Abstract
Background: Diseases caused by gammaherpesviruses continue to be a challenge for human health and antiviral treatment. Most of the commonly used antiviral drugs are directed against viral gene products. However, the emergence of drug-resistant mutations ma limit the effectiveness of these drugs. Since viruses require a host cell to propagate, the search for host cell targets is an interestin alternative. Methods: In this study, we infected three different cell types (fibroblasts, endothelial precursor cells and macrophages with a murine gammaherpesvirus and analysed the host cell response for changes either common to all or unique to a particular cell type using oligonucleotide microarrays. Results: The analysis revealed a number of genes whose transcription was significantly up- or down-regulated in either one or two of the cell types tested. After infection, only two genes, Lman1 (also known as ERGIC53) an synaptobrevin-like 1 (sybl1) were significantly up-regulated in all three cell types, suggestive for a general role for the virus life cycl independent of the cell type. Both proteins have been implicated in cellular exocytosis and transport of glycoproteins through the secre tory pathway. To test the significance of the observed up-regulation, the functionality of these proteins was modulated, and the effect on virus replication was monitored. Inhibition of either Lman1 or sybl1 resulted in a significant reduction in virus production. Conclusions: This suggests that proteins of the secretory pathway which appear to be rate limiting for virus production may represent new targets for intervention.
Keywords: gammaherpesvirus, MHV-68, microarray, virus productivity, secretory pathway, lytic infection
Introduction
Diseases caused by gammaherpesviruses continue to be a challenge for human health as well as for antiviral treatment. Most of the commonly used antiviral drugs are directed against viral gene products but the emergence of drug-resistant mutations may limit their effectiveness. Because viruses require a host cell to propagate, the search for cellular targets is an interesting alternative. Studies with the human gammaherpesviruses Epstein-Barr virus (EBV) and Human Herpesvirus 8 (HHV-8, also called KSHV) are hampered by the lack of readily available cell culture systems capable of supporting productive replication. In contrast, murine gammaherpesvirus 68 (MHV-68) efficiently replicates in various cells in tissue culture and thus provides a model to study gammaherpesvirus infection. In particular, it allows the systematic comparison of the global response of different cell types to infection. MHV-68 is a natural pathogen of wild rodents [1]. The nucleotide sequence of MHV-68 is similar to EBV, but it is more closely related to KSHV [2]. Thus, MHV-68 is used as a model for infections with human gammaherpesviruses [3–9].
It is generally assumed that genes conserved among different organisms or different species/strains within the same group of organisms usually are of functional importance [10, 11]. The same holds true for viruses. For example, all herpesvirus genomes share a set of conserved genes to accomplish viral replication [12]. In addition, the genomes also contain genes which facilitate the specific life style of a herpesvirus in the specific host species or in a specific cell type of the host. To tune a cell to its specific needs, the virus modulates functions of the host cell. To match the specific requirements, the virus may regulate one and the same cellular gene differentially in different cell types. Nevertheless, some important functions have to be accomplished in most of the host cell types. Thus, we hypothesized that cellular genes targeted by a given virus in all tested host cell types should be important for the viral life cycle and could therefore reflect new targets for antiviral therapy. In particular, cellular genes being expressed at low level in uninfected cells and up-regulated only during infection could indicate rate limiting functions. Global monitoring of cellular gene expression using microarrays after infection of different cell types with MHV-68 allows to search for such cellular genes important for gammaherpesvirus replication.
In this study, we infected three different cell types (fibroblasts, endothelial precursor cells and macrophages) with MHV-68 and analysed the host cell response for changes either common to all or unique to a particular cell type using DNA microarrays covering 12,488 probe sets. The analysis revealed a number of genes whose transcription was significantly up- or down-regulated in either one or two of the cell types tested. The transcription of two genes was found to be regulated in all three cell types, suggesting an important role for the virus life cycle independent of the cell type. Lman1 (also known as ERGIC53) and synaptobrevin-like 1 (sybl1) were significantly up-regulated after infection in all three cell types. Both proteins are implicated in cellular exocytosis by transport of glycoproteins through the secretory pathway. Inhibition of these proteins resulted in a significant reduction in virus production.
