Abstract
The solute carrier (Slc) superfamily is a major group of membrane transport proteins present in mammalian cells. Although Slc transporters play essential and diverse roles in the central nervous system, the localization and function of the vast majority of Slc genes in the mammalian brain are largely unknown. Using high-throughput in situ hybridization data generated by the Allen Brain Atlas, we systematically and quantitatively analyzed the spatial and cellular distribution of 307 Slc genes, which represent nearly 90% of presently known mouse Slc genes, in the adult C57BL/6J mouse brain. Our analysis showed that 252 (82%) of the 307 Slc genes are present in the brain, and a large proportion of these genes were detected at low to moderate expression levels. Evaluation of 20 anatomical brain subdivisions demonstrated a comparable level of Slc gene complexity but significant difference in transcript enrichment. The distribution of the expressed Slc genes was diverse, ranging from near-ubiquitous to highly localized. Functional annotation in 20 brain regions, including the blood-brain and blood-cerebral spinal fluid (CSF) barriers, suggests major roles of Slc transporters in supporting brain energy utilization, neurotransmission, nutrient supply, and CSF production. Furthermore, hierarchical cluster analysis revealed intricate Slc expression patterns associated with neuroanatomical organization. Our studies also revealed Slc genes present within defined brain microstructures and described the putative cell types expressing individual Slc genes. These results provide a useful resource for investigators to explore the roles of Slc genes in neurophysiological and pathological processes.
Transporters are integral membrane proteins whose primary function is to facilitate small molecule exchange across cell membranes. Because of its central role in neurotransmission and its high energy demand, the brain employs numerous membrane transport proteins to supply nutrients, facilitate energy production, control ion and pH balance, regulate neurotransmitter levels and activity, and remove metabolic byproducts (Blakely et al., 1994; Kusuhara and Sugiyama, 2004; Simpson et al., 2007). Many transporters, such as those within the solute carrier (SLC) 6 family of sodium and chloride-dependent neurotransmitter transporters, are involved in the genesis and development of brain disorders (Garland et al., 2002; Beart and O'Shea, 2007). Several transporters, such as the serotonin and norepinephrine reuptake transporters (SLC6A4 and SLC6A2, respectively) and the high-affinity glutamate transporters [e.g., excitatory amino acid transporter 2 (EAAT2)/SLC1A2], are important drug targets for the treatment of various psychiatric and neurological diseases (Torres et al., 2003; Rothstein et al., 2005). In addition, transporters expressed at the blood-brain interfaces, including the blood-brain barrier (BBB) and blood-cerebral spinal fluid barrier (BCSFB), are of great pharmaceutical importance because they are important determinants for drug disposition and targeting into the brain (Smith et al., 2004; Pardridge, 2007).
The SLC (or Slc) superfamily is a major group of membrane transport proteins that control cellular uptake and efflux of nutrients, neurotransmitters, metabolites, drugs, and toxins (Hediger et al., 2004). Approximately 360 Slc genes have been identified from the human and rodent genomes and assigned into 46 gene families (Hediger et al., 2004; refer to the HUGO Gene Nomenclature Committee Solute Carrier Family Series, http://www.bioparadigms.org/slc/menu.asp). Few of these transporters have been characterized fully in the central nervous system (CNS), and the biological and neurological functions of the vast majority of the Slc genes in the mammalian brain are largely unknown. In particular, the expression and distribution of most of the Slc genes in various brain microstructures and cell types have not been characterized. In addition, the physiological substrates of many Slc transporters in the brain remain unknown. Because specific physiological, behavioral, and higher order cognitive functions and many CNS disease processes are frequently associated with distinct brain regions and cell groups, deciphering the spatial and cellular expression patterns of Slc genes in the brain will provide crucial information regarding their neurophysiological and pathological functions in the CNS.
The Allen Brain Atlas (ABA) is a newly developed, genome-wide digital atlas of gene expression in the adult mouse brain (Bhattacharjee, 2006; Lein et al., 2007; Sunkin and Hohmann, 2007). Using highly standardized, automated high-throughput in situ hybridization (ISH) procedures, anatomically comprehensive expression patterns of over 16,000 genes were obtained at cellular resolution in the 56-day-old male C57BL/6J mouse brain (Lein et al., 2007). The primary goal of this study was to construct a comprehensive brain expression map for mouse Slc genes using high-quality ISH data generated by the ABA. Original ISH photomicrographs were manually analyzed for the expression of 307 Slc genes, corresponding to nearly 90% of all presently known mouse Slc genes, in 20 anatomically comprehensive brain areas, including two previously overlooked structures: the BBB and the BCSFB (i.e., choroid plexus). The putative cell types expressing each Slc gene were evaluated based on cell morphology and with reference to cellular marker genes. Unsupervised hierarchical cluster analysis was then employed to elucidate the distributional and potential functional relatedness of the Slc genes across the brain.
Materials and Methods
Databases and Analysis Tools. The primary database used in our study was the Allen Brain Atlas (http://www.brain-map.org). The construction and validation of the ABA was described previously (Lein et al., 2007). The NCBI databases (Genbank, UniGene), the Human Genome Organization Nomenclature Committee Database, the SLC genomic database at Bioparadigms, which includes more than 40 transporter families of the Slc gene series (available online at http://www.bioparadigms.org/slc/menu.asp) (Hediger et al., 2004), and PubMed were used to query individual Slc genes and to obtain information related to their substrates, tissue distribution, and other functions.
ISH Data Analysis. Image series from the Allen Brain Atlas were retrieved by entering the term “Slc” into the search function. Incomplete transcripts and pseudogenes were excluded from the data set. An initial search from the first ABA data release in 2006 returned ISH photomicrograph series for 298 Slc genes. Additional mouse Slc genes were further identified in GenBank, and corresponding probe sets were synthesized and utilized in automated high-throughput ISH at the Allen Institute for Brain Science. The ISH photomicrographs were visually inspected at multiple magnification levels. The sagittal image series near midline (lateral 0.5-1.5 mm) was primarily used for annotation, and when available, coronal sections were also evaluated for image and gene expression consistency. If apparent hybridization artifacts were seen, adjacent images or images from cross-sections were used for primary scoring. For each gene, expression density and level were subjectively scored with reference to the ABA expression heat mask in 20 anatomical subdivisions: olfactory bulb; cortex; striatum; pallidum; dentate gyrus; hippocampal fields CA1, CA2, and CA3; retrohippocampal formation; choroid plexus; thalamus; hypothalamus; superior and inferior colliculi (evaluated as one region); ventral tegmental area; substantia nigra; raphe nucleus; locus coeruleus; pons; medulla; and cerebellum. Brain microvessel expression was analyzed in regions enriched in vasculature, such as the thalamus, and expression level was semiquantitatively evaluated independently from the other brain regions.
