Abstract
Antigen expressed as MHC Class I glycoprotein (pMHCI) complexes on dendritic cells is the primary driver of CD8+ T cell clonal expansion and differentiation. As we seek to define the molecular differences between acutely stimulated cytotoxic T lymphocyte (CTL) effectors and long-lived memory T cells, it is essential that we understand the duration of in vivo pMHCI persistence. Although infectious influenza A virus is readily cleared by mammalian hosts, that does not necessarily mean that all influenza antigen is totally eliminated. An exhaustive series of carefully controlled adoptive transfer experiments using 3 different carboxy fluorescein diacetate succinimidyl ester–labeled T cell receptor–transgenic CTL populations and a spectrum of genetically engineered and wild-type influenza A viruses provided no evidence for pMHCI persistence over the 30–60-d interval after virus challenge. Molecular profiles identified in antigen-specific T cells at this time may thus be considered to reflect established immunologic memory and not recent CTL activation from a persistent pMHCI pool.
Virus-specific CD8+ T cell–mediated immunity is a critical component of the host response. Naive CD8+ T cells recognize virus-derived peptides presented in the context of MHC Class I glycoproteins (pMHCI). Encountering these pMHCI complexes on professional antigen-presenting cells (APCs) in draining lymph nodes (1) triggers CD8+ T cell proliferation and differentiation (2, 3) to mediate cytotoxic T lymphocyte (CTL) effector function, a primary mechanism of virus clearance (4). Other clonally expanded CD8+ T cells survive to establish a stable memory pool (2). Although it is possible to probe the molecular events that underlie these events using in vitro culture systems, what happens to T cells during and after an infectious challenge is optimally determined by minimizing the extent of manipulation after lymphocyte separation from the intact host. For example, we have recently demonstrated that a minority of influenza-specific memory CTLs recovered directly ex vivo can maintain cytotoxic gene expression for up to 1 year after infection (5). Understanding what this means in a molecular sense requires a clear picture of the likely stimulatory environment. Defining the importance of antigen load through the establishment and maintenance phases of CD8+ T cell immunity is also essential if we are to take rational approaches to vaccine design.
Some viruses that first establish an acute, lytic infectious process have evolved mechanisms that allow them to persist at a low level (or in a latent form) for the life of the host. This is not the case for influenza in immunologically competent mice or, so far as we are aware, in other mammalian species. Numerous lines of evidence based on (i) feeding the thymidine analogue bromodeoxyuridine to monitor antigen-specific T cell expansion (6), (ii) isolating dendritic cells (DCs) to probe for pMHCI expression (7), and (iii) PCR to detect viral genome in the recovered lung (8–10) support the view that the antigenic footprints of influenza virus infection cannot be detected for more than 16 d or so after the initial exposure. This conclusion has, however, recently been challenged by published data indicating that influenza A virus pMHCI complexes can, when probed by the adoptive transfer of naïve carboxy fluorescein diacetate succinimidyl ester (CFSE)-labeled T cell receptor (TCR) transgenic (Tg) CD8+ T cells, be maintained (or generated) for months after infection (8, 9, 11). Given the utility of the influenza A virus mouse model for comparing effector and memory responses by a spectrum of pMHCI-specific CD8+ T cells, it is obviously important to have a clear picture of acute and persistent antigen load in this infectious process. The demonstration that antigen can activate naïve T cells months after viral clearance has the potential to change current dogma regarding how virus-specific responses are initiated and maintained. It is paramount to test the validity of such findings using extensive analyses because we may have to reinterpret previous results in light of these findings. For example, maintenance of CD8+ T cell memory is currently considered not to require pMHCI persistence (12–14). Is this indeed the case?
Here, we investigated pMHCI presentation in the respiratory tract and regional lymph nodes for weeks after the resolution of acute influenza A virus infection. This comprehensive search for the long-term (LT) maintenance of pMHCI antigen used multiple assays of T cell function and investigated a panel of influenza-derived epitopes. No support whatsoever was found for the contention that influenza A virus pMHCI complexes either persist or are generated LT after the clearance of infectious virus.
Results
The protocol used throughout to probe both acute antigen load and the duration of pMHCI persistence after primary (1°) exposure was to infect naïve B6 mice intranasally (i.n.) with a nonlethal dose of one or other WT or engineered (peptide in the NA stalk) influenza A virus. In secondary (2°) challenge experiments, variants of non–cross-neutralizing H1N1 and H3N2 influenza A viruses were given i.n. at least 30 d after primary infection. The TCR Tg CD8+ T cells that were labeled with CFSE, then used to detect the presence of antigen, were recovered directly from the lymph nodes of naïve mice and injected i.v. into virus-primed recipients at intervals after infection. The viruses used were the H1N1 isolates A/WS/N/33 (WSN) and A/PR/8/34 (PR8), or the H3N2 A/NT/68 (NT68) and A/HK/x31 (X31) strains. The peptides engineered into the various PR8, WSN, and X31 viruses were derived from the ovalbumin (OVA257–264) and herpes simplex virus gB (gB489–505) proteins. The TCR Tg CD8+ T cells specific for the KbOVA257, KbgB489–505, and NT68 DbNP366–374 epitopes are designated OT-I, gBT, and F5, respectively. The WT nucleoprotein (NP) DbNP366–374 epitope recognized by responding CD8+ T cells is the same for PR8 (ASNENMETM) and X31 (a reassortant with all of the PR8 internal proteins), whereas the NT68 peptide (ASNENMDAM) that targets the F5 TCR Tg CD8+ T cells is different.
Kinetics of KbOVA257 Presentation After 1° and 2° X31-OVA Infection.
