Abstract
The structure of the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase in complex with cognate DNA was determined at 1.89 Å resolution in the presence of S-adenosyl-l-homocysteine. DNA recognition and the dynamics of base-flipping were studied by site-directed mutagenesis, DNA methylation kinetics and fluorescence stopped-flow experiments. Our data illustrate the mechanism of coupling of DNA recognition and base-flipping. Contacts to the non-target strand in the second (3′) half of the GATC site are established by R124 to the fourth base-pair, and by L122 and P134 to the third base-pair. The aromatic ring of Y119 intercalates into the DNA between the second and third base-pairs, which is essential for base-flipping to occur. Compared to previous published structures of bacteriophage T4 Dam, three major new observations are made in E. coli Dam. (1) The first Gua is recognized by K9, removal of which abrogates the first base-pair recognition. (2) The flipped target Ade binds to the surface of EcoDam in the absence of S-adenosyl-l-methionine, which illustrates a possible intermediate in the base-flipping pathway. (3) The orphaned Thy residue displays structural flexibility by adopting an extrahelical or intrahelical position where it is in contact to N120.
Keywords: Dam methylation, GATC recognition, base flipping, bacterial virulence factor
Introduction
DNA methylation plays important roles in a large variety of biological processes, including gene regulation, genomic (parental) imprinting, DNA repair and host defense.1 DNA from most higher eukaryotes contains 5-methylcytosine (5mC); in bacterial DNAs, 6-methyladenine (N6mA) and 4-methylcytosine (N4mC) are observed as well. These modifications are introduced after DNA replication by DNA methyltransferases (MTases), which catalyze methyl group transfer from the donor S-adenosyl-l-methionine (AdoMet), producing S-adenosyl-l-homocysteine (AdoHcy) and methylated DNA.2 Generally, DNA MTases recognize specific nucleotide sequences and utilize a so-called base-flipping mechanism3 to rotate the target base-out of the DNA helix and into the active-site pocket of the enzyme.4
While most prokaryotic DNA MTases are components of restriction–modification systems and function as part of a host defense mechanism, some MTases are not associated with a restriction enzyme; e.g. the Escherichia coli DNA adenine MTase (EcoDam), which methylates the exocyclic amino nitrogen (N6) of the Ade in GATC.5,6 Dam MTase gene orthologs are widespread among γ-proteobacteria and their bacteriophages.7 DNA-adenine methylation at specific GATC sites plays a pivotal role in the regulation of bacterial gene expression, DNA replication, and mismatch repair, and is essential for bacterial virulence of many Gram-negative bacteria.8 For example, there is a cluster of GATC sites near the origin of replication of E. coli and Salmonella typhimurium, all of which are conserved between the two species. It is the hemimethylated GATC sites, produced immediately following DNA replication, that regulate the timing and targeting of a number of cellular functions.9 Dam methylation is essential in the post-replicative mismatch repair system, where the methylation mark present on one DNA strand after replication is used to distinguish between parental and daughter DNA strands to allow for a directed repair of base mismatches.10 In addition, Dam methylation regulates the expression of certain genes in E. coli.11,12 For example, the expression of pyelonephritis-associated pili (Pap) in uropathogenic E. coli is epigenetically controlled by the methylation state of the two GATC sites in the Pap regulon,13 which determines, in part, the phase variation of pili formation.
The involvement of Dam in pathogenicity was first described for Salmonella enterica serovar Typhimurium where a dam− mutant was out-competed by wild-type in establishing fatal infections in mice; moreover, mice previously infected with the dam− mutant were less susceptible to superinfection by the wild-type.14,15 Recently, it was shown that inactivation of Dam MTase attenuates Haemophilus influenzae virulence,16 and a role for Dam as a virulence factor has been observed for a growing list of bacterial pathogens.17 In fact, a Dam deletion strain of Salmonella used as live attenuated vaccines was able to raise cross-protective immunity.18,19 An understanding of the mechanism of Dam methylation would pave the way for the rational design of specific inhibitors of this enzyme. This might provide the basis for the development of a new class of antibiotics that could have broad antimicrobial action because humans, as well as other higher eukaryotes, do not have detectable adenine methylation.
Results and Discussion
Here, we report the crystal structure of EcoDam bound to cognate DNA in the presence of the coenzyme product AdoHcy. Together with parallel biochemical studies, we verified structural predictions and reconciled the effect of site-directed mutations on DNA binding, target-sequence specificity and base-flipping. DNA recognition is mediated by a hairpin loop that is positioned similar to that observed in the T4Dam–DNA structure reported earlier.20 The contacts of EcoDam to the third (by L122 and P134) and fourth base-pair (by R124) are analogous to those observed in T4Dam. As in T4Dam, two residues are intercalated into the DNA (Y119 and N120). In contrast, the first base-pair is contacted by K9 in EcoDam, whereas R130 interacts with the Gua1 base in T4Dam. Mutational studies and methylation kinetics were performed to investigate the functional role of K9 in Gua1 recognition. By combining structural, site-directed mutagenesis and rapid kinetics data we investigate the role of each of the various enzyme–DNA contacts for base-flipping. The biochemical and structural data demonstrate that in the absence of AdoMet in EcoDam the flipped target Ade does not enter the active site pocket but binds to an alternative site on the surface of the enzyme, which might resemble an intermediate in the base-flipping pathway. The orphan Thy can adopt an intrahelical or an extrahelical position, which illustrates the structural flexibility of the DNA in complex with the MTase.
Overall structure of EcoDam
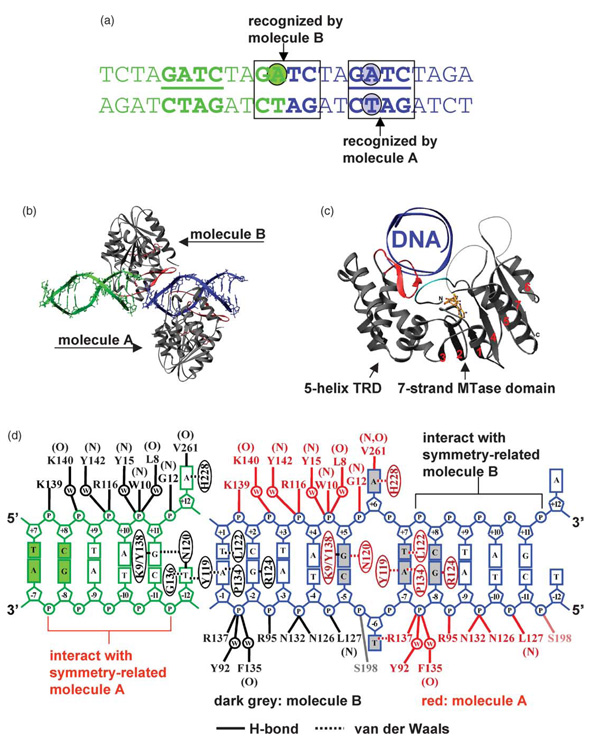
We crystallized a ternary complex containing EcoDam, AdoHcy, and a 12-mer oligodeoxynucleotide duplex containing a single, centrally located GATC target site. The end sequence of the duplex was chosen such that the sequence at the joint of two molecules mimics a GATC target site if the DNA duplexes are stacked head-to-tail (Figure 1(a)). The resulting crystals diffracted X-rays to 1.89 Å resolution (Table 1).
