Abstract
Current global phylogenies are built predominantly on rRNA sequences. However, an experimental system for studying the evolution of rRNA is not readily available, mainly because the rRNA genes are highly repeated in most experimental organisms. We have constructed an Escherichia coli strain in which all seven chromosomal rRNA operons are inactivated by deletions spanning the 16S and 23S coding regions. A single E. coli rRNA operon carried by a multicopy plasmid supplies 16S and 23S rRNA to the cell. By using this strain we have succeeded in creating microorganisms that contain only a foreign rRNA operon derived from either Salmonella typhimurium or Proteus vulgaris, microorganisms that have diverged from E. coli about 120–350 million years ago. We also were able to replace the E. coli rRNA operon with an E. coli/yeast hybrid one in which the GTPase center of E. coli 23S rRNA had been substituted by the corresponding domain from Saccharomyces cerevisiae. These results suggest that, contrary to common belief, coevolution of rRNA with many other components in the translational machinery may not completely preclude the horizontal transfer of rRNA genes.
One of the most intriguing hypotheses in modern biology is that all organisms are descendants of an ancient, self-replicating entity that depended on RNA both for its genetic material and for catalysis (1). The notion of an “RNA world” has gained substantial support from the remarkable discovery of catalytic RNAs (1). Particularly striking is the demonstration that the peptidyl transferase step of protein synthesis is catalyzed in vitro by rRNA with few or no proteins present (2–4). This result is consistent with the highly conserved primary structure of rRNA in all living organisms (5, 6). This evolutionary conservation of rRNA has had a significant impact on several fields of study in biology. For systematic biology this conservation has been instrumental in revolutionizing taxonomy and in the construction of major phylogenetic trees (5–7). The identification, classification, and placement of many organisms into evolutionary schemes now heavily depend on the determination of the sequences of their rRNA genes. The rapid phylogenetic placement of even unculturable microorganisms is now possible once their rRNA genes have been sequenced (7–10). Although the validity of using a single gene for the analysis of organism history has been questioned (11), the omnipresence of rRNA and its seemingly slow pattern of evolutionary change have resulted in many successful applications in the practical sciences of clinical and environmental microbiology, for example. The comparative analysis of rRNA sequences also has played a major role in identifying many important structural and functional features of rRNA (12, 13).
Each species’ rRNA has evolved in a particular context of ribosomal proteins (r-proteins), tRNAs, and factors that must interact with rRNA at particular steps in its synthesis, assembly, and function. An experimental system to examine rRNA in a foreign setting, forcing it to interact and succeed in an evolutionarily distant environment therefore would provide a powerful method of studying the evolution of fundamental features of rRNA and other factors involved in protein synthesis. Development of such a system has been difficult, however, primarily because most organisms carry multiple, essentially identical rRNA genes. The repetitive nature of the genes also has restricted the mutational analysis of rRNA and the in vivo production of pure mutant ribosome populations that are necessary for a variety of in vitro assays (discussed in ref. 14). Homogeneous mutant ribosomes usually are obtained by the in vitro reconstitution of ribosomes (15, 16), but the drawback to this technique is that the activity of reconstituted ribosomes is generally low (14). The use of organisms and organelles carrying only one copy of an rRNA operon on their genomes has not been aggressively pursued to solve these problems because genetic and molecular biological techniques are not well developed in these systems (14). In this work we describe the construction of a mutant strain of E. coli in which all seven chromosomal rRNA operons have been deleted and replaced with a single rRNA operon carried by a plasmid, with the aim of developing a simple genetic system for expressing homogeneous rRNA in vivo. A similar system developed in yeast previously was reported but has not yet been broadly used (some applications of the yeast system were reported by Liebman and coworkers in refs. 17–21). The advantages of the E. coli system are that both classical genetics and recombinant DNA techniques are highly advanced in this organism and that the vast majority of biochemical, structural, and mutational studies of rRNA have been done on E. coli ribosomes, providing a broad base of information on which to add new observations (14, 22–24). We report here an application of our system to the experimental analysis of rRNAs from foreign species that are laterally transferred into E. coli, studies that essentially bypass normal evolutionary processes.
MATERIALS AND METHODS
Bacterial Growth Conditions.
Cells were grown at 37°C in Luria–Bertani broth. Overnight cultures were incubated without shaking and subcultured into fresh medium. The initial cell density of all subcultures was adjusted to 5 Klett units with a Klett-Summerson photoelectric colorimeter using a red filter and incubated with aeration by shaking. Antibiotics were added at the following concentrations when required: 100 μg/ml ampicillin (Ap), 50 μg/ml kanamycin (Km), 30 μg/ml chloramphenicol, 12.5 μg/ml tetracycline, and 40 μg/ml spectinomycin (Spc).