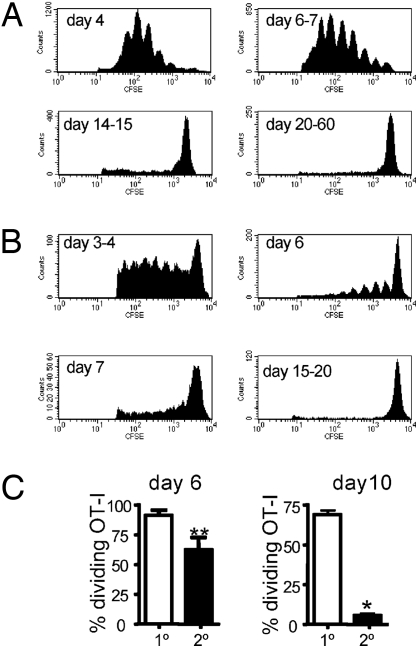
The first experiments looked at pMHCI persistence after 1° respiratory exposure (Fig. 1A). Naïve B6 mice were infected i.n. with the X31-OVA virus, then given 2 × 106 CFSE-labeled OT-I T cells at intervals thereafter. The regional, mediastinal lymph nodes (MLNs) were sampled at ≈64 h after transfer, and the CFSE profiles were assessed to determine the extent of cell cycling. By this measure it was apparent that there was abundant antigen present at Day 4 (d4) and d6–7 after infection, some residual KbOVA257 on d14–15, and nothing detectable by d20–60 (Fig. 1A). We then repeated the analysis after 2° infection (Fig. 1B). The B6 mice were first exposed i.n. to X31-OVA, then challenged i.n. with PR8-OVA after a further 30–60 d (Fig. 1B). The CFSE dilution results for the transferred OT-Is indicated that, as might be expected for previously primed mice, viral antigen was cleared more quickly. Peak antigen load detected by the transferred OT-Is was now seen at d3–4, cycling was much reduced by d6–7, and there was no evidence of antigen persistence at d15–20 (Fig. 1B). The significant difference in the duration of antigen expression after 1° and 2° challenge was confirmed in a further experiment in which we made a contemporary comparison (Fig. 1C). Overall, the findings are in accord with the well-established kinetics of infectious virus clearance from the lung after 1° or 2° virus challenge (6, 15).
Fig. 1.
Kinetics of KbOVA257 presentation after 1° and 2° X31-OVA infection. (A) Antigen-driven CD8+ T cell proliferation at intervals after primary infection: the Ly5.2+ B6 mice were given 1 × 104 PFU X31-OVA i.n., followed by 2 × 106 CFSE-labeled Ly5.1+ OT-I CD8+ T cells i.v. at 4 d, 6 to 7 d, 14 to 15 d, or 20–60 d later. The MLNs were harvested ≈64 h after transfer, and OT-I division was assessed by flow cytometry. Histograms display CD8+ Ly5.1+ CFSE+ cells. The data are representative of 2–4 independent experiments with 2 to 3 mice per group. (B) Antigen persistence after 2° challenge: the B6 mice were primed i.n. with 1 × 104 PFU X31-OVA >30 d previously, then challenged i.n. with 50 PFU PR8-OVA, and the experiment described in A was repeated. Data are representative of a minimum of 3–7 mice per time-point. (C) Contemporary comparison of pMHCI presentation on d6 and d10 after 1° or 2° virus challenge. Bars represent mean ± SEM. *P < 0.05; **P < 0.01.
Probing Influenza pMHCI Persistence with LT CFSE Transfer Assay.
The analysis in Fig. 1 did not confirm the conclusions from recent studies demonstrating that influenza pMHCI complexes persist LT after virus clearance from the lung (8, 9, 11). However, Zammit et al. (8) emphasized that it is necessary to allow the CFSE-labeled T cells a much longer interval (9 d) than the 64 h we used in Fig. 1 if they are to adequately “sample” the antigen environment of the host. With this LT CFSE assay, we were concerned that exposing the transferred CD8+ T cells to previously infected environments for such protracted intervals might enhance “homeostatic” proliferation independent of specific pMHCI availability. We thus included both naïve (uninfected) mice and hosts infected with viruses lacking the cognate immunogenic peptides as controls to ensure that any CD8+ T cell division was indeed antigen specific.
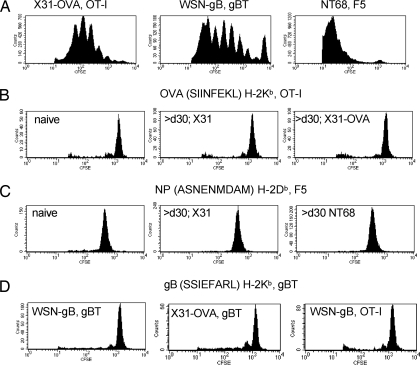
The first necessity was to confirm that the 3 sets of TCR Tg CD8+ T cells that we intended to use in this analysis indeed proliferate when exposed to their cognate antigen in acutely infected hosts. Fig. 2A thus repeats the 64-h time point (Fig. 1) after transferring CFSE-labeled OT-I, gBT, and F5 CD8+ T cells into separate groups of B6 mice infected i.n. with the cognate X31-OVA, WSN-gB, or NT68 viruses 3 to 4 d previously. As can be seen, all CD8+ T cells responded with ample division (Fig. 2A).
Fig. 2.