Figure 1. Structure of the EcoDam–AdoHcy–12mer DNA complex.
(a) Two DNA duplexes (green and blue) are stacked head-to-end, with one GATC site in the middle of each duplex and one in the joint of two duplexes. The nucleotides in extrahelical positions are in shaded circles. (b) Molecule A binds to the GATC site in the middle of each DNA duplex, while EcoDam molecule B binds to the joint of two DNA duplexes. (c) EcoDam contains two domains: a seven-stranded catalytic domain that harbors the binding site for AdoHcy (in stick model) and a DNA-binding domain consisting of a five-helix bundle and a β-hairpin (red) that is conserved in the family of GATC-related MTase orthologs. The N-terminal residues 7 to 10, colored in cyan, also interact with the DNA (see (d)). (D) Summary of the protein–DNA contacts of molecule A (red) and molecule B (grey). Backbone-mediated interactions are indicated with main chain amide (N) or carbonyl (O). For simplicity, only single water molecule (w)-mediated interactions are shown. Focusing on a single DNA duplex (blue), 20 out of 22 phosphate groups interact with three EcoDam molecules (A, B, and symmetry-related molecule B). Thus, the choice of the length (12 base-pairs) and the end sequence of the oligonucleotide used for crystallization optimally maximized the DNA–protein interactions and DNA-mediated protein–protein interactions in the crystal lattice of packing. The only two phosphate groups that are not involved in EcoDam interactions are the 5′ phosphate groups of the two Thy of the central GATC site, which are the phosphate groups missing from the joint GATC site.
Table 1.
Crystallographic data and refinement statistics
| Crystal | EcoDam–DNA (cognate)–AdoHcy |
|---|---|
| PDB code | 2G1P |
| Beamline (wavelength Å) | APS 22-ID (0.97179) |
| Space group | P21 |
| Unit cell dimensions | |
| a (Å) | 46.2 |
| b (Å) | 71.3 |
| c (Å) | 97.8 |
| α (deg.) | 90 |
| β (deg.) | 90.03 |
| γ (deg.) | 90 |
| Resolution range (Å) | 33.6–1.89 (1.92–1.89) |
| Measured reflections | 377,638 |
| Unique reflections | 51,126 |
| 〈I/σ〉 | 9.1 |
| Completeness (%)a | 99.9 (98.7) |
| Rlineara | 0.102 (0.575) |
| R-factora | 0.188 (0.278) |
| Rfree (5% data)a | 0.216 (0.287) |
| Non-hydrogen atoms | |
| Protein | 3996 (2 EcoDam) |
| DNA | 486 (1 DNA duplex) |
| Heterogen | 52 (2 AdoHcy) + 6 (1 glycerol) |
| Water | 321 |
| Thermal B values (Å2) | |
| Overall mean B value | 29.8 |
| From Wilson plot | 18.5 |
| rmsd from ideality | |
| Bond lengths (Å) | 0.010 |
| Bond angles (deg.) | 1.4 |
| Dihedral (deg.) | 22.0 |
| Improper (deg.) | 1.0 |
| Estimated coordinate error (Å) | |
| From Luzzati plot | 0.20 |
| From σA | 0.19 |
Values in parentheses are for the highest resolution shell.
Rlinear = ∑|I – 〈I〉|/ ∑〈I〉, R-factor = ∑|Fo – Fc|/∑|Fc|
Two EcoDam monomers (molecules A and B) and one DNA duplex are contained in the crystallographic asymmetric unit. EcoDam molecule A binds primarily to a single DNA duplex, while EcoDam molecule B binds to the joint between the two DNA duplexes (Figure 1(b)). EcoDam contains two domains: a seven-stranded catalytic domain (residues 1–56 and 145–270) harboring the binding site for AdoHcy and a DNA binding domain (residues 57–144) consisting of a five-helix bundle and a β-hairpin loop (residues 118–139, red in Figure 1(b) and (c)) that is conserved in the family of GATC-related MTase orthologs.21 The two protein molecules are highly similar (see Materials and Methods). Three regions are disordered in both molecules (Figure 1(b) and (c)): residues 188–197 immediately after the active-site D181-P-P-Y184 motif (after strand β4), residues 247–259 between strands β6 and β7, and residues 271–278 at the C terminus.
EcoDam–DNA interactions
The EcoDam molecule spans ten base-pairs, four base-pairs on the 5′ side and five on the 3′ side of the flipped-out target Ade (Figure 1(d)), whether they are froma single DNA duplex (EcoDam molecule A) or the joint between two 12-mer DNA duplexes (EcoDam molecule B). Five phosphate groups 5′ to the Ade residues in both strands are in contact with a single EcoDam molecule. Among the side-chains making direct interactions with the phosphate groups, four conserved residues (R95, N126, N132, and R137) interact with three consecutive phosphate groups flanking the Gua of the fourth GATC base-pair of the non-target strand (Figure 1(d)).
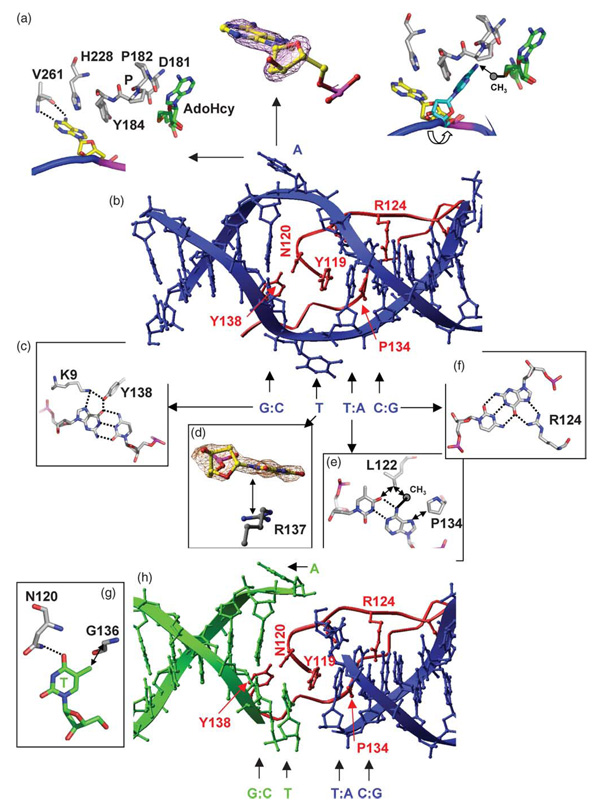
The methylation target, the Ade of the second base-pair in GATC (Ade2), flips out from the DNA helix (Figure 2(a)). The specific interactions with the remaining bases of the site occur in the DNA major groove. The amino acid residues from the β-hairpin (red in Figure 2(b)) make the majority of base-specific interactions, but K9 from the N-terminal loop also forms a base contact (cyan in Figure 1(c)). These two regions, the β-hairpin and the N terminal loop, are connected tightly together through many intramolecular interactions, including hydrogen binding of the main chain amide nitrogen atom and the carbonyl oxygen of K9 with the N115 side-chain carbonyl and amide groups, respectively (not shown). In the following sections we describe EcoDam recognition of the first, third and fourth base-pair, and its interaction with the target base-pair, including protein side-chain intercalation and DNA base-flipping.
