caulobacter motility structures are found at the cell poles and proteins that mediate cell division are found near mid-cell; therefore, cell motility and cell division proteins were expected to be enriched among localized proteins. Other over-enriched classes such as signal transduction, trafficking and secretion, and membrane and cell wall biogenesis were less expected. Although individual proteins with such functions have been reported to be localized, these data suggest that spatial organization may be of general significance for these processes. Together, the over-enriched GO classes suggest that localized proteins tend to be involved in the spatial and temporal regulation and sorting of cellular activities. Consistent with this result, we found that the genes whose transcription had been found to be cell-cycle-regulated (20), were also over-represented among our localized proteins (Fig. 2A).
Fig. 2.
Functions of localized Caulobacter proteins. (A) The enrichment of localized proteins was examined for each of the 21 GO-term functional subcategories and for a previously generated list of genes whose transcripts are cell-cycle-regulated (20). The dotted blue lines represent a P = 0.05 significance threshold. The names and values of over-enriched categories are shown in red and the under-enriched categories are shown in green. (B–D) The CckA, ChpT, and CtrA proteins that function together also localize together in Caulobacter. For each protein, an overlay of phase contrast and mCherry fluorescence images is shown on the Left and the fluorescence image alone is shown on the Right. Arrows point to fluorescent foci found at the stalked pole. (Scale bars represent 2 μm.)
Proteins that function together might be expected to localize together. To illustrate a connection between localization and function, we examined the localization of a protein that has been described by genetic and biochemical methods as functioning at a specific cellular location. ChpT (CC3470) is a histidine phosphotransferase that links the CckA hybrid 2-component kinase to its downstream CtrA response regulator (22). Both CckA and CtrA are found at the cell pole (23, 24), such that although ChpT localization has not been examined, it too was predicted to localize to the cell pole. The Caulobacter localizome data verified that ChpT indeed localized to the stalked pole with both CckA and CtrA (Fig. 2 B–D). ChpT only exhibited polar localization as an N-terminal fusion. Interestingly, the N terminus of ChpT is poorly conserved and predicted to be unstructured, whereas the C-terminal 200 aa of ChpT contain a well-conserved domain (COG5383). This correlation supports the hypothesis that proteins localized only as fusions to a single terminus may reflect and possibly reveal the presence of conserved structural motifs important for protein function or localization.
Quantitative Analysis of Large-Scale Protein Localization Data.
Quantitation of large-scale datasets has proven to be an important aspect of genomic studies. However, the quantitative analysis of protein localization data has not been routinely performed in high throughput. We recently developed a software suite termed PSICIC that enables automated image analysis of protein localization and intensity (7). PSICIC directly addresses 2 major obstacles to quantitating Caulobacter localization data: It uses interpolated contours to achieve accurate and precise subpixel resolution and generates an internal coordinate system for each cell that allows data from multiple cells to be directly compared regardless of variabilities in cell geometry (7). To reduce the complexity associated with interpreting heterogeneously distributed proteins, we excluded the “patchy/spotty” localized proteins from this initial analysis.
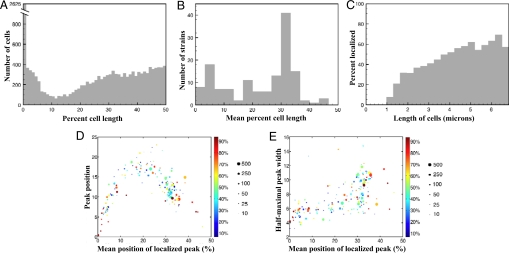
To generally survey Caulobacter protein localization, we first examined the frequency at which proteins are localized along the cell length by generating a histogram of the positions of peak fluorescence intensity in each localized cell (Fig. 3A). We found a very large enrichment of proteins localized to the extreme cell pole (<1% of cell length) (Fig. 3A). In addition, a region of the cell near the poles (5–25% of cell length) was noticeably depleted of localized proteins (Fig. 3A). Such a zone of reduced protein localization, or “localization coldspot,” has not been previously reported in bacterial cells and determining its molecular basis will prove interesting for future studies.