Probing pMHCI persistence for a variety of influenza epitopes. (A) Antigen-driven proliferation of TCR Tg CD8+ T cells transferred 64 h previously into mice that had been infected for 3 to 4 d with viruses expressing the cognate peptide. (B) Lack of differential OT-I cycling at 11–14 d after cell transfer into naïve B6 mice or into mice infected >30 d previously with a virus that did (X31-OVA) or did not (X31) express the SIIINFEKL peptide. (C) Lack of differential F5 cycling in naïve mice or in mice infected >30 d previously with viruses that did (NT68) or did not (X31) express the immunogenic ASNENMDAM peptide. (D) Reciprocal transfer of gBT and OT-I CTLs into mice infected >30 d previously with viruses that did or did not carry the cognate peptide. All recipient mice were infected i.n. with 1 × 104 PFU X31-OVA, 1 × 104 PFU X31, 50 PFU WSN-gB, or 1 × 104 PFU NT68, with the variation in virus doses reflecting differences in virulence. The Ly5.2+ B6 mice were then transferred i.v. at different times after infection with 2 × 106 CFSE-labeled Ly5.1+ OT-I, Ly5.1+ gBT, or F5 CD8+ T cells and sampled at 64 h (A) or 11–14 d (B–D). Data are representative of a minimum of 2–11 mice per experimental group from 2–4 independent experiments.
In the next experiment, we transferred OT-I T cells into naïve mice or mice that had been infected 30–60 d previously with X31 or X31-OVA, then left the CD8+ T cells in situ for at least 11 d (Fig. 2B). There was evidence of some homeostatic turnover for the 3 groups, but the profiles were identical for the OT-I sets recovered from naïve recipients or from those that had been given viruses that do (X31-OVA) or do not (X31) contain the SIINFEKL peptide (Fig. 2B). This was also the case when we transferred F5 CD8+ T cells in a comparably controlled experiment (Fig. 2C). Similarly, gBT CD8+ T cells showed no evidence of differential proliferation in mice that had been primed i.n. with WSN-gB or X31-OVA >30 d previously, and they behaved no differently than OT-I T cells in WSN-gB–infected mice (Fig. 2D). In short, we found no evidence that any of the pMHCI complexes examined persisted through to the 30–60-d interval after primary influenza A virus infection.
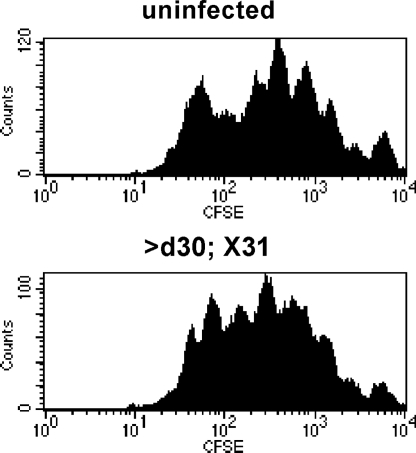
Pulmonary DC Status in Naïve and Previously Infected Hosts.
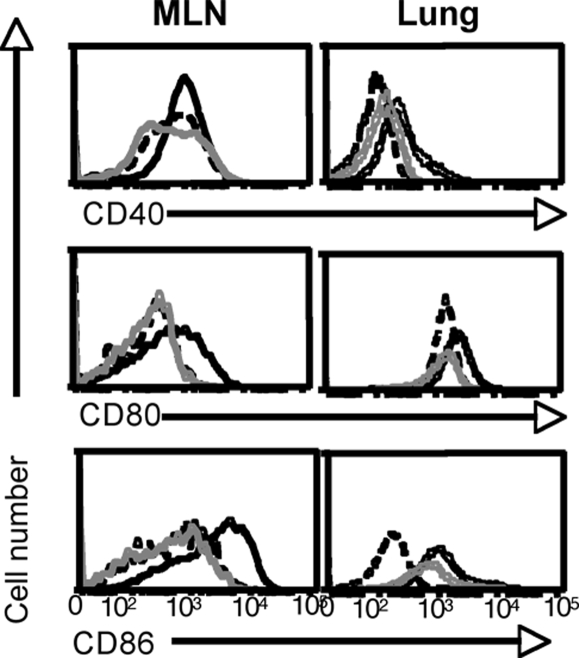
The strictly time-limited capacity of epitope-specific CD8+ T cells to divide in mice that had previously been infected with influenza A viruses (Figs. 1 and 2) could be thought to reflect that the APC environment is in some way compromised after the acute phase of this viral pneumonia is resolved (16, 17). To exclude that possibility, we examined the status of the regional lymph node/pulmonary DC network. The MLNs of naïve SPF mice enlarge greatly (and remain bigger) after virus challenge, but whereas more DCs were obtained from the MLNs of recovered mice, the CD11c+ DCs were at comparable prevalence in MLN and lung populations from previously uninfected and LT-exposed (>30 d) hosts (Fig. S1). Using the influenza model, Dahl et al. (16) reported that lung DCs from recovered mice display sustained, increased levels of costimulatory markers. Given that DC activation has the potential to impair antigen presentation (18), this could explain the failure to detect evidence of pMHCI expression beyond 1 week or so after infectious virus clearance (Figs. 1 and 2). The CD11c+ DCs recovered from the MLNs of recently infected (4 d) mice expressed increased levels of CD80 and CD86, but not CD40, although none of these markers were elevated on comparable cells recovered at the LT 30–60-d interval (MLN; Fig. 3). The lung CD11c+ DCs upregulated all 3 costimulatory molecules after short-term (ST) infection, and CD86 was marginally higher in the LT hosts (Fig. 3). Overall, though, the highest levels of DC activation (Fig. 3) were seen at the early time point when there was evidence of ample pMHCI expression and maximal CD8+ T cell stimulation (Figs. 1 and 2).
Fig. 3.
Phenotypic analysis of the CD11c+ DC populations. Activation phenotypes of CD11c+ DCs recovered from the MLN and lung. The mice were uninfected (dashed line) or infected i.n. with 1 × 104 PFU X31 3 to 4 d (black line) or 30–60 d (gray line) previously. The flow cytometry histograms display CD11c+ cells. Data are representative of 2 to 3 independent experiments in which organs were pooled from groups of 2–6 mice.