Figure 2. EcoDam–DNA base interactions.
(a) The target Ade is bound in an alternative nucleotide-binding site, on the outside edge of the active-site pocket formed by the DPPY motif (left panel). The target Ade is superimposed with an omit (base and ribose) electron density map contoured at 3.5σ above the mean (middle panel). Large rotations about the P–O5′ bond of the DNA backbone drive the insertion of Ade into the active site (right panel). The transferable methyl group, modeled onto the sulfur atom of AdoHcy, would lie out of the plane of the Ade base, consistent with the target nitrogen lone pair deconjugated and positioned for an in-line direct methyl group transfer (indicated by an arrow), as seen in the M.TaqI–DNA complex.31 (b) The hairpin loop of molecule A (red) in the major groove of the blue DNA duplex with a central GATC site. (c) Interaction with the first base-pair (G:C) of GATC. Dotted lines indicate hydrogen bonds. (d) The flipped orphan Thy, superimposed with an omit electron density map contoured at 3.5σ above the mean, stacked with the side-chain of R137. (e) Interaction with the third base-pair (T:A) of GATC. A methyl group is modeled onto the exocyclic amino nitrogen N6 atom of the Ade in the non-target strand. Double arrows indicate van der Waals contacts. (f) Interaction with the fourth base-pair (C:G) of GATC. (G) The orphan Thy–N120 interaction in the joint of two DNA duplexes. The Thy–N120 interaction is similar to other protein side-chain-orphaned base interactions of base-flipping enzymes, such as those for Thy–S112 of T4Dam20 and Gua-Q237 of M.HhaI.3 (h) The hairpin loop of molecule B (red) in the joint of two DNA molecules (green and blue). The interactions with the first, third, and fourth bases-pairs are identical with that of molecule A (see (b)).
Recognition of the first base-pair by N-terminal K9
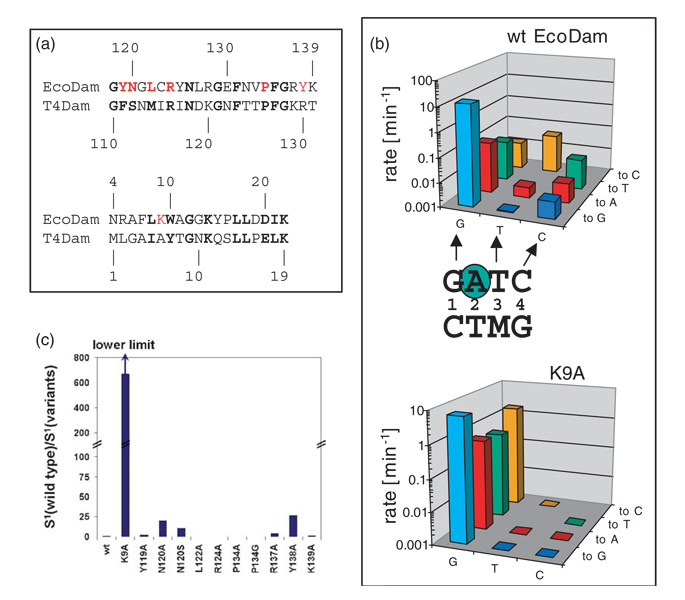
The recognition of the first base-pair is an interesting deviation between T4Dam and EcoDam. In the T4Dam structure, the first Gua of the GATC site is contacted by R130 with bifurcated hydrogen bonds to the N7 and O6 atoms of Gua1.20 R130 is located at the end of the β-hairpin, but it is not conserved among the Dam-related MTases (Figure 3(a)). The EcoDam structure shows that Gua1 interacts, via the N7 and O6 atoms respectively, with two side-chains, K9 and Y138 (Figure 2(c)). At the position corresponding to EcoDam K9, T4Dam has an Ala (Figure 3(a)) whose β-carbon atom points towards the DNA, but does not contact it. Previously, we examined the involvement of EcoDam Y138 and the two flanking residues, R137 and K139, in the recognition of the first base-pair; however, changing these residues to Ala did not affect the DNA recognition of EcoDam strongly.20 In response to the EcoDam–DNA structure described here, we changed K9 to alanine and investigated its role in DNA recognition. The EcoDam K9A variant shows slightly reduced catalytic activity (~60% of wild-type) (comparing the light blue bars in Figure 3(b)) and DNA binding (~70%) (data not shown).
Figure 3. Recognition of the first base-pair by N-terminal K9.
(a) Pair-wise sequence alignment of EcoDam and T4Dam in two regions: the β hairpin loop and the N-terminal loop. The residues colored in red were targets for site-directed mutagenesis. (b) Specificity profile of EcoDam wild-type (top panel) and the K9A variant (bottom panel). For easy comparison, we duplicate the wild-type panel.20 The single-turn-over methylation rates of the wild-type and the K9A variant are given for the cognate, hemimethylated GATC substrate (light blue bars) as well as for all nine near-cognate hemimethylated substrates. On the horizontal axis, the three positions of the GATC site that are mutated are given (G=GATC, T=GATC, C=GATC, M=N6 mA). The new base introduced at each position is specified on the right-hand axis. The methylation rates of the respective pair of enzyme and substrate are given on the vertical axis (note the logarithmic scale). (c) Specificity factor (defined in Materials and Methods) of EcoDam variants for recognition of the first position of the GATC sequence (S1). The values are given as relative changes with respect to the wild-type. Because no activity could be detected at near-cognate sites modified at the third or fourth base-pair of GATC with the K9A variant, the S1 factor given here is a lower limit, indicated by the arrow. The specificity factors of wild-type EcoDam and the K9A were calculated using the data given in (b) and (c); the data for all other variants were taken from Horton et al.20
To study DNA recognition by EcoDam, we compared the rates of DNA methylation of the canonical versus variant duplexes, all containing a single hemimethylated target (Figure 3(b)) to ensure that only one strand of the DNA was subject to methylation. The variant duplexes contained a single base substitution at either the first, third, or fourth base-pair of the target sequence; these variant sites are designated here as “near-cognate” sites (a total of nine) (Figure 3(b)). The results showed that relative to GATC, near-cognate substrates that carry a base-pair substitution at the first position were methylated by wild-type EcoDam at a 100 to 1000-fold reduced rate (Figure 3(b)). In contrast, the K9A variant showed a loss of specificity at the first base-pair; because relative to GATC the rate of methylation of CATC was only fourfold lower, and AATC and TATC methylation was reduced tenfold (Figure 3(b), bottom panel). In addition, the K9A variant was unable to methylate any of the near-cognate sites, carrying a substitution in the third or fourth base-pair, demonstrating an increased discrimination for these positions. This is probably due to the fact that, in K9A–GAXC and K9A–GATX complexes (where X denotes a nucleotide exchange in the third or fourth base-pair of the GATC sequence), the interaction of the enzyme and DNA is disrupted at two base-pairs: at the first by the K9A exchange and at the third or fourth base-pair by mutation of the DNA sequence.
