Abstract
Background & Aims
The function of microRNA (miRNA) in liver development is unknown. To address this issue, we characterized miRNA expression in the embryonic mouse liver, performed functional miRNA analysis in zebrafish larvae, and identified novel hepatic miRNA targets.
Methods
Hepatic RNA isolated from mice at embryonic days 15.5, 18.5, and post-natal day 2 was hybridized to a mouse miRNA microarray. The microarray results were confirmed by Northern blot hybridization and quantitative RT-PCR. The spatial distribution of selected miRNAs was determined by in situ hybridization. Functional analysis of miR-30a was performed in zebrafish using antisense-mediated miRNA knockdown. Targets of miR-30a were identified by microarray analysis of gene expression following knockdown in cultured cells.
Results
A set of 38 differentially expressed fetal hepatic miRNAs was identified. Several of these miRNAs were found to exhibit distinct temporal and spatial patterns of expression in hepatocytes, cholangiocytes, and non-epithelial cells within the liver. Two (miR-30a and miR-30c) are the first examples of ductal plate and bile duct-specific hepatic miRNAs. Knockdown of miR-30a in the zebrafish larva results in defective biliary morphogenesis. Several newly identified targets of miR-30a are known regulators of liver development and function.
Conclusions
We have identified miRNAs whose spatial and temporal patterns of expression are suggestive of functional roles in hepatic development and/or function. One of these, the biliary miRNA miR-30a, is required for biliary development in zebrafish. This is the first demonstration of a functional role for miRNA in hepatic organogenesis.
Introduction
The past decade has seen an increasing recognition of the widespread importance of small noncoding RNA molecules in regulating gene expression in a wide variety of organisms.1, 2 The most abundant of these are microRNAs (miRNA), 21-23nt single-stranded RNAs found in both plants and animals. One miRNA can regulate many genes; given the existence of hundreds of miRNAs per vertebrate genome, the regulatory potential of these molecules is very large.
The functional importance of miRNA in development has been borne out in worms, flies, and plants. 3-6 Less is known regarding miRNA function in vertebrate development. An essential role for miR-375 in pancreas development has been demonstrated using antisense-mediated knockdown in zebrafish.7 Loss of miR-1-2 function has profound effects on cardiac development,8 while conditional loss of miR-155 has broad effects on the immune system 9 However, it is not known if any miRNAs regulate the process of hepatobiliary development.
While previous studies have documented liver-expressed miRNA, these have generally surveyed the static miRNA content of the adult or fetal liver.10 To date, changes in miRNA expression during embryonic liver development have not been explored. Furthermore, the regional specificity of liver-specific miRNA has not been examined. We speculated that miRNA plays a role in the establishment of hepatocyte and cholangiocyte cell fates occurring between embryonic day 15.5 (E15.5) and the neonatal period.11 This process is intimately tied to the appearance and remodeling of the ductal plate, a transient sheet of CK19-expressing hepatoblasts that gives rise to mature bile ducts.11 Failure of ductal plate morphogenesis and remodeling is characteristic of several human diseases, including congenital hepatic fibrosis, autosomal recessive and dominant polycystic kidney diseases, and Caroli syndrome.12-18 It is associated with approximately 25% of cases of biliary atresia, the most common indication for pediatric liver transplantation.19
In this study we have obtained a global profile of changes in the liver miRNA transcriptome during the critical phase of biliary development from late gestation to the perinatal period. Two miRNAs identified by this approach are miR-30a miR-30c, which are the first biliary-specific hepatic miRNAs to be described. Functional analysis in zebrafish indicates that miR-30a is required for normal hepatobiliary development, suggesting a similar critical role in mammalian hepatogenesis. This is supported by our identification of several miR-30a target genes that are known or suspected regulators of hepatic differentiation, growth, or function.
Materials and Methods
Mice
FVB/N mice were obtained from Charles River Laboratory. All mice were housed, handled, and euthanized in accordance with federal and institutional guidelines under the supervision of the Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee.
Reagents
3’ digoxygenin-labeled miRNA antisense probes were purchased from Exiqon. Tyramide amplification was performed using the TSA Plus Fluorescence systems kit (Perkin Elmer).
RNA isolation
Fetal livers were dissected in PBS, and tissue was lysed immediately for RNA isolation. RNA was purified using the mirVana™ miRNA Isolation Kit (Ambion). Sample quantification and quality assurance were performed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), and an Agilent 2100 Bioanalyzer (Agilent Technologies).
Microarray analysis of miRNA expression
Total liver RNA was labeled and hybridized to the locked nucleic acid (LNA) based miRcury™ 8.0 microarray at Exiqon A/S, (Vedbaek, Denmark). Three biological replicates of each time point (E15.5, E18.5, P2) were analyzed. Hybridization intensities were scored relative to a common control consisting of diluted, pooled RNA from all of the samples. Raw intensity data were normalized and analyzed using the SAM add-in for Microsoft Excel.20 MiRNAs exhibiting a fold-change of greater than 1.5 at a false discovery rate of 10% were chosen for further study. The microarray data have been deposited at the NCBI GEO repository under the accession number GSE10602.
Microarray analysis of mRNA expression
RNA from BMELs transfected with either control or miR-30a ASO was prepared and 100 ng of total RNA per sample was amplified and biotinylated using the MessageAmp Premier kit (Ambion). Samples (n=5 for each) were hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 Arrays in the Children’s Hospital of Philadelphia Nucleic Acids Core Facility and analyzed with the assistance of the Penn Bioinformatics Core. Probe intensities were normalized using the GCRMA method 21 and the significance of the log2-transformed, GCRMA-normalized signal intensities was determined using SAM20. The microarray data have been deposited at the NCBI GEO repository under the accession number GSE12908.
