Abstract
Cryptocyanin, a copper-free hexameric protein in crab (Cancer magister) hemolymph, has been characterized and the amino acid sequence has been deduced from its cDNA. It is markedly similar in sequence, size, and structure to hemocyanin, the copper-containing oxygen-transport protein found in many arthropods. Cryptocyanin does not bind oxygen, however, and lacks three of the six highly conserved copper-binding histidine residues of hemocyanin. Cryptocyanin has no phenoloxidase activity, although a phenoloxidase is present in the hemolymph. The concentration of cryptocyanin in the hemolymph is closely coordinated with the molt cycle and reaches levels higher than hemocyanin during premolt. Cryptocyanin resembles insect hexamerins in the lack of copper, molt cycle patterns of biosynthesis, and potential contributions to the new exoskeleton. Phylogenetic analysis of sequence similarities between cryptocyanin and other members of the hemocyanin gene family shows that cryptocyanin is closely associated with crustacean hemocyanins and suggests that cryptocyanin arose as a result of a hemocyanin gene duplication. The presence of both hemocyanin and cryptocyanin in one animal provides an example of how insect hexamerins might have evolved from hemocyanin. Our results suggest that multiple members of the hemocyanin gene family—hemocyanin, cryptocyanin, phenoloxidase, and hexamerins—may participate in two vital functions of molting animals, oxygen binding and molting. Cryptocyanin may provide important molecular data to further investigate evolutionary relationships among all molting animals.
Keywords: phenoloxidase, Ecdysozoa
Hemocyanin, the blue copper-containing oxygen-transport molecule, usually is described as the predominant protein in the hemolymph of many crustaceans. It has been reported to contribute >90% of the total hemolymph protein. The properties of this large extracellular oligomer, found in three classes of Arthropoda (Crustacea, Chelicerata, and Myriapoda), have been extensively studied for many years, as have those of the structurally distinct but functionally similar molluscan hemocyanin (for review, see refs. 1–3). Cancer magister, the Dungeness crab, contains a typical arthropod hemocyanin. Its six heterogeneous subunits, ranging from ≈82 to ≈67 kDa, assemble into two distinct populations of molecules, 16S hexamers and 25S two-hexamers (4, 5), that are not in association–dissociation equilibria with one another. Each subunit contains one binuclear copper site and combines reversibly with one molecule of oxygen.
Insects lack a functional hemocyanin. This class of Arthropoda does have other hemolymph proteins, the hexamerins, whose quaternary structures resemble hexameric hemocyanin (for review, see refs. 6 and 7). Some hexamerins are also known as larval serum proteins or storage proteins because of their high concentrations in larval stages and incorporation into new body structures, including cuticle, during metamorphosis or nonfeeding periods of adult development (6, 8, 9). Although these insect hemolymph proteins lack a binuclear copper-binding site, a common ancestral molecule for the copper-containing hemocyanins and the copperless hexamerins has been postulated based on sequence similarities, subunit size, hexameric shape, and a conserved exon–intron boundary (7, 10–14).
We have found a protein in crustacean blood that reaches hemolymph concentrations even greater than hemocyanin during each molt cycle of C. magister and disappears from the hemolymph during intermolt. It is so structurally similar to hemocyanin that early studies often overlooked it. Because this protein masquerades as hemocyanin in structure but neither combines reversibly with oxygen nor has the 340 nm absorbance maximum characteristic of oxyhemocyanin, we have named it cryptocyanin. A “nonrespiratory protein” had been noted previously in hemolymph of several crustaceans and chelicerates (15–21) but has not been fully characterized. A large protein like cryptocyanin present in such high concentrations in the circulation must have profound effects on many aspects of arthropod physiology including cardiovascular function and ion regulation. It may also participate in forming the new exoskeleton during the molt, similar to some insect hexamerins. Could this crustacean protein resolve the evolutionary link between the oxyhemocyanins and the insect hexamerins?
Another arthropod protein, phenoloxidase, has recently been shown to have a close structural relationship to hemocyanin (22–25). Arthropod prophenoloxidases contain the copper A (CuA) and copper B (CuB) binding sites of arthropod hemocyanins, and they share sequence similarities with arthropod hemocyanins and hexamerins. Proteins with phenoloxidase (tyrosinase) activity are widely distributed among animals, plants, fungi, and prokaryotes (26). They incorporate oxygen into other molecules, converting monophenols to o-diphenols and diphenols to the corresponding o-quinones. In arthropods, phenoloxidases function in defense responses and in exoskeleton sclerotization at ecdysis. Molluscan hemocyanins have phenoloxidase activity (27, 28), as do arthropod hemocyanins, although the latter must be activated by partial unfolding or proteolysis (refs. 29 and 30; N.B.T., H. Decker, and M.R., unpublished data). Some crustaceans and insects have a separate prophenoloxidase in hemolymph and/or blood cells, which must be activated to phenoloxidase via a series of proteolytic reactions (31, 32). Does cryptocyanin exhibit any phenoloxidase activity?