To compare the contribution of recognition of Gua1 more quantitatively, a specificity factor (S1) fxor the recognition of Gua1 was calculated for wild-type EcoDam, K9A and other EcoDam variants from the β-hairpin. Since a high specificity of the recognition of the first base-pair results in a low rate of methylation of near-cognate substrates modified at the first base-pair, S1 is defined by the average methylation rate of all near-cognate substrates carrying a base-pair exchange at position 3 or 4 divided by the average of the methylation rates of all near-cognate substrates carrying an alteration at the first base-pair. On the basis of S1, in comparison to wild-type EcoDam, K9A has an at least 800-fold reduced recognition of the first base-pair (Figure 3(c)), whereas all other variants displayed only minor effects. These results demonstrate that the EcoDam K9–Gua1 contact (via the N7 atom) is important for the recognition of the first base-pair in GATC. Y138A, which loses its interaction with the O6 atom of Gua1 (Figure 2(c)), and N120A, which loses its π-stacking with Gua1 (Figure 2(b)), also show small changes in specificity factor S1, indicating that S1 correctly identified all three side-chains that are in the vicinity of Gua1.
Our data for EcoDam and T4Dam demonstrate that the recognition of Gua1 is not mediated by conserved structural elements, although the overall enzyme structures are highly similar. Since it is difficult to imagine that one of the enzymes would lose its specificity for Gua1 during molecular evolution and later re-establish the same specificity using another amino acid residue, this finding suggests that T4Dam and EcoDam were derived from an enzyme that recognized only the ATC part of the recognition sequence and they developed their specificity for Gua1 of the GATC sequence independently. Our finding indicates also that interaction via the N7 atom is more important for the Gua1 recognition.
Interaction with the target Ade and base-flipping
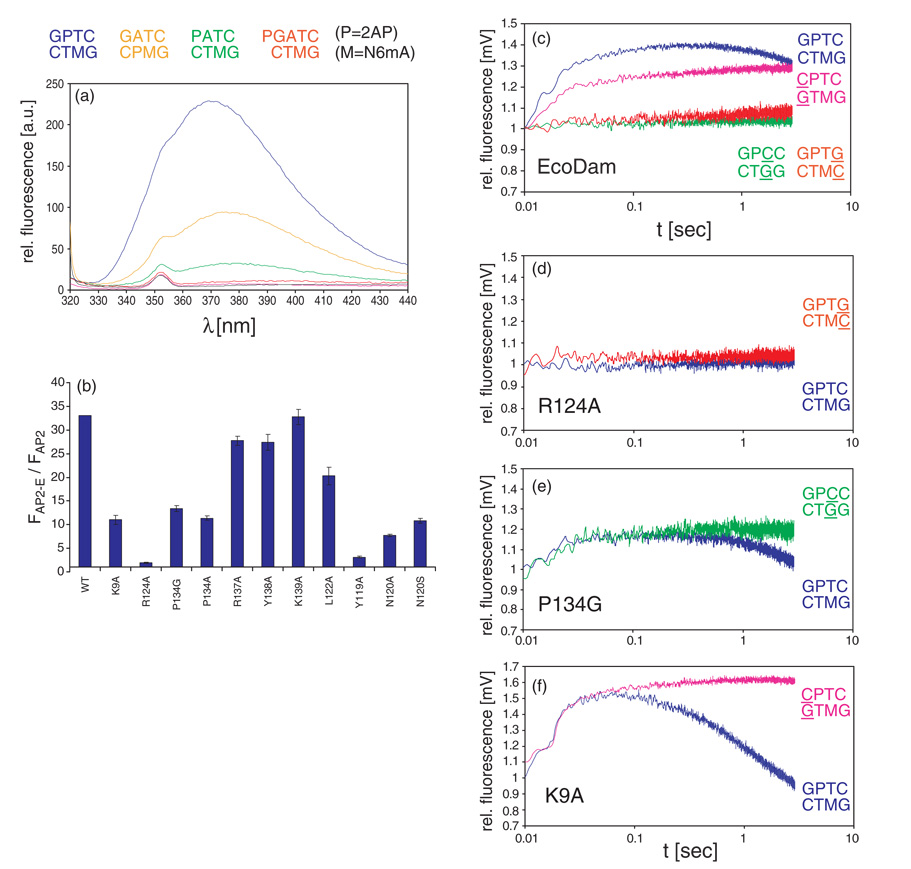
Incorporation of the nucleotide analog 2-aminopurine (2AP) into synthetic oligodeoxynucleotide duplexes has been used extensively to probe DNA conformational changes, such as base-flipping,22–25 because 2AP fluorescence increases dramatically when it is removed from the stacking environment of double helical DNA.26 Fluorescence changes of a hemimethylated G-2AP-TC substrate, which carries 2AP at the position of the target Ade, were correlated with base-flipping by EcoDam.27 Base-flipping by EcoDam comprises two steps: (i) flipping of the target base out of the DNA helix, and (ii) binding of the flipped base into the active site pocket of the enzyme (formed by the D181-P-P-Y184 motif). Target base-flipping leads to a complete loss of the stacking interactions of the Ade with the neighboring bases, which causes a strong increase in fluorescence. During binding of the flipped base into the active site pocket, it stacks with aromatic residue(s), which leads to a reduction of 2AP fluorescence during this step of trapping.27 However, binding of the flipped target base into the active site pocket does not occur in the presence of AdoHcy or if no AdoMet is bound to the enzyme.27
In agreement with these observations, in the current structure formed in the presence of AdoHcy, the flipped target Ade lies against the protein surface (side-chains of Y184 and H228) outside the active-site pocket (Figure 2(a), left panel). The imidazole ring of H228 stacks edge-to-face on the Ade ring. In addition, the ring nitrogen atom N1 and the exocyclic amino nitrogen N6 atom of the Ade form a hydrogen bond with the main-chain amide nitrogen atom and carbonyl oxygen atom of V261, respectively. Comparison of the DNA conformation with that in the T4Dam-specific complex20 reveals that a simple but large rotation (~90°) around only one dihedral bond (P–O5′) would drive insertion of the flipped Ade into the active-site pocket (Figure 2(a), right panel). We postulate that this movement could take place only after AdoMet binding when the unstructured loop (amino acid residues 188–197) following the active-site residues becomes ordered and enables the formation of a closed active site. Future studies are required to uncover the mechanism by which binding of AdoMet triggers insertion of the flipped target base into the active site pocket.