Fig. 3.
Quantitative analysis of fluorescent protein localization. (A) A histogram of the location of the fluorescent peak in individual cells, broken into 1% bins, shows high enrichment for polar localization and a region, between ≈5% and 20% of the cell length, where the focus is relatively rarely found. (B) A histogram of the mean position of localization for each strain, broken into 3.3% bins, shows enrichment for localization near the pole and at ≈1/3 the cell length (only strains with >30 cells exhibiting a fluorescent focus are shown). (C) The frequency of a cell displaying localization increases with cell length. (D) Strains exhibiting localization patterns near the pole or midcell have tighter distributions of localization than those at the midcell. The x axis is the mean position of each localized peak as a percentage of cell length. The y axis is the standard deviation of each peak position as a percentage of cell length. Each point represents a single strain, point size represents the number of localized cells present, and point color represents the percentage of cells in that strain exhibiting localization. (E) The mean diffuseness of the localized focus increases as the mean localization moves toward the midcell, with a large jump nearest midcell. The x axis is the mean position of each localized peak as a percentage of cell length. The y axis is the mean half-maximal width of each peak as a percentage of cell length. Point size and color as in D.
Examining the distribution of the mean position of each type of fluorescent fusion confirmed the enrichment of polar proteins and depletion of juxta-polar proteins (Fig. 3B). This analysis also revealed a second site of protein localization enrichment near the cell middle, at ≈30–40% of cell length (Fig. 3B), suggesting that the asymmetric division site of Caulobacter cells represents a second landmark or hotspot for protein localization. The mid-cell localization hotspot appears to be obscured in the single-cell data by the relatively noisy localization of these proteins, as detailed below. This noise likely also explains the slight shift of the polar peak in the averaged data. An analysis of the percentage of cells with localized proteins as a function of cell length revealed that protein localization becomes more prevalent with increasing cell length (Fig. 3C). This finding suggests that protein localization plays fewer functions early in the cell cycle, potentially because of the later development of the division-associated localization hotspot.
Quantitation of the Caulobacter localized protein library also enabled the analysis of how reproducibly and accurately proteins are localized in both cell populations and individual cells. To examine how reproducibly a specific type of fluorescent protein fusion is localized, we analyzed the standard deviation of the peak intensity positions of all of the localized cells for each strain (Fig. 3D). Near the cell poles, a low standard deviation of peak intensity position suggests that polar proteins are localized in a highly reproducible fashion. From the cell pole to the quarter-cell position, protein localization becomes less and less accurate, but then increases again in accuracy toward mid-cell. The increased prevalence and accuracy of polar and mid-cell proteins suggests that the majority of the active protein targeting mechanisms direct proteins to these hotspots. Strains with mean localization in other positions could reflect either rarer and less tightly regulated localization mechanisms or the averaged localization of proteins with bimodal distributions. To examine how accurately or tightly proteins are localized in individual cells, we calculated the distance between the peak intensity and its half-maximal value. By this metric, polar proteins are the most accurately distributed and both the range and level of localization distribution increases steadily as a function of the distance from the pole (Fig. 3E). The finding that the reproducibility of localization position within a cell population increases at mid-cell but the accuracy of protein distribution at the single-cell level does not, indicates that polar and mid-cell proteins may be localized by fundamentally different types of targeting mechanisms.
Discussion
New Resources for Studying Protein Localization and Functional Genomics.