In this study, we have characterized purified cryptocyanin of C. magister, determined its cDNA sequence, and compared its derived amino acid sequence with those of arthropod hemocyanins, hexamerins, and prophenoloxidases to explore their structural, functional, and evolutionary relationships. We have also begun to investigate the expression of cryptocyanin throughout the molt cycle of juvenile and adult crabs. We find a close synchrony between stage of molt cycle and hemolymph levels of cryptocyanin that suggests that cryptocyanin is a hormonally regulated molting protein. Thus, cryptocyanin and related molecules may provide important molecular data to further investigate the monophyly of the newly proposed clade of molting animals, the Ecdysozoa (33), in addition to enhancing our understanding of the molecular phylogeny of the hemocyanins.
MATERIALS AND METHODS
Animals.
Adult and juvenile C. magister (Dana) were collected from Coos Bay, Oregon, and maintained in running seawater at ambient temperature and salinity at the Oregon Institute of Marine Biology.
Protein Purification.
Fresh hemolymph was purified by BioGel A-1.5m column chromatography as in ref. 19. Fractions were assayed by absorbance at 280 nm and 340 nm and by using pH 7.4 PAGE. Cryptocyanin was further purified by using ion exchange chromatography on a DEAE BioGel A column (1.5 × 26 cm) equilibrated with 0.05M Tris⋅HCl (pH 7.5) containing 5 mM MgCl2 and 5 mM CaCl2. Proteins were eluted with a linear gradient of 0–0.2 M NaCl.
Electron Microscopy.
Transmission electron microscopy was done on negatively stained samples as in ref. 19 by using a Philips CM12 TEM/STEM operated at 100 kV.
Sedimentation Velocity.
Studies were carried out in a model E analytical ultracentrifuge. Beckman Scanner optics were used at 280 nm, with either 12 mm or 30 mm double-sector cells. All experiments were carried out at temperatures near 20°C.
Copper Analysis.
BioGel A-1.5m and DEAE BioGel A-purified samples of 25S hemocyanin and cryptocyanin were concentrated and rinsed in A-1.5m column buffer in Centricon 100 tubes. Filtrates (used to measure free copper levels in the buffer) and protein samples were analyzed for copper content by using atomic absorption spectroscopy. Localization of fluorescence quenching was done on pH 7.4 PAGE gels (34).
Electrophoresis.
Nondissociating, nondenaturing PAGE at pH 7.4 was done to identify and isolate native hemolymph components and purified proteins (19). Proteins were stained with Coomassie blue, whereas glycoproteins were detected by periodic acid/Schiff reagent staining (35). Prophenoloxidase activity after electrophoresis was demonstrated according to ref. 36. Dissociating, nondenaturing PAGE at pH 8.9 in EDTA (37) was also carried out. Apparent molecular weights and purification of protein subunits were determined by using SDS/PAGE in the presence of DTT (38). Two-step electrophoresis of isolated bands from pH 7.4 PAGE to SDS/PAGE was done as in ref. 19. Purified proteins were either electroeluted from gel slices in a Schleicher & Schuell Elutrap or electroblotted onto Immobilon poly(vinylidene difluoride) membranes.
Western Blots.
Monoclonal antibodies were produced by the University of Oregon Monoclonal Antibody Facility against C. magister 25S hemocyanin and cryptocyanin purified by column chromatography and assayed by pH 7.4 PAGE and SDS/PAGE. For Western blots, proteins were electroblotted from SDS/PAGE onto Immobilon membranes. Primary antibodies were detected by using goat-anti-mouse-biotin secondary antibodies, strepavidin–alkaline phosphatase and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP; Zymed).
Amino Acid Sequence Analysis.
Cryptocyanin was further purified after BioGel A-1.5m chromatography by C4 reverse phase HPLC. HPLC-purified cryptocyanin and two cyanogen bromide fragments were submitted to automated Edman degradation on an Applied Biosystems Model 477A modified gas-phase sequenator equipped with an on-line Applied Biosystems 120A PTH amino acid analyzer (Worcester Foundation Protein Sequencing Center). SDS/PAGE-purified cryptocyanin band 1 was excised from Immobilon membranes after electroblotting and sequenced on an Applied Biosystems Model 470A liquid-phase protein sequenator and an Applied Biosystems Model 120 PTH Analyzer (University of Oregon Biotechnology Laboratory).
PCR Amplification of Cryptocyanin cDNA.
Hypodermis, muscle, and hepatopancreas tissues from C. magister were dissected and immediately frozen in liquid nitrogen. RNA was isolated by using the SNAP total RNA isolation kit (Invitrogen). Reverse transcription–PCR was carried out as in ref. 39. Degenerate PCR primers were designed based on the cryptocyanin N-terminal and CNBr fragment amino acid sequences. The sense primer was 5′-CGG-ATC-CGA-T/CGA-G/ACC-A/G/T/CGA-T/CGG-A/G/T/CG-3′; the antisense primer was 5′-GGA-ATT-CCG-AG/A/CA-G/AA/GG-GGA-TGA-AC/A/GA-C-3′. These primers allowed the amplification of a single 2,000-bp fragment from hepatopancreas cDNA. It was gel purified and cloned into a pCRII vector by using the TA Cloning Kit (Invitrogen) and sequenced in both directions, initially by using T7 and SP6 primers and subsequently by using primers designed according to the resulting sequences. Sequencing was done at the University of Oregon DNA Sequencing Facility.