Construction of rrn-Deletion and rrn+ Strains.
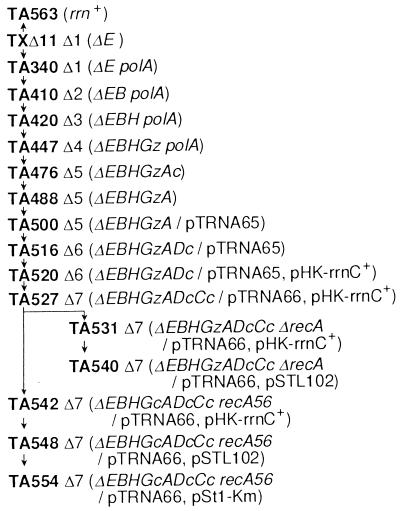
Details of strain construction will be published elsewhere. Briefly, starting with strain TXΔ11 carrying a deletion of rrnE (25), four operons (rrnB, rrnH, rrnG, and rrnA) were sequentially inactivated without using antibiotic resistance genes by a gene replacement technique. [The rrnG operon was inactivated with the lacZ coding region (ΔrrnG∷lacZ+).] The remaining rrn operons (rrnD and rrnC) were inactivated by P1-transducing deletion/insertion mutations (ΔrrnD∷cat+ and ΔrrnC∷cat+) in which the deleted regions were replaced with the cat (chloramphenicol resistance) gene (26).
Strain TA531 was constructed from TA527 by P1-transducing a recA-deletion mutation, Δ(srlR-recA)306, which was linked to srlR∷Tn10. Another recA derivative of TA527, TA542, was constructed by mating TA527 with an Hfr strain carrying the recA56 mutation linked to srlC∷Tn10. The Hfr strain also contained the ΔrrnG∷cat+ mutation and, during the mating, the ΔrrnG∷lacZ+ mutation in TA527 was replaced with ΔrrnG∷cat+.
An rrn+ strain, TA563, was constructed from TXΔ11 by introducing rrnE+ with Hfr mating. The donor strain was CAG5052 (27), and the exconjugants were screened for Pur+ and rrnE+.
Construction of pTRNA Plasmids.
The following tRNA-containing fragments (shown in Fig. 1A) were placed downstream from the tac promoter, generating pTRNA65. A DNA fragment (226 bp) carrying the tRNA genes for Asp-1 and Trp was amplified from the rrnC operon by PCR with the following primers: 5′-GCCGGTCATAAAATCGATGGTTG-3′, 5′-CCTTAGCTGTCGACAAGGATGAT-3′. DNA fragments containing the tRNA genes for Ile-1 and Ala-1B and the tRNA gene for Glu-2 were obtained by SmaI and HpaI digestion of the rrnD and rrnB operons, respectively. A fragment with the tRNA gene for Thr-1 and the rRNA transcription terminators was prepared by digesting the rrnD operon with XmnI and EcoRI. pTRNA66 was spontaneously generated from pTRNA65 by deletion of the gene for tRNA Glu-2.
Figure 1.
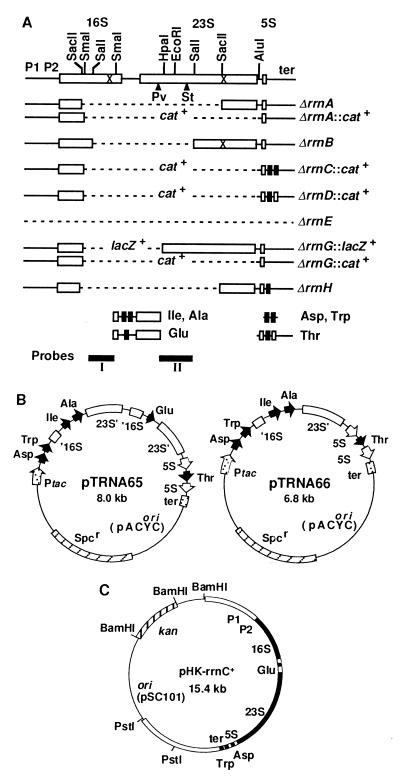
(A) Deletion mutations introduced into the chromosome. Open and filled rectangles are rRNA and tRNA genes, respectively. Deleted regions are shown by broken lines. cat+ and lacZ+ indicate the deleted regions are replaced with the coding regions of these genes. The rRNA promoters (P1P2) and terminators (ter) also are shown. The crosses in the 16S and 23S rRNA genes indicate the positions of the Spc and erythromycin resistance mutations, respectively. ▴ indicates the position of an intervening sequence in the 23S rRNA gene of S. typhimurium (St) or P. vulgaris (Pv). The four DNA fragments with tRNA genes cloned in pTRNA65 are shown below the rRNA operons. (B) pTRNA plasmids. Relative positions and orientation of the tac promoter, tRNA genes and the 5S rRNA gene are indicated by arrows. The rRNA transcription terminators and the truncated 16S and 23S rRNA genes also are shown. Truncation of the genes is indicated by a prime. The plasmids are not drawn to scale. (C) An rRNA plasmid carrying the wt rrnC operon. Filled and open boxes indicate stable RNA genes and their flanking sequences, respectively. The size of the BamHI–PstI fragment containing the rrnC operon is 8.4 kb.