Northern blot analysis
Northern blot analysis was carried out using antisense LNA probes directed against the mature microRNA sequence. 10 pmol 3’-digoxygenin labeled LNA probe was 5’ phosphorylated with 5μCi γ-32P-ATP using polynucleotide kinase (New England Biolabs) according to the manufacturer’s instructions. Hybridization was carried as described.22 A probe directed against the Valyl-tRNA served as a loading control.
Characterization of miRNA spatial expression
The spatial expression of miRNAs was characterized by in situ hybridization on frozen liver sections using 3’ digoxygenin-labeled antisense LNA probes using a modified version of a published protocol.23 To ensure detection of miRNA with low expression a tyramide signal amplification step was incorporated (see Supplementary Methods).
In situ hybridization and immunofluorescence double detection
For simultaneous detection of miRNA and protein, incubation with anti-digoxygenin antibody-HRP conjugate was performed simultaneously with either anti-CK19 (1:1000, lab collection) for mouse tissue, anti-CK19 (1:1000, Dako) for human tissue, or anti-elastin (1:400, Cedarlane Laboratories Limited). Tyramide signal amplification was performed prior to secondary antibody incubation.
In situ hybridization in zebrafish
In situ hybridization on whole 5dpf zebrafish embryos was performed as described24 using a 3’ digoxygenin-labeled probe against dre-miR-30a (Exiqon).
Zebrafish injections and immunostaining
Wild-type TLF zebrafish were injected with scrambled ASO or ASO directed against zebrafish miR-30a (Exiqon) at 2 days post fertilization (dpf). Consistent with previous studies,25 we injected 2 dpf rather than 1-cell stage embryos with the aim of specifically disrupting biliary development, which begins around that time.26 Larvae were exposed to PED6 at 5 dpf and examined for gallbladder fluorescence, then collected and fixed in methanol, and stained for cytokeratin as described previously.26, 27 Schematic projections were constructed by tracing over bile ducts in Adobe Photoshop as described.28 Several 5 dpf larvae were also fixed in buffered glutaraldehyde and examined under electron microscopy as described previously.26
Cell culture
The bipotential mouse embryonic liver (BMEL) cell line, 9A1 was cultured in an undifferentiated state29. The NIH3T3 cell line was obtained from the ATCC and cultured under standard conditions. Cholesterol-tagged LNA oligonucleotides were obtained from Sigma-Proligo. Transient transfections were performed using Oligofectamine (Invitrogen). Following a 4-hour transfection in the absence of serum, cells were grown in the presence of serum for 24 hours before harvesting.
Dual luciferase assays
NIH3T3 cells were co-transfected with psiCHECK reporters with either control or inhibitory ASOs as described above. Luciferase assays were performed using the Dual-Luciferase® Reporter Assay System (Promega), on a Turner Biosystems 1000 luminometer. Relative light unit values were normalized to the values for the empty psiCHECK-2 vector. P-values were calculated using Student’s t-test, comparing the normalized relative light units between empty vector and each candidate reporter, co-transfected with control or miR-30 ASO.
Results
Statistical analysis of microarrays defines the miRNA transcriptome of the embryonic liver
Total RNA was extracted from embryonic day 15.5 (E15.5), E18.5, and postnatal day 2 (P2) mouse liver and hybridized to a locked nucleic acid microarray comprising probes to all known mouse miRNAs. Initial analysis identified a subset of miRNAs whose expression levels were altered at least 1.5-fold between samples. Consistent with previous studies, this group includes miR-122a, which is estimated to account for approximately 70% of the total miRNA content in adult liver,30, 31 as well as known liver-expressed miRNAs of lower abundance such as miR-194 and let7b.31, 32
We then performed pair-wise comparisons of the E15.5, E18.5, and post-natal time points using the Significance Analysis for Microarrays (SAM) software package. SAM analysis identified 38 known or predicted miRNA transcripts with statistically significant alterations in probe hybridization between the E15.5 and E18.5 samples (Supplementary Table 1). Of this group only miR-331 was detected at lower levels at E18.5 than at E15.5. For the remaining 37 miRNAs there was a remarkable concordance in the inferred level of gene expression; in almost all cases, expression peaked in the E18.5 samples and remained stable or decreased slightly by the P2 time point.
As shown in Figure 1, the results of the microarray analysis were in close agreement with Northern blot detection. Beyond the time points examined by microarray, some transcripts continue to accumulate in the adult liver, while others are expressed only during the development of the organ and are not present in adult tissue.
Figure 1. Fetal and post-natal expression of hepatic miRNA.