Because of the extreme labor involved, previous large-scale protein localization studies have focused on imaging and subjectively analyzing a single set of C-terminal fusions. To address these limitations, we developed a series of high-throughput methods for generating, imaging, and quantitatively analyzing fluorescent protein fusions. In addition to localization efforts, these methods should benefit other types of functional genomic studies. The in vivo LR method for constructing protein fusions can be directly applied to any species and can be easily modified for the purposes of other types of genomic experiments such as 2-hybrid analysis, heterologous expression, or overexpression. Because the current study included the assembly of a modular Caulobacter ORFeome library, these other types of genomic studies will now also be feasible for Caulobacter. In addition, both the pedestal slide system for high-throughput high-resolution microscopy and the PSICIC software suite for quantitating localization data can be directly applied to other model systems, including other prokaryotes and both unicellular and multicellular eukaryotes.
By combining these methods into a pipeline for high-throughput localization analysis, we were able to examine the localization of the Caulobacter proteome as both N- and C-terminal fusions. In addition to enabling the rapid identification of localized proteins, the ability to rapidly reimage these existing fusion libraries should enable the future examination of how proteome localization responds to changes in growth conditions, mutant backgrounds, or pharmacological treatments. These methods thus provide a powerful postgenomic platform for global studies of protein localization.
The ability to readily examine protein localization promises to significantly advance our ability to understand the functions of individual proteins of interest. In addition, quantitating protein distributions enables the systems-level characterization of proteome localization. For example, our findings that there is a juxta-polar localization coldspot and that the polar and mid-cell localization hotspots may be mediated by different targeting mechanisms represent emergent properties that could not be readily determined by traditional methods. In the future it will be interesting to determine how such quantitative localization data can be most productively integrated with other genomic datasets.
Caulobacter Localizome.
In the past decade, the emerging field of bacterial cell biology has identified multiple examples of the use of subcellular localization to execute or regulate spatially restricted processes (4, 5). Our genomic analysis of protein localization in Caulobacter crescentus demonstrates that such localization is not exclusive to a small number of specific proteins, but rather is a common feature of the bacterial proteome, with >10% of proteins examined exhibiting a nonuniform subcellular distribution. These localized proteins assume a number of specific patterns that can be combined in different ways to achieve a larger number of possible localizations. This arrangement suggests that Caulobacter could use a relatively small number of targeting mechanisms to distribute a large number of proteins, and the identification of proteins restricted to the stalk indicates the existence of previously unappreciated localization pathways. Protein localization (at least at the resolution detectable by light microscopy) does not appear to play a role in the metabolism of rapidly diffusing small molecules. Localization does appear to be a general feature of cellular processes including cell division, motility, and signal transduction that involve spatial and temporal dynamics.
In addition to identifying a large number of new localized proteins and describing the types of proteins that tend to be localized, the fact that we repeated our analysis with both N- and C-terminal fusions allows us to generate insight into the types of fusions that can be localized. The localizome data demonstrate that many proteins cannot tolerate fusions to one terminus but are localized when the reporter is fused to the opposite terminus. The differences in these fusion proteins may result from disruptions in important structural or interaction motifs such that this dataset may provide future insight into specific protein localization mechanisms. Although the validation of individual localization patterns will require additional protein-specific experiments, global analysis suggests that our dataset contains relatively few false negatives or inclusion body-mediated false positives. These data thus empirically demonstrate the importance of studying genomic fusion libraries in multiple ways to achieve high confidence data.
Experimental Procedures
Entry Vector Library and Gateway Destination Vector Construction.
A detailed description of the methods used is contained in SI Experimental Procedures. Entry vector cloning into the donor vector pDONR223 (8) was performed for 3,763 annotated Caulobacter ORFs (CB15; National Center for Biotechnology Information) by using methods and reagents from the Invitrogen Gateway cloning system. Correct size and identity of the ORF contained in each entry vector was verified by PCR by using primers flanking the insert region followed by sequencing of each entry vector insert. Two destination vectors were engineered to allow xylose-inducible expression in Caulobacter of each ORF-encoded protein fused to mCherry at either the C terminus (gXRC) or N terminus (gXRN). Expression vectors were created by using an “in vivo LR” procedure described in SI Experimental Procedures. High-throughput conjugation was then used to transfer each expression vector from E. coli to Caulobacter.