Plasmid Replacement.
TA531 contains pHK-rrnC+ and is resistant to Km. This strain was transformed to Ap resistance with pSTL102 (28), and the transformants were grown to saturation in the absence of Km. The cultures were diluted and plated on Ap plates, and colonies on the plates were screened for sensitivity to Km. In this experiment, ≈20% of Ap-resistant transformants were Km-sensitive. The efficiencies of plasmid replacement in the other experiments were similar to this value.
Primer extension was carried out essentially as described (29).
Determination of rRNA/Protein Ratios.
The amount of total RNA was obtained by measuring the absorption at 260 nm of RNA hydrolysates as described in refs. 30 and 31. The amount of rRNA in stable RNA then was determined from the molar ratio of rRNA to tRNA. To obtain this ratio, total RNA was prepared from cells as described in ref. 32, and 5S rRNA and tRNA (4S) were fractionated by polyacrylamide gel electrophoresis (4% gel, see ref. 33), and the intensity of each band was determined with the IS-1000 Digital Imaging System (Alpha Innotech, San Leandro, CA). The ratio of rRNA to tRNA thus obtained with wild-type (wt) cells should represent the known ratio, i.e., one 5S rRNA molecule per nine tRNA molecules (30). The rRNA/tRNA ratios in other strains were calculated based on this assumption. The total size of rRNA and the average size of tRNAs used to obtain the amount of rRNA in total RNA were 4,566 bp and 80 bp, respectively. The amount of total protein was determined as described in ref. 31 with BCA Protein Assay Reagent (Pierce).
RESULTS
Deletion of All Seven E. coli Chromosomal rRNA Operons.
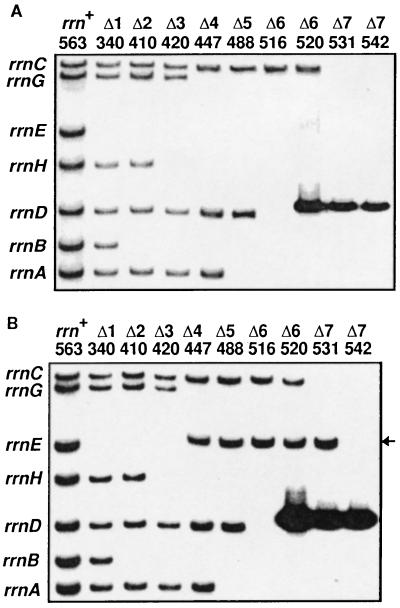
Each of the seven E. coli rRNA (rrn) operons contains three (16S, 23S, and 5S) rRNA genes (Fig. 1A). Ellwood and Nomura (25) were the first to construct several E. coli strains in which all three rRNA genes of the rrnE operon were completely deleted. We chose one of their strains (TXΔ11) for the starting material and sequentially introduced deletion mutations into the 16S and 23S rRNA genes of the remaining six operons (see Materials and Methods). The deletions are shown in Fig. 1A, and the relevant strains and the order of deletions introduced are summarized in Fig. 2. The lack of intact 16S and 23S rRNA genes in these strains (termed here Δ1, Δ2, Δ3, etc.) was verified by Southern blot analysis, and a typical result is shown in Fig. 3.
Figure 2.
The pedigree of rrn-deletion strains. Inactivated rRNA operons are indicated by capital letters derived from their specific operon names (for example, A for rrnA). When the inactivation was carried out by a deletion/insertion mutation, a capital letter is followed by a lowercase letter c or z representing the inserted gene cat+ or lacZ+, respectively (for example, Ac for rrnA∷cat+). pTRNA, pHK-rrnC+, and pSTL102 contain Spc, Km, and Ap resistance markers, respectively. All rRNA operons cloned in the plasmids shown contain the gene for tRNA Glu-2.
Figure 3.