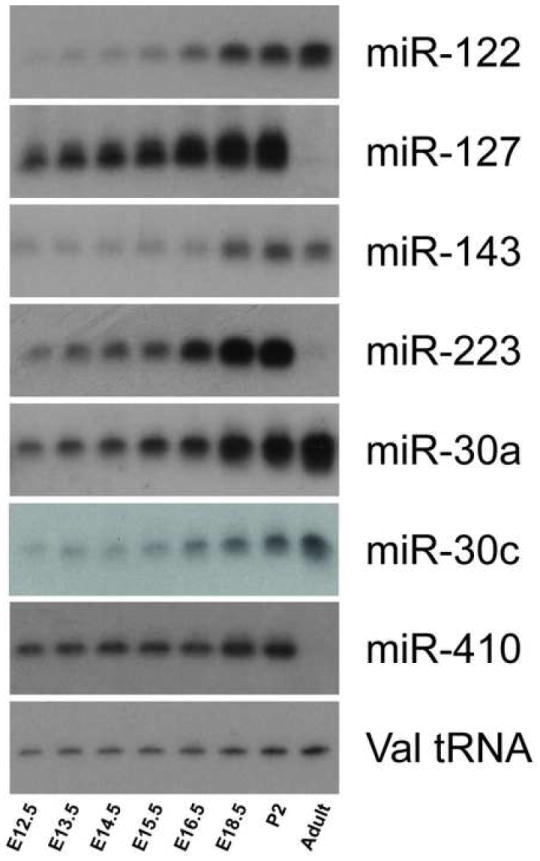
The expression patterns of hepatic miRNAs identified by microarray analysis were confirmed by Northern blot hybridization to 5 micrograms of total RNA isolated from mouse liver from mid-gestational embryonic, neonatal, and adult stages. The exposures times for the blots shown were: miR-122, 10 minutes; miR-127, 4 hours; miR-143, miR-223, miR-30a, 24 hours; miR-30c, 5 hours; miR-410, 16 hours; Val-tRNA, 5 minutes.
miRNAs are expressed in regionally restricted patterns within the developing liver
To identify miRNAs whose spatial and temporal expression patterns suggest a role in the establishment and/or maintenance of the biliary system, we performed in situ hybridization analysis on embryonic, perinatal, and adult mouse liver sections. The results of this analysis are illustrated in Supplementary Figure 1 and Supplementary Table 1. The miRNAs analyzed fell into five classes comprising miRNAs specific to (i) hepatocytes, (ii) hepatoblasts of the developing ductal plate and mature cholangiocytes, (iii) the peri-portal region, (iv) scattered blood cells throughout the lobule, and (v) very low or undetectable levels throughout the lobule.
miR-30a and miR-30c are associated with the developing ductal plate
Initially, miR-223 and miR-30a were observed to have grossly similar patterns of expression in cells surrounding branches of the portal vein in a pattern reminiscent of the developing ductal plate (Fig. 2). Double labeling with antibodies directed against cytokeratin CK19 indicated that the cell populations expressing miR-223 and miR-30a were distinct; there was little overlap between CK19 expression and that of miR-223 (Fig. 2A). To investigate the identity of the miR-223 expressing cells we examined co-expression with other cellular markers, including elastin (Fig. 2B), smooth muscle actin, fibroblast secretory protein, and myeloperoxidase (data not shown). These results indicated that the miR-223 expressing cells were granulocytes preferentially distributed to the periportal area. While the expression of miR-223 in granulocytes has been described,33 it is not known if this distribution within the fetal liver is of functional significance.
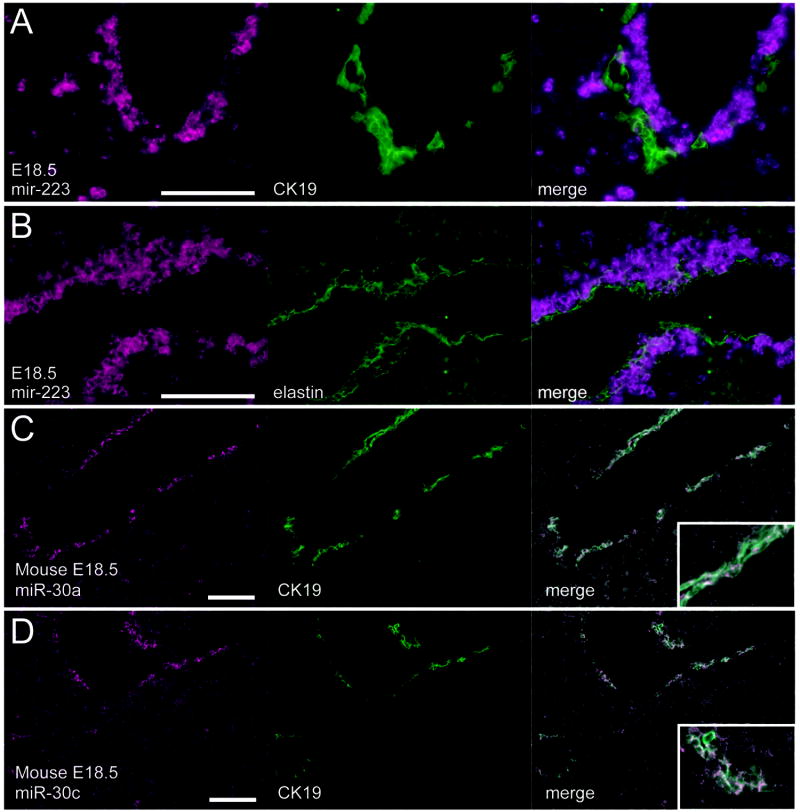
Figure 2. MicroRNAs associated with the ductal plate.
Simultaneous detection of miR-223 RNA (A, B), miR-30a (C), or miR-30c (D) by in situ hybridization and either CK19 (A, C, D), or elastin (B) proteins by immunofluorescence reveals that the peri-portal miR-223 expressing cells are juxtaposed between the developing ductal plate and the vascular endothelium. In contrast, miR-30a and miR-30c are predominantly expressed in the ductal plate itself. Scale bars: 100μm.