Materials and methods
Cell culture, virus stocks and plaque assays
The original stock of MHV-68 (clone G2.4) was obtained from Dr. A. Nash (University of Edinburgh, Edinburgh, UK). Cell culture, virus stocks and plaque assays were performed essentially as described [13]. Briefly, working stocks of virus were prepared on BHK-21 cells (ATCC CCL10), which were maintained in Glasgow modified Eagle's medium (GIBCO) supplemented with 5% newborn calf serum, 5% tryptose phosphate broth, penicillin (100U/ml) and streptomycin (100μg/ml). The BHK-21 cells were infected at a multiplicity of infection (MOI) of 0.1, and virus stocks were prepared when the cytopathic effect (cpe) was complete by 2x freezing and thawing the cells. Cellular debris were removed by cen-trifugation, and the supernatants were stored in aliquots at -80°C. Virus titres were determined by plaque assay on BHK-21 cells. Briefly, tenfold dilutions of virus were adsorbed onto BHK-21 cells. After 90 min. at 37°C, the inoculum was removed and fresh medium containing 1.5% car-boxymethylcellulose was added. Cells were stained after 4–5 days with 0.1% crystal violet solution to determine the number of plaques. Primary mouse embryonal fibroblasts (MEFs) were prepared from embryos of C57BL/6 and Balb/c mice. MEFs were cultured in Dulbecco's modified Eagle medium supplemented with 10% newborn calf serum, penicillin (100U/ml), streptomycin (100μg/ml) and 1% L-glutamine. The C57BL/6-derived macrophage cell line ANA-I [14] (kindly provided by Dr. K. Pfeffer University of Düsseldorf, Düsseldorf, Germany) was cultured in RPMI 1640 medium supplemented with 10% newborn calf serum, penicillin (100U/ml), streptomycin (100μg/ml) and 1% L-glutamine. Mouse embryonic endothelial progenitor cells (eEPCs) were isolated and propagated as described before [15, 16]. Vero cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% newborn calf serum, penicillin (100U/ml), streptomycin (100μg/ml) and 1% L-glutamine. A clinical isolate of HSV-1 was kindly provided by Dr. J. Haas (Max von Pettenkofer-Institute, Munich, Germany). A virus stock was prepared as clarified lysate of infected Vero cells. Briefly, Vero cells were infected at low multiplicity, and virus stock was prepared after removing the supernatant by two rounds of freezing and thawing of the infected cell pellets in culture medium. The clarified lysate was aliquoted and stored at -80°C until used. Viral titres were determined by standard plaque assay on Vero cells as described previously [17].
Infection of cells and RNA-isolation for microarray analysis
In order to analyse the host cell response for common or unique changes in response to MHV-68 infection, three different cell types including fibroblasts, endothelial precursor cells and macrophages were infected with MHV-68. In pilot experiments, we first determined the optimal infection conditions for each cell type using a recombinant MHV-68 expressing the green fluorescent protein (gfp) [13]. To allow for comparison of the results obtained from the different cell types, the conditions for infection were adjusted to result in a similar number of gfp-expressing cells at 48 hrs after infection in all three cell types. This adjustment included the MOI used for each cell type as well as using centrifugal enhancement [18] for the infection of macrophages. The MOI determined to be necessary to achieve a 100% infection rate 48 hrs after infection was 10, 50 and 40 for fibroblasts, endothelial precursor cells and macrophages, respectively. After establishing the infection conditions for each cell type, the cells were infected with wild-type MHV-68 (clone G2.4) for microarray analysis. Untreated fibroblasts and endothelial precursor cells or macrophages treated with an equal amount of heat-inactivated (by boiling for 30 min.) virus were used as uninfected controls. Total RNA was isolated 48 hrs after infection from infected or uninfected cells using the TRI-REAGENT (SIGMA, Saint Louis, Missouri, USA), according to the instructions of the manufacturer. Three independent experiments were performed with each cell type.
Microarray analysis
Total RNA was processed and hybridized to the mouse expression array MG-U74Av2 (Affymetrix, Santa Clara, USA) according to the manufacturer's protocols. Subsequently microarrays were scanned and analysed using Affymetrix Microarray Suite V5.0 (MAS 5.0) software. For each condition (C57BL/6 or Balb/c derived MEF cells, ANA-1 macrophages or eEPCs, mock-treated and infected, respectively) three biological replicates were used, resulting in total of 24 microarrays. ANA-1 and eEPCs derived cell intensity file (CEL) files were normalized separately for each cell line by using dCHIP 1.3 (http://www.dCHIP.org) [19] and expression values were generated using the same program and the PM/MM model. Arrays derived from MEF cells were normalized to the probe sets AFFX-BioB-3_at, AFFX-BioC-3_at, AFFX-BioDn-3_at, and AFFX-CreX-3_at, with a mean target value of 1000 using the MAS 5.0 software. These probe sets represent the signals of four pre-labelled controls, which were spiked routinely into the hybridization cocktail. The Present Call percentages were obtained using the affy package for R1 (http://www.R-project.org).
Significance analyses between mock-infected and MHV-68 infected cells were performed with the significance analysis of microarrays (SAM) two-class paired algorithm (http://www-stat.stanford.edu/~tibs/SAM/) [20]. Probe sets were considered as significantly regulated if the median false discovery rate (FDR) was below 1% and the transcript was induced or repressed at least twofold. Clustering was performed with Genesis software, with the Euclidean distance used as the similarity distance measurement [21]. Further filtering and list comparison was done using Spotfire DecisionSite (http://www.spotfire.com). Analysis of overrepresentation of distinct gene categories was done with the program EASE [22].