Autoradiograms of Southern blot hybridization showing the inactivation of chromosomal rRNA operons. DNA was analyzed as described in ref. 26. The number of operons inactivated and strain numbers are shown on the top. The rRNA operon carried by each fragment is shown on the left. (A) Inactivation of the 16S rRNA genes. The SalI–SmaI fragment (probe I, Fig. 1A) of the 16S rRNA gene was labeled with 32P and used for hybridization. TA520, 531, and 542 contain an rRNA plasmid, pHK-rrnC+ and thus give an additional band (8.4 kb) carrying the plasmid-borne rrnC operon (see Fig. 1C) just above the rrnD band (8.1 kb). (B) Inactivation of the 23S rRNA genes. Probe I was removed from the membrane shown in A, and the cellular DNA was rehybridized with 32P-labeled probe II carrying the DNA sequence between the HpaI and the SalI sites in the 23S rRNA gene (Fig. 1A). TA520, 531, and 542 again gave a plasmid-derived band. The ΔrrnG∷lacZ+ construct contains the HpaI–SalI region of the gene (Fig. 1A). Therefore, in TA447, 488, 516, 520, and 531 in which the rrnG operon was inactivated with this construct, the rrnG+-containing band (15.5 kb) disappeared and a new band with the expected size (11.6 kb, indicated by an arrow) appeared just above the rrnE band (11.2 kb). This band disappeared in TA542 in which ΔrrnG∷lacZ+ was replaced with ΔrrnG∷cat+.
Each rRNA operon contains at least one tRNA gene between the 16S and 23S rRNA genes; rrnB, rrnC, rrnE, and rrnG contain the tRNA gene for Glu-2, whereas the tRNA genes for Ile-1 and Ala-1B are found in rrnA, rrnD, and rrnH (34). These tRNA (spacer tRNA) genes are encoded only in the rRNA operons. Because introduction of our deletions ultimately removes all spacer tRNA genes, we have cloned these genes, as well as other tRNA (distal tRNA) genes encoded in the rRNA operons, into a derivative of pACYC184 carrying the Spc resistance marker (resulting in plasmids pTRNA65 and 66, Fig. 1B). The presence of one of these plasmids is essential for the viability of TA516, in which only the rrnC operon is left on the chromosome, and derivatives of this strain. Similarly, a plasmid producing active 16S and 23S rRNAs (termed here prrn) is essential for strains carrying no intact rRNA operons on the chromosome. We used a derivative of pSC101 containing the wt rrnC operon, pHK-rrnC+ (Fig. 1C), for the construction of the first Δ7 prrn strain, TA527.
Expression of Homogeneous rRNA in Δ7 prrn: Manipulation of rRNA Species by Plasmid Replacement.
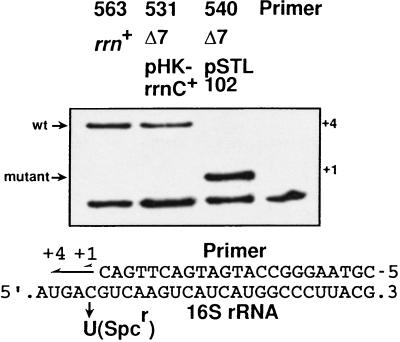
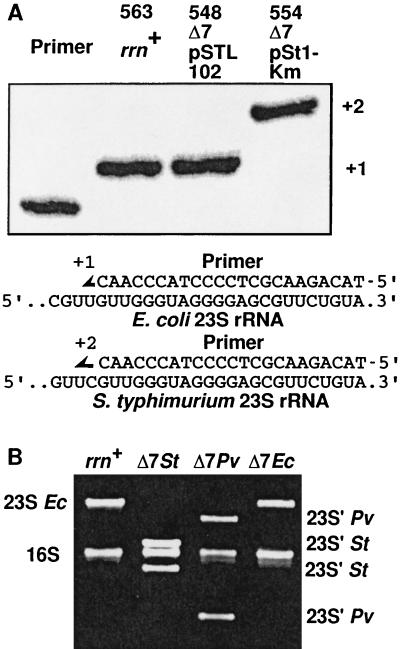
Pure ribosome populations containing rRNA molecules from a single operon are optimal for detailed analyses of rRNA functions in vitro. The Δ7 prrn strain should provide such a population of homogeneous ribosomes as it contains only a single intact rRNA operon on a plasmid. To confirm that this homogeniety was the case, we replaced the rRNA-producing plasmid, pHK-rrnC+, in a recA derivative of Δ7 prrn (TA531, Fig. 2) with a plasmid producing rRNA from a different operon, pSTL102 (plasmid replacement, see Materials and Methods). The resulting strain is TA540. Plasmid pSTL102 is a derivative of pBR322 carrying a mutant, but functional, rrnB operon of E. coli (28). It contains single base substitutions in the 16S rRNA gene at position 1192, conferring resistance to Spc, and in the 23S rRNA gene at position 2058, conferring resistance to erythromycin (Fig. 1A; ref. 35). Either of these base substitutions can be distinguished from the wt base by a modified primer extension method developed by Morgan and coworkers (29). As shown in Fig. 4, wt 16S molecules were not detected in a total RNA sample prepared from TA540, suggesting that 16S rRNA molecules in Δ7 prrn strains were indeed homogeneous. By using an rRNA plasmid carrying the S. typhimurium rrnD operon, we also showed that 23S rRNA molecules in Δ7 prrn strains were homogeneous (see below). We conclude that both 16S and 23S rRNA molecules in Δ7 prrn strains are homogeneous and that the plasmid replacement technique provides a powerful tool for manipulating rRNA species in Δ7 prrn strains.