The existence of a conformation in which the flipped Ade is not bound to the active site suggests that the base-flipping occurs through a series of intermediates.27–29 The Ade-binding mode observed here in the EcoDam complex could mimic an intermediate in the Ade-flipping pathway in which the Ade is flipped but not yet inserted into the active-site pocket. Such an intermediate could occur during processive DNA methylation, when after one turnover the coenzyme product AdoHcy is still bound to the enzyme, but the next target site has already been located and base-flipping is initiated. Alternatively, since N6-methyl-Ade could replace the target Ade in the outer binding conformation without unfavorable constraints, the conformation could represent an intermediate formed immediately after release of the methylated base from the active-site pocket.
Interaction with the orphan Thy: a double base-flipping
The conformation of the orphan Thy (opposite the flipped Ade) represents a major difference between the EcoDam–DNA complexes formed by molecule A versus molecule B. Unexpectedly, the orphan Thy in the center in molecule A is also flipped out of the DNA helix, where it is stabilized by the π-stacking interactions with the guanidino group of R137 (Figure 2(d)). In contrast, the orphan Thy in molecule B hydrogen bonds with the amide side-chain of N120 (Figure 2(g)). There, N120 inserts its side-chain into the helical space vacated by the flipped Ade. The immediate flanking phosphate groups of the orphan Thy have either no interaction (5′ phosphate) or only a weak interaction with higher thermal B-factor (3′ phosphate with S198, the first ordered residue after the unstructured loop comprising residues 188–197). The less constrained conformation allows bond rotations about the DNA backbone at the orphaned site, which moves the Thy to an extrahelical position and disrupts the Thy–N120 interaction.
These results illustrate that the orphan Thy can adopt at least two different conformations. Therefore, we examined whether Thy-flipping occurs in solution. To this end, we used a substrate that has 2AP substituted in place of the Thy to be orphaned (GATC/GA-2AP-C). Under equilibrium binding conditions in the presence of AdoHcy, a sixfold fluorescence increase was observed (Figure 4(a)), which indicates a significant loss of 2AP stacking interactions. Because in the intrahelical conformation of the Thy observed in molecule B there is no obvious change in the stacking interaction of the Thy and no changes in the DNA conformation (bending or unwinding), this protein-induced change in fluorescence intensity of 2AP is most likely due to base-flipping. A 2AP fluorescence change with the GATC/GA-2AP-C substrate was observed with the R137A variant as well (data not shown), indicating that docking of the flipped 2AP to R137 (Figure 2(d)) is not required for the process, and that there might be other alternative extrahelical conformations for this base.
Figure 4. Base flipping by EcoDam and its variants.
(a) Fluorescence intensities of several DNA substrates in the presence of EcoDam. The Figure displays the fluorescence of 2AP (=P in the labels) at the position of target Ade (blue curve), the orphan Thy (orange curve), the Gua1 position of the first pair (green curve), and the immediate 5′ position to the GATC (red curve). The pink curve displays free DNA (the hemimethylated G-2AP-TC) as a control and the black curve is for free enzyme. (b) Changes of relative fluorescence of hemimethylated G-2AP-TC during binding of EcoDam and its variants. (c) Stopped-flow studies of base-flipping using substrates containing the 2AP at the target position (blue curve) and with three near-cognate substrates that carry a single base-pair substitution at the first (pink curve), third (green curve) or fourth base-pair (red curve) of the recognition site. (d)–(f) Stopped-flow studies of base-flipping with EcoDam variants with various substrates: (d) R124A, (e) P134G, and (f) K9A.
Previously, local conformational changes of the orphan base have been observed in the M.HaeIII–DNA (base repairing),30 and M.TaqI–DNA structures (base-shifting toward the center of helix).31 Double base-flipping has been observed in the structure of the DNA repair enzyme endonuclease IV and its DNA substrate32 as well as in the stopped-flow fluorescence studies with the MutY adenine DNA glycosylase using 2AP-containing DNA.33 However, in the structure of the MutY–DNA lesion-containing complex, the oxoG lesion lies completely in the DNA complex, while the Ade flipped out.34 Taken together, these studies suggest that the oxoG, like the orphan Thy bound with EcoDam, can be in either an intrahelical or an extrahelical location. Flexibility of the conformation of the partner base might be important for the initiation of base-flipping of the target base, where the Watson–Crick base-pair is disrupted. Also it might contribute to the back-flipping of the Ade base after methylation, when the amino acid residues inserted into the DNA have to be removed to free the space for the Ade.
Y119 intercalation is necessary for base-flipping
The Y119 aromatic ring intercalates into the DNA duplex and stacks between the third base-pair of GATC and the Thy:N120 base-amino acid pair in molecule B (Figure 2(h)) or the side-chain of N120 in molecule A (Figure 2(b)), resulting in a local doubling in helical rise. The helical expansions in the middle and end of the DNA duplex effectively increase the length of the DNA such that it corresponds to 14 base-pairs, matching the crystal a axis with the length of ~46 Å. A similar intercalation by F111 is observed in T4Dam.20
Previously, we showed that substitution of Y119 by Ala led to a strong reduction in catalytic activity.20 Fluorescence studies now reveal an almost complete loss of detectable base-flipping in the presence of AdoHcy (Figure 4(b)) or AdoMet (data not shown). Therefore, intercalation of Y119 into the DNA is a necessary event for base-flipping, possibly by expanding the helical rise distance between the base-pairs, reminiscent of the recent finding of flipping of two bases at the junction between B and Z DNA,35 or by preventing rapid back-flipping. In contrast, the invasion of N120 into the DNA helix where it contacts the orphaned Thy in its intrahelical conformation is of less importance; its substitution by Ala reduced catalytic activity20 and 2AP-flipping only about fourfold (Figure 4(b)). This difference might be explained by the strong stacking interactions of the aromatic Y119 to the adjacent base-pairs.