High-Throughput Imaging and Image Scoring.
Induced Caulobacter strains were imaged on 48-pedestal slides (1% agarose) and fields of ≈50–200 cells were imaged with a Nikon90i epifluorescent microscope equipped with a 100 × 1.4 NA objective (Nikon), Rolera XR cooled CCD camera (QImaging), and NIS Elements software (Nikon). Phase contrast and fluorescent images were taken of each field by using an automated acquisition and file-handling script. The resulting images were scored by several individuals using custom MATLAB (MathWorks) scripts. Strains containing localized proteins were reimaged and sequencing was performed to verify the expected gene product.
Statistical Analysis and Quantitative Image Analysis.
Quantitative attributes of the localized Caulobacter proteins were compared with those of all Caulobacter proteins by using a 2-sample T test (Microsoft Excel) when appropriate. The overlaps between protein subsets were compared by using the hypergeometric distribution (MATLAB; MathWorks). Cell outlines were identified, cell length was measured, and 1-dimensional intensity profiles were calculated for each cell by using the PSICIC software toolkit, as described in ref. 7.
Supplementary Material
Acknowledgments.
We thank the members of the Gitai lab and Bonnie Bassler, Leonid Kruglyak, and Coleen Murphy for helpful discussions and input on the manuscript. We also thank Greg Phillips (Iowa State University, Ames IA), Michael Kahn (Washington State University, Pullman, WA), Martin Thanbichler (Max Planck Institute for Terrestrial Microbiology, Marburg, Germany), and M. R. Alley (Anacor Pharmaceuticals, Palo Alto, CA) for materials, and Denis Dupuy and the rest of Marc Vidal's lab for their help in constructing the Caulobacter ORFeome entry library. J.N.W is supported by a postdoctoral fellowship, Grant 1F32AI073043–01A1, from the National Institute of Allergy and Infectious Diseases; J.G. is supported by a fellowship from the Burroughs Wellcome Fund; and Z.G. is supported by funding from Grant DE-FG02-05ER64136 from the U.S. Department of Energy Office of Science (Biological and Environmental Research).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901781106/DCSupplemental.
References
- 1.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 2.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 3.Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 4.Gitai Z. The new bacterial cell biology: Moving parts and subcellular architecture. Cell. 2005;120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 5.Collier J, Shapiro L. Spatial complexity and control of a bacterial cell cycle. Curr Opin Biotechnol. 2007;18:333–340. doi: 10.1016/j.copbio.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell JH, Keiler KC. Screen for localized proteins in Caulobacter crescentus. PLoS ONE. 2008;3:e1756. doi: 10.1371/journal.pone.0001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: Noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput Biol. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walhout AJ, et al. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 9.Brasch MA, Hartley JL, Vidal M. ORFeome cloning and systems biology: Standardized mass production of the parts from the parts-list. Genome Res. 2004;14:2001–2009. doi: 10.1101/gr.2769804. [DOI] [PubMed] [Google Scholar]
- 10.Shaner NC, et al. Improved monomeric red, orange, and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 11.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 12.Meisenzahl AC, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rey S, et al. PSORTdb: A protein subcellular localization database for bacteria. Nucleic Acids Res. 2005;33:D164–D168. doi: 10.1093/nar/gki027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen RB, Wang SC, Shapiro L. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 2001;20:4952–4963. doi: 10.1093/emboj/20.17.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 16.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briegel A, et al. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. [DOI] [PubMed] [Google Scholar]
- 18.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 22.Biondi EG, et al. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 23.Ryan KR, Huntwork S, Shapiro L. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc Natl Acad Sci USA. 2004;101:7415–7420. doi: 10.1073/pnas.0402153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs C, Domian IJ, Maddock JR, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.