Figure 4.
Expression of homogeneous rRNA in Δ7 prrn strains. 16S rRNA molecules in the rrn+ strain and in Δ7 prrn strains carrying either wt (pHK-rrnC+) or mutant (pSTL102) rRNA plasmid were analyzed by primer extension. DNA primers annealed to 16S rRNA molecules were extended in the presence of one dideoxynucleotide (ddATP) and three deoxynucleotides (dGTP, dCTP, and dTTP). Because mutant 16S rRNA produced from pSTL102 contains a C to U change, primers hybridized to wt and mutant 16S molecules are extended by four nucleotides and one nucleotide, respectively.
Expression of Foreign rRNA in the Δ7 prrn Strain.
As a first step toward the experimental analysis of rRNA evolution, we constructed a strain producing homogeneous hybrid ribosomes in vivo. These ribosomes contain S. typhimurium 16S and 23S rRNAs and E. coli r-proteins. Unlike E. coli rRNAs, the 23S rRNAs of S. typhimurium contain at least one intervening sequence that is excised by RNase III during rRNA maturation (36). This cleavage results in 23S rRNA molecules that are fragmented, but nevertheless functional in the bacterium. pSt1 is a derivative of pBR322 carrying the entire rrnD operon (including regulatory regions) of S. typhimurium (36). The 23S gene in this operon contains a single intervening sequence in helix 45 beginning at position 1164 (Fig. 1A). We inserted the Km resistance gene into pSt1 within the vector sequence and the resulting plasmid, pSt1-Km, was introduced into a Δ7 prrn strain TA548, replacing pSTL102 and generating TA554 (Fig. 2). To prove that this E. coli strain was growing uniquely with S. typhimurium 16S and 23S rRNAs, we did the following experiments showing that only S. typhimurium rRNA was expressed. First, no E. coli 23S rRNA molecules could be detected by primer extension on a total RNA preparation isolated from TA554 (Fig. 5A). Second, all of the 23S rRNA molecules visible in this strain on agarose gels were fragmented, characteristic of Salmonella rRNA; no intact 23S rRNAs could be seen (Fig. 5B). Third, the 500-bp EcoRI–SalI fragment present in all E. coli 23S genes was not detected by Southern blot of a total DNA preparation isolated from TA554, using probe II (shown in Fig. 1A), specific for early 23S sequences. Instead, we detected a larger fragment, characteristic of Salmonella operons, the result of the presence of the ≈90-bp intervening sequence (data not shown; ref. 36). Fourth, the sequence of cDNA molecules amplified by PCR from 16S rRNA in TA554 corresponded uniquely to S. typhimurium rRNA (data not shown). From these results, we conclude that S. typhimurium 16S and 23S rRNAs can form functional ribosomes in vivo, which, in turn, can interact correctly with the other components of the E. coli translational machinery. We detected very little difference in either growth rate or rRNA/protein ratio between TA548, containing E. coli ribosomes, and TA554, containing hybrid ribosomes (Table 1). Because a ribosome defect or deficiency would be predicted to lower the growth rate of the cells (30), these results suggest that translation efficiency and fidelity of the hybrid ribosomes are similar to those of E. coli ribosomes.
Figure 5.
Expression of foreign rRNA in Δ7 prrn strains. (A) Primer extension analysis. 23S rRNA molecules in the rrn+ strain and in Δ7 prrn strains carrying either E. coli (pSTL102) or S. typhimurium (pSt1-Km) rRNA plasmid were analyzed by primer extension as described in Fig. 4. The primer used in this experiment hybridizes to a common sequence in E. coli and S. typhimurium 23S rRNAs but gives different extension products because of a sequence difference between the two molecules. (B) Fragmentation of 23S rRNA. Total cellular RNA prepared from the rrn+ strain and Δ7 prrn strains carrying plasmids with E. coli (Ec, pSTL102), S. typhimurium (St, pSt1-Km), or P. vulgaris (Pv, pPM2) rRNA is shown. Fragmented 23S rRNA molecules are indicated by a prime.
Table 1.