RT-PCR
1.0 μg RNA was reverse transcribed using the First Strand cDNA Synthesis Kit for RT-PCR (AMV) (Roche Diagnostics GmbH, Mannheim, Germany) according to the instructions of the manufacturer. 2 μl of the resulting cDNA were used as template for PCR amplification of selected genes. The primer sequences were as follows:
| GAPDH: | 5′primer: CTCACTCAAGATTGTCAGCAATG |
| 3′primer: GAGGGAGATGCTCAGTGTTGG | |
| CD14: | 5′primer: CCTGGAATACCTTCTAAAGCGTGTG |
| 3′primer: CCCTTGTTGCCCACGACACGTTGC | |
| TGFβR3: | 5′primer: GTCTTCTCTGTGGCAGAGAATGAGC |
| 3′primer: CTAGAGAGGTGCAGGCGTCGTCAGG | |
| Sybl1: | 5′primer: TGGAAACCACCAAACTTATGACCTG |
| 3′primer: CAGTTCAACTTGGTAATGGATAAAC | |
| CCL3: | 5′primer: TTCTCAGCGCCATATGGAGCTGACAC |
| 3′primer: GAGATGGAGCTATGCAGGTGGCAGG | |
| Wisp1: | 5′primer: TACACATCAAGGCAGGGAAGAAATG |
| 3′primer: CATGGAACTTTACCCTGAGCCACAC | |
| Lman1: | 5′primer: GGTGCAGACAGTGTTGTTCGTTGG |
| 3′primer: GTGCTGTGACACTTCTAGGATAAATGC | |
| Irg1: | 5′primer: CAATATCTAAACGAGTTGCACTGG |
| 3′primer: GTGTATTTCATAGGGGTACAGTC |
The PCR reactions were performed using Titanium Taq DNA polymerase (CLONTECH, Palo Alto, CA, USA) according to the instructions of the manufacturer.
siRNA experiments
For siRNA experiments, the C57BL/6-derived MEF cell line MEF-BL/6–1 (ATCC SCRC-1008), NIH3T3 cells (ATCC CRL1658) and VERO cells (ATCC CCL81) were used. Cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% newborn calf serum, penicillin (100U/ml) streptomycin (100μg/ml) and 1% L-glutamine. Cells were plated in 24-wel plates, and after overnight culture, transfected with 50 pmoles of the appropriate siRNAs. siRNAs directed against murine sybl1 (synaptobrevin like 1 NM_011515) and siRNA libraries as negative control were designed and purchased from EUROGENTEC (EUROGENTEC, Seraing, Belgium) and transfected using jetSI-ENDO (EUROGENTEC) as recommended by the manufacturer. The sequences of the siRNAs directed against murine sybl1 are as follows: siRNA1: 5'-CCACUAUCCUUGCCAAACA99–3' and siRNA2 5'-GCAUCACUCUGAGAAUAAG99–3'. The siRNAs directed against murine Lman1 (also called ERGIC53; NM_027400) were purchased from Qiagen (Qiagen, Hilden, Germany) (siRNAQ12: HP GenomWide siRNA 1027400 Mm_Lman1_2_HP siRNA and siRNAQ14: Mm_Lman1_-4_HP siRNA) and transfected using HiPerfect transfection reagent (Qiagen) as recommended by the manufacturer. 24 hrs after transfection, cells were infected with MHV 68 or HSV-1 at an MOI of 1, unless otherwise indicated. After 60 min. a 37°C, the inoculum was removed and fresh medium was added. Cells and supernatants were harvested 48 hrs after infection, unless otherwise indi cated, and virus titres were determined by plaque assay after two rounds o freezing and thawing. To determine whether transfection with siRNAs migh have any effect on cell proliferation or viability, cell numbers and viability were determined either using trypan blue or 3-(4,5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazotiumbromide (MTT) after harvesting the cells from paralle cultures. To demonstrate the ability of the siRNAs to inhibit the expression of the target genes, total RNA from parallel cultures was isolated and analysed by RT-PCR as described above.
Treatment of infected cells with ONO-RS-082
NIH3T3 cells in 6-well plates were pre-incubated at 4°C for 30 min. and infected with MHV-68 at an MOI of 0.1. After an incubation period of 60 min at 4°C to allow virus adsorption, inocula were removed and the temperature was shifted to 37°C in the presence or absence of 50 μM of the phospholi pase A2 (PLA2) inhibitor 2-(p-amylcinnamoyl)amino- 4-chlorobenzoic acid (ONO-RS-082) (Biomol, Hamburg, Germany), dissolved in ethanol. Viral titres were determined after 48 hrs by plaque assay on BHK-21 cells.
Results
Global changes of gene transcription profiles in different cell types
For a comprehensive analysis of the changes in the steady state levels of the cellular transcription program during lytic MHV-68 replication, we performed a microarray screen. Three cell types, Ana-1 macrophages (ANA), embryonic endothelial progenitor cells (eEPCs, also labelled ENDO) and primary MEF, all derived from the C57BL/6 (BL/6) background, were studied. In addition, MEF cells derived from the Balb/c background were used. In all infection experiments, the same time-point (48 hrs) was chosen, and for each condition, three biological replicates were analysed, resulting in 24 microarrays altogether. The time-point 48 hrs after infection was chosen since studies with KSHV during later time points of lytic infection are difficult due to the lack of cell culture systems that support KSHV replication. In contrast, MHV-68 efficiently replicates in various tissue culture cells and thus allowed us to analyse pathways most dramatically modulated when lytic replication is fully ongoing. This should reflect how cellular genes are regulated to aid production of virus particles. We used Affymetrix murine MG-U74Av2 oligonucleotide arrays, measuring 12,488 transcripts.