Recognition of the third base-pair: discrimination of unmethylated and hemimethylated DNA
The third base-pair of GATC makes van der Waals contacts with two hydrophobic side-chains of L122 and P134 (Figure 2(e)). P134 has been shown to be central for the discrimination of the TA base-pair. 20 However, in vivo hemimethylated GATC sites produced during DNA replication are the natural substrates for the Dam MTase. Therefore, we were interested to investigate the interaction of EcoDam with hemimethylated DNA. In the mismatch repair enzyme mutH, which interacts also with hemimethylated GATC sites, the methyl group directs the binding of the nuclease to the DNA in an orientation that leads to cleavage of the unmethylated DNA strand. A recent structure of mutH in complex with hemimethylayed GATC DNA demonstrates that P185 of MutH closely contacts the methyl group of methylated adenine.36
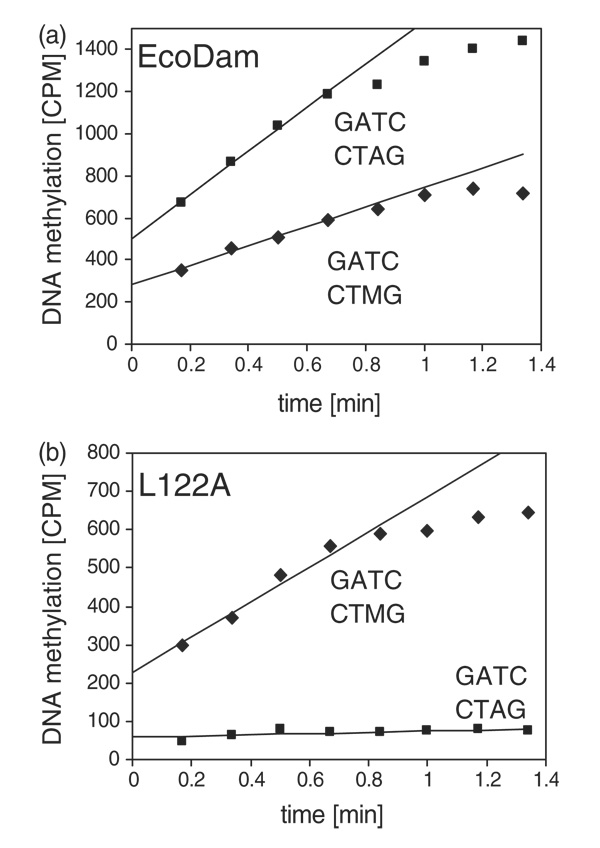
We modeled a methyl group onto the exocyclic amino nitrogen N6 atom of Ade3 (Figure 2(e)): the methyl group sits between the side-chains of L122 and P134, but the L122–CH3 contact distance (~3.6 Å) is much shorter than that of P134–CH3 (~4.9 Å). Thus, we studied the influence of residue L122 on EcoDam interaction with hemimethylated DNA duplexes. As shown in Figure 5(a), the rate of methyl transfer with the unmethylated substrate was roughly twice as fast as with the hemimethylated substrate. Three, not mutually exclusive, lines of argument might explain this finding: (I) the unmethylated substrate has twice the number of target sites as the hemimethylated one. (II) If the initial EcoDam binding is random with respect to the two strands, 50% of the binding events with the hemimethylated substrate will be unproductive, because EcoDam will be positioned such that the methylated Ade would be at the target position. (III) The methyl transfer step might be faster with unmethylated DNA. However, the L122A variant showed a drastically altered behavior (Figure 5(b)); viz. it was almost inactive on unmethylated DNA, while modifying the hemimethylated substrate at a rate similar to that of wild-type EcoDam.
Figure 5. Discrimination between unmethylated and hemimethylated DNA.
Methylation of unmethylated (squares) and hemimethylated (diamonds) oligonucleotide substrates by the (a) EcoDam (WT) and (b) L122A variant. These experiments were performed in 50 mM Hepes (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.5 mM DTT and 0.2 µg/µl of BSA using 0.5 µM DNA, 0.25 µM enzyme and 0.76 µM labeled AdoMet.
The mechanism of this pronounced change in the catalytic properties of L122A is not clear. We speculate that Ala at position 122 interacts with the methyl group of methylated Ade3, which might compensate for the loss of the contact between L122 and Thy3 (see Figure 2(e)). It is interesting to note that a single-point mutation (L122A), which reduced the size of an aliphatic hydrocarbon side-hain, was sufficient to convert EcoDam into a bona fide maintenance MTase with pronounced preference for hemimethylated DNA. In this respect, the L122A variant resembles the mammalian maintenance MTase Dnmt1, which has a high preference for hemimethylated CpG sites over unmethylated CpG sites,37,38 and plays a central role in the propagation of CpG methylation patterns in mammals.39
Recognition of the fourth base-pair by R124
The Gua in the fourth base-pair of GATC interacts via its O6 and N7 atoms with the guanidino group of R124 in a bifurcated hydrogen bonding pattern (Figure 2(f)). This contact is identical with that observed in T4Dam, and we showed previously that R124 has a critical role in the recognition of the fourth base-pair.20 The R124A variant had an overall reduction in catalytic activity but methylated two near-cognate substrates (GATT and GATG) faster than the canonical GATC, demonstrating that the interaction of R124 and Gua4 is required to activate the enzyme for catalysis.20
Coupling of base-flipping and DNA recognition
A central mystery of DNA methylation concerns the mechanism by which DNA MTases cause flipping of the target base within their recognition sequences. To study the coupling of base-flipping and DNA recognition, we investigated base-flipping by EcoDam variants with an altered specificity, viz. P134G and P134A (which show reduced discrimination at the third base-pair), R124A (which shows altered recognition at the fourth base-pair) and K9A (which has relaxed recognition of the first base-pair).
As shown in Figure 4(c), wild-type EcoDam shows no detectable change in 2AP fluorescence with substrates containing sequence changes at the third or fourth base-pair (green and redlines in Figure 4(c)), whereas base-flipping occurs with substrates containing a base substitution in the first base-pair. Conversely, no base-flipping signal was detected with the R124A variant (Figure 4(d)),which correlates with its pronounced reduction in catalytic activity.
In agreement with their high catalytic activities, P134G (Figure 4(e)) and P134A (data not shown) exhibited only a small reduction in the amplitude of the fluorescence change, but no detectable changes in the kinetics of base-flipping. However, both variants induced base-flipping of the substrate with altered sequence at the third base-pair (green line in Figure 4(e)), which did not occur with wild-type EcoDam (green line in Figure 4(c)). K9A behaved in a similar fashion: base-flipping of substrates carrying a base-pair substitution at the first position of the target site was more efficient than with wild-type EcoDam (compare the cyan lines in Figure 4(f) and (c)). We conclude that the change in specificity of these variants is based on a change in their flipping Ade in near-cognate sites.
Interestingly, in the interaction of K9A and P134G with the modified C-2AP-TC and G-2AP-CC substrates, respectively, the insertion of the flipped base into the base-binding pocket is still delayed. This may explain, in part, the difference in the methylation rates if the GATC substrate is compared to CATC and GACC. One cannot rule out the possibility that the insertion of 2AP as an artificial target base influences this step in the rapid kinetics experiments. Better understanding of this step awaits the availability of an EcoDam–DNA complex with the flipped base fully inserted into the active-site pocket, and development of additional spectroscopic tools to study the interaction of EcoDam and DNA.