To assess further whether lung APC capacity is in some way depressed after i.n. exposure >30 d previously, we took LT mice that had been given the WT X31 virus then challenged them (and naïve controls) i.n. with OVA protein. Transferred CFSE-labeled OT-I T cells given 4 d later divided to an equivalent extent in the OVA-pulsed uninfected and LT mice, indicating that the levels of KbOVA257 expression were comparable (Fig. 4). There is thus no reason to think that persistent influenza virus pMHCI complexes are in some way hidden in the LT mice by compromised DC function.
Fig. 4.
Prior experience of influenza pneumonia does not compromise APC function. Uninfected or B6 mice infected i.n. with 1 × 104 PFU X31 30–60 d previously were given OVA protein i.n. 1 day before i.v. transfer of 2 × 106 CFSE-labeled Ly5.1+ OT-I CTLs. The MLN was harvested ≈64 h after transfer. The dose of OVA protein varied from 0.05 mg (displayed) to 0.5 mg, with similar results. The histograms display CD8+ Ly5.1+ CFSE+ cells, and the data are representative of 2 independent experiments with 2 to 3 mice per group.
Do CD8+ T Cell Numbers or Phenotypes Indicate pMHCI Persistence?
So far, we found no evidence supporting the idea of persistent pMHCI expression in mice that had cleared influenza A virus infection. However, with our previous analyses (Fig. 2), slow turnover of a small number of cells beyond the normal “homeostatic” cycling would be hard to distinguish, given the presence of the large undivided CFSEhi population. We thus monitored both CD8+ T cell activation phenotypes and cell counts to determine whether there was any indication that they were encountering antigen.
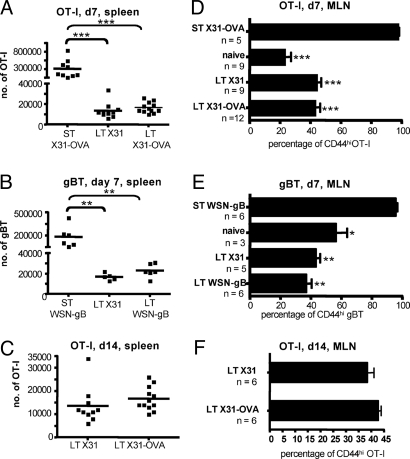
First, we examined the expansion and accumulation of transferred CD8+ T cells. In these experiments, 105 unlabeled OT-I T cells were transferred into hosts infected 3 to 4 d (ST) or 30–60 d previously with X31-OVA (LT X31-OVA) or a control virus lacking OVA257 expression (LT X31). As expected, substantial accumulation of OT-I CD8+ T cells was observed on d7 (compare naïve and ST, Fig. 5A) after transfer into the ST X31-OVA–infected hosts where KbOVA257 is abundant (Figs. 1 and 2). These ST counts were significantly higher than those from the LT X31 (P < 0.001, 22-fold) or X31-OVA (P < 0.001, 18-fold) mice, but there was no difference between the 2 LT groups (Fig. 5A). The same result was observed for a comparable experiment with the gBT Tg/WSN-gB system (Fig. 5B). To exclude the possibility that expansion rates after CD8+ T cell encounter of LT pMHCI “depots” could be slower than those observed in response to high pMHCI levels in the ST situation, CTL expansion was also assessed 14 d after OT-I transfer into the LT mice. Again, there was no significant difference in OT-I numbers for the LT X31 and X31-OVA hosts (Fig. 5C), although in all 3 LT experiments (Fig. 5 A–C) the mean values for the T cells transferred into a situation in which there is the possibility of pMHCI persistence were slightly higher but not statistically different.
Fig. 5.
CD8+ T cell numbers and activation status after transfer into infected hosts. The experiments used mice that had been infected short term (ST, 3 to 4 d) or long term (LT, 30–60 d) with different influenza A viruses before the transfer of 105 TCR Tg CTLs. The histograms show spleen (A–C) and MLN (D–F) results for samples taken 7 d (A, B, D, E) or 14 d (C and F) after cell transfer. TCR Tg CD8+ T cell number (A–C) and the percentage of CD44hi CD8+ TCR Tg T cells (D–F) were determined. The virus infections and the transferred cell populations are described in greater detail in the legends to Figs. 1 and 2 and in Materials and Methods. For each experiment, data are pooled from 2 to 3 independent infections; in A–C, each square represents an individual mouse and the line designates the mean; in D–F, the bar represents mean ± SEM. **P < 0.01; ***P < 0.001.
Virus-specific CD8+ T cell activation induces the upregulation of CD44 and CD69 and the downregulation of CD62L. We next measured the prevalence of CD44hi TCR Tg CD8+ T cells at 7d (Fig. 5 D and E) or 14 d (Fig. 5F) after transfer into naïve, ST-infected, or LT-infected mice. Again, antigen-specific CD44 upregulation was apparent for the ST group, reflecting stimulation by the cognate pMHCI complex, but there were no significant differences between the findings in naïve recipients and in those primed LT with a virus that did, or did not, express the cognate peptide (Fig. 5 D–F). The levels of CD62L and CD69 did change on the transferred OT-Is but, because the profiles were equivalent for the Tg T cells recovered from the LT X31 and X31-OVA recipients, the effect was not antigen specific (data not shown).
Might pMHCI Complexes Persist Somewhere in the Infected Lung?