Sequence Comparisons.
A search of the European Molecular Biology Laboratory (EMBL) and SWISS-PROT databases by using fasta3(40) yielded best scores against hemocyanin, insect hexamerins, and prophenoloxidases. The derived amino acid sequence of cryptocyanin was aligned with sequences for members of these protein groups by using the clustal-w 1.7 program (41). The alignment is in excellent agreement with the alignments and conserved residues published in ref. 7. Neurospora crassa tyrosinase, representing an ancestral CuB site, was used as the outgroup and aligned by hand, based on CuB site similarities (42, 43). Phylogenetic analysis was performed with the paup program (44). The single most parsimonious tree was obtained through a heuristic search algorithm, treating gaps as missing data. Bootstrap analysis was used to assess confidence (45); 500 replicates were done by using the 50% majority-rule consensus.
RESULTS
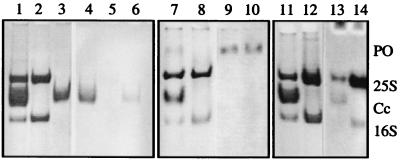
Cryptocyanin was present in premolt hemolymph from megalopa, juvenile, and adult male and female C. magister. It was readily apparent as a single band with an electrophoretic mobility midway between C. magister 25S two-hexamer hemocyanin and 16S one-hexamer hemocyanin when hemolymph was analyzed by pH 7.4 PAGE (Fig. 1, lane 1). The amount of cryptocyanin fluctuated in synchrony with the phase of the animal’s molt cycle. Levels of cryptocyanin peaked in late premolt, usually higher than hemocyanin levels, plummeted just before ecdysis, and were not detectable in intermolt adult crabs (Fig. 1, lane 2). Periodic acid/Schiff reagent staining of hemolymph on a pH 7.4 gel gave a strongly positive reaction for cryptocyanin, indicating it was a glycoprotein (Fig. 1, lanes 4 and 6). Hemocyanin showed only a weak periodic acid/Schiff reagent reaction (Fig. 1, lanes 4 and 5), consistent with the low carbohydrate content reported for other crustacean hemocyanins (46). Cryptocyanin migrated much faster than lipovitellin, a protein present in hemolymph of female crabs during vitellogenesis (see ref. 21, plate 2), and it showed no reaction to Sudan black staining for the presence of lipids. Prophenoloxidase analysis of C. magister hemolymph after pH 7.4 PAGE showed reactivity within 10 min of incubation; a slowly migrating band distinct from both hemocyanin and cryptocyanin bands was present (Fig. 1, lanes 9 and 10).
Figure 1.
Cancer magister hemolymph proteins, nondissociating, nondenaturing PAGE, pH 7.4. CB, Coomassie blue stain. Lane 1, hemolymph with cryptocyanin, CB; lane 2, hemolymph lacking cryptocyanin, CB; lane 3, purified cryptocyanin, CB; lanes 4, 5, 6, same as 1, 2, 3 with PAS reaction; lane 7, hemolymph with cryptocyanin, CB; lane 8, hemolymph lacking cryptocyanin, CB; lanes 9, 10, same as 7, 8 with phenoloxidase reaction; lane 11, hemolymph with cryptocyanin, CB; lane 12, hemolymph lacking cryptocyanin, CB; lanes 13, 14, same as 11, 12 with fluorescence quenching after bathocuproine reaction. PO, phenoloxidase activity; 25S, 25S hemocyanin; Cc, cryptocyanin; 16S, 16S hemocyanin.
When hemolymph from pre-molt crabs was chromatographed on BioGel A-1.5m, cryptocyanin coeluted with the trailing side of the second peak (16S hemocyanin) and had an apparent molecular mass of 430–450 kDa. Analysis of the A-1.5m fractions by pH 7.4 PAGE confirmed this elution pattern. Peak 2 had a higher 280/340 nm absorbance ratio than purified hemocyanin or peak 1 (25S hemocyanin) consistent with the presence of both a hemocyanin and a copper-free protein in peak 2. Cryptocyanin could be separated from 16S hemocyanin by using ion exchange chromatography on DEAE BioGel A, and the purified cryptocyanin electrophoresed as a single band on pH 7.4 PAGE (Fig. 1, lane 3).
Atomic absorption spectroscopy of purified cryptocyanin showed that the protein did not contain copper. Furthermore, the protein lacked the absorbance maximum at 340 nm characteristic of copper-containing oxyhemocyanin. Cryptocyanin did show slight fluorescence quenching on pH 7.4 PAGE (Fig. 1, lane 13), a phenomenon that has been attributed to the presence of copper in protein (34). An equivalent concentration of hemocyanin gave a strong positive quenching reaction (Fig. 1, lane 14), whereas bovine serum albumin, ovalbumin, and horse myoglobin gave negative results. The source of the quenching by cryptocyanin was unknown.