In a first quality screen of the pre-processed data, unusually high-scaling factors for the microarrays derived from RNA samples from MEF cells infected with virus were observed, which pointed to a lower median intensity for these arrays. As shown in Figure 1A, the average present calls from three microarrays per cell type and condition for all cell types was higher than 40%, except for infected MEF cells were the average present call percentage was 30.7 (± 4.5). This indicated a general shut down of the host RNA transcription, frequently seen after viral infection of mammalian cells [23–25], that would make the application of standard global normalization procedures of microarray data obsolete. Therefore, we normalized the MEF-derived data to spike-in control RNAs, which are routinely added to the cRNA after the labelling process (see Materials and methods). A box plot analysis of expression values normalized to spike-in controls indeed revealed a global down-regulation of cellular transcription at 48 hrs after infection with MHV-68 (Fig. 1B). Only the virus-infected MEF samples showed a dramatically decreased median signal compared to the mock samples. This effect of overall decrease in signal intensity was even more pronounced in MEF cells derived from the Balb/c background (data not shown).
Fig 1.
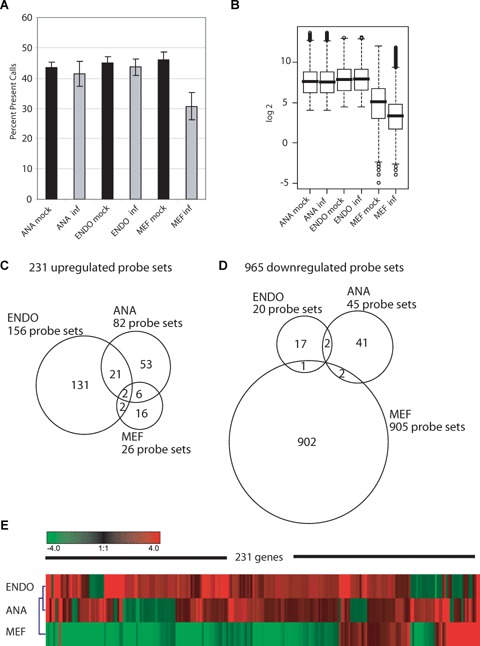
Overview of the general changes in expression values upon murine gammaherpesvirus 68 (MHV-68) infection. (A) Barplot showing the mean percentage of Affymetrix present calls for the three arrays of one condition, respectively. The present call is an indication for the reliability of detection of a probe set. Standard deviation is shown based on three array replicates. (B) Boxplots representing the normalized log2 expression values of all 12,488 transcripts present on the arrays, averaged per condition, for the three BL/6-derived cells, mock and MHV-68 infected, respectively. (C) Venn diagram describing the partitioning and overlap of the 231 significantly up-regulated probe sets among the three cellular systems. (D) Diagram as in C for the 965 significantly down-regulated probe sets. (E) Hierarchical Clustering of the average fold-changes induced by the viral infection. The 231 transcripts clustered are significantly different in at least one cell type with a fold-change >2 compared to the respective mock-treated cells. Red colour indicates up-regulation, green colour down-regulation after virus infection. ANA, Ana-1 macrophages; ENDO, embryonic endothelial progenitor cells or eEPCs; MEF, primary mouse embryonic fibrob-lasts; all cell types are of BL/6 genetic background.
We found that MHV-68 infection up-regulated 231 transcripts and down-regulated 965 probe sets. In order to define a core response to MHV-68 infection, we compared the genes from the three cell types for up- and down-regulated transcripts. In Figure 1C, the distribution and the overlap of concordant induction is summarized in a Venn diagram. The cell types did not react to the same extent to the viral infection. Endothelial progenitor cells showed the greatest change in gene induction with 156 probe sets. Ana-1 macrophages gene expression was with 82 probe sets intermediate, while the embryonal fibroblast cells showed the weakest induction in this set with just 26 probe sets. Despite differences in the amount of up-regulation, there was an overlap between cell types with regard to the genes found up-regulated (overlap of ANA/ENDO 28%, ANA/MEF 9.8%, MEF/ANA 30.8%, MEF/ENDO 15.4%, ENDO/ANA 14.7% and ENDO/MEF 2.6%). Two genes were induced in all three cell types: Lectin, mannose binding 1 (Lman1, probe set 161622_f_at) and synaptobrevin like 1 (sybl1, probe set 162113_r_at), both assigned to be involved in intracellular protein trafficking. The up-regulation of Lman1 was 122.5, 16.1 and 4.2-fold in fibroblasts, endothelial precursor cells and macrophages, respectively. The corresponding changes of sybl1 were 13.1, 7.8 and 5.0-fold. It has to be mentioned that the probe set we refer to Lman1 is not automatically annotated, since just five out of the nine oligos representing this probe set matches to the 3´ UTR of the transcript as deposited in the ENSEMBL database (http://www.ensembl.org/-Mus_musculus/index.html). A second probe set (160270_at), which is matching 100% to the 3´ UTR of Lman1, was not induced in the three cell types. Nevertheless, Lman1 induction could be confirmed by an independent RT-PCR as described later. By looking at the distribution of the 965 down-regulated transcripts (Fig. 1D), the remarkable repression of mRNA transcription in MEF cells after virus infection that was already assumed by the drop in present calls, with 905 probe sets down-regulated, could be confirmed. ANA and ENDO cells down-regulated only 45 and 20 probe sets, respectively. With regard to down-regulation, the overlap of the gene lists was much weaker than seen before for up-regulated transcripts. ANA and MEF cells share two (two times lymphocyte specific 1, LSP1), MEF and ENDO cells just one (coagulation factor III, F3) and ENDO and ANA cells two probe sets (Zinc finger protein of the cerebellum 2, Zic2 and Necdin, Ndn). Interestingly, Ndn was also down-regulated in MEF cells (15.1-fold), however, it did not pass the significance threshold.