Quantitative Analysis of Slc Gene Expression. Within a defined brain region, the tremendous cellular diversity and heterogeneity present a challenge for quantitative analysis of gene expression from histological ISH data. Previously, Lein et al. (2007) showed that relative density (D) and expression level (L) are the two most salient variables for quantitative analysis of brain ISH data. D is defined as the proportion of positively labeled cells in a defined area, and L is a measure correlated with the classic transcript intensity. The product, D × L, was proposed as the metric to quantify gene expression in a defined area (Lein et al., 2007). The number of labeled cells in a defined brain structure was found previously to be the best predictor of whether or not a gene is considered to be “expressed” in that structure by colormetric ISH detection. A minimal cutoff of >1.5% of all 8 × 8-pixel regions containing segmented cell body was defined as the detection threshold (Lein et al., 2007). The same paradigm was used in our analysis. In brief, each ISH image was visually analyzed, and the numerical values for relative density (D, 0-4) and expression level (L, 0-5) were scored for each brain region with reference to the ABA expression heat mask. The D and L values were then multiplied to obtain the expression factor (E = D × L). The mean and S.D. of the E values in the 20 individual brain regions were computed to obtain average expression factor (Ê) for the whole brain. This average expression factor is a global assessment of transcript expression across the brain, whereas the S.D. reflects the degree of heterogeneous expression across the 20 anatomical regions. By definition, the upper limit of expression factor, in average or in individual areas, is 20, and the lower limit is 0. Based on the E value, five expression categories were assigned: “not detected” (E = 0), “low” (0 < E < 4), “moderate” (4 ≤ E < 10), “intermediate to high” (10 ≤ E < 14), and “very high” (E ≥14). Putative cell type annotation of neurons, astrocytes, oligodendrocytes, brain microvessels, and choroid plexus epithelia was performed by morphological examination and by reference to cell type-specific markers, according to Lein et al. (2007).
Unsupervised Hierarchical Cluster Analysis. To investigate the potential anatomical and functional relationship of Slc gene expression in the brain, regional expression factor data were analyzed by unsupervised, two-way hierarchical cluster analysis using MATLAB R2007a software (Mathworks Inc., Natick, MA). Cluster analysis was first evaluated by running multiple combinations of distance and linkage models, and the quality of the various clustering solutions was then assessed by comparing the cophenetic distance coefficients for each model. It was found that the Euclidean distance followed by Ward's linkage (on the x-axis) and single linkage (on the y-axis) produced the highest cophenetic correlation coefficient (>0.9) and, therefore, was used in hierarchical cluster analysis. Because genes with uniform expression interfere with clustering and pattern identification, such genes, characterized by a S.D. of 0 for their average brain expression factor values, were filtered out of the data set, leaving a 235-gene matrix for analysis. The resulting cluster analysis was displayed as a graphic depicting a heat map of gene expression, flanked by dendrograms on the x and y axes.
Results
Global Expression of Slc Transcripts in the Mouse Brain. To identify Slc transporter genes analyzed by the ABA, the term “Slc” was used in the query function in the ABA. This search initially returned acceptable ISH data sets for 298 independent Slc genes. A family-based search in the NCBI Genbank and UniGene databases further identified 29 putative mouse Slc genes, which were either not assayed by the ABA in their first data release or had failed ISH assay. Probe sets corresponding to these genes were synthesized and included in additional runs of automated high-throughput ISH at the Allen Institute for Brain Sciences. Of these probe sets, nine generated acceptable ISH data, whereas the remainder failed quality control. Therefore, the total number of Slc genes included in our final analysis was 307, which corresponds to 87% of the 351 full-length mouse Slc open reading frames existing in the NCBI database.
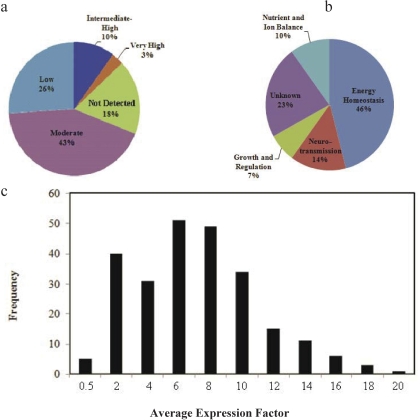
ISH data sets for each of the 307 Slc genes were retrieved and visually inspected in 20 brain anatomical subdivisions. For each gene, expression density (D) and level (L) were scored in individual brain areas, and the expression factor (E) in each area was calculated. The average expression factor (Ê) across the brain was calculated as the mean of the E values from 20 individual areas. Based on the Ê values, the 307 analyzed Slc genes were classified into five categories. Approximately 18% of the 307 Slc genes had anÊ value of 0, and was considered not detected in the brain (Fig. 1a). Twenty-six percent of Slc genes was expressed at low levels (0 <Ê < 4), and 43% was expressed at moderate levels (4 ≤Ê < 10). Ten percent was expressed at intermediate to high levels (10 ≤Ê < 14), and only 3% was expressed at very high levels (Ê ≥ 14) (Fig. 1a). Based on the known and proposed functions and substrates of human and rodent Slc genes, 46% of the 252 Slc genes present in the brain is involved in energy production and consumption, 14% is dedicated to neurotransmitter regulation, 10% mediates nutrient and ion balance, and 7% seems to play a role in cell growth and regulatory pathways (Fig. 1b). It is notable that approximately 23% of the Slc genes encodes orphan transporters with no identifiable substrates. The 252 Slc genes demonstrated a normal distribution of average expression factor values, but the resulting histogram was slightly skewed leftward because of a significant representation of genes expressed at low to intermediate levels (Ê falling between 2 and 10 units) (Fig. 1c).
Fig. 1.
Global expression of Slc transcripts in the mouse brain. a, pie chart shows the percentage of Slc genes in five expression categories based upon their average expression factor values (Ê) across the brain. Based onÊ values, five expression categories were assigned: not detected (Ê = 0), low (0 < Ê < 4), moderate (4 ≤Ê < 10), intermediate to high (10 ≤Ê < 14), and very high (E ≥14). b, pie chart displays the functional categories of Slc genes with detectable expression in the brain (Ê > 0). c, histogram shows the distribution of average expression factor values of Slc genes expressed in the brain.
The 39 Slc genes with intermediate-high to very high average expression factor (Ê ≥ 10) in the brain are shown in Table 1. These genes, representing 20 different Slc families, are generally highly expressed and widely distributed in the brain. The Slc25 family of mitochondrial carriers, the largest Slc family with at least 46 isoforms in man (Palmieri, 2004; Haitina et al., 2006), has nine isoforms represented in this category, accounting for nearly one quarter of the high expressers in the brain. Four of them (Slc25a1, Slc25a4, Slc25a12, Slc25a23) are expressed at the highest level (Ê ≥ 14), with a single isoform, the ADP/ATP carrier 1 (AAC1/Slc25a4), showing the highestÊ value (Ê = 20) among all analyzed Slc genes. Other transporters that are highly expressed and widely distributed in the brain include transporters involved in nutrient (e.g., sugar, amino acids, fatty acids) transport, ion and pH balance, and trace metal (e.g., copper, zinc) homeostasis (Table 1). Several neurotransmitter transporters, including the EAAT2 (Slc1a2), a widely expressed glial transporter responsible for the majority of glutamate uptake in the brain (Kanai and Hediger, 2004; Tian et al., 2007), the plasma membrane monoamine transporter (Slc29a4), a newly identified brain biogenic amine transporter (Engel et al., 2004; Dahlin et al., 2007), the GABA transporter 3 (Slc6a11), and the vesicular glutamate transporter 1 (Slc17a7), are also associated with highÊ values. Several Slc genes in this high expression/wide distribution category are orphan transporters whose substrates have not been identified yet (Table 1).
TABLE 1.