The sedimentation coefficient of purified cryptocyanin, S20,w = 15.95, was similar to that of 16S hemocyanin. When examined by transmission electron microscopy, negatively stained cryptocyanin molecules appeared as single hexagons or squares. They were indistinguishable in size and shape from 16S hemocyanin molecules, consistent with the molecular mass and sedimentation velocity data.
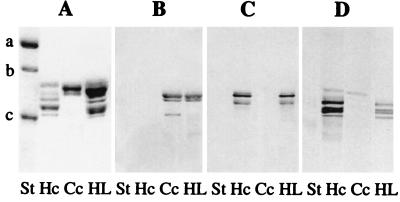
Cryptocyanin electrophoresed as a single slowly migrating component at pH 8.9 in the presence of EDTA, whereas hemocyanin dissociated into rapidly migrating individual subunits, typical of most arthropod hemocyanins. By using SDS/PAGE in the presence of DTT, purified cryptocyanin hexamers dissociated into one major and two minor polypeptide chains with apparent molecular masses of approximately 88, 85, and 74 kDa (Fig. 2).
Figure 2.
Immunological specificity of C. magister cryptocyanin and hemocyanin. (A) SDS 7.5% polyacrylamide gel, Coomassie blue stain. (B) Western blot, anti-cryptocyanin antibody 23A1. (C) Western blot, anti-hemocyanin antibody BB2E4. (D) Western blot, anti-hemocyanin Ab CC4. St, Molecular mass standards, Bio-Rad (a = 116, 250, b = 97, 400, c = 66,200 Da); Hc, hemocyanin; Cc, cryptocyanin; HL, hemolymph containing both hemocyanin and cryptocyanin.
To further explore the relatedness between cryptocyanin and hemocyanin, we developed monoclonal antibodies against the column-purified proteins. Western blot analysis indicated that most antibodies against cryptocyanin did not crossreact with hemocyanin and vice versa, whereas some anti-hemocyanin antibodies showed a slight reaction to cryptocyanin (Fig. 2). These experiments indicated that the two proteins from C. magister are distinctive but related and probably have shared epitopes.
With a size and shape similar to 16S hemocyanin but with no copper, cryptocyanin could be a hemocyanin precursor awaiting its copper, an apohemocyanin, or perhaps a degradation product of hemocyanin. Conversely, it could be a unique member of the hemocyanin gene family, one that has lost its ability to bind copper. To differentiate between these alternatives, we used reverse transcription–PCR in combination with cryptocyanin-specific primers and C. magister hepatopancreas tissue RNA and were able to amplify and sequence a 2,000-bp cDNA product. No product was obtained using hypodermis or muscle RNA, suggesting that the primary site of synthesis is hepatopancreas tissue. The three cryptocyanin peptide sequences aligned identically with the derived amino acid sequence of the 2,000-bp PCR product, indicating that we had amplified cryptocyanin mRNA. The sequence showed remarkable similarity with C. magister hemocyanin subunit 6 (43). Especially intriguing, however, was the discovery that the cryptocyanin sequence contained only two of the three highly conserved histidines of the arthropod hemocyanin CuA binding site and only one of the three conserved histidines in the CuB site. The N-terminal portion was distinctly different from those of all six C. magister hemocyanin subunits (39). Thus, cryptocyanin is a distinct gene product and neither an apo- nor a degraded hemocyanin.
We aligned the cryptocyanin sequence with sequences of other crustacean and chelicerate hemocyanins, insect hexamerins representative of the widely diverse hexamerin types (9, 47), crustacean and insect prophenoloxidases, and an insect embryonic protein—all potential members of the hemocyanin gene family and displaying apparent sequence similarities to one another. The average percentage of cryptocyanin amino acid residues identical to crustacean hemocyanins was 48% (range 40–51%), chelicerate hemocyanins 33% (32–34%), insect hexamerins 27.5% (26–33%), and prophenoloxidases 31% (29–32%). Between any two crustacean hemocyanins, one finds 60% identity, and between chelicerate Hcs, 53%. The high overall sequence similarity between cryptocyanin and these proteins was particularly pronounced at the CuA and CuB sites (Fig. 3). In those proteins that bind copper, hemocyanins and prophenoloxidases, the CuA and CuB histidines were all conserved, whereas cryptocyanin had three of the six and insect hexamerins had only two or less of the histidines.
Figure 3.
Sequence conservation in copper-binding sites CuA and CuB of Cancer magister cryptocyanin and other hemocyanin-type proteins. ∗ indicates conserved histidine, acting as copper ligand in hemocyanin. Residues conserved in more than three taxa and identical to cryptocyanin are shaded. Sequences are identified in Fig. 4.