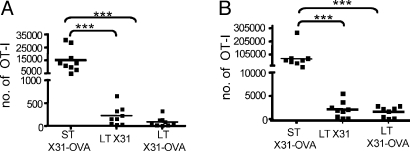
In mice, the requirement for a trypsin-like enzyme to cleave the viral HA molecule effectively limits productive influenza A virus infection to the superficial epithelial layer of the respiratory tract. Although we seem to have excluded the draining MLN as a site of pMHCI persistence (Figs. 1–5), could this be occurring in the lung? As expected, transferring OT-I T cells into mice infected for 3 to 4 d with X31-OVA led to greatly increased numbers in the population recovered 10 d later by BAL (ST, Fig. 6A) or by disruption of the lung parenchyma (ST, Fig. 6B). However, there was no difference in the OT-I T cell counts from either site in mice that had been infected i.n. with X31 or X31-OVA 30–60 d before cell transfer (LT, Fig. 6 A and B). In both cases, the OT-I numbers were significantly lower than those found for the acutely infected hosts (P > 0.001). If pMHCI complexes are maintained in such mice, then they do not function either to retain T cells in the infected lung or to facilitate their recruitment into the airways.
Fig. 6.
Antigen-specific CTL migration into the infected respiratory tract. Transferred OT-I T cells (105 i.v.) were enumerated 10 d later in cell populations from the BAL (A) and lung (B) after ST (3 to 4 d) or LT (30–60 d) infection with X31 (LT) or X31-OVA (ST and LT). Data are pooled from 3 independent experiments; each square represents an individual mouse, and the line designates the mean. ***P < 0.001.
Could the Provision of Inflammatory Signals Reveal Persistent pMHCI Complexes?
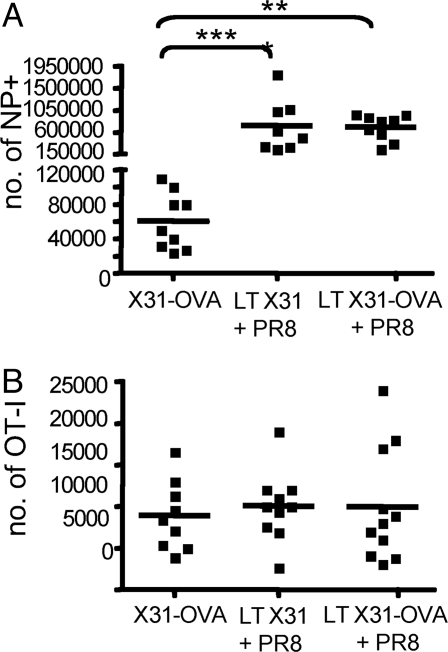
The failure to detect antigen-specific CD8+ T cell division, expansion, activation, and migration in LT-recovered mice (Figs. 1–6) suggested that influenza A virus pMHCI complexes are completely absent from those infected more than 2 to 3 weeks previously. A further possibility is, however, that CD8+ T cell proliferation is compromised when pMHCI antigen is encountered in the absence of inflammatory mediators and/or virus-associated danger signals. The repaired respiratory tract of the LT mice would not be expected to provide such a milieu. Mice that had been infected i.n. 30–60 d previously (LT) with X31 or X31-OVA were thus challenged i.n. with PR8 influenza A virus to restore that “inflammatory” environment. The expansion of the endogenous CD8+DbNP366+ CTL memory set was monitored by tetramer staining of spleen cells on d10 after exposure to the PR8 virus, showing a mean 10-fold increase in number over the unchallenged (X31-OVA) controls (Fig. 7A). However, the OT-I T cells that were given on d3 of PR8 challenge showed no evidence of KbOVA257-specific proliferation 10 d later (Fig. 7B). Providing the cytokine/chemokine milieu associated with active infection thus fails to reveal the presence of a “cryptic” KbOVA257+ APC pool in recovered, LT mice.
Fig. 7.
Provision of virus-associated inflammation does not uncover pMHCI depots. OT-I (105 i.v.) were given to B6 mice that had been infected i.n. with 1 × 104 PFU X31 or X31-OVA 30–60 d previously (X31-OVA), then challenged i.n. with 50 PFU PR8 3 d before cell transfer. Spleens were taken 7 d later, and flow cytometry was used to measure (A) the endogenous CD8+DbNP366+ response, determined by tetramer staining; and (B) the CD8+Ly5.1+OT-I T CTL counts. Data are pooled from 3 independent experiments; each square represents an individual mouse, and the line designates the mean. *P < 0.05; ***P < 0.001.
Discussion
Correlating the molecular profiles of ex vivo–isolated CD8+ T cells with particular differentiation states, especially the memory phase (5, 19–21), is potentially confounded by the possibility of pMHCI persistence and continued TCR ligation subsequent to the control of the infectious process. Early experiments indicated that pMHCI complexes survive in only the very short term after infectious influenza A virus clearance (6, 22), leading to the conclusion that the maintenance of influenza-specific CD8+ T cell memory is antigen independent. Recent findings have suggested that influenza pMHCI complexes may persist for 1 month or more after the active infection is controlled (8, 9, 11, 23). If this is indeed the case, it is incumbent on us to reinterpret the nature of influenza A virus-specific CD8+ T cell memory.
Persistent pMHCI expression beyond the termination of influenza A virus replication has been described by 2 groups (8, 9, 11). The most convincing evidence describes LT pMHCI depots after infection with the E61–13-H17 (NT68) influenza A virus. In these studies, the readout for pMHCI presentation was the division of CFSE-labeled F5 CD8+ T cells transferred into hosts that had been infected 30 d or 60 d previously (8, 11). This experiment is repeated here, with the finding that there is no difference in the minimal rates of proliferation for F5 Tg CD8+ T cells transferred into naïve mice or those infected >30 d previously with the “cognate” NT68 virus or the “irrelevant” PR8 virus that, nonetheless, induces comparable lung pathology. The same result was found for OT-I and gBT-I TCR Tg CD8+ T cells when we used influenza A viruses that had, or had not, been engineered to express the immunogenic peptide. A second report argued that pMHCI depots persist after infection of BALB/c mice with PR8 virus, although these fail to elicit CD8+ T cell priming or memory development (9). Using T cell proliferation, population size, and activation status as readouts, we were unable to demonstrate that influenza pMHCI complexes are maintained much beyond 15 d after infection and, despite considerable efforts to “reveal” possible cryptic pMHCI pools, none of our experiments showed such effects, which were seen repeatedly when we used recipient mice that were still supporting virus replication.