Slc genes with high average expression factor values (Ê ≥ 10) in the brain
| Gene Symbol | Protein Name | Substrates | Average Expression Factor |
|---|---|---|---|
| Slc1a2 | EAAT2 | L-Glutamate, D-/L-aspartate | 11.38 |
| Slc2a12 | GLUT12 | Glucose | 10.71 |
| Slc6a11 | GAT3 | GABA | 10.33 |
| Slc6a17 | NTT4 | Unknown | 13.86 |
| Slc7a4 | Arginine, lysine, and ornithine | 11.19 | |
| Slc7a6 | y+ LAT2 | Cationic and large neutral L-amino acids | 13.48 |
| Slc8a1 | NCX1 | Na+, Ca2+ | 10.14 |
| Slc8a2 | NCX2 | Na+, Ca2+ | 10.81 |
| Slc9a1 | NHE1 | Na+, H+, Li+, NH4+ | 11.43 |
| Slc9a3r1 | NHERF | NHE3-regulatory factor | 12.90 |
| Slc9a7 | NHE7 | Na+, K+, H+, Li+ | 12.52 |
| Slc12a5 | KCC2 | K+, Cl− | 10.90 |
| Slc17a7 | VGLUT1 | Glutamate | 10.05 |
| Slc22a7 | OAT2 | Organic anions, polyspecific | 12.90 |
| Slc22a17 | BOIT | Unknown | 16.05 |
| Slc23a2 | SVCT2 | L-Ascorbic acid | 10.05 |
| Slc24a2 | NCKX2 | Na+, Ca2+, K+ | 15.33 |
| Slc24a3 | NCKX3 | Na+, Ca2+, K+ | 13.90 |
| Slc25a1 | CIC | Citrate, malate, PEP | 15.19 |
| Slc25a3 | PiC | Phosphate | 11.76 |
| Slc25a4 | AAC1 | ADP, ATP | 20.00 |
| Slc25a5 | AAC2 | ADP, ATP | 12.43 |
| Slc25a11 | OGC | Oxoglutarate, malate | 12.00 |
| Slc25a12 | AGC1 | Aspartate, glutamate | 17.24 |
| Slc25a23 | APC2 | ATP, ADP, AMP, and Pi | 14.24 |
| Slc25a27 | UCP4 | H+ | 13.62 |
| Slc25a36 | Unknown | 13.90 | |
| Slc27a1 | FATP1 | Long-chain fatty acids | 10.71 |
| Slc29a3 | ENT3 | Purine and pyrimidine nucleosides, nucleobases | 10.05 |
| Slc29a4 | PMAT | Biogenic amines, adenosine | 10.48 |
| Slc30a2 | ZNT2 | Zinc | 16.57 |
| Slc30a9 | ZNT9 | Unknown | 10.00 |
| Slc31a2 | Ctr2 | Copper | 11.10 |
| Slc35a1 | CST | CMP-sialic acid | 14.52 |
| Slc35b4 | Unknown | 12.90 | |
| Slc38a2 | SNAT2 | Alanine, asparagine, cysteine, glutamine, glycine, histidine, methionine, proline, serine | 12.62 |
| Slc39a7 | KE4 | Manganese | 12.29 |
| Slc43a2 | LAT4 | Neutral amino acids | 10.29 |
| Slc43a3 | EEG1 | Unknown | 11.00 |
Among the 307 Slc genes analyzed, 55 showed ISH signals not significantly above background in all 20 brain regions (Ê = 0) and were classified as not detected in the brain (Supplemental Table 1). A number of these transporters, such as the Na+-taurocholate-cotransporting polypeptide (Slc10a1), the organic anion-transporting polypeptide 1a1 (OATP1a1/Slco1a1), and the urate transporter 1 (Slc22a12), are known to be expressed in a tissue-specific manner in liver and/or kidney, where they mediate the excretion or reabsorption of circulating metabolites (e.g., bile acids, bile salts, urate, etc.) (Hagenbuch and Meier, 1994, 2004; Enomoto et al., 2002). Many others genes not detected in the brain seem to have equivalent isoforms that transport similar categories of substrates in the brain (Table 1; Supplemental Table 1). One gene, Slc1a7 (EAAT5), is exclusively expressed in the retina (Arriza et al., 1997), an area not included in the ABA ISH assay.
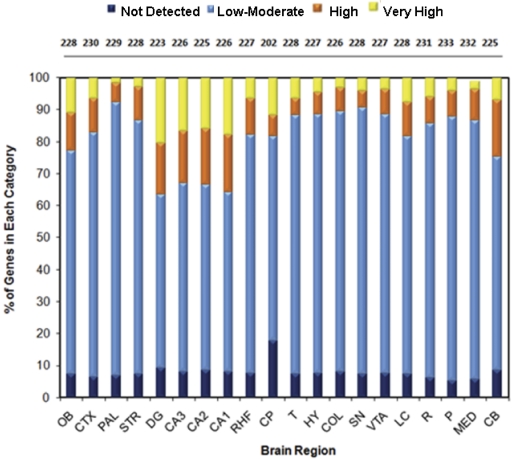
Gene Expression Patterns and Regional Enrichment. To investigate the regional distribution of Slc genes across the brain, the number of genes present in a specific anatomical region was determined, and the percentage of these genes in each expression factor category was calculated (Fig. 2). In terms of gene diversity (i.e., gene numbers), all regions, with the exception of the choroid plexus, exhibited a similar level of complexity, expressing 223 to 233 different Slc genes (mean, 226 ± 6), corresponding to ∼90% of the 252 Slc genes present in the brain. The choroid plexus contained slightly fewer genes (202 genes). No single brain region expressed all 252 genes. With respect to expression factor, significant regional differences were observed. In structures containing dense populations of cell bodies relative to nerve terminals, such as the hippocampal areas (dentate gyrus and fields CA1, CA2, and CA3), cerebellum, and olfactory bulb, a larger proportion of Slc genes (>20%) was observed at high expression levels (E ≥10). Other regions contained a smaller percentage of highly expressed Slc genes. For example, within the pallidum, which is densely innervated by neural processes and contains fewer cell bodies, the smallest fraction (8%) of highly expressed Slc genes was found (Fig. 2).
Fig. 2.
Gene diversity and distribution of expression factor in individual brain regions. The numbers above each bar indicate the total number of genes present (E > 0) in the corresponding brain region. The percentage of Slc genes within each expression factor category, indicated by color-coded bars [dark blue, not detected (E = 0); light blue, low to moderate (0 < E ≤ 9); orange, intermediate to high (9 < E ≤ 13); yellow, very high (13 < E ≤20)] for each brain area is shown on the y-axis. Labels on x-axis correspond to brain regions: OB, olfactory bulb; CTX, cerebral cortex; PAL, pallidum; STR, striatum; DG, dentate gyrus; CA3, CA3 hippocampal field; CA2, CA2 hippocampal field; CA1, CA1 hippocampal field; RHF, retrohippocampal formation; CP, choroid plexus; T, thalamus; HY, hypothalamus; COL, colliculi; SN, substantia nigra; VTA, ventral tegmental area; LC, locus coeruleus; R, raphe; P, pons; MED, medulla; and CB, cerebellum.