Conclusions
Our findings demonstrate the contacts of the β-hairpin loop with second half of the recognition sequence (the third and fourth base-pairs) are required to position the enzyme on the target sequence. In particular, we conclude that the Gua4 base contact by R124, and its flanking phosphate contacts by conserved residues (R95, N126, N132, and R137; see above), positions EcoDam on the DNA duplex such that other residues involved in base-flipping (such as Y119, the second most important residue after R124 in base-flipping (Figure 4(b)) and DNA recognition (such as L122 and P134) resemble the closure of a zipper from the fourth to the first base-pair. With the Gua1–K9 interactions (an unique interaction for EcoDam) firmly engaged, the flipped Ade positions on the outside edge of the active site (Figure 2(a), left panel), a distinct intermediate of Ade flipping observed for EcoDam. A flexible conformation of the orphan Thy could have a role in the dynamics of the base-flipping process. Only after these events have taken place (some common between EcoDam and T4Dam and some specific for EcoDam, including the unique Gua1–K9 interaction, the distinct intermediate of Ade flipping, the alternative intra or extra-helical conformation of the orphan Thy), and the target Ade has entered the active site, does catalysis of methyl transfer from AdoMet take place (see Figure 2(a), right panel).
Materials and Methods
Crystallography
His-tagged EcoDam was expressed in HMS174(DE3) cells using autoinduction procedures,40 and purified utilizing Ni2+-affinity, UnoS, and S75 Sepharose sizing columns. A 0.5 l autoinduced culture yielded ~7 mg of purified enzyme. In the last purification step and during concentration, AdoHcy was added to the protein at a molar ratio of approximately 2:1. Concentrated binary complexes were mixed with oligonucleotide duplex (synthesized by New England Biolabs, Inc) at a protein to DNA molar ratio of ~2:1 and kept on ice for at least 2 h before crystallization; the final concentration of protein for crystallization trials was ~15 mg/ml. In hanging drop crystallization trials with AdoHcy, the ternary complex crystals appeared under these conditions: 100 mM buffer (Mes or Hepes) pH 6.6–7.4, 100 mM KCl, 10 mM MgSO4, 5–15% (w/v) PEG400 (Table 1). Crystals, acquired with a nylon loop (Hampton), were transferred quickly to mother liquor containing either 25% (v/v) glycerol or ethylene glycol before being flash-frozen directly in liquid nitrogen or in a stream of nitrogen gas at 100 K.
Structural determination of the cognate ternary complex proceeded by molecular replacement with the program REPLACE,41 using protein coordinates of a DpnM monomer structure (PDB 2DPM).42 The DpnM model was modified on the basis of a pair-wise sequence alignment of EcoDam with DpnM; differing amino acid residues in DpnM were changed to those in EcoDam and visually given the best rotamer using the program O,43 and some residues were deleted in the loop regions. DNA molecules were built manually into densities of difference maps. Refinement proceeded with the program CNS.44
The crystal of EcoDam–DNA (cognate)-AdoHcy can be indexed in the space group P212121 with the same cell dimensions but one EcoDam molecule and one half DNA duplex in the asymmetric unit. However, we were not able to distinguish whether the molecule bound at the central GATC site or at the joint GATC of two DNA duplexes. The DNA sequences surrounding both GATC sites are almost identical, except that the joint GATC lacks phosphate between the A and T. This difference is averaged out by the crystallographic orthorhombic 2-fold. To distinguish the two sites, we indexed the diffraction data in a lower symmetry space group, P21, with the asymmetric unit cell containing two EcoDam molecules and one complete DNA duplex. This enabled us to assign one molecule bound to the central GATC site (molecule A) and the other at the joint of two DNA duplexes (molecule B). Furthermore, Thy2 evidently adopts different conformations in molecule A (extrahelical) and in molecule B (intrahelical). Another important difference between the A and B molecules is the presence of one glycerol molecule bound to the hinge region between the DNA and the catalytic domains of molecule B, but not molecule A. To reduce the number of free parameters to fit the diffraction data in the P21 space group, we initially imposed non-crystallographic symmetry (NCS) restraints for the protein component during the refinement. Because of high resolution and an excess number of unique reflections (51,126) over the number of atomic parameters (4861 atoms × 4 = 19,444), the NCS was released in the final cycles of the refinement without adding or deleting any atoms, resulting in nearly no change of refinement statistics. It is evident that the two protein molecules (A and B) are highly similar, and the ~0.1 Å root-mean-square deviation (rmsd) between them (comparing 241 pairs of Cα atoms) is probably not due to real differences, just a reflection of errors in the data (a fact that the data could be indexed in a higher symmetry space group) and the finite precision of the refinement process at the 1.89 Å resolution.
Biochemical experiments
Site-directed mutagenesis was performed as described.45 EcoDam wild-type and its variants were purified as described.20 DNA binding was analyzed using surface plasmon resonance in a BiaCore X instrument as described.20 Methylation of oligonucleotide substrates (purchased from Thermo Electron, Dreieich, Germany in purified form) was carried out as described.20 Methylation experiments with the near-cognate substrates were performed in 50 mM Hepes (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 0.2 µg/µl of bovine serum albumin (BSA) containing 0.76 µm [methyl-3H]AdoMet (NEN) at 37 °C as described,46 using single-turnover conditions with 0.5 µM oligonucleotide substrate and 0.6 µM enzyme for specificity analysis (Figure 3(b) and (c)) and 0.25 µM enzyme for the study of interaction with hemimethylated DNA (Figure 5). The single turnover rate constants determined under these conditions incorporate the effects of DNA binding and catalysis. The sequence of the 20-mer oligonucleotide substrate was a duplex of
5′-GCGACAGTGATCGGCCTGTC-3′
5′-GACAGGCCGMTCACTGTCGC-3′
where M is N6-methyl-Ade. In addition, nine substrates with near-cognate sites, differing in one base-pair from GATC at the first, third or fourth position, were used. To compare the recognition of the first position of the target sequence by different variants, a specificity factor was defined as the ratio between the rates of methylation of all near-cognate sites modified at other positions and the rates of methylation of substrates modified at the first position, viz.:
To measure equilibrium base-flipping, the fluorescence change of oligonucleotides containing 2AP was detected in the absence and in the presence of EcoDam using 2 µM enzyme and 0.5 µM DNA in 50 mM Hepes (pH7.5), 50 mM NaCl containing 100 µM AdoHcy at ambient temperature (Figure 4(a)). The 2AP fluorescence was excited at 313 nm in an F2810 spectrofluorimeter (Hitachi). Emission spectra were recorded between 320 nm and 500 nm. Emission and excitation slits were set to 2.5 nm, and the data were analyzed by integration of the fluorescence peak after subtraction of the background signal from the buffer sample alone. The kinetics of base-flipping were investigated by stopped-flow experiments performed in an SF-3 stopped-flow device (BioLogic, Claix, France) as described,27 using enzyme and DNA at equal concentrations (350 nM) at ambient temperature. The enzyme was pre-incubated in buffer containing 50 mM Hepes (pH 7.5), 50 mM NaCl and 10 µM AdoMet, and mixed rapidly with DNA in the same buffer (Figure 4(c)–(f)). The 2AP fluorescence was excited at 313 nm and emission was observed using a 340 nm cutoff filter. The dead-time of the experiments was 3.1 ms. Typically, 15–20 shots were accumulated, averaged and analyzed. All experiments were carried out at least in triplicate, with no significant differences in the results. The data shown represent examples of individual experiments.