In contrast to miR-223, miR-30a was expressed in CK19-positive ductal plate cells (Fig. 2C). Based on this finding, we quantified the expression of the entire miR-30a family by Taqman RT-PCR (Supplementary Fig. 2), which revealed that miR-30a, miR-30b, and miR-30c are each highly induced at E18.5, whereas the expression of miR-30d and mIR-30e is lower and less highly induced. We then analyzed the spatial expression of the miR-30 family; of these, only miR-30c exhibited restricted expression in the developing ductal plate, with a pattern of expression indistinguishable from miR-30a (Fig. 2D). The other miR-30 family members (miR-30b, d, and e) were expressed in a more diffuse pattern within the hepatic parenchyma (Supplementary Fig. 3). Despite the observation by Northern analysis that miR-30a transcripts continue to increase in abundance in adult mouse liver, cholangiocyte accumulation of miR-30a and miR-30c substantially decreased in mouse juvenile bile ducts (between P2 and P14) and was not observed in adult mouse bile ducts. Rather it seems that the higher level of miR-30a was due to moderately increased expression in hepatocytes throughout the lobule (data not shown).
miR-30a and miR-30c are expressed in human cholangiocytes
To determine if murine miR-30a and miR-30c are also expressed in human liver, we performed in situ hyridization to human liver samples (Figure 3). As was observed in the mouse, both miRNAs were predominantly expressed in bile ducts. Furthermore, we found that the proliferating bile ducts formed in a chronic cholestatic state (in this case, biliary atresia) express miR-30a and miR-30c, as shown in Figure 4.
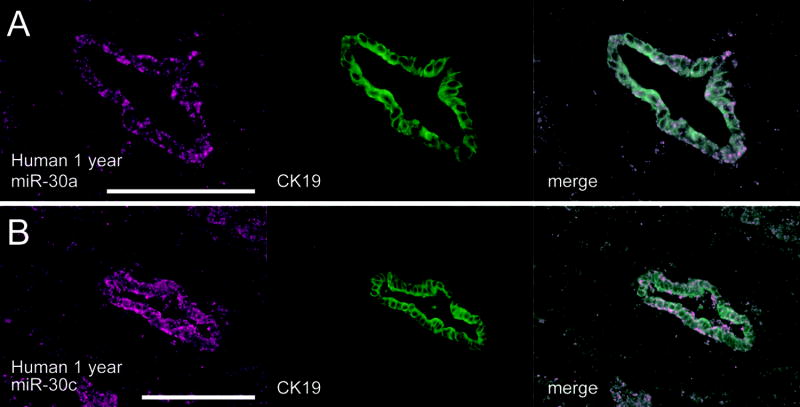
Figure 3. MiR-30a and miR-30c are expressed in cholangiocytes in normal human tissue.
Simultaneous detection of miR-30a (A) or miR-30c (B) and CK19 protein in human liver reveals that both are expressed in bile ducts in human liver (B). The expression of miR-30c is indistinguishable from that of miR-30a. Scale bars: 100μm.
Figure 4. MiR-30a and miR-30c expression is expanded in biliary atresia samples.
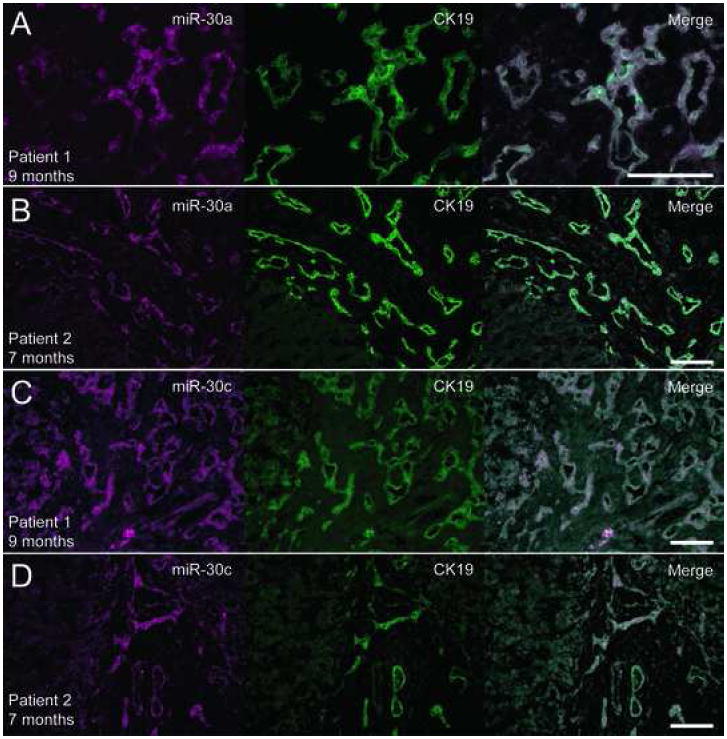
Double detection of miR-30a (A, B) or miR-30c (C, D) RNA and CK19 protein in liver explants obtained from two patients with biliary atresia at the time of liver transplantation. Both miR-30a and miR-30c are highly expressed in the proliferating bile ductules typically seen in this disorder. Scale bars: 100μm.
miR-30a is required for bile duct development in zebrafish
To test the function of miR-30a in vertebrate liver development, we used ASO-mediated knockdown of zebrafish miR-30a. Using in situ hybridization, we verified that as in mice, miR-30a is expressed in the zebrafish liver (Fig. 5). In addition to the relative ease of genetic knockdown (via antisense oligonucleotides) in zebrafish, this model also has the advantage of a functional test of the biliary system using the fluorogenic lipid reporter PED6.26, 27 The action of intestinal lipases on PED6 liberates a fluorophore, which is taken up by the liver and concentrated in the gall bladder, allowing for the easy detection of disruptions of the biliary system.34
Figure 5. The zebrafish miR-30a ortholog is expressed in liver.