The present analysis focuses primarily on the adoptive transfer of naïve T cells as a readout for antigen persistence. Others have suggested that there is preferential retention of influenza A virus-specific memory CTLs within the draining lymph node for >30 d after infection (11). Although we could find no evidence for LT maintenance of pMHCI complexes at levels sufficient to drive proliferation or activation, there remains the formal possibility that very small amounts of pMHCI influence memory T cell localization patterns. However, in the same analysis that suggested this requirement for antigen to “hold” memory T cells in the lymph node, naïve CD8+ CTLs were able to respond to the putative LT pMHCI depot (11), an observation that we have not been able to reproduce in this series of rigorously controlled experiments. A further possibility is that CD8+ T cells primed in a particular site develop an inducible “cell surface language” of integrins and so forth that favors their return to/retention in the anatomic niche where they encountered antigen. A population of influenza A virus-specific memory CD8+ T cells does seem to be resident in the lung parenchyma (24, 25), although we have little understanding of the rate of T cell turnover between blood, tissue, and lymph for this, or for any other, site of former pathology. Our efforts to reveal “cryptic” pMHCI in the recovered lung met with no success.
Other, less-direct evidence also calls into question the idea that pMHCI persistence is in some way required for the maintenance of influenza virus-specific CD8+ T cell memory. One is that such memory, once established, is remarkably stable. The profiles of TCR usage characteristic of the acute response are maintained, and there is no suggestion that periodic “bursts” of proliferation as a consequence of random encounters between individual clonotypes and scarce pMHCI+ APCs skew the memory TCR profile in unpredicted ways (26). Additionally, with time, memory CD8+ T cells shift progressively to the less-activated CD62Lhi phenotype. Furthermore, if the putative “persistent” pMHCI complexes are thought to come from, say, antigen–antibody complexes on the surface of follicular DCs (27), it would be expected that proteins made in great abundance (like NP) should be more likely to persist at high levels than, say, the low abundance (28) acid polymerase (PA). What the evidence shows, though, is that the larger, antigen-driven clonal expansions that result in a bigger CTL memory pool specific for DbNP366 tend, with time, to converge in size to be more like those recognizing DbPA224 (29).
Why is there such a substantial difference in findings between the 3 established immunology groups that have looked seriously at this issue? One possibility is that evidence of pMHCI persistence could reflect a failure of complete virus clearance due to some immunosuppressive effect, perhaps mediated via a concurrent, subclinical, and unrelated disease. Additionally, our experiments have been done with B6 mice that have been breeding for some time in Australia and were not sourced recently from any of the major supply houses. Otherwise, we can think of no reasonable explanation for the fact that such different results have been achieved with what look to be essentially identical experimental systems and readouts.
Although it is essentially impossible to prove the absence of something we have tried, without prejudice and using every reasonable approach we could think of, to investigate the claim that influenza A virus pMHCI complexes are maintained well beyond the stage of infectious virus clearance. Although the experiments have been done with great care, evidence for pMHCI persistence has been completely lacking. At least for mice infected with influenza A viruses under the conditions described here, it is valid to argue that influenza A virus–specific CD8+ T cell memory is maintained in the absence of further TCR ligation by the inducing pMHCI epitope
Materials and Methods
Mice.
Female C57BL/6J (B6), OT-I × B6.SJL-PtprcaPep3b/BoyJ (Ly5.1 × OT-I), and gBT-I.1 × B6.SJL-PtprcaPep3b/BoyJ (Ly5.1 x gBT) mice were bred and housed in the Department of Microbiology and Immunology animal facility at The University of Melbourne. Female B6 and F5 TCR Tg mice were bred and housed at The Walter and Eliza Hall Institute for Medical Research animal facility. OT-I (30), gBT (31), and F5 (32) CD8+ TCR Tg mice are specific for the H-2Kb restricted OVA-derived epitope OVA257–264 (KbOVA257), the H-2Kb restricted gB-derived epitope gB498–505 (KbgB498), and the H-2Db restricted NT68 virus NP-derived epitope NP366–374 (DbNP366), respectively.
Viruses.
The recombinant A/HKx31-OVA (X31-OVA), A/PR8-OVA (PR8-OVA), and WSN-gB influenza A viruses have been described previously (20, 33, 34). The X31-OVA and PR8-OVA express the OVA257–264 peptide, whereas WSN-gB expresses gB498–505. Naïve B6 mice (6–8 weeks) were lightly anesthetized and infected via i.n. administration of virus. Secondary challenge with PR8 or PR8-OVA was performed by i.n. infection of mice that had been given X31-OVA more than 30 d previously. Mice received 1 × 104 plaque-forming units (PFU) of HKx31 (X31), 1 × 104 PFU of X31-OVA, 50 PFU of WSN-gB, 50 PFU of PFU PR8-OVA, or 1 × 104 PFU of A/NT/60/68 (NT68).
CFSE Labeling and Transfer.