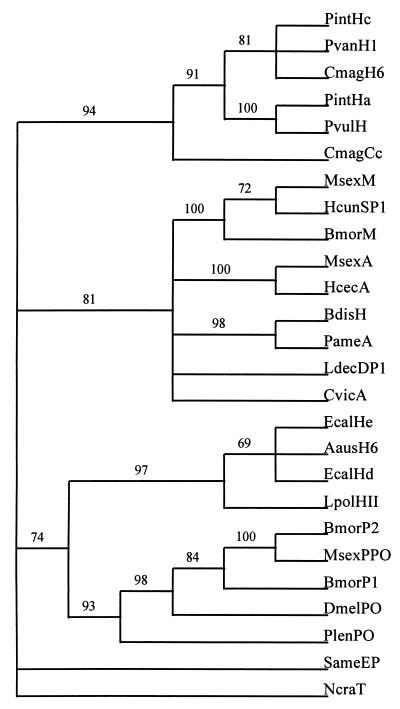
Phylogenetic analysis of the aligned sequences shows that cryptocyanin is clearly associated with the crustacean hemocyanins (Fig. 4). The arthropod hemolymph proteins sort into four monophyletic groups: cryptocyanin and the crustacean hemocyanins, the insect hexamerins, the chelicerate hemocyanins and the prophenoloxidases. The hemocyanin/hexamerin/phenoloxidase pattern is generally consistent with other phylogenies of the hemocyanin gene family (7, 43, 47–49). Grasshopper (Schistocerca americana) embryonic protein (50) does not strongly align with any of the others in our analysis. These conclusions are supported by very robust nodes in the phylogenetic tree indicated by bootstrap values well over 50%. Before bootstrap analysis, the single most parsimonious tree showed the grasshopper embryonic protein grouped with either cryptocyanin/crustacean hemocyanins or insect hexamerins, depending on the choice of hexamerin sequences aligned. After bootstrap analysis, the associations collapsed, illustrating the poorly supported topology of these trees. When prophenoloxidases are used as the outgroup, cryptocyanin/crustacean hemocyanins and insect hexamerins form a monophyletic group and chelicerate hemocyanins form another group. Conversely, when chelicerate hemocyanins are used as the outgroup, cryptocyanin/crustacean hemocyanins and insect hexamerins form one group and phenoloxidases form another group. However, the close relationship between the phenoloxidases and the chelicerate hemocyanins is more obvious when N. crassa tyrosinase is used as the outgroup.
Figure 4.
Single most parsimonious phylogenetic tree of aligned sequences of cryptocyanin and other potential members of hemocyanin gene family. Gaps are treated as missing data. Total tree size: 6,569 substitutions. Bootstrap values (500 replicates) are indicated above branches. PintHc = Panulirus interruptus hemocyanin subunit c, SwissProt accession no. (SP) P80096; PvanH1 = Penaeus vannamei hemocyanin subunit 1, GenBank accession no. (GB) X82502; CmagH6 = Cancer magister hemocyanin subunit 6, GB U48881; PintHa = P. interruptus hemocyanin subunit a, SP P04254; PvulH = Palinurus vulgaris hemocyanin, SP P80888; CmagCc = C. magister cryptocyanin subunit 1, GB AF091261; MsexM = Manduca sexta methionine-rich storage protein, GB L07609; HcunSP1 = Hyphantria cunea storage protein 1, GB U60988; BmorM = Bombyx mori sex-specific storage protein-1, GB X12978; MsexA = M. sexta arylphorin a, GB M28396; HcecA = Hyalophora cecropia arylphorin, GB AF032396; BdisH = Blaberus discoidalis hexamerin, GB U31328; PameA = Periplaneta americana Cr-PI allergen, GB L40818; LdecDP1 = Leptinotarsa decemlineata diapause protein 1, GB X76080; CvicA = Calliphora vicina arylphorin, GB M76480; EcalHe = Eurypelma californica hemocyanin subunit e, GB X16650; AausH6 = Androctonus australis hemocyanin subunit 6, SP P80476, EcalHd = E. californica hemocyanin subunit d, SP P02241; LpolHII = Limulus polyphemus hemocyanin subunit II, SP P04253; BmorP2 = B. mori prophenoloxidase 2, GB D49371; MsexPPO = M. sexta prophenoloxidase, GB L42556; BmorP1 = B. mori prophenoloxidase 1, GB D49370; DmelPO = Drosophila melanogaster prophenoloxidase, GB D45835; PlenPO = Pacifastacus leniusculus prophenoloxidase, GB X83494; SameP = Schistocerca americana embryonic hemolymph protein, GB AF038569; NcraT = N. crassa tyrosinase, GB M33271.