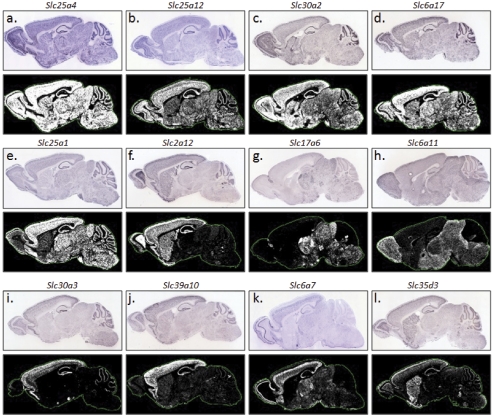
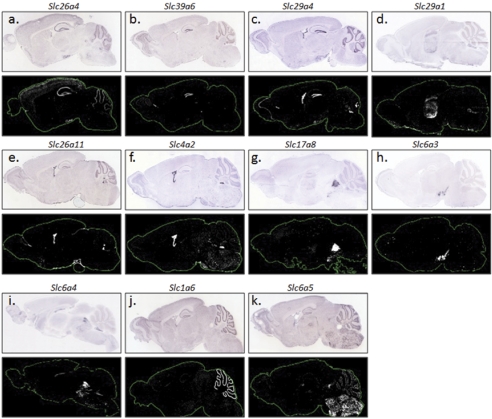
The 252 Slc genes present in the brain revealed diverse expression patterns. The vast majority of Slc genes showed heterogeneous expression; of these, many were enriched in distinct brain regions. In contrast, a small number of Slc genes were uniformly or near uniformly expressed in the brain. Figure 3 provides visual illustrations of Slc genes that are either ubiquitously distributed or regionally enriched in the cerebral cortex, striatum, and brain stem. A number of Slc genes were highly localized to specific fine structures (Fig. 4). These include known highly localized genes, such as the dopamine transporter (DAT/Slc6a3) in the substantia nigra and ventral tegmental area (Torres et al., 2003), the serotonin transporter (SERT/Slc6a4) in the median and dorsal raphe nuclei (Blakely et al., 1994), EAAT4 (Slc1a6) in the Purkinje cell layer of the cerebellum (Kanai and Hediger, 2004), and the anion exchanger 2 (AE2/Slc4a2) in the choroid plexus (Lindsey et al., 1990) (Fig. 4). Highly localized or uniquely enriched patterns were revealed for the first time for a number of other Slc genes. For example, we found that the anion exchanger gene Slc26a4 (pendrin), which is mutated in congenital sensorineural deafness and thyroid goiter (Pendred syndrome) (Scott et al., 1999), is present in the brain and highly enriched in the hippocampal areas (dentate gyrus, CAI-III) (Fig. 4a). In addition, Slc39a6 (Liv-1), a newly described zinc transporter first identified as an estrogen-regulated gene in breast cancer (Taylor et al., 2007), is highly localized to the dentate gyrus of the hippocampus, with minimal presence in CA fields or other brain areas (Fig. 4b). The newly discovered plasma membrane monoamine transporter (Slc29a4) is particularly enriched in the dentate gyrus and choroid plexus (Fig. 4c). The equilibrative nucleoside transporter 1 (Slc29a1), the principal adenosine uptake transporter (Kong et al., 2004), is uniquely enriched in the thalamus (Fig. 4d). The gene encoding Slc26a11, a putative sulfate transporter, is highly localized to the choroid plexus (Fig. 4e). The vesicular glutamate transporter 3 (Slc17a8) is highly localized to the dorsal and medial raphe nuclei (Fig. 4g). The highly localized expression patterns of these genes may suggest novel and specialized functions of these transporters in the neurochemical network. Based upon the specificity of their cellular expression and fine structure distribution, these Slc genes may also serve as potential markers for specific cell groups and microstructures.
Fig. 3.
Examples of expression patterns for ubiquitously and regionally expressed Slc genes. a through e, typical ISH results for ubiquitously and near-ubiquitously expressed genes. Regionally localized Slc genes are shown in f through l. Genes enriched in the cortex are shown in i through l. For each gene, the top represents the ISH image, and the bottom is the expression mask generated from the ISH image.
Fig. 4.
Expression patterns observed for some highly localized Slc genes. In the forebrain, genes that are highly localized in the hippocampal formation, dentate gyrus and thalami are shown in a through d. e and f, choroid plexus-enriched genes. Slc genes that were highly localized to the midbrain (g-i), cerebellum (j), and medulla (k) were also observed. For each gene, the top represents the ISH image, and the bottom is the expression mask generated from the ISH image.
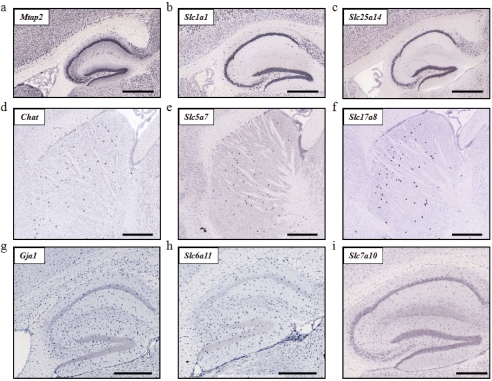
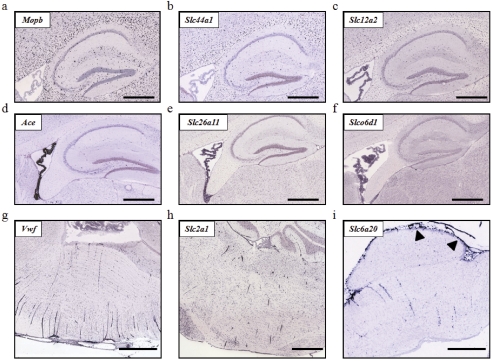
Enriched Expression in Major Cell Types. The expression of Slc genes in major brain cell types is largely unknown. The fine resolution achieved by the ABA ISH data permits expression analysis at the cellular level. Thus, an important goal of this study was to evaluate the putative cell types of Slc expression in neurons, astrocytes, oligodendrocytes, choroid plexus epithelia, and microvessel endothelia. The putative cell types expressing individual Slc genes are summarized in Supplemental Figs. 1 to 4. Figures 5 and 6 provide examples of the observed cellular expression patterns for selected Slc genes. Brain sections showing distinct expression patterns for established cellular markers of neurons (Mtap2), interneurons (Chat), astrocytes (Gja1), oligodendrocytes (Mopb), choroid plexus epithelia (Ace) and brain microvessels (Vwf) were used as references for comparison (Figs. 5 and 6). In general, most Slc genes seem to have neuronal or mixed cellular expression patterns. Many Slc genes showed distinctly neuronal expression (e.g., Slc1a1 and Slc25a14 in Fig. 5, b-c), of which some were clearly present in interneurons (e.g., Slc5a7 and Slc17a8 in Fig. 5, e-f). A number of genes, such as Slc6a11 (Fig. 5h), Slc1a2, and Slc1a3 (Fig. 5i), demonstrated enriched expression in cells that are morphologically similar to astrocytes. A small number of genes, including the choline transporter-like protein 1 (CLT1/Slc44a1), were present in oligodendrocytes as demonstrated by their characteristic distribution in the white matter (e.g., corpus callosum) and by similarity to the distribution pattern of Mopb, an oligodendrocyte-specific gene marker (Fig. 6, a-c).
Fig. 5.
Cellular expression patterns of Slc genes within neurons and astrocytes. Neuron-specific marker Mtap2 (shown in a) was used as a reference for genes with neuronal expression patterns [e.g., Slc1a1 (b) and Slc25a14 (c) in the hippocampus]. The interneuron-restricted Chat [expressed in striatal interneurons in (d)] was used as a marker for genes with interneuron expression [Slc5a7 (e) and Slc17a8 (f) in the striatum]. In h and i, Slc transporters demonstrate cellular expression patterns similar to that of the astrocyte marker, Gja1 (g).