Lymph nodes were harvested from OT-I, gBT, or F5 TCR Tg mice, and suspensions of 107 cells per milliliter in PBS containing 1% bovine albumin (Invitrogen) were incubated with 5 μM of CFSE (Invitrogen) at 37 °C for 10 min. Mice were injected i.v. with 105 to 2 × 106 CFSE-labeled CD8+ TCR Tg T cells and sampled 64 h or 11–14 d later in the different experiments. Cell suspensions from the MLN were prepared by tissue dissociation using forceps, then stained with fluorescently conjugated anti-CD8α (53–6.7; BD PharMingen), anti-CD45.1 (A20; BD PharMingen), or anti-Vβ11 TCR (RR3–15; BD PharMingen) antibodies in PBS containing 5% bovine albumin and 0.02% sodium azide (Sigma-Aldrich) for 30 min at 4 °C. The dilution of CFSE by CD8+ Ly5.1+ (OT-I and gBT) or CD8+ Vβ11 TCR+ (F5) cells was assessed by flow cytometry (FACSCalibur; BD Biosciences). The analysis used CellQuest or FlowJo software.
CD8+ T Cell Adoptive Transfer, Tissue Sampling, and Analysis.
Lymph nodes were harvested from OT-I and gBT mice, and cell suspensions were prepared and stained with anti-CD8α (53–6.7; BD PharMingen) and anti-CD45.1 (A20; BD PharMingen) to estimate the proportion of CD8+ TCR Tg T cells. Mice were injected i.v. with 105 to 2 × 106 OT-I or gBT cells as above, then spleen, MLN, BAL, and lung samples were taken at intervals. For spleen, cell suspensions were prepared using a 40–70-μm nylon cell strainer (BD Falcon; BD Biosciences), followed by treatment with red cell lysis buffer (0.14 M NH4Cl and 0.017 M Tris). MLN cell suspensions were prepared as above, and BAL samples were treated with red cell lysis buffer. Lung tissue was digested in the presence of 2 mg/mL of collagenase A (Roche) at 37 °C for 30 min, followed by dissociation through a 40–70-μm nylon sieve (BD Falcon) and treatment with red cell lysis buffer. Cell suspensions were stained with fluorescently conjugated anti-CD8α (53–6.7; BD PharMingen) and anti-CD45.1 (A20; BD PharMingen) antibodies in PBS containing 5% BSA and 0.02% sodium azide for 30 min at 4 °C. The frequency of CD8+Ly5.1+ cells was determined by flow cytometry (FACSCalibur). Upregulation of CD44 was assessed by staining with conjugated anti-CD44 (IM7; BD PharMingen). Analysis used CellQuest or FlowJo software. Cell counts were performed using trypan blue to exclude nonviable cells.
Detection of Endogenous Influenza A Virus–Specific CD8+ T Cells.
Spleen cell suspensions were stained with PE-conjugated DbNP366 tetramer for 1 h at room temperature. The frequency of DbNP366 specific CD8+ T cells was determined by flow cytometry (FACSCalibur).
DC Enrichment.
For DC enrichment, organs were harvested from PBS-perfused mice. Lung or MLN samples were dissociated and digested for 20 min at room temperature with 1 mg/mL Type II collagenase (Worthington Biochemical) and 0.0014% (wt/vol) DNase (Roche Molecular Biochemicals). Any T cell–DC complexes were disrupted by treatment for 5 min with 0.1M EDTA (Gibco BRL). The DCs were enriched by depleting with anti-CD3 (KT3), anti-Thy1 (T24, 31.7), anti-CD19 (ID3), anti-GR-1 (RB6–8C5), and anti-erythrocyte (TER-119) in combination with antirat Ig-coupled magnetic beads (Dynabeads; Dynal). Cells were stained for CD11c, CD86, CD80, CD40, and IA/E.
Statistical Analysis.
All graphing and statistical analysis used the Prism graphing program (GraphPad). P values were calculated using a nonparametric, Mann-Whitney T test.
Supplementary Material
Acknowledgments.
We thank Drs. Nicole La Gruta and Katherine Kedzierska for helpful discussion. J.D.M. was supported by Australian National Health and Medical Research Project Grant 508905 and The University of Melbourne C.R. Roper Fellowship. P.C.D. was supported by National Health and Medical Research Council Program Grant 299907 and National Institutes of Health Grant AI170251. S.J.T. was supported by a Pfizer Senior Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901128106/DCSupplemental.