Doubling times and rRNA/protein ratios of Δ7 prn cells expressing E. coli or hybrid ribosomes
| rRNA-producing plasmid | rRNA
species
|
Doubling time, min | rRNA/protein ratio, relative value | |
|---|---|---|---|---|
| 16S | 23S | |||
| pSTL102 | Ec | Ec | 47.9 ± 1.5 | 1.00 ± 0.02 |
| pSt1-Km | St | St | 52.6 ± 2.4 | 0.99 ± 0.02 |
| pPM2 | Pv | Pv | 55.6 ± 1.1 | 0.97 ± 0.02 |
| pWM1 | Ec | Ec/yeast | 61.8 ± 1.6 | 0.93 ± 0.05 |
Ec, E. coli; St, S. typhimurium; Pv, P. vulgaris; Ec/yeast, E. coli/yeast hybrid.
We used the same approach to replace the E. coli rRNAs with those from one of the most distantly related enteric bacteria, Proteus vulgaris (37). Although the sequence of the 23S rRNA gene of P. vulgaris is unknown, we found that the 23S gene cloned in pPM2 (38) contained a large (200 bp) intervening sequence in helix 25 beginning at position 533 (Fig. 1A; D.Z. and C.L.S., unpublished data). As shown in Fig. 5B, we were able to replace all of the E. coli 23S rRNA molecules with the corresponding P. vulgaris rRNAs. When cDNA amplified from 16S rRNA in a Δ7 prrn strain carrying pPM2 was sequenced, only P. vulgaris rRNA was detected (data not shown). Although the rRNA/protein ratio was unchanged, the doubling time of the cells expressing P. vulgaris rRNA was slightly larger than that of E. coli (Table 1). We conclude that P. vulgaris rRNA can function effectively with E. coli translational components, but the slightly lowered growth rate suggests a somewhat lowered ribosome efficiency. In contrast, an rRNA operon from Pseudomonas aeruginosa, which is more distantly related to E. coli than P. vulgaris, failed to replace an E. coli operon. We have used the expression and processing regions of the Pseudomonas operon, perhaps these signals do not function adequately in E. coli. Experiments are in progress to determine the specific characteristics of the P. aeruginosa operon that prevent it from functioning in E. coli.
Thompson et al. (39) replaced the r-protein L11 binding domain comprising the GTPase center of E. coli 23S rRNA with the equivalent region from yeast 26S rRNA. A plasmid (pWM1) expressing such a hybrid 23S rRNA, along with wt E. coli 16S rRNA, had no effect on the growth rate of wt E. coli cells, despite the fact that more than 75% of the ribosome population was mutant. Because the secondary, but not the primary, structure of this domain is extremely conserved between E. coli and yeast rRNAs, the result suggested that only the secondary structure is crucial for rRNA-L11 interaction in vivo. The hybrid particles did, however, show a somewhat reduced rate of GTP hydrolysis in vitro compared with wt ribosomes (39). We were interested to see whether E. coli could survive with 100% mutant ribosomes. We successfully replaced the plasmid-borne E. coli rRNA operon in the Δ7 prrn strain with the E. coli/yeast hybrid operon, showing that a pure population of hybrid ribosomes can support growth of E. coli. However, unlike the result obtained by Thompson et al. (39), the doubling time of Δ7 prrn cells containing the hybrid ribosomes was increased by 29% (Table 1). This increase might be related to the decreased GTPase activity of the hybrid ribosomes. We believe that the use of the Δ7 prrn strain has allowed the in vivo unmasking of an otherwise subtle phenotype. In contrast to the doubling time, the rRNA/protein ratio in Δ7 prrn cells expressing the hybrid ribosomes remained similar to that in cells containing E. coli ribosomes (Table 1). We therefore conclude that although the E. coli/yeast hybrid 50S subunits are functional, their efficiency is significantly decreased relative to E. coli ribosomes. There are 20 single base differences between the E. coli and yeast sequence in this domain (39), so it is possible that the primary structure is also important for its interaction with r-protein L11 and other translational components.
DISCUSSION
Early studies by Nomura and coworkers (40) demonstrated that functional 30S subunits can be reconstituted in vitro from the 16S rRNA of one species of bacteria and the r-proteins of a distantly related species. They concluded that only certain regions of rRNA are evolutionary conserved and directly involved in the specific interaction with r-proteins in the assembly of ribosomal particles. Comparison of various rRNA sequences and localization of r-protein binding sites subsequently have confirmed their conclusions (12, 13, 23, 24), providing the validity of using hybrid ribosomes for studying the evolution of rRNA and r-proteins. Such an approach has not been extensively exploited, however, probably because the ribosome is a large and complex ribonucleoprotein particle and reconstitution of ribosomes is technically difficult. In vivo analysis of rRNA evolution also has been hindered by the repetitive nature of the rRNA genes. Formation of hybrid ribosomes in vivo first was reported by Stackebrandt’s group (38). They expressed P. vulgaris rRNA in E. coli from a plasmid-borne gene and found that ≈25% of total ribosomes contained Proteus 16S rRNA (38). This group also introduced an entire rRNA operon of P. vulgaris into the E. coli chromosome and showed that the fraction of hybrid ribosomes was ≈5% (41). In the study reported here, we have succeeded in generating 100% homogeneous hybrid ribosomes in vivo. This system therefore should provide a powerful tool for the study of rRNA evolution.