Fig. 6.
Cellular expression patterns of Slc genes within oligodendrocytes, choroid plexus epithelia, and brain microvessel endothelial cells. Cell type-specific markers for oligodendrocytes (a), choroid plexus epithelia (d), and brain microvessel endothelia (g) were used to distinguish the cellular expression patterns of Slc genes. Examples of Slc genes expressed in oligodendrocytes (b and c), choroid plexus (e and f), and brain microvessel (h and i) and pia mater membrane [darts (i)] are shown.
Expression at the Blood-Brain Barrier. Transporters at the BBB are thought to play central roles in supplying vital nutrients to neurons and other cells within the brain (Allen and Geldenhuys, 2006; Simpson et al., 2007). Here, we semiquantitatively analyzed Slc gene expression in brain microvessels by morphology and in reference to the blood vessel endothelial cell marker, Vwf (Fig. 6g). Distinct microvessel expression patterns are apparent for 36 Slc genes, and examples of typically observed BBB expression patterns are shown in Fig. 6, h and i. The 36 Slc genes include transcripts not previously observed in the BBB in addition to well characterized BBB genes such as GLUT1 (Slc2a1), Oatp1c1 (Slco1c1), LAT1 (Slc7a5), and OAT3 (Slc22a8) (Lee et al., 2001; Li et al., 2001; Enerson and Drewes, 2006). These genes, their transported substrates, relative expression levels at the BBB, and average brain expression factor values were summarized in Table 2. Consistent with the role of the BBB in supplying essential nutrients to the brain, we found that 25 of the 36 (∼70%) Slc genes present at the BBB are engaged in the transport of nutrients or nutrient supplements, including transporters for simple sugars (Slc2a1, Slc2a5, Slc2a10, Slc2a13); amino acids (Slc1a2, Slc3a1, Slc7a5, Slc6a6, Slc6a20, Slc7a11, Slc38a3, Slc38a5); oligopeptides (Slc15a2, Slc15a3); long-chain fatty acids (Slc27a5); mono-, di-, and tricarboxylates (Slc13a3, Slc13a5, Slc16a1); folates/thiamines (Slc19a1, Slc19a2); l-ascorbic acids (Slc23a1, Slc23a2); carnitine (Slc22a5); and thyroid hormones (Slc16a2, Slco1c1). Other Slc genes found at the BBB include a urea transporter (Slc14a1) and five polyspecific organic cation or anion transporters (Slco1a4, Slco1a6, Slc22a3, Slc22a7, Slc22a8), which may mediate the efflux of various metabolic waste products from the brain. The remaining BBB-expressed genes include two trace metal transporters (Slc11a2, Slc30a1), a phosphate transporter (Slc20a2), a sulfate transporter (Slc13a4), and a scaffold protein (Slc9a3r2). The majority of genes present in brain microvessels were also expressed at low to moderate levels in other brain regions (between 2 and 10 units). However, several genes, including the facilitative glucose transporters 1, 5, and 10 (GLUT1/Slc2a1, GLUT5/Slc2a5, GLUT10/Slc2a10), the Na+-imino acid transporter (Slc6a20), the cystine/glutamate exchanger (Slc7a11), the sodium-dicarboxylate cotransporter 2 (Slc13a3), and OATP1a6 (Slco1a6), are specifically enriched at the BBB because they showed zero or very low average brain expression factor values (Table 2).
TABLE 2.
Slc genes that are significantly expressed in the BBB
| Gene Symbol | Protein Name | Substrates | Relative BBB Expression Levela | Average Expression Factor in Brain |
|---|---|---|---|---|
| Slc 1a2 | GLT-1 EAAT2 | l-Glu, d-/l-Asp | + | 11.38 |
| Slc2a1 | GLUT1 | Glucose, galactose, mannose, glucosamine | +++ | 0 |
| Slc2a5 | GLUT5 | Fructose | ++ | 1 |
| Slc2a10 | GLUT10 | Glucose, galactose | + | 1 |
| Slc2a13 | HMIT | Myo-inositol | + | 8.9 |
| Slc3a1 | rBAT | Slc7a9 heavy subunit | + | 7.43 |
| Slc6a6 | TAUT | Taurine | ++ | 6.81 |
| Slc6a20 | SIT1 | Imino acids | +++ | 4.33 |
| Slc7a5 | LAT1 | Large neutral amino acids, T3, T4, l-dopa | + | 6.52 |
| Slc7a11 | xCT | Cystine, l-glutamine | +++ | 0.57 |
| Slc9a3r2 | NHERF-2 | NHE3 | + | 9.38 |
| Slc11a2 | DMT1 | Fe2+, Cd2+, Co2+, Cu1+, Mn2+ | + | 3.67 |
| Slc13a3 | NaC2 SDCT2 | Succinate, citrate, α-ketoglutarate | ++ | 0.1 |
| Slc13a4 | NaS2 | Sulfate | + | 7 |
| Slc13a5 | NaC3 | Citrate | + | 2.38 |
| Slc14a1 | UT-B1 | Urea | + | 5.19 |
| Slc15a2 | PEPT2 | Di- and tripeptides | + | 4.19 |
| Slc15a3 | PHT2 | Histidine and di- and tripeptides | + | 2.67 |
| Slc16a1 | MCT1 | Monocarboxylates | + | 6.10 |
| Slc16a2 | MCT8 | T3, T4 | + | 7.9 |
| Slc19a1 | RFT | N5-Methyltetrahydrofolate | + | 9.52 |
| Slc19a2 | ThTr1 | Thiamine | + | 4.9 |
| Slc20a2 | PiT-2 | Pi, Na+ | + | 8.57 |
| Slco1a4 | Oatp1a4 | Digoxin, bile salts, organic anions, organic cations | ++ | 6 |
| Slco1a6 | Oatp1a6 | Organic anions | + | 0 |
| Slco1c1 | Oatp1c1 | T4, rT3, BSP | + | 6.81 |
| Slc22a3 | Oct3 | Organic cations | ++ | 6.19 |
| Slc22a5 | OCTN2 | l-Carnitine, organic cations | ++ | 6.19 |
| Slc22a7 | Oat2 | Organic anions | ++ | 12.9 |
| Slc22a8 | Oat3 | Organic anions | ++ | 5.67 |
| Slc23a1 | SVCT1 | l-Ascorbic acid | + | 3.14 |
| Slc23a2 | SVCT2 | l-Ascorbic acid | + | 10.05 |
| Slc27a5 | FATP5 | Long-chain fatty acids | + | 5.38 |
| Slc30a1 | ZNT1 | Zinc | + | 1.71 |
| Slc38a3 | SNAT4 | Neutral amino acids | + | 8.9 |
| Slc38a5 | SNAT5 | Neutral amino acids | + | 4.29 |
+, significant expression; ++, strong expression; +++, very high expression.
Relative expression level in the BBB.