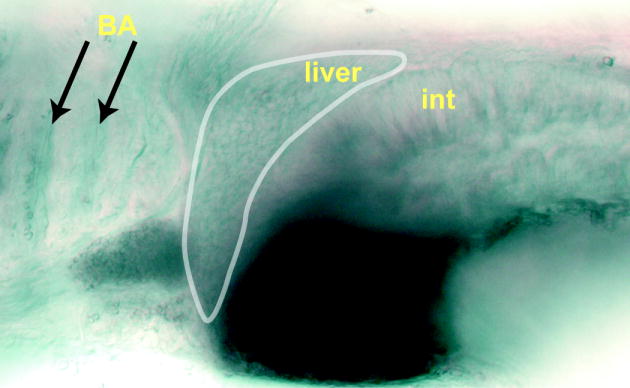
Colorimetric in situ hybridization of the zebrafish miR-30a ortholog reveals a reticular staining pattern in the liver (outlined) consistent with cholangiocyte expression, as well as expression in the branchial arches (BA, indicated by the arrows) and intestine (int). Dorsal is up, anterior left.
Both the control and miR-30a ASO-injected fish appeared grossly normal at 5 dpf, but zebrafish injected with the miR-30a ASO demonstrated less gallbladder uptake of PED6 compared to controls, suggesting an impairment of biliary function (Fig. 6A). Cytokeratin immunostaining of these fish revealed abnormal intrahepatic bile ducts (Fig. 6B-C), with fewer terminal ductules, which is consistent with defects in the later stages of biliary development similar to that previously reported for zebrafish larvae with ASO-induced deficiency of the biliary transcription factor hnf6.27 Similarly, staining with antibodies to the canalicular marker Mdr indicated a substantial reduction in the number of terminal ductules (data not shown). Ultrastructural analysis of liver tissue was consistent with these findings (Fig. 6D-E). Canaliculi appeared enlarged in the miR-30a ASO injected fish, as might be expected if bile excretion were impaired or obstructed. A similar change was observed in hnf6 morphants.27 These results demonstrated that ASO-mediated knockdown of miR-30a leads to abnormal intrahepatic biliary and/or canalicular development in zebrafish.
Figure 6. Intrahepatic bile duct and canalicular defects in zebrafish injected with an ASO directed against zebrafish miRNA 30a.
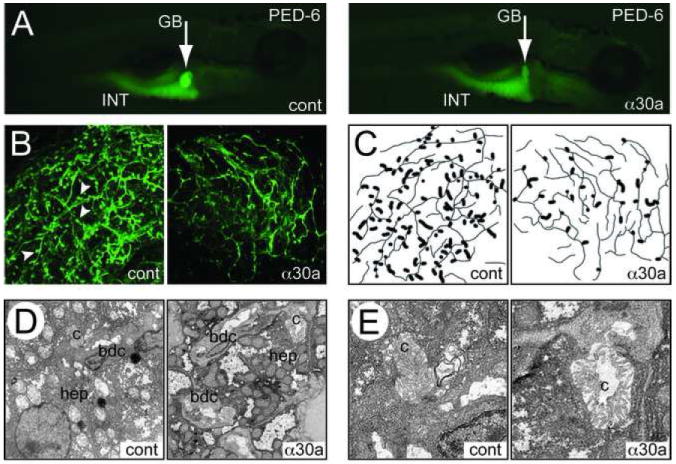
(A) PED6 uptake in 5 dpf larvae injected with an control oligonucleotide (cont) or with an ASO against miR-30a (α30a) GB, gall bladder; INT, intestine. (B) Cytokeratin immunostaining on control and α30a-injected 5 dpf larvae (Magnification 600x). (C) Schematic representation of (B) reveals a decrease in the number of terminal ductules (arrowheads) in the ASO-injected fish, indicated in the schematic as the wider lines. (D) Low-power (2500x) electron micrographs of control and α30a ASO-injected 5 dpf larvae showing bile duct cells (bdc) and hepatocytes (hep). Note the relatively similar overall appearance. (E) Higher-power (10000x) views showing a representative canaliculus (c), which is dilated and abnormal-appearing in the α30a ASO-injected larva.
Identification of miR-30a target genes
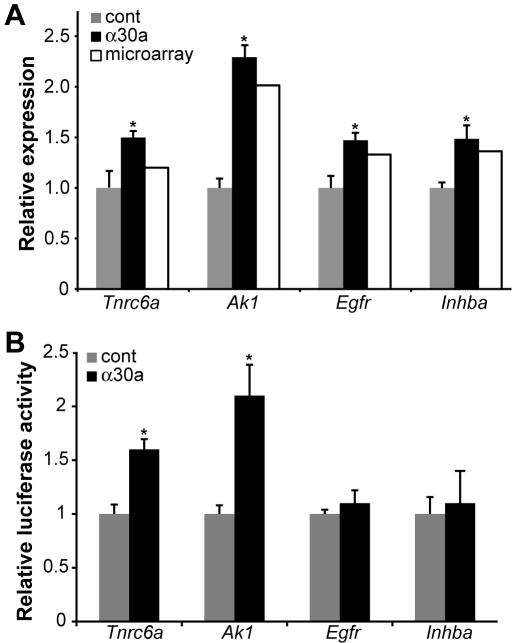
To elucidate the mechanism of miR-30a function in mammalian biliary development, we pursued the identification of miR-30a targets in cultured bipotential murine embryonic liver (BMEL cells)29. Control or miR-30a ASO oligonucleotides were transfected into BMEL cells, and specific knockdown of miR-30a was confirmed by Taqman RT-PCR (35% of control levels; p < 0.05). Using a microarray approach, we identified 305 genes whose expression was perturbed in the miR-30a ASO-treated cells by greater than 10% at a false discovery rate of 5% or less (Supplementary Table 2). With rare exception, miRNA-mediated regulation leads to repression of the target gene. Consistent with this, increases in gene expression comprised 75% (230/305) of the altered mRNA levels. The results of the microarray were validated by quantitative RT-PCR of several mRNA targets, selected on the basis of their relevance to liver biology, their degree of expression change in response to the miR-30a ASO, and the presence of miR-30a binding sites as predicted by Targetscan (Fig. 7A).35
Figure 7. Identification of miR-30a targets in BMEL cells.