DISCUSSION
The discovery that cryptocyanin shows remarkable similarity in amino acid sequence as well as patterns of molecular size and shape to other arthropod hemolymph proteins provides significant evidence that cryptocyanin is a member of the arthropod hemocyanin gene family. Cryptocyanin most closely resembles hemocyanin in quaternary structure, especially in comparison to the phenoloxidases. The latter exist as monomers, dimers, trimers or larger depending on ionic strength (51), but to date, no one has reported a clearly hexameric prophenoloxidase or phenoloxidase. The similarity in appearance with transmission electron microscopy between cryptocyanin and hemocyanin is interesting, because calliphorin, an insect hexamerin from the blowfly, Calliphora vicuna, has a distinctly triangular shape in electron micrographs (13). Whether the ultrastructure of calliphorin is typical of insect hexamerins awaits publication of micrographs of other hexamerins. The incomplete copper-binding sites of cryptocyanin are consistent with its inability to bind oxygen reversibly as a hemocyanin or to carry out the phenoloxidase reaction. Even after an extended reaction time of up to 1 hour, when purified 25S hemocyanin and 16S hemocyanin from C. magister had a slight positive phenoloxidase reaction, similar to other arthropod hemocyanins, cryptocyanin did not (N.B.T., H. Decker, and M.R., unpublished data). Thus cryptocyanin is neither a hemocyanin nor a phenoloxidase. Cryptocyanin resembles the insect hexamerins in its lack of copper as well as absence of histidines in the CuA and CuB binding sites, but the histidines are more conserved in cryptocyanin than in the insect hexamerins.
The presence of two related proteins, cryptocyanin and hemocyanin, within the hemolymph of one organism, C. magister, provides an ideal system for studying the structure, function, and evolution of these proteins from a common ancestral molecule and strengthens the hypothesis that insect hexamerins are derived from hemocyanin. We hypothesize that gene duplications in an ancestral arthropod hemocyanin allowed loss of the copper-binding capability in one of the gene products. Having lost its role as an oxygen-transport molecule, the resulting cryptocyanin could assume new functions related to the molt cycle and formation of the new exoskeleton. Whereas recent phylogenetic inferences based on 18S rRNA sequences support monophyly of arthropods and chelicerates, the relationship between crustaceans and insects is not robust (52). Crustaceans may be ancestral to insects. If so, cryptocyanin could be ancestral to the insect hexamerins; its presence in crustaceans along with hemocyanin suggests this possibility. Alternatively, crustaceans and insects might have evolved independently from a common ancestor, and the molecular phylogeny of cryptocyanin and hexamerins could parallel this pattern. Common ancestry of two sequences can be a result of either a speciation event or a gene duplication event, and only the former can be used to infer phylogeny of species.
A separate hemocyanin gene duplication, occurring before the divergence of the chelicerate and crustacean hemocyanins, would have led to arthropod phenoloxidase, a molecule that retained its copper-binding sites but shifted function totally from oxygen transport to oxygen incorporation into phenolic compounds with resultant exoskeleton hardening. The persistent grouping of the chelicerate hemocyanins with the insect and crustacean phenoloxidases (Fig. 4) is consistent with this idea. Extant crustaceans like C. magister express all three members of the hemocyanin gene family—hemocyanin, phenoloxidase and cryptocyanin—whereas the air-breathing insects utilize phenoloxidase and the cryptocyanin-like hexamerins.
Cryptocyanin is a molting protein. The increase in hemolymph concentration during premolt and the precipitous decline as the animal goes into ecdysis, as well as cryptocyanin’s absence during intermolt is similar to a pattern of hexamerin levels seen in some insects (9). Hemocyanin levels fluctuate during a crab’s molt cycle also, but the decrease at ecdysis is more moderate, and hemocyanin levels quickly return to premolt conditions. Only during intermolt, when cryptocyanin disappears, does hemocyanin contribute 90% of the hemolymph protein; cryptocyanin dominates during premolt. The differences between the two C. magister proteins reflect separate patterns of regulation. Whether cryptocyanin is ubiquitous among all crustaceans is not yet known, but in a preliminary survey of eastern north Pacific crustaceans, six brachyuran and three anomuran crabs all contained a cryptocyanin-like protein (N.B.T. and S. Johnson, unpublished data). A nonhemocyanin protein whose hemolymph concentrations varied with molt stage is also present in the green crab Carcinus maenas (15, 16); its electrophoretic behavior is similar to cryptocyanin (N.B.T., unpublished data).
Initial functional studies indicate similarities between cryptocyanin and those hexamerins that increase during larval stages, disappear from hemolymph during pupation, and become incorporated into the new cuticle, as in Manduca sexta, for example (8). The hypodermis of a crab synthesizes a new exoskeleton during premolt. Molt cycle fluctuations of cryptocyanin and tissue-specific expression of cryptocyanin mRNA suggest that cryptocyanin is synthesized in cells in or around the hepatopancreas, secreted into the blood vessels, and transported via the hemolymph to the hypodermal cells. Cryptocyanin could also bind and transport molecules such as hormones, ions, or catechols in the hemolymph. During late premolt, some or all of the cryptocyanin molecule may be transferred across the cell into the extrahypodermal space along with other cuticular proteins (53), prophenoloxidase (54), and possibly hemocyanin. Immediately postecdysis, the prophenoloxidase cascade activates the prophenoloxidases in the extrahypodermal space (54), and the process of sclerotization, or hardening of the newly formed exoskeleton, begins. Western blots of hypodermis from early postmolt crabs showed high levels of cryptocyanin whereas other tissues did not; this localization of cryptocyanin had disappeared by 4–5 days postmolt (N.B.T. and C. Otoshi, unpublished data). The entire hemocyanin gene family—hemocyanin, cryptocyanin, prophenoloxidase and hexamerins—may participate to varying degrees in these two vital functions of molting animals (oxygen binding and molting). The newly proposed clade of Ecdysozoa, suggested by Aguinaldo et al. (33), would incorporate all molting animals, including nematodes, onychophorans, and rotifers, into one evolutionary grouping based on sequence comparisons of 18S ribosomal RNA. Cryptocyanin and its relatives may be key molecules to further trace the relationships of the Ecdysozoa.