S. typhimurium and P. vulgaris differ from E. coli in the 16S coding sequence by 3.2% and 6.8%, respectively. Although one thinks of E. coli and S. typhimurium as rather close relatives, Ochman and Lawrence (37) estimate, based on 16S rRNA sequence differences, that they diverged from a common ancestor about 120–140 million years ago. Thus, their lineages probably have been separated since the advent of mammals and flowering plants. Also based on 16S rRNA sequence differences, it has been suggested that E. coli diverged from P. vulgaris about 350 million years ago (42, 43). The rRNAs of Salmonella and Proteus have evolved independently within the constraints of their own specific protein and RNA factors. For example, the interaction sites mapped for E. coli r-proteins S8, S9, S10, S19, and S20 have different sequences in S. typhimurium. A similar analysis of the P. vulgaris 16S rRNA sequence suggests that the S8, S9, S10, and S14 interaction sites are altered. The stem-loop structure between positions 1130 and 1140 of E. coli 16S rRNA has been shown to be important for interaction with S9 and S10 (23). Four of five bases in this loop region are different in P. vulgaris, whereas two are changed in S. typhimurium. It therefore is surprising that Salmonella and Proteus rRNAs are able to form functional hybrid ribosomes with E. coli r-proteins and translation factors. It is even more surprising that E. coli cells containing only these hybrid ribosomes grow nearly as well as those containing pure E. coli ribosomes. As we have done with the E. coli/yeast hybrid, studies in which the rRNA genes of even more distantly related organisms are placed into E. coli should provide considerable information about specific interactions within the translational machinery, elucidating the limits to which these interactions can be extended. Furthermore, the fate of heterologous rRNA genes that are transferred into E. coli from different sources can be experimentally evaluated with the rrn-deletion strains. This work should help to understand the significance of horizontal gene transfer (44), one of the most debated questions in the field of molecular evolution and one that recently has become the focus of much attention (11).
In addition to experimental evolutionary biology, the rrn-deletion strains can be used for many other studies of rRNA. For example, one can directly test the hypothesis that the downstream box enhancer element in some mRNAs requires base pairing to a specific region of the 16S rRNA by simply changing that region of the 16S sequence (45). Replacement of the plasmid-encoded wt rRNA operon in a Δ7 prrn strain with one in which the 16S gene was altered in the antidownstream box was predicted to result in a cell in which mRNAs with a downstream box base pairing are not selectively translated. However, preliminary data suggest that the downstream box does not form the proposed base-pairing interaction with mRNA and indicate that enhancement of translation by the downstream box does not involve rRNA-mRNA base pairing (M. O’Connor, C.L.S., and A. E. Dahlberg, unpublished work).
In conclusion, the rrn-deletion strains described here should find broad fundamental and practical application and prove to be exceptionally useful for the experimental analysis of many aspects of rRNA evolution, regulation, physiology, structure, and function.
Acknowledgments
We thank N. R. Pace for pSt1; E. Stackebrandt, R. Söller, and R. K. Hartmann for pPM2; W. Ludwig and R. K. Hartmann for a plasmid containing a P. aeruginosa rRNA operon; A. E. Dahlberg for pWM1; A. E. Dahlberg and all members of his laboratory for helpful discussions; C. Condon, A. E. Dahlberg, P. Dennis, W. F. Doolittle, R. K. Hartmann, M. O’Connor, A. Wright, and C. Yanofsky for comments on the manuscript; and Binghua Shen for technical assistance. This work was supported by National Institutes of Health Grant GM24751 to C.L.S.
ABBREVIATIONS
- r-protein
ribosomal protein
- Ap
ampicillin
- Km
kanamycin
- Spc
spectinomycin
- wt
wild type
Footnotes
A Commentary on this article begins on page 1820.