(A) Confirmation of microarray results by quantitative RT-PCR of RNA from transfected BMEL cells. Genes identified in the microarray analysis as significantly altered by α30a transfection were selected. Candidate gene expression was normalized to Hprt expression, and the results are expressed relative to a value of 1 in the control ASO-treated cells. (B) Luciferase reporter assays to test for regulation of candidate gene 3’-UTRs by miR-30a. The indicated 3’-UTRs were sub-cloned into a dual luciferase reporter plasmid. Luciferase activity was normalized for both transfection efficiency and for effects on a reporter plasmid lacking the 3’-UTR. The results are expressed relative to a value of 1 in the control ASO-treated cells. *p < 0.05
Many of the miR-30a target genes are of relevance to liver development and/or function. For example, two are known to control hepatocyte growth and differentiation, namely: Egfr (the EGF receptor) and Inhba (the TGFβ agonist Activin). A third, Abcb1b, encodes the hepatocyte canalicular transporter Mdr1. A fourth, Tnrc6a, encodes the GW182 protein, a component of the RNA interference silencing complex, suggesting that miR-30a may participate in the broad control of miRNA-mediated gene regulation. Ak1 encodes an isoform of the enzyme adenylate kinase which is predominantly restricted to muscle, brain, and erythrocytes; our results suggest that miR-30a promotes this tissue-specific expression by repressing Ak1 in the liver.
Ak1 and Tnrc6a are direct targets of miR-30a mediated regulation
To determine if genes with increased expression in the microarray data were direct targets of miR-30a regulation, we cloned selected 3’UTRs into a luciferase reporter plasmid and co-transfected the reporter and control or miR-30a ASO into BMEL cells. The 3’UTRs of two genes Ak1 and Tnrc6a conferred regulation by miR-30a. While there was a modest increase in the case of the Egfr and Inhba 3’UTRs, these did not reach statistical significance (Figure 7B). These results support the use of ASO-induced gene expression changes as a means of detecting direct miRNA target genes.
Discussion
This study is the first description of the miRNA transcriptome during the critical period of hepatobiliary development that spans the end of gestation to the early neonatal period. We have identified a core set of 38 miRNAs whose expression levels change during the period of hepatobiliary specification. When we examined the spatial expression patterns of selected miRNAs we found additional evidence for complex, independent regulation of their expression. We have also demonstrated regionally restricted expression of miRNAs within subsets of cells of both parenchymal and non-parenchymal origin within the liver. As is to be expected given the hematopoietic nature of the fetal liver, several of the miRNAs identified in our study were found to be restricted to developing blood cells. However, both hepatocyte and cholangiocyte-specific miRNAs were also identified.
The finding that cells expressing the granulocyte-specific miRNA miR-223 are interposed between the developing ductal plate and the portal vein was unexpected. Previous studies have implicated the Notch signaling pathway in normal biliary development,36, 37 and it has been hypothesized that direct signaling between the portal vein or periportal mesenchyme and the ductal plate is a critical component of biliary differentiation.38 Our work indicates that during the period between E18.5 and the neonatal period, these populations of cells are not in direct intercellular contact, as would be required for Notch signaling.
Any attempt to identify cholangiocyte-specific gene products is complicated by the fact that these cells comprise only a small proportion of the total mass of the liver. Since the appearance and remodeling of the ductal plate involves only a small number of cells and occurs in a relatively brief developmental period, the search for miRNA transcripts specific to these cells by large-scale cloning strategies would have been unlikely to be successful. Our discovery of ductal plate-specific miRNA transcripts validates the approach of investigating the subset of miRNAs that are dynamically changing in abundance in late gestation. In the case of the mouse miR-30a and miR-30c transcripts, the cholangiocyte-specific expression is temporally restricted to the period between E18.5 and the early neonatal period, suggesting that these miRNAs function in the final differentiation of cholangiocytes.
Given the highly similar pattern of the miR-30a and miR-30c positive cells, and given the genomic location of the genes encoding miR-30a and miR-30c2 (less than 20kb apart), it seems likely that the two transcripts are co-regulated and that miR-30c2 rather than miR-30c1 is the source of the mature miR-30c transcript; however, we have not formally tested this hypothesis.
Since functional studies of miR-30a and miR-30c in mice will require the generation of conditional knockout strains, we have used ASO technology to reduce expression of the zebrafish ortholog of miR-30a as a rapid, preliminary assay. While bile duct development in the zebrafish does not occur through a ductal plate intermediate, the regulatory molecules which function in mammals (the Onecut factors, HNF1β, Jagged/Notch) play analogous roles in the fish.26, 27 Our findings in the zebrafish model provide the first evidence that miRNA plays a critical role in the development of the biliary system.