Expression at the Blood-CSF Barrier. The choroid plexus epithelial cells in the brain ventricles form the second largest brain-blood interface, the BCSFB. A major function of the choroid plexus is to secret CSF, which is accomplished by active transport of small ions (e.g., Na+, Cl-, bicarbonate) across the choroidal epithelium. Besides CSF production, the choroid plexus may also supply the brain with certain nutrients, hormones, and metal ions and remove metabolites and toxins from the CSF (Miller, 2004; Smith et al., 2004). By annotation, 202 Slc genes were found in the choroid plexus (Fig. 2). The majority (∼80%) of the Slc genes in the choroid plexus are expressed at low to moderate levels, whereas 27 Slc genes are expressed at the highest levels, with E ≥ 16 (Table 3). These highly expressed genes include several electrolyte transporters involved in CSF secretion, such as NKCC1 (Slc12a2), AE2 (Slc4a2), and NCBE (Slc4a10). The Slc25 family of mitochondrial carriers was also well represented, with six isoforms highly expressed in the choroid plexus. This observation is consistent with mitochondrial enrichment and high Na+/K+ ATPase activity in the choroid plexus, which provides the ion gradient and driving force for CSF production (Smith et al., 2004). Other highly expressed Slc genes include transporters for nutrients, thyroid hormones, sulfate, and trace metals. Most transporters expressed in the choroid plexus were coexpressed in other brain regions and cell types; however, a very small number of genes, e.g., the sodium-independent sulfate transporter Slc26a11 (Fig. 6e; Table 3), seemed to be expressed only in the choroidal epithelia. Although Slc26a11 is reportedly expressed at high levels in the placenta, kidney, brain, and high endothelial venules (Vincourt et al., 2003), this is the first report of its localization to a specific brain structure. The role of Slc26a11 in the choroid plexus epithelia is presently unknown but may be potentially related to sulfate utilization in the brain.
TABLE 3.
Slc genes with highest expression values (E ≥ 16) in the choroid plexus (CP)
| Gene Symbol | Protein Name | Substrates | CP Expression Factor | Average Brain Expression Factor |
|---|---|---|---|---|
| Slc2a12 | GLUT12 | Glucose | 20 | 11 |
| Slc4a2 | AE2 | Cl−, 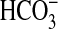
|
20 | 5.8 |
| Slc4a10 | NCBE | Na+, Cl−,
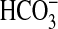
|
16 | 8.5 |
| Slc5a3 | SMIT | Myo-inositol | 20 | 6.1 |
| Slc6a20 | SIT1 | Imino acids | 16 | 4.5 |
| Slc7a4 | CAT-4 | Unknown | 16 | 11 |
| Slc7a6 | y1 LAT2 | Large, neutral l-amino acids and cationic amino acids | 16 | 14 |
| Slc12a2 | NKCC1 | Sodium, potassium, chloride | 16 | 5.4 |
| Slc13a4 | NaS2 | Sulfate | 20 | 7.1 |
| Slc16a2 | MCT8 | T3, T4 | 20 | 8 |
| Slc19a1 | RFT | N5-Methyltetrahydrofolate | 16 | 9.6 |
| Slco1c1 | Oatp1c1 | T4, rT3, BSP | 20 | 6.9 |
| Slc22a17 | BOIT | Unknown | 20 | 16 |
| Slc23a2 | SVCT2 | l-Ascorbic acid | 20 | 10 |
| Slc24a5 | NCKX5 | Na+, Ca2+ | 16 | 1.8 |
| Slc25a3 | PiC | Phosphate | 16 | 12 |
| Slc25a4 | AAC1 | ADP, ATP | 16 | 20 |
| Slc25a5 | AAC2 | ADP, ATP | 16 | 12 |
| Slc25a11 | OGC | Oxoglutarate, malate | 16 | 12 |
| Slc25a12 | ACG1 | Aspartate, glutamate | 16 | 17 |
| Slc25a17 | ANC1 | ATP | 16 | 8 |
| Slc26a11 | Sulfate | 16 | 0.76 | |
| Slc27a1 | FATP1 | Long-chain fatty acids | 16 | 11 |
| Slc29a4 | PMAT | Biogenic amines, adenosine | 20 | 10 |
| Slc31a1 | Ctr1 | Copper | 16 | 7.7 |
| Slc38a3 | SNAT4 | System A amino acids | 20 | 8.9 |
| Slc39a12 | Zinc | 16 | 2.1 |
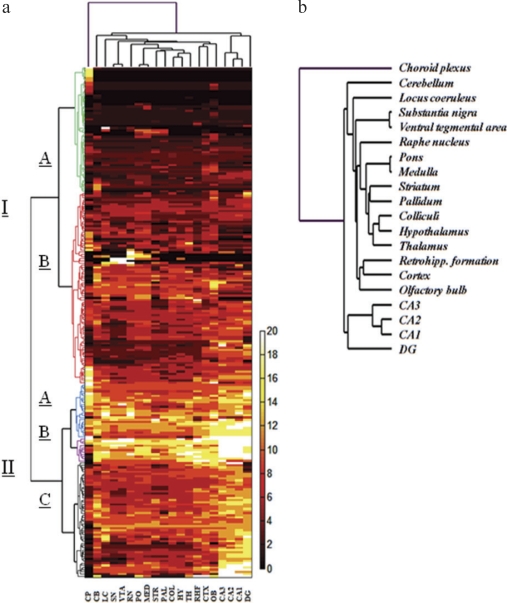
Clustering of Correlated Gene Expression. To explore potential functional relatedness of the Slc genes and possible anatomic association with brain microstructures, we implemented unsupervised, two-way hierarchical cluster analysis to reveal the expression patterns of Slc genes in 20 brain regions. The results of this analysis are shown in Fig. 7. On the x-axis, the resulting dendrogram ordered the 20 evaluated brain regions into two major clusters, with the choroid plexus comprising a third cluster. The other 19 brain areas were ordered into several branches and subbranches within the two dominant clusters. Anatomically proximal regions were closely coclustered (Fig. 7b), illustrating that functionally and anatomically related structures share a higher degree of overlapping Slc expression profiles.
Fig. 7.
Unsupervised hierarchical cluster analysis (UHCA) of Slc gene expression across the brain. Clustergram shown in a demonstrates results of UHCA of E values for Slc genes. UHCA produced two major clusters (labeled I and II, left), divided into five subclusters (dendrogram is color-coded for ease of identification). Label bar at the right of clustergram indicates the range of color-coded expression factor values presented in the heat map (from 0 to 20). In b, a magnified image of the dendrogram of clustered brain regions [shown as x-axis labels (a)] is shown. OB, olfactory bulb; CTX, cerebral cortex; PAL, pallidum; STR, striatum; DG, dentate gyrus; CA3, CA3 hippocampal field; CA2, CA2 hippocampal field; CA1, CA1 hippocampal field; RHF, retrohippocampal formation; CP, choroid plexus; T, thalamus; HY, hypothalamus; COL, colliculi; SN, substantia nigra; VTA, ventral tegmental area; LC, locus coeruleus; R, raphe; P, pons; MED, medulla; CB, cerebellum.