References
- 1.Gesteland R F, Atkins J F, editors. The RNA World. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 2.Noller H, Hoffarth V, Zimniak L. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 3.Nitta I, Ueda T, Watanabe K. RNA. 1998;4:257–267. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Nitta I, Kamada Y, Noda H, Ueda T, Watanabe K. Science. 1998;281:666–669. doi: 10.1126/science.281.5377.666. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K H. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 6.Woese C R. In: Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Biosynthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC Press; 1996. pp. 23–48. [Google Scholar]
- 7.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong E F, Wu K Y, Prezelin B B, Jovine R V M. Nature (London) 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 10.Barns S M, Delwiche C F, Palmer J D, Pace N R. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennisi E. Science. 1998;280:672–674. doi: 10.1126/science.280.5364.672. [DOI] [PubMed] [Google Scholar]
- 12.Woese C R, Pace N R. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 91–117. [Google Scholar]
- 13.Gutell R R, Larsen N, Woese C R. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green R, Noller H F. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 15.Nomura M, Held W A. In: Ribosomes. Nomura M, Tissieres A, Lengyel P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. pp. 193–223. [Google Scholar]
- 16.Nierhaus K H. In: Ribosomes: Structure, Function, and Genetics. Chambliss G, Craven G R, Davies J, Davis K, Kahan L, Nomura M, editors. Baltimore: University Park Press; 1980. pp. 267–294. [Google Scholar]
- 17.Chernoff Y O, Vincent A, Liebman S W. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liebman S W, Chernoff Y O, Liu R. Biochem Cell Biol. 1995;73:1141–1149. doi: 10.1139/o95-123. [DOI] [PubMed] [Google Scholar]
- 19.Liu R, Liebman S W. RNA. 1996;2:254–263. [PMC free article] [PubMed] [Google Scholar]
- 20.Chernoff Y O, Newman G P, Liebman S W. Proc Natl Acad Sci USA. 1996;93:2517–2522. doi: 10.1073/pnas.93.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velichutina I V, Rogers M J, McCutchan T F, Liebman S W. RNA. 1998;4:594–602. doi: 10.1017/s1355838298980049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson K S, Noller H F. Cell. 1998;92:337–349. doi: 10.1016/s0092-8674(00)80927-7. [DOI] [PubMed] [Google Scholar]
- 23.Noller H F, Nomura M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: ASM Press; 1996. pp. 167–186. [Google Scholar]
- 24.Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Biosynthesis. Boca Raton, FL: CRC Press; 1996. [Google Scholar]
- 25.Ellwood M, Nomura M. J Bacteriol. 1980;143:1077–1080. doi: 10.1128/jb.143.2.1077-1080.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Condon C, Philips J, Fu Z-Y, Squires C, Squires C L. EMBO J. 1992;11:4175–4185. doi: 10.1002/j.1460-2075.1992.tb05511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singer M, Baker T, Schnitzler G, Deischel S, Goel M, Dove M, Jaacks K, Grossman A, Erikson J, Gross C. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triman K, Becker E, Dammel C, Katz J, Mori H, Douthwaite S, Yapijakis C, Yoast S, Noller H F. J Mol Biol. 1989;209:645–653. doi: 10.1016/0022-2836(89)92000-7. [DOI] [PubMed] [Google Scholar]
- 29.Sigmund C D, Ettayebi M, Borden A, Morgan E A. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- 30.Bremer H, Dennis P. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 31.Brunschede H, Dove T L, Bremer H. J Bacteriol. 1977;129:1020–1033. doi: 10.1128/jb.129.2.1020-1033.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emilsson V, Kurland C. EMBO J. 1990;9:4359–4366. doi: 10.1002/j.1460-2075.1990.tb07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peacock A C, Dingman C W. Biochemistry. 1967;6:1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- 34.Komine Y, Adachi T, Inokuchi H, Ozeki H. J Mol Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- 35.Sigmund C D, Ettayebi M, Morgan E A. Nucleic Acids Res. 1984;12:4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgin A B, Parados K, Lane D J, Pace N R. Cell. 1990;60:405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- 37.Ochman H, Lawrence J G. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: ASM Press; 1996. pp. 2627–2637. [Google Scholar]
- 38.Niebel H, Dorsch M, Stackebrandt E. J Gen Microbiol. 1987;133:2401–2409. doi: 10.1099/00221287-133-9-2401. [DOI] [PubMed] [Google Scholar]
- 39.Thompson J, Musters W, Cundliffe E, Dahlberg A E. EMBO J. 1993;12:1499–1504. doi: 10.1002/j.1460-2075.1993.tb05793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomura M, Traub P, Bechmann H. Nature (London) 1968;219:793–799. doi: 10.1038/219793b0. [DOI] [PubMed] [Google Scholar]
- 41.Stackebrandt E, Niebel H, Söller R. J Gen Appl Microbiol. 1991;37:141–146. [Google Scholar]
- 42.Ochman H, Wilson A. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 43.Dennis P P, Shimmin L C. Microbiol Mol Biol Rev. 1997;61:90–104. doi: 10.1128/mmbr.61.1.90-104.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith M W, Feng D-F, Doolittle R F. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 45.Sprengart M L, Porter A G. Mol Microbiol. 1997;24:19–28. doi: 10.1046/j.1365-2958.1997.3161684.x. [DOI] [PubMed] [Google Scholar]