At the inception of this study, none of the computationally predicted targets of miR-30a or miR-30c had been validated. A recent study has identified 100 targets of miR-30a using proteomic methods by transfection of a miR-30a mimic into HeLa cells39. Of these, only two (Slc12a4 and Ptrh1) were also identified in our screen. There are several potential explanations for this small overlap. The use of miR-30a over-expression carries the risk of false positives due to supra-physiologic levels of the miRNA. Furthermore, since the regulatory function of miRNA involves binding of miRNAs to target mRNAs, a particular miRNA may modulate expression of distinct targets in different cell types. Our study was performed by decreasing the levels of endogenous miR-30a in BMELs, a hepatoblast model cell line, and is thus more likely to identify liver-specific target mRNAs.
Our study provides the first links between miR-30a and two well-described regulators of liver development: Inhba (a component of the TGFβ agonist, Activin) and Egfr (the epidermal growth factor receptor). The expression of both of these in the fetal liver has already been described40, 41. Clotman and colleagues have shown that a gradient of TGFβ activity is essential for the normal differentiation of hepatoblasts into cholangiocytes and hepatocytes.42 Our study suggests that miR-30a expression in the ductal plate promotes the formation of this gradient by inhibiting Activin production. Egfr is a growth factor receptor expressed in the ductal plate which is down-regulated in the mature cholangiocyte; conversely, over-expression is Egfr is frequently observed in cholangiocarcinoma. Our findings suggest that miR-30a participates in the control of Egfr expression in normal cholangiocytes and loss of miR-30a may promote excess Egfr expression in cancer. A third miR-30a target identified in this study is the canalicular marker Abcb1b. In the context of embryonic biliary development, miR-30a may inhibit Abcb1b expression in nascent cholangiocytes in the ductal plate as they undergo terminal differentiation from hepatoblasts.
Neither the Inhba nor the Egfr 3’-UTR is targeted by miR-30a in our reporter assay, although both are predicted to contain miR-30a binding sites by Targetscan. This may be because the luciferase gene is expressed at high levels under the control of the SV40 promoter; this strong expression may mask the effects of targeting by miR-30a. It is also possible that the regulation of Inhba and Egfr requires both the 3’-UTR and other elements within the mature transcript. Finally, the regulation of Inhba and Egfr by miR-30a may be indirect.
In summary, these results provide the first links between miRNA and hepatogenesis, particularly biliary development. Future studies will be directed to testing the function of miR-30a and miR-30c in the mouse by genetic means and to further establishing links between these regulators and other pathways relevant to liver ontology and function.
Supplementary Material
Simultaneous detection of selected miRNAs by in situ hybridization and immunofluorescent antibody staining for CK19. (A) At E18.5 miR-200a is expressed in scattered cells presumed to be hematopoietic and uniformly throughout the lobule, with no alteration in expression close to the developing ductal plate. (B) Expression of miR-410 at E18.5 persists in scattered cells that are morphologically consistent with hematopoietic and endothelial cells. (C) Lack of overlap of the miRNA probe and CK19 signals indicate that while miR-122 is highly expressed in hepatocytes, it is neither expressed in CK19-expressing cells nor in non-parenchymal cells. (D) miR-27b is expressed throughout the lobule at E18.5, with higher expression surrounding portal tracts. At P14, miR-27b expression is retained in vascular tissue in the portal tract. (E) miR-127 is expressed in hepatocytes at E18.5, with slightly elevated peri-portal expression. By P1 there is no longer any discernible asymmetry in the expression of miR-127 adjacent to the portal tract.
Quantitative RT-PCR was used to measure the expression of all miR-30a family members in mouse liver at the indicated time points. Expression was normalized to that of Hprt.
In situ hybridization was performed on E18.5 mouse liver using the indicated LNA probes. MiR-30b and miR-30e are present in hepatocytes, whereas only a faint signal is detected for miR-30d. None of these three miR-30a family members is expressed in a biliary pattern. Scale bars, 100 μm.
Acknowledgments
Anti-elastin antibody was a kind gift from Rebecca G. Wells. The authors wish to thank K. Kaestner and L. Greenbaum for critical reading of the manuscript, and A. Silahtaroglu for technical suggestions regarding the in situ hybridization protocol.
Grant support: NIH K08DK070881 (JRF), NIH K08DK68009 (RPM), Children’s Digestive Health and Nutrition Foundation (JRF), The Fred and Suzanne Biesecker Pediatric Liver Center (JRF, RPM).