References
- 1.Hickman HD, et al. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat Immunol. 2008;9:155–165. doi: 10.1038/ni1557. [DOI] [PubMed] [Google Scholar]
- 2.Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: Initial antigen encounter triggers a developmental program in naive cells. Nat Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Stipdonk MJ, et al. Dynamic programming of CD8+ T lymphocyte responses. Nat Immunol. 2003;4:361–365. doi: 10.1038/ni912. [DOI] [PubMed] [Google Scholar]
- 4.Jones CM, et al. Herpes simplex virus type 1-specific cytotoxic T-lymphocyte arming occurs within lymph nodes draining the site of cutaneous infection. J Virol. 2000;74:2414–2419. doi: 10.1128/jvi.74.5.2414-2419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenkins MR, Kedzierska K, Doherty PC, Turner SJ. Heterogeneity of effector phenotype for acute phase and memory influenza A virus-specific CTL. J Immunol. 2007;179:64–70. doi: 10.4049/jimmunol.179.1.64. [DOI] [PubMed] [Google Scholar]
- 6.Flynn KJ, Riberdy JM, Christensen JP, Altman JD, Doherty PC. In vivo proliferation of naive and memory influenza-specific CD8(+) T cells. Proc Natl Acad Sci USA. 1999;96:8597–8602. doi: 10.1073/pnas.96.15.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence CW, Braciale TJ. Activation, differentiation, and migration of naive virus-specific CD8+ T cells during pulmonary influenza virus infection. J Immunol. 2004;173:1209–1218. doi: 10.4049/jimmunol.173.2.1209. [DOI] [PubMed] [Google Scholar]
- 8.Zammit DJ, Turner DL, Klonowski KD, Lefrancois L, Cauley LS. Residual antigen presentation after influenza virus infection affects CD8 T cell activation and migration. Immunity. 2006;24:439–449. doi: 10.1016/j.immuni.2006.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jelley-Gibbs DM, et al. Persistent depots of influenza antigen fail to induce a cytotoxic CD8 T cell response. J Immunol. 2007;178:7563–7570. doi: 10.4049/jimmunol.178.12.7563. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton-Easton A, Eichelberger M. Virus-specific antigen presentation by different subsets of cells from lung and mediastinal lymph node tissues of influenza virus-infected mice. J Virol. 1995;69:6359–6366. doi: 10.1128/jvi.69.10.6359-6366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khanna KM, et al. In situ imaging reveals different responses by naive and memory CD8 T cells to late antigen presentation by lymph node DC after influenza virus infection. Eur J Immunol. 2008;38:3304–3315. doi: 10.1002/eji.200838602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullbacher A. The long-term maintenance of cytotoxic T cell memory does not require persistence of antigen. J Exp Med. 1994;179:317–321. doi: 10.1084/jem.179.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou S, Hyland L, Ryan KW, Portner A, Doherty PC. Virus-specific CD8+ T-cell memory determined by clonal burst size. Nature. 1994;369:652–654. doi: 10.1038/369652a0. [DOI] [PubMed] [Google Scholar]
- 14.Lau LL, Jamieson BD, Somasundaram T, Ahmed R. Cytotoxic T-cell memory without antigen. Nature. 1994;369:648–652. [PubMed] [Google Scholar]
- 15.Flynn KJ, et al. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 16.Dahl ME, Dabbagh K, Liggitt D, Kim S, Lewis DB. Viral-induced T helper type 1 responses enhance allergic disease by effects on lung dendritic cells. Nat Immunol. 2004;5:337–343. doi: 10.1038/ni1041. [DOI] [PubMed] [Google Scholar]
- 17.Didierlaurent A, Goulding J, Hussell T. The impact of successive infections on the lung microenvironment. Immunology. 2007;122:457–465. doi: 10.1111/j.1365-2567.2007.02729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson NS, et al. Systemic activation of dendritic cells by Toll-like receptor ligands or malaria infection impairs cross-presentation and antiviral immunity. Nat Immunol. 2006;7:165–172. doi: 10.1038/ni1300. [DOI] [PubMed] [Google Scholar]
- 19.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 20.Mintern JD, Guillonneau C, Carbone FR, Doherty PC, Turner SJ. Cutting edge: Tissue-resident memory CTL down-regulate cytolytic molecule expression following virus clearance. J Immunol. 2007;179:7220–7224. doi: 10.4049/jimmunol.179.11.7220. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar S, et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belz GT, et al. Distinct migrating and nonmigrating dendritic cell populations are involved in MHC class I-restricted antigen presentation after lung infection with virus. Proc Natl Acad Sci USA. 2004;101:8670–8675. doi: 10.1073/pnas.0402644101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner DL, Cauley LS, Khanna KM, Lefrancois L. Persistent antigen presentation after acute vesicular stomatitis virus infection. J Virol. 2007;81:2039–2046. doi: 10.1128/JVI.02167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogan RJ, et al. Activated antigen-specific CD8+ T cells persist in the lungs following recovery from respiratory virus infections. J Immunol. 2001;166:1813–1822. doi: 10.4049/jimmunol.166.3.1813. [DOI] [PubMed] [Google Scholar]
- 25.Wiley JA, Hogan RJ, Woodland DL, Harmsen AG. Antigen-specific CD8(+) T cells persist in the upper respiratory tract following influenza virus infection. J Immunol. 2001;167:3293–3299. doi: 10.4049/jimmunol.167.6.3293. [DOI] [PubMed] [Google Scholar]
- 26.Turner SJ, Diaz G, Cross R, Doherty PC. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity. 2003;18:549–559. doi: 10.1016/s1074-7613(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 27.Gray D, Kosco M, Stockinger B. Novel pathways of antigen presentation for the maintenance of memory. Int Immunol. 1991;3:141–148. doi: 10.1093/intimm/3.2.141. [DOI] [PubMed] [Google Scholar]
- 28.La Gruta NL, et al. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc Natl Acad Sci USA. 2006;103:994–999. doi: 10.1073/pnas.0510429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall DR, et al. Measuring the diaspora for virus-specific CD8+ T cells. Proc Natl Acad Sci USA. 2001;98:6313–6318. doi: 10.1073/pnas.101132698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 31.Mueller SN, Jones CM, Smith CM, Heath WR, Carbone FR. Rapid cytotoxic T lymphocyte activation occurs in the draining lymph nodes after cutaneous herpes simplex virus infection as a result of early antigen presentation and not the presence of virus. J Exp Med. 2002;195:651–656. doi: 10.1084/jem.20012023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moskophidis D, Kioussis D. Contribution of virus-specific CD8+ cytotoxic T cells to virus clearance or pathologic manifestations of influenza virus infection in a T cell receptor transgenic mouse model. J Exp Med. 1998;188:223–232. doi: 10.1084/jem.188.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins MR, Webby R, Doherty PC, Turner SJ. Addition of a prominent epitope affects influenza A virus-specific CD8+ T cell immunodominance hierarchies when antigen is limiting. J Immunol. 2006;177:2917–2925. doi: 10.4049/jimmunol.177.5.2917. [DOI] [PubMed] [Google Scholar]
- 34.Stock AT, Jones CM, Heath WR, Carbone FR. CTL response compensation for the loss of an immunodominant class I-restricted HSV-1 determinant. Immunol Cell Biol. 2006;84:543–550. doi: 10.1111/j.1440-1711.2006.01469.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.