Acknowledgments
We thank Kristin O’Brien and Clete Otoshi for their dedicated technical assistance. We also thank Karen Miller and Kensal van Holde (Oregon State University) for sedimentation velocity analysis and fruitful discussion, Eric Schabtach (University of Oregon) for electron microscopy, and Ninian Blackburn (Oregon Graduate Institute) for atomic absorption spectroscopy. This work was supported by National Science Foundation Grants IBN 92-17530 and IBN 96-05321 to N.B.T.
ABBREVIATIONS
- CuA and CuB
copper binding sites in arthropod hemocyanins
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF091261).
References
- 1.van Holde K E, Miller K I. Adv Protein Chem. 1995;47:1–81. doi: 10.1016/s0065-3233(08)60545-8. [DOI] [PubMed] [Google Scholar]
- 2.Markl J, Decker H. In: Advances in Comparative and Environmental Physiology. Mangum C P, editor. Vol. 13. Berlin: Springer; 1992. pp. 325–376. [Google Scholar]
- 3.Terwilliger N B. J Exp Biol. 1998;201:1085–1098. doi: 10.1242/jeb.201.8.1085. [DOI] [PubMed] [Google Scholar]
- 4.Ellerton H D, Carpenter D E, van Holde K E. Biochemistry. 1970;9:2225–2232. doi: 10.1021/bi00813a002. [DOI] [PubMed] [Google Scholar]
- 5.Larson B A, Terwilliger N B, Terwilliger R C. Biochim Biophys Acta. 1981;667:294–302. doi: 10.1016/0005-2795(81)90195-1. [DOI] [PubMed] [Google Scholar]
- 6.Telfer W H, Kunkel J G. Annu Rev Entomol. 1991;36:205–228. doi: 10.1146/annurev.en.36.010191.001225. [DOI] [PubMed] [Google Scholar]
- 7.Beintema J J, Stam W T, Hazes B, Smidt M P. Mol Biol Evol. 1994;11:493–503. doi: 10.1093/oxfordjournals.molbev.a040129. [DOI] [PubMed] [Google Scholar]
- 8.Webb B A, Riddiford L M. Dev Biol. 1988;130:671–681. doi: 10.1016/0012-1606(88)90359-4. [DOI] [PubMed] [Google Scholar]
- 9.Haunerland N H. Insect Biochem Mol Biol. 1996;26:755–765. doi: 10.1016/s0965-1748(96)00035-5. [DOI] [PubMed] [Google Scholar]
- 10.Telfer W H, Massey H C., Jr . In: Molecular Entomology. Law J H, editor. New York: Liss; 1987. pp. 305–314. [Google Scholar]
- 11.Sakurai H, Fujii T, Ixumi S, Tamino S. J Biol Chem. 1988;263:7876–7880. [PubMed] [Google Scholar]
- 12.Willot E, Wang X-Y, Wells M A. J Biol Chem. 1989;264:19052–19059. [PubMed] [Google Scholar]
- 13.Markl J, Burmester T, Decker H, Savel-Niemann A, Harris J R, Suling M, Naumann U, Scheller K. J Comp Physiol B. 1992;162:665–680. doi: 10.1007/BF00301616. [DOI] [PubMed] [Google Scholar]
- 14.Voll W, Voit R. Proc Natl Acad Sci USA. 1990;87:5312–5316. doi: 10.1073/pnas.87.14.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busselen P. Comp Biochem Physiol. 1970;37:73–82. [Google Scholar]
- 16.Truchot J P. Arch Zool Exp G. 1978;119:265–282. [Google Scholar]
- 17.Markl J, Markl A, Schartau W, Linzen B. J Comp Physiol. 1979;130:283–292. [Google Scholar]
- 18.Markl J, Hofer A, Bauer G, Markl A, Kempter B, Brenzinger M, Linzen B. J Comp Physiol. 1979;133:167–175. [Google Scholar]
- 19.Terwilliger N B, Terwilliger R C. J Exp Biol. 1982;221:181–191. [Google Scholar]
- 20.Wache S, Terwilliger N B, Terwilliger R C. J Exp Zool. 1988;247:23–32. [Google Scholar]
- 21.Terwilliger N B. In: Crustacean Egg Production. Wenner A, Kuris A, editors. Rotterdam: A. A. Balkema; 1991. pp. 31–36. [Google Scholar]
- 22.Aspán A, Huang T-S, Cerenius L, Söderhäll K. Proc Natl Acad Sci USA. 1995;92:939–943. doi: 10.1073/pnas.92.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall M, Scott T, Sugumaran M, Söderhäll K, Law J H. Proc Natl Acad Sci USA. 1995;92:7764–7768. doi: 10.1073/pnas.92.17.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimoto K, Okino N, Kawabata S, Iwanaga S, Ohnishi E. Proc Natl Acad Sci USA. 1995;92:7769–7773. doi: 10.1073/pnas.92.17.7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawabata T, Yasuhara Y, Ochiai M, Matsuura S, Ashida M. Proc Natl Acad Sci USA. 1995;92:7774–7778. doi: 10.1073/pnas.92.17.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Gelder C W, Flurkey W H, Wichers J. Phytochemistry. 1997;45:1309–1323. doi: 10.1016/s0031-9422(97)00186-6. [DOI] [PubMed] [Google Scholar]