Abbreviations
- ASO
antisense oligonucleotide
- dpf
days post fertilization
- E
embryonic day
- LNA
locked nucleic acid
- miRNA
microRNA
- P
post-natal day
Footnotes
Financial disclosures: No conflicts of interest exist.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116:S89–92. doi: 10.1016/s0092-8674(04)00035-2. 1 p following S96. [DOI] [PubMed] [Google Scholar]
- 3.Poethig RS, Peragine A, Yoshikawa M, Hunter C, Willmann M, Wu G. The function of RNAi in plant development. Cold Spring Harb Symp Quant Biol. 2006;71:165–70. doi: 10.1101/sqb.2006.71.030. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–97. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–50. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 6.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci U S A. 2005;102:18017–22. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 9.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–58. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 11.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 12.Akhan O, Karaosmanoglu AD, Ergen B. Imaging findings in congenital hepatic fibrosis. Eur J Radiol. 2007;61:18–24. doi: 10.1016/j.ejrad.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Adeva M, El-Youssef M, Rossetti S, Kamath PS, Kubly V, Consugar MB, Milliner DM, King BF, Torres VE, Harris PC. Clinical and molecular characterization defines a broadened spectrum of autosomal recessive polycystic kidney disease (ARPKD) Medicine (Baltimore) 2006;85:1–21. doi: 10.1097/01.md.0000200165.90373.9a. [DOI] [PubMed] [Google Scholar]
- 14.Shneider BL, Magid MS. Liver disease in autosomal recessive polycystic kidney disease. Pediatr Transplant. 2005;9:634–9. doi: 10.1111/j.1399-3046.2005.00342.x. [DOI] [PubMed] [Google Scholar]
- 15.Kahn E. Biliary atresia revisited. Pediatr Dev Pathol. 2004;7:109–24. doi: 10.1007/s10024-003-0307-y. [DOI] [PubMed] [Google Scholar]
- 16.Tamiolakis D, Arvanitidou V, Nikolaidou S, Barbagadaki S, Avgidou K, Boglou P, Papadopoulos N. Caroli’s syndrome. A case report and review of the literature. Minerva Gastroenterol Dietol. 2004;50:179–81. [PubMed] [Google Scholar]
- 17.Terada T, Nakanuma Y. Congenital biliary dilatation in autosomal dominant adult polycystic disease of the liver and kidneys. Arch Pathol Lab Med. 1988;112:1113–6. [PubMed] [Google Scholar]
- 18.Desmet VJ. Congenital diseases of intrahepatic bile ducts: variations on the theme “ductal plate malformation”. Hepatology. 1992;16:1069–83. doi: 10.1002/hep.1840160434. [DOI] [PubMed] [Google Scholar]
- 19.Sokol RJ, Shepherd RW, Superina R, Bezerra JA, Robuck P, Hoofnagle JH. Screening and outcomes in biliary atresia: summary of a National Institutes of Health workshop. Hepatology. 2007;46:566–81. doi: 10.1002/hep.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F. A Model Based Background Adjustment for Oligonucleotide Expression Arrays. Johns Hopkins University Department of Biostatistics Working Papers; 2004. [Google Scholar]
- 22.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silahtaroglu AN, Nolting D, Dyrskjot L, Berezikov E, Moller M, Tommerup N, Kauppinen S. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat Protoc. 2007;2:2520–8. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 24.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–9. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 25.Stenkamp DL, Frey RA. Extraretinal and retinal hedgehog signaling sequentially regulate retinal differentiation in zebrafish. Dev Biol. 2003;258:349–63. doi: 10.1016/s0012-1606(03)00121-0. [DOI] [PubMed] [Google Scholar]
- 26.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–66. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 27.Matthews RP, Lorent K, Russo P, Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–59. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 28.Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev Dyn. 2008;237:124–31. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- 29.Strick-Marchand H, Weiss MC. Inducible differentiation and morphogenesis of bipotential liver cell lines from wild-type mouse embryos. Hepatology. 2002;36:794–804. doi: 10.1053/jhep.2002.36123. [DOI] [PubMed] [Google Scholar]
- 30.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 31.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 32.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 33.Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, McCoy JP, Jr, Sloand EM, Kajigaya S, Young NS. Hematopoietic-specific microRNA expression in human cells. Leuk Res. 2006;30:643–7. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Farber SA, Pack M, Ho SY, Johnson ID, Wagner DS, Dosch R, Mullins MC, Hendrickson HS, Hendrickson EK, Halpern ME. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292:1385–8. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- 35.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–51. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 37.Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–42. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 38.Kodama Y, Hijikata M, Kageyama R, Shimotohno K, Chiba T. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology. 2004;127:1775–86. doi: 10.1053/j.gastro.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 40.Peng M, Palin MF, Veronneau S, LeBel D, Pelletier G. Ontogeny of epidermal growth factor (EGF), EGF receptor (EGFR) and basic fibroblast growth factor (bFGF) mRNA levels in pancreas, liver, kidney, and skeletal muscle of pig. Domest Anim Endocrinol. 1997;14:286–94. doi: 10.1016/s0739-7240(97)00025-8. [DOI] [PubMed] [Google Scholar]
- 41.Carver RS, Stevenson MC, Scheving LA, Russell WE. Diverse expression of ErbB receptor proteins during rat liver development and regeneration. Gastroenterology. 2002;123:2017–27. doi: 10.1053/gast.2002.37060. [DOI] [PubMed] [Google Scholar]
- 42.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–54. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Simultaneous detection of selected miRNAs by in situ hybridization and immunofluorescent antibody staining for CK19. (A) At E18.5 miR-200a is expressed in scattered cells presumed to be hematopoietic and uniformly throughout the lobule, with no alteration in expression close to the developing ductal plate. (B) Expression of miR-410 at E18.5 persists in scattered cells that are morphologically consistent with hematopoietic and endothelial cells. (C) Lack of overlap of the miRNA probe and CK19 signals indicate that while miR-122 is highly expressed in hepatocytes, it is neither expressed in CK19-expressing cells nor in non-parenchymal cells. (D) miR-27b is expressed throughout the lobule at E18.5, with higher expression surrounding portal tracts. At P14, miR-27b expression is retained in vascular tissue in the portal tract. (E) miR-127 is expressed in hepatocytes at E18.5, with slightly elevated peri-portal expression. By P1 there is no longer any discernible asymmetry in the expression of miR-127 adjacent to the portal tract.
Quantitative RT-PCR was used to measure the expression of all miR-30a family members in mouse liver at the indicated time points. Expression was normalized to that of Hprt.
In situ hybridization was performed on E18.5 mouse liver using the indicated LNA probes. MiR-30b and miR-30e are present in hepatocytes, whereas only a faint signal is detected for miR-30d. None of these three miR-30a family members is expressed in a biliary pattern. Scale bars, 100 μm.