The resulting y-axis dendrogram consisted of two major branches (Fig. 7). The upper branch (cluster I) contained the low- and variable-expression factor clusters, whereas the lower branch (cluster II) included the high expression factor clusters. Cluster I was further divided into subclusters, IA and IB. Cluster IA contained Slc genes that tended to have a localized distribution pattern (Supplemental Fig. 1). A group of these genes, including Slc39a12, Slc24a5, Slco6d1, and Slc26a11, were highly expressed and localized in the choroid plexus. Another group of genes, including Slc41a3, Slco4a1, and Slc9a3, were highly enriched in the cerebellum. Cluster IB, the largest subcluster, contained genes that were characterized generally by wide distribution and low to moderate expression factor values (Supplemental Fig. 2). However, a minority of cluster IB genes were highly localized. Genes in the second major branch, cluster II, possessed two consistent traits: widespread distribution and intermediate to high levels of expression in multiple regions (Fig. 7). Another interesting observation of cluster II was the marked enrichment of Slc genes in the hippocampal areas (Fig. 7). Within cluster II, three subbranches, IIA, IIB, and IIC, were present. Cluster IIA contained genes that seem to be high expressers in choroid plexus, but are also highly expressed in a few other areas including the hippocampus (Supplemental Fig. 3). The smallest subbranch, cluster IIB, contained only 11 genes, which generally demonstrated high expression in nearly all brain regions but were low or not detected in the choroid plexus (Supplemental Fig. 3). Cluster IIC, the largest subbranch in cluster II, contained genes that showed variable expression across the brain but were particularly enriched in the hippocampal region. Like cluster IIB, genes in cluster IIC were also absent or expressed at low levels in the choroid plexus (Supplemental Fig. 4).
Discussion
In this study, we comprehensively and systematically analyzed the expression patterns of 307 Slc genes, representing nearly 90% of presently known mouse Slc genes, in the mouse brain. To our knowledge, this is the first study in which a detailed expression analysis was performed for a single gene superfamily in the mammalian brain. This is also the first study in which mammalian Slc genes were comprehensively characterized in a distinct organ structure.
Across the brain, the average expression factor for the majority of Slc genes was low to moderate (0 <Ê < 10), suggesting that many genes are either expressed at low levels throughout the brain or localized to a few brain areas. However, 39 Slc genes showed relatively high average expression factor values (Ê ≥ 10). Nine isoforms within the Slc25 mitochondrial carrier family are represented in this category, and four of them (Slc25a1, Slc25a4, Slc25a12, Slc25a23) are expressed at the highest level among all genes evaluated in this analysis. The abundant expression and wide distribution of Slc25 mitochondrial transporters across the brain is consistent with the brain's high energy demand, which consumes 25% of total body energy (Clark and Sokoloff, 1999). Located in the inner membrane of mitochondria, these proteins transport ADP/ATP and tricarboxylic acid cycle intermediates and are essential for ATP production and mitochondrial energy metabolism, an activity that fuels all basic and higher order cellular activities (Palmieri, 2004). Other highly expressed genes include transporters for nutrients, ions, and neurotransmitters (Table 1). These findings, together with the functional categorization of the 252 Slc genes present in the brain (Fig. 1b), suggest that Slc transporters play crucial roles in energy metabolism, nutrient supply, and neurotransmitter regulation in the brain.
The mammalian brain is anatomically complex, containing diverse cell types and distinct microstructures. These microstructures are associated with unique functional tasks and are specifically targeted by disease processes. Previous histological studies have only mapped the expression of a limited number of Slc genes in brain microstructures. In this study, we systematically and quantitatively analyzed the spatial and cellular distribution of 307 Slc genes in 20 anatomically comprehensive brain regions. Our analysis revealed complex and diverse expression patterns for the Slc genes (Figs. 3, 4, 5, 6; Supplemental Figs. 1, 2, 3, 4), suggesting important and diverse roles for this gene superfamily in the brain. Moreover, the identification of highly localized patterns and cell type-specific expression of certain Slc genes may provide important clues to their physiological functions within various neurochemical networks. For example, our study found that the low-affinity, high-capacity CTL1 (Slc44a1), a novel and less characterized Slc transporter (O'Regan et al., 2000), is uniquely enriched in oligodendroglial cells (Fig. 6b). In contrast, the Na+-dependent, high-affinity choline transporter 1 (Slc5a7) is known to be mainly expressed in cholinergic neurons, where it plays a critical role in sustaining synaptic cholinergic signaling (Bazalakova and Blakely, 2006). The distinct cellular distribution of CTL1 suggests a fundamentally different role of this transporter in the brain. Because choline is also an essential precursor of membrane lipids (e.g., phosphatidylcholine, shingomylin) (Michel et al., 2006), CTL1 may function in the synthesis of biomembranes (including the myelin sheath) in oligodendroglial cells.
An important contribution of our study is the annotation of Slc gene expression at the BBB and choroid plexus, two important barriers that protect the brain from circulating xenobiotics and blood-borne pathogens. Several studies have attempted to characterize the gene expression profiles at the BBB using microarray or polymerase chain reaction-based serial analysis of gene expression approaches (Li et al., 2001; Enerson and Drewes, 2006). This study is the first where BBB expression of the Slc genes is systematically analyzed using ISH histological data. Here, we clearly identified 36 Slc genes expressed at the BBB, including many novel transcripts in addition to genes with known localization in this structure. Compared with the large number (202-233) of Slc genes identified in the choroid plexus and other brain subdivisions (Fig. 2), this number is relatively small, suggesting that our method is not sensitive enough to detect genes with relatively low expression at the BBB. Consistent with the role of the BBB in supplying essential nutrients to the brain, ∼70% of the 36 Slc genes found at the BBB is involved in the transport of nutrients, vitamins, hormones, and trace metals (Table 2). On the other hand, genes involved in electrolyte transport and ATP production are well represented at the choroid plexus, consistent with its role in CSF production (Table 3). It is interesting that a few genes, including the thyroid hormone transporters Slc16a2 and Slco1c1, the sulfate transporter Slc13a4, the reduced folate transporter Slc19a1, the l-ascorbic acid transporter Slc23a2, and the system A amino acid transporter Slc38a3, are coexpressed at high levels at both the BBB and the choroid plexus. These genes may play coordinated roles at the two blood-CNS interfaces to regulate brain homeostasis of nutrients and hormones.
Unsupervised, two-way hierarchical cluster analysis revealed that Slc gene expression patterns are associated with the neuroanatomical organization of the brain. In general, anatomically adjacent and functionally related structures share overlapping Slc expression profiles (Fig. 7b). It is interesting that marked enrichment of Slc genes was observed in the hippocampal areas (dentate gyrus, fields CA1, CA2, and CA3) in cluster II (Fig. 7; Supplemental Figs. 3-4). The highly expressed genes in the hippocampus include many mitochondrial carriers in the Slc25 family (Slc25a1, Slc25a5, Slc25a11, Slc25a12, Slc25a16, Slc25a23, Slc25a27, Slc25a36) and Golgi nucleoside-sugar transporters in the Slc35 family (Slc35a1, Slc35a4, Slc35b1, Slc35b2, Slc35b3, Slc35e2), which may suggest high volume of energy consumption and protein processing activities in this area.
In conclusion, we have identified the major Slc transporters expressed in defined brain structures and revealed the complex expression patterns represented by this gene superfamily. These studies will ultimately serve as a helpful resource for delineating the roles of Slc genes in the mammalian brain.
Supplementary Material
Acknowledgments
We thank Dr. Michael Hawrylycz for assistance in bioinformatic analysis and critical reading of the manuscript.
This work was supported by the National Institutes of Health [Grants R01-GM066233, T32-GM007750].
doi:10.1124/jpet.108.149831.