- 27.Salvato B, Jori G, Piazzese A, Ghiretti F, Beltramini M, Lerch K. Life Chem Rep Suppl Ser. 1983;1:313–317. [Google Scholar]
- 28.Nakahara A, Suxuki S, Kino J. Life Chem Rep Suppl Ser. 1983;1:319–322. [Google Scholar]
- 29.Zlateva T, Di Muro P, Salvato B, Beltramini M. FEBS Lett. 1996;384:251–254. doi: 10.1016/0014-5793(96)00326-2. [DOI] [PubMed] [Google Scholar]
- 30.Decker H, Rimke T. J Biol Chem. 1998;273:25889–25892. doi: 10.1074/jbc.273.40.25889. [DOI] [PubMed] [Google Scholar]
- 31.Ashida M, Yamazaki H I. In: Molting and Metamorphosis. Ohnishi E, Ishizaki H, editors. Tokyo: Japan Sci. Soc. Press; 1990. pp. 239–265. [Google Scholar]
- 32.Söderhäll K, Cerenius L. Curr Opin Immunol. 1998;10:23–28. doi: 10.1016/s0952-7915(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 33.Aguinaldo A M, Turbeville J M, Linford L S, Rivera M C, Garey J R, Raff R A, Lake J A. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 34.Bruyninckx W J, Gutteridge S, Mason H S. Anal Biochem. 1978;89:174–177. doi: 10.1016/0003-2697(78)90738-8. [DOI] [PubMed] [Google Scholar]
- 35.Kapitany R A, Zebrowski E J. Anal Biochem. 1973;56:361–369. doi: 10.1016/0003-2697(73)90202-9. [DOI] [PubMed] [Google Scholar]
- 36.Nellaiappan K, Vinayakam A. Biotech Histochem. 1993;68:193–195. doi: 10.3109/10520299309104697. [DOI] [PubMed] [Google Scholar]
- 37.Davis B. Ann N Y Acad Sci. 1964;12:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli H K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Durstewitz G, Terwilliger N B. J Biol Chem. 1997;272:4347–4350. doi: 10.1074/jbc.272.7.4347. [DOI] [PubMed] [Google Scholar]
- 40.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huber M, Lerch K. In: Invertebrate Oxygen Carriers. Linzen B, editor. Berlin: Springer; 1986. pp. 265–276. [Google Scholar]
- 43.Durstewitz G, Terwilliger N B. Mol Biol Evol. 1997;14:266–276. doi: 10.1093/oxfordjournals.molbev.a025762. [DOI] [PubMed] [Google Scholar]
- 44.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1993. , Version 3.1. [Google Scholar]
- 45.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 46.Van Kuik J A, Kamerling J P, Vliegenthart J F G. In: Invertebrate Dioxygen Carriers. Préaux G, Lontie R, editors. Leuven, Belgium: Leuven Univ. Press; 1990. pp. pp.157–163. [Google Scholar]
- 47.Burmester T, Massey H C, Jr, Zakharkin S O, Benes H. J Mol Evol. 1998;47:93–108. doi: 10.1007/pl00006366. [DOI] [PubMed] [Google Scholar]
- 48.Jamroz R C, Beintema J J, Wytze S T, Bradfield J Y. J Insect Physiol. 1996;42:115–124. [Google Scholar]
- 49.Burmester T, Scheller K. J Mol Evol. 1996;42:713–728. doi: 10.1007/BF02338804. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez D, Ganfonina M D, Gutierrez G, Bastiani M J. Mol Biol Evol. 1998;15:415–426. doi: 10.1093/oxfordjournals.molbev.a025938. [DOI] [PubMed] [Google Scholar]
- 51.Jiang H, Wand Y, Ma C, Kanost M R. Insect Biochem Mol Biol. 1997;27:835–850. doi: 10.1016/s0965-1748(97)00066-0. [DOI] [PubMed] [Google Scholar]
- 52.Turbeville J M, Pfeifer D M, Field K G, Raff R A. Mol Biol Evol. 1991;8:669–686. doi: 10.1093/oxfordjournals.molbev.a040677. [DOI] [PubMed] [Google Scholar]
- 53.Sass M, Kiss A, Locke M. J Insect Physiol. 1994;40:561–575. [Google Scholar]
- 54.Ashida M, Brey P T. Proc Natl Acad Sci USA. 1995;92:10698–10702. doi: 10.1073/pnas.92.23.10698. [DOI] [PMC free article] [PubMed] [Google Scholar]