Acknowledgements
We thank Dr Zhe Yang for discussion, Dr Richard J. Roberts for the oligonucleotides used for crystallization, and Drs Stanley Hattman, Robert M. Blumenthal, and Richard I. Gumport for comments on the manuscript. Data for this study were measured at the beamline SERCAT-22 of the Advanced Photon Source at Argonne National Laboratory. Financial support for the beamline operation of Emory’s shares comes from the Dean’s Office of Emory University School of Medicine. Figures were drawn using the program Pymol, a user-sponsored molecular modeling system with an OPEN-SOURCE foundation. These studies were supported, in part, by US public Health Services grants GM068680 and GM049245 and the Georgia Research Alliance to X.C. and grants of the German BMBF (BioFuture programme) and DFG (JE 252/2) to A.J.
Abbreviations used
- MTase
methyltransferase
- AdoMet
S-adenosyl-l-methionine
- AdoHcy
S-adenosyl-l-homocysteine
- EcoDam
E. coli DNA adenine MTase.
References
- 1.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 2.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 3.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 4.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucl. Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacks S, Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J. Mol. Biol. 1977;114:153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- 6.Hattman S, Brooks JE, Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J. Mol. Biol. 1978;126:367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- 7.Hattman S, Malygin EG. Bacteriophage T2Dam and T4Dam DNA-[N6-adenine]-methyltransferases. Prog. Nucl. Acid Res. Mol. Biol. 2004;77:67–126. doi: 10.1016/S0079-6603(04)77003-8. [DOI] [PubMed] [Google Scholar]
- 8.Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 2005;8:154–160. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Messer W, Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988;54:735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- 10.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 11.Oshima T, Wada C, Kawagoe Y, Ara T, Maeda M, Masuda Y, et al. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 2002;45:673–695. doi: 10.1046/j.1365-2958.2002.03037.x. [DOI] [PubMed] [Google Scholar]
- 12.Lobner-Olesen A, Marinus MG, Hansen FG. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc. Natl Acad. Sci. USA. 2003;100:4672–4677. doi: 10.1073/pnas.0538053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernday AD, Braaten BA, Low DA. The mechanism by which DNA adenine methylase and PapI activate the pap epigenetic switch. Mol. Cell. 2003;12:947–957. doi: 10.1016/s1097-2765(03)00383-6. [DOI] [PubMed] [Google Scholar]
- 14.Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Del Portillo F, Pucciarelli MG, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl Acad. Sci. USA. 1999;96:11578–11583. doi: 10.1073/pnas.96.20.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson ME, Jarisch J, Smith AL. Inactivation of deoxyadenosine methyltransferase (dam) attenuates Haemophilus influenzae virulence. Mol. Microbiol. 2004;53:651–664. doi: 10.1111/j.1365-2958.2004.04140.x. [DOI] [PubMed] [Google Scholar]
- 17.Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 2001;69:7197–7204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dueger EL, House JK, Heithoff DM, Mahan MJ, Salmonella DNA. adenine methylase mutants elicit protective immune responses to homologous and heterologous serovars in chickens. Infect. Immun. 2001;69:7950–7954. doi: 10.1128/IAI.69.12.7950-7954.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dueger EL, House JK, Heithoff DM, Mahan MJ. Salmonella DNA adenine methylase mutants prevent colonization of newly hatched chickens by homologous and heterologous serovars. Int. J. Fd Microbiol. 2003;80:153–159. doi: 10.1016/s0168-1605(02)00152-6. [DOI] [PubMed] [Google Scholar]
- 20.Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng X. Transition fromnonspecific to specific DNAinteractions along the substrate-recognition pathway of dam methyltransferase. Cell. 2005;121:349–361. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Horton JR, Zhou L, Zhang XJ, Dong A, Zhang X, et al. Structure of the bacteriophage T4 DNAadenine methyltransferase. Nature Struct. Biol. 2003;10:849–855. doi: 10.1038/nsb973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allan BW, Reich NO. Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry. 1996;35:14757–14762. doi: 10.1021/bi9615708. [DOI] [PubMed] [Google Scholar]
- 23.Allan BW, Beechem JM, Lindstrom WM, Reich NO. Direct real time observation of base flipping by the EcoRI DNA methyltransferase. J. Biol. Chem. 1998;273:2368–2373. doi: 10.1074/jbc.273.4.2368. [DOI] [PubMed] [Google Scholar]
- 24.Holz B, Klimasauskas S, Serva S, Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucl. Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stivers JT. 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucl. Acids Res. 1998;26:3837–3844. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 27.Liebert K, Hermann A, Schlickenrieder M, Jeltsch A. Stopped-flowandmutational analysis of base flipping by the Escherichia coli DamDNA-(adenine-N6)-methyltransferase. J. Mol. Biol. 2004;341:443–454. doi: 10.1016/j.jmb.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 28.Horton JR, Ratner G, Banavali NK, Huang N, Choi Y, Maier MA, et al. Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase. Nucl. Acids Res. 2004;32:3877–3886. doi: 10.1093/nar/gkh701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 30.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base-pairing. Cell. 1995;82:143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 31.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nature Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 32.Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 33.Bernards AS, Miller JK, Bao KK, Wong I. Flipping duplex DNA inside out: a double base-flipping reaction mechanism by Escherichia coli MutY adenine glycosylase. J. Biol. Chem. 2002;277:20960–20964. doi: 10.1074/jbc.C200181200. [DOI] [PubMed] [Google Scholar]
- 34.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 35.Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 36.Lee JY, Chang J, Joseph N, Ghirlando R, Rao DN, Yang W. MutH complexed with hemiand unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell. 2005;20:155–166. doi: 10.1016/j.molcel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol. 2001;309:1189–1199. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- 38.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 39.Grace Goll M, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 40.Studier FW. Protein production by autoinduction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Tong L, Rossmann MG. Rotation function calculations with GLRF program. Methods Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 42.Tran PH, Korszun ZR, Cerritelli S, Springhorn SS, Lacks SA. Crystal structure of the DpnMDNA adenine methyltransferase from the DpnII restriction system of streptococcus pneumoniae bound to S-adenosylmethionine. Structure. 1998;6:1563–1575. doi: 10.1016/s0969-2126(98)00154-3. [DOI] [PubMed] [Google Scholar]
- 43.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallog. sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 44.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography &NMR system: a new software suite for macromolecular structure determination. Acta Crystallog. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Jeltsch A, Lanio T. Site-directed mutagenesis by polymerase chain reaction. Methods Mol. Biol. 2002;182:85–94. doi: 10.1385/1-59259-194-9:085. [DOI] [PubMed] [Google Scholar]
- 46.Roth M, Jeltsch A. Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol. Chem. 2000;381:269–272. doi: 10.1515/BC.2000.035. [DOI] [PubMed] [Google Scholar]