In Figure 1E, a hierarchical cluster of the 231 up-regulated probe sets, based on the fold-changes, is depicted. The homology plot reveals a stronger similarity between ENDO and ANA cells, compared to MEF cells, because most transcripts induced upon infection in ENDO and ANA cells appear to be strongly down-regulated in MEF cells due to the general decrease in median signal levels in MEF cells compared to the mock samples. However, despite the overall similarity, a number of gene clusters were identified in ANA and ENDO cells, which were regulated in a cell type-specific manner.
Cell type specific activation programs
For a thorough analysis of possible functional relationships among the induced sequences, the list of 231 transcripts was subjected to a term analysis with the EASE program to detect overrep-resentation of distinct gene classification units (e.g. GO terms) hidden in the list. Nearly all significantly overrepresented gene categories were functionally connected to the immune response. On the contrary, gene categories like apoptosis, cell cycle arrest or transcriptional repression were not represented. A subsequent similar analysis for each individual cell type did not yield any cell type specific categories. Depicted in Table 1A is a selection of overrepresented categories with their probabilities (P-values). Genes from the GO category defense responewere induced with high significance (P-value = 7*10−13), including the more specific sub-categories MHCI and Interferon induction.
Table 1.
Functional analysis of the 231 significantly up-regulated transcripts upon MHV-68 infection. (A) Analysis of overrepresentation of gene categories for induced transcripts. Gene categories were assigned to the list of 231 up-regulated genes with the program EASE. The probability of seeing the number of ‘List Hits' in the ‘List Total’, given the frequency of ‘Population Hits' in the ‘Population Total’, is calculated by the Fisher exact probability and the P-values are shown in the last column. (B) Selected genes were manually assigned to functional biological categories. For each of the three cell lines, the average fold-change (FC) is shown.
| A | ||||||
|---|---|---|---|---|---|---|
| System | Gene category | List hits | List total | Population hits | Population total | Probability |
| GO Biological process | Defense response | 45 | 168 | 629 | 7516 | 7*10−13 |
| GO Biological process | Innate immune response | 17 | 168 | 146 | 7516 | 2*10−08 |
| GO Biological process | Chemotaxis | 13 | 168 | 95 | 7516 | 2*10−07 |
| SwissProt keywordinte | Interferon induction | 7 | 130 | 27 | 5351 | 3*10−06 |
| GO Molecular function | Chemokine receptor binding activity | 7 | 170 | 30 | 7550 | 3*10−06 |
| GO Molecular function | Cytokine activity | 15 | 170 | 177 | 7550 | 1*10−05 |
| SwissProt keywordinte | MHC I | 4 | 130 | 10 | 5351 | 6*10−05 |
| GO Molecular function | Antiviral response protein activity | 5 | 170 | 24 | 7550 | 1*10−04 |
| B | ||||||
|---|---|---|---|---|---|---|
| Classification | Probe set | Gene symbol | Gene title | MEF FC | ANA FC | ENDO FC |
| Antigen presentation | 9754_f_at | H2-D1 | Histocompatability 2, D region locus 1 | −2.1 | 1.1 | 2.1 |
| 99378_f_at | H2-D1 | Histocompatability 2, Q region locus 1 | −1.4 | 1.3 | 2.0 | |
| 98472_at | H2-T23 | Histocompatability 2, T region locus 23 | −1.3 | 2.4 | −1.1 | |
| 99379_f_at | LOC676689 | Similar to H-2 class I histocompatability antigen. L-D a chain precursor | −1.6 | 1.3 | 2.1 | |
| 93085_at | Psmb9 | Proteosome (prosome, macropain) subunit, β type 9 | 1.0 | 1.1 | 2.4 | |
| 100154_at | Tapbp | TAP binding protein | −1.9 | 1.4 | 3.1 | |
| Chemokines | 102736_at | Ccl2 | Chemokine (C-C motif) ligand 2 | −6.0 | 3.8 | −1.0 |
| 94146_at | Ccl4 | Chemokine (C-C motif) ligand 4 | 7.7 | 3.0 | 1.2 | |
| 94761_at | Ccl7 | Chemokine (C-C motif) ligand 7 | −24.7 | 4.8 | −1.1 | |
| 96953_at | Cxcl14 | Chemokine (C-X-C motif) ligand 14 | −7.7 | 1.1 | 3.7 | |
| 101160 at | Cxcl2 | Chemokine (C-X-C motif) ligand 2 | 2.2 | 2.9 | −1.1 | |
| Cytokine and other Immune modulators | 98240_at | ll12rb1 | Interleukin 12 receptor, β 1 | −1.6 | 2.7 | 2.4 |
| 94755_at | ll1a | Intrleukin 1 α | 10.3 | 1.0 | 1.0 | |
| 103486_at | ll1b | Interleukin 1 β | 11.3 | 1.6 | 1.5 | |
| 93871_at | ll1m | Interleukin, receptor antagonist | −1.1 | 3.1 | 1.6 | |
| 102629_at | Tnf | Tumour necrosis factor | 5.1 | 1.2 | −1.0 | |
| Innate immunity | 98088_at | Cd14 | CD14 antigen | 1.6 | 2.7 | 5.5 |
| 94747_at | Csf2rb1 | Colony stimulating factor 2 receptor β 1 | 7.1 | 1.4 | 1.1 | |
| 101800_at | Fpr-rs2 | Formyl peptid receptor, related sequence 2 | 7.2 | 1.0 | −1.2 | |
| 99387_at | Fpr1 | Formyl peptide receptor 1 | 3.1 | 1.0 | 1.1 | |
| 95585_at | Proc | Protein C | −1.5 | 2.2 | 1.6 | |
| 98018_at | procr | Protein C receptor, endothelial | 3.4 | 2.2 | −1.1 | |
| Interferon associated | 98406_at | Ccl5 | Chemokine (C-C motif) ligand 5 | 3.6 | 6.7 | −1.0 |
| 98822_at | Isg15 | ISG15 ubiquitin-like modifier | −4.1 | 1.7 | 6.6 | |
| 98466_r_at | Ifi204 | Interferon activated gene 204 | −2.9 | 3.1 | 1.1 | |
| 100013_at | Ifi35 | Interferon-induced protein 35 | −2.6 | 1.7 | 3.8 | |
| 100981_at | Ifit1 | Interferon-induced protein with tetratricopeptide repeats 1 | −6.0 | 1.2 | 7.9 | |
| 93956-at | Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | −2.5 | 1.3 | 3.8 | |
| 160253_at | Ifitm3 | Interferon induced transmembrane protein 3 | −1.9 | 1.6 | 3.3 | |
| 161699_i_at | Irf6 | Interferon regulatory factor 6 | 1.0 | 1.1 | 2.2 | |
| 104669_at | Irf7 | Interferon regulatory factor 7 | 1.2 | 6.9 | 1.5 | |
| 97409-at | Irgm | immunity-related GTpase family, M | −6.1 | 2.0 | 2.7 | |
| 103432_at | Isg20 | Interferon-stimulated protein | −4.1 | −1.0 | 6.0 | |
| 102717_at | Oas1g | 2'5' oligoadenylate synthetase 1G | −1.4 | 2.3 | 3.9 | |
| 101465_at | Stat1 | single transducer and activator of transcription 1 | −3.9 | 1.4 | 2.4 | |
| Scavenger receptor activity | 97507_at | Lgals3bp | Lectin, galactoside-binding, soluble, 3 binding pro-tein | −2.2 | 2.4 | 2.1 |
| 102974_at | Marco | macrophage receptor with collagenous structure | 6.2 | 1.1 | −1.1 | |
With the help of the gene category annotation derived from the EASE output, we manually assigned up-regulated transcripts to the functional classes as shown in Table 1B. In this manner, a number of previously unrecognized cell type specific differences in gene expression became apparent. For example, MHC class I expression in response to virus infection was induced in both ENDO and ANA cells, whereas antigen processing molecules, like the proteosome subunit, β type 9 or the TAP-binding protein were solely induced in ENDO cells. MEF cells did not show any induction of genes associated with antigen presentation.
Chemokines as a group were induced in all cases, but there was little overlap of individual members among the three cell types. In brief, Ccl2 (MCP1) and Ccl7 (MCP3), both capable to chemoattract macrophages, were induced only in ANA cells and the only chemokine induced in ENDO cells was Cxcl14 (Mip2g). Ccl4 (Mip1b), Ccl5 (Rantes) and Cxcl2 (Mip2a) were all found induced in MEF and ANA cells. Interleukin 1 α and β and tumour necrosis factor α are highly induced solely in infected MEF cells. This might reflect the immune modulatory function of fibroblasts in the infection process. Interestingly, no cytokine was found to be induced in either ANA macrophages or endothelial cells.
Examining genes related to innate immunity, it is surprising that four out of 26 probe sets were induced in MEF cells (15%), namely Csf2rb, Fpr-rs2, Fpr1 and Procr, to a high extent. The induction of interferon-associated genes was, with 10 genes, highest in ENDO cells. It should be mentioned that no induction of interferon itself was detected in this screen.
Transcriptome comparison between BL/6 and Balb/c derived MEF cells after infection
To search for possible genotype-specific patterns of gene expression changes after MHV-68 infection, we compared the response of BL/6- and Balb/c-derived embryonic fibroblasts (MEF). Similar to the BL/6 results, there was only a weak general induction of transcription in Balb/c MEFs. As shown in Figure 2A, 16 probe sets were induced, with a reasonable overlap of five transcripts that were also induced in BL/6 cells. Of note, Lman1 and Sybl1, which were found as part of the core response in all three cell types of BL/6 background, were also induced in Balb/c MEF. The remaining three probe sets induced in both MEF cell types could not be assigned to any known gene. One annotated transcript, which showed concordant induction in both cell types, although not significant for the BL/6 background, was the major intrinsic protein of eye lens fibre (Mip).
Fig 2.
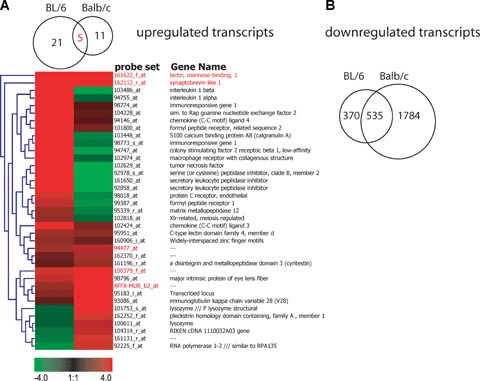
Common and unique changes in gene expression comparing BL/6 and Balb/c-derived MEF cells. (A) Venn diagram and heat map showing the 36 transcripts that are significantly up-regulated in either BL/6 or Balb/c MEF cells. The heat map is based on fold-changes. Red colour indicates up-regulation, green colour down-regulation after MHV-68 infection. (B) Venn diagram of the 2698 transcripts that are significantly down-regulated in MEF cells.
Strikingly, none of the chemokines, cytokines and genes associated with innate immune function, which were induced in the BL/6 cells, was up-regulated in Balb/c MEF cells, suggesting that the genetic background might play a fundamental role in fibrob-last-associated immune response.
MEF cells from the BL/6 background showed a remarkable shutdown of transcription (905 transcripts), a process even more pronounced in MEFs from Balb/c mice (2319 probe sets repressed). 535 probe sets were repressed in both cell types indicating that the type of gene down-regulation is not incidental (Fig 2B).
Validation of microarray data by RT-PCR analysis
To verify the microarray data, we performed RT-PCR analysis on a subset of representative genes. The RT-PCR analysis indicated a pattern of up- or down-regulation of transcription which correlated well with the microarray data (Fig. 3).
Fig 3.
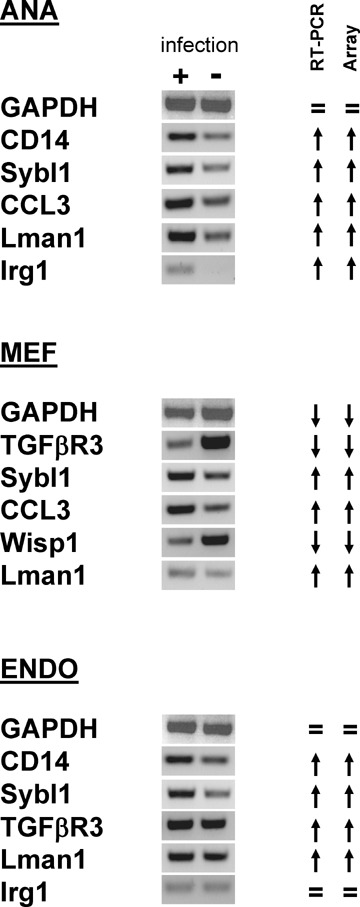
Validation of microarray data by RT-PCR. Total RNA derived from infected or non-infected cells was reverse-transcribed, and the resulting cDNA was amplified using gene-specific primers. RT-PCR products were separated by electrophoresis in 2% agarose gels and visualized by ethidium bromide staining. Upward- and downward-pointing arrows indicate up- and down-regulation, whereas ‘=’ indicates that the expression was unchanged. In all cases, RT-PCRs were consistent with the microarray data. ANA, Ana-1 macrophages; ENDO, embryonic endothelial progenitor cells or eEPCs; MEF, primary mouse embryonic fibroblasts
Down-regulation of both Lman1 or sybl1 by siRNA reduces virus productivity
Because Lman1 and sybl1 were significantly up-regulated after infection in all three cell types, we tested whether up-regulation of these proteins is important for lytic virus replication. Lman1 and sybl1, respectively, were down-regulated by siRNA technology. We first showed that selected gene-specific siRNAs blocked efficiently the expression of the respective target genes (Fig. 4A). Next, we analysed the effect of two different siRNAs directed against sybl1 on MHV-68 lytic replication in MEF cells. As shown in Figure 4B, down-regulation of sybl1 by two different siRNAs resulted in a significant reduction in virus yield 48 hrs after infection. Unrelated effects of siRNAs on cell numbers or viability could be excluded by either trypan blue exclusion test or MTT assay (data not shown). To exclude interferon induced by the siRNA transfection as the mediator of the effect, we performed the same experiments in NIH3T3 cells, which are hyporesponsive to interferon [26–28]. As in MEF cells, down-regulation of sybl1 by two different siRNAs resulted in a significant reduction in virus yields (Fig. 4C). Consistent with these results, we failed to detect the induction of interferon (IFN-β) after siRNA transfection of NIH3T3 cells both at the level of transcription, as analysed by RT-PCR, and at the protein level, as analysed by ELISA in supernatants of transfected cells (data not shown). Similar to the results with sybl1, we were able to demonstrate a significant reduction in virus yield using two different siRNAs directed against Lman1 (Fig. 4D). To further rule out that the drop in virus yields is due to interferon effects, we performed an experiment in Vero cells, which have a defect in the interferon system [29, 30]. One of the siRNAs directed against murine sybl1 was predicted by computer analysis to target the monkey sybl1 sequence. Using RT-PCR, we could indeed demonstrate the ability of this siRNA to inhibit the expression of sybl1 in Vero cells (data not shown). Consistent with the results in the other cell lines, down-regulation of sybl1 in Vero cells resulted in a significant reduction in MHV-68 virus yield (Fig. 4E). Finally, we tested the siRNA1 effect at a later time point. For this purpose, NIH3T3 cells were transfected with siRNA1, infected 72 hrs after transfection with either an MOI of 1 (Fig. 4F) or 0.1 (Fig. 4G), and virus titres were determined 72 hrs after infection. In this experiment, down-regulation of sybl1 by siRNA treatment resulted in a strong (99.75% and 96%, respectively) reduction in MHV-68 virus yield (Fig. 4F and G).
Fig 4.
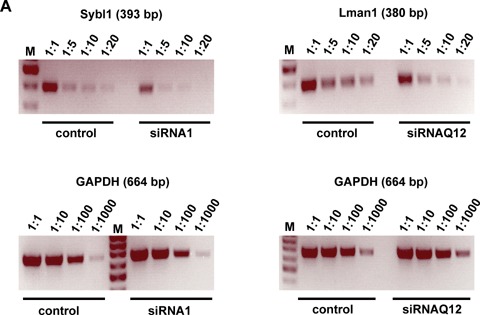
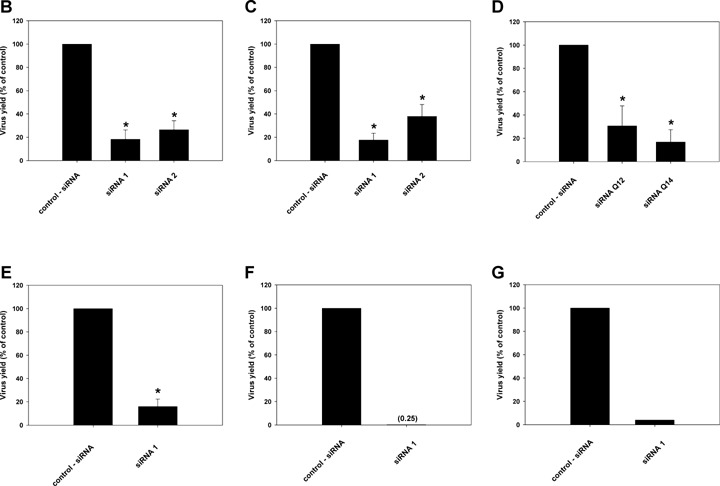
Effect of siRNA-mediated gene silencing on MHV-68 yield. NIH3T3 cells (A, C, D, F and G), MEF cells (B) and Vero cells (E) were transfected with 50 pmoles of the indicated siRNAs. 24 hrs (A-E) or 72 hrs (F and G) after transfection, cells were infected with MHV-68 with an MOI of 1 (A-F) or 0.1 (G). After 60 min. at 37°C, the inoculum was removed and fresh medium was added. Cells and supernatants were either harvested 48 hrs (A-E) or 72 hrs (F and G) after infection, and RNA was isolated and analysed by RT-PCR (A). Virus titres were determined by plaque assay (B-G). In panel A, down-regulation of synaptobrevin-like 1 (sybl1) by siRNA1 directed against sybl1 is shown on the left, and down-regulation of Lman1 by siRNAQ12 directed against Lman1 is shown on the right. Different concentrations of cDNA template, as indicated on top of each panel, were amplified by PCR with primer pairs specific for sybl1 and Lman1, respectively. A primer set for the housekeeping gene GAPDH was used in parallel for PCR amplification as a control. In panels B-G titres are expressed as percentages of the virus yield, where 100% represents the titre of virus grown in wells transfected with control siRNA. In panels B-G, values are means ± SD from three independent experiments. The asterisks indicate a statistically significant difference with P<0.003 (Student's t-test). In panels F and G, data from single experiments are shown.
Inhibition of the retrograde transport of Lman1 results in a significant reduction in virus production
To confirm the important role of Lman1 in the process of viral infection, we blocked Lman1 activity with an alternative method using a phospholipase A2 (PLA2) antagonist. PLA2 enzymes are involved in regulating membrane trafficking events, and thus, PLA2 antagonists can be used to inhibit constitutive retrograde membrane traffic to the endoplasmic reticulum [31]. Since Lman1 is a transmembrane lectin, which cycles constitutively between the ER, ERGIC and cis-Golgi elements, it can be inhibited by PLA2 antagonists [31]. For our experiments, we selected the well-described PLA2 inhibitor 2-(p-amylcinnamoyl)amino-4-chloroben-zoic acid (ONO-RS-082) [31]. Inhibition of Lman1 by treatment with ONO-RS-082 resulted in a significant reduction in virus yield (Fig. 5). Again, this reduction was not due to effects of ONO-RS-082 on cell numbers or viability, which were similar in all samples (data not shown). Thus, pharmacological evidence further supports the notion that inhibition of retrograde membrane traffic results in reduced virus production.
Fig 5.
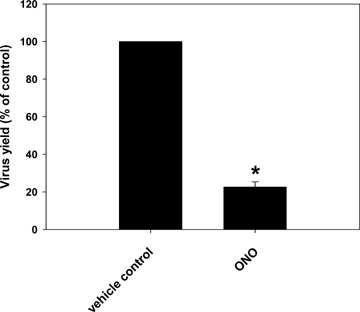
Effect of pharmacological phospholipase A2 (PLA2) inhibition on MHV-68 yield. NIH3T3 cells in 6-well plates were pre-incubated at 4°C for 30 min. and infected with MHV-68 at an multiplicity of infection