ABBREVIATIONS: Slc, solute carrier; EAAT, excitatory amino acid transporter 2; BBB, blood-brain barrier; BCSFB, blood-cerebrospinal fluid barrier; CNS, central nervous; ABA, Allen Brain Atlas; ISH, in situ hybridization; NCBI, National Center for Biotechnology Information; OATP, organic anion-transporting polypeptide; CLT, choline-like transporter; GLUT, glucose transporter; CSF, cerebrospinal fluid; UHCA, unsupervised hierarchical cluster analysis.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
References
- Allen DD and Geldenhuys WJ (2006) Molecular modeling of blood-brain barrier nutrient transporters: in silico basis for evaluation of potential drug delivery to the central nervous system. Life Sci 78 1029-1033. [DOI] [PubMed] [Google Scholar]
- Arriza JL, Eliasof S, Kavanaugh MP, and Amara SG (1997) Excitatory amino acid transporter 5, a retinal glutamate transporter coupled to a chloride conductance. Proc Natl Acad Sci U S A 94 4155-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazalakova MH and Blakely RD (2006) The high-affinity choline transporter: a critical protein for sustaining cholinergic signaling as revealed in studies of genetically altered mice. Handb Exp Pharmacol 175 525-544. [DOI] [PubMed] [Google Scholar]
- Beart PM and O'Shea RD (2007) Transporters for l-glutamate: an update on their molecular pharmacology and pathological involvement. Br J Pharmacol 150 5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee Y (2006) Neuroscience: “Google of the brain”: atlas maps brain's genetic activity. Science 313 1879. [DOI] [PubMed] [Google Scholar]
- Blakely RD, De Felice LJ, and Hartzell HC (1994) Molecular physiology of norepinephrine and serotonin transporters. J Exp Biol 196 263-281. [DOI] [PubMed] [Google Scholar]
- Clark DD and Sokoloff L (1999) in Basic Neurochemistry: Molecular, Cellular and Medical Aspects (Siegel GJ, Agranoff BW, Albers RW, Fisher SK, and Uhler MD eds) pp 637-670, Lippincott, Philadelphia, PA.
- Dahlin A, Xia L, Kong W, Hevner R, and Wang J (2007) Expression and immunolocalization of the plasma membrane monoamine transporter in the brain. Neuroscience 146 1193-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerson BE and Drewes LR (2006) The rat blood-brain barrier transcriptome. J Cereb Blood Flow Metab 26 959-973. [DOI] [PubMed] [Google Scholar]
- Engel K, Zhou M, and Wang J (2004) Identification and characterization of a novel monoamine transporter in the human brain. J Biol Chem 279 50042-50049. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Kimura H, Chairoungdua A, Shigeta Y, Jutabha P, Cha SH, Hosoyamada M, Takeda M, Sekine T, Igarashi T, et al. (2002) Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature 417 447-452. [DOI] [PubMed] [Google Scholar]
- Garland EM, Hahn MK, Ketch TP, Keller NR, Kim CH, Kim KS, Biaggioni I, Shannon JR, Blakely RD, and Robertson D (2002) Genetic basis of clinical catecholamine disorders. Ann N Y Acad Sci 971 506-514. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B and Meier PJ (1994) Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest 93 1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B and Meier PJ (2004) Organic anion transporting polypeptides of the OATP/SLC21 family: phylogenetic classification as OATP/SLCO superfamily, new nomenclature and molecular/functional properties. Pflugers Arch 447 653-665. [DOI] [PubMed] [Google Scholar]
- Haitina T, Lindblom J, Renström T, and Fredriksson R (2006) Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics 88 779-790. [DOI] [PubMed] [Google Scholar]
- Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, and Bruford EA (2004) The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch 447 465-468. [DOI] [PubMed] [Google Scholar]
- Kanai Y and Hediger MA (2004) The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch 447 469-479. [DOI] [PubMed] [Google Scholar]
- Kong W, Engel K, and Wang J (2004) Mammalian nucleoside transporters. Curr Drug Metab 5 63-84. [DOI] [PubMed] [Google Scholar]
- Kusuhara H and Sugiyama Y (2004) Efflux transport systems for organic anions and cations at the blood-CSF barrier. Adv Drug Deliv Rev 56 1741-1763. [DOI] [PubMed] [Google Scholar]
- Lee G, Dallas S, Hong M, and Bendayan R (2001) Drug transporters in the central nervous system: brain barriers and brain parenchyma considerations. Pharmacol Rev 53 569-596. [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445 168-176. [DOI] [PubMed] [Google Scholar]
- Li JY, Boado RJ, and Pardridge WM (2001) Blood-brain barrier genomics. J Cereb Blood Flow Metab 21 61-68. [DOI] [PubMed] [Google Scholar]
- Lindsey AE, Schneider K, Simmons DM, Baron R, Lee BS, and Kopito RR (1990) Functional expression and subcellular localization of an anion exchanger cloned from choroid plexus. Proc Natl Acad Sci U S A 87 5278-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel V, Yuan Z, Ramsubir S, and Bakovic M (2006) Choline transport for phospholipid synthesis. Exp Biol Med (Maywood) 231 490-504. [DOI] [PubMed] [Google Scholar]
- Miller DS (2004) Confocal imaging of xenobiotic transport across the choroid plexus. Adv Drug Deliv Rev 56 1811-1824. [DOI] [PubMed] [Google Scholar]
- O'Regan S, Traiffort E, Ruat M, Cha N, Compaore D, and Meunier FM (2000) An electric lobe suppressor for a yeast choline transport mutation belongs to a new family of transporter-like proteins. Proc Natl Acad Sci U S A 97 1835-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch 447 689-709. [DOI] [PubMed] [Google Scholar]
- Pardridge WM (2007) Drug targeting to the brain. Pharm Res 24 1733-1744. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Patel S, Regan MR, Haenggeli C, Huang YH, Bergles DE, Jin L, Dykes Hoberg M, Vidensky S, et al. (2005) Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature 433 73-77. [DOI] [PubMed] [Google Scholar]
- Scott DA, Wang R, Kreman TM, Sheffield VC, and Karniski LP (1999) The Pendred syndrome gene encodes a chloride-iodide transport protein. Nat Genet 21 440-443. [DOI] [PubMed] [Google Scholar]
- Simpson IA, Carruthers A, and Vannucci SJ (2007) Supply and demand in cerebral energy metabolism: the role of nutrient transporters. J Cereb Blood Flow Metab 27 1766-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Johanson CE, and Keep RF (2004) Peptide and peptide analog transport systems at the blood-CSF barrier. Adv Drug Deliv Rev 56 1765-1791. [DOI] [PubMed] [Google Scholar]
- Sunkin SM and Hohmann JG (2007) Insights from spatially mapped gene expression in the mouse brain. Hum Mol Genet 16 R209-R219. [DOI] [PubMed] [Google Scholar]
- Taylor KM, Morgan HE, Smart K, Zahari NM, Pumford S, Ellis IO, Robertson JF, and Nicholson RI (2007) The emerging role of the LIV-1 subfamily of zinc transporters in breast cancer. Mol Med 13 396-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Lai L, Guo H, Lin Y, Butchbach ME, Chang Y, and Lin CL (2007) Translational control of glial glutamate transporter EAAT2 expression. J Biol Chem 282 1727-1737. [DOI] [PubMed] [Google Scholar]
- Torres GE, Gainetdinov RR, and Caron MG (2003) Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci 4 13-25. [DOI] [PubMed] [Google Scholar]
- Vincourt JB, Jullien D, Amalric F, and Girard JP (2003) Molecular and functional characterization of SLC26A11, a sodium-independent sulfate transporter from high endothelial venules. FASEB J 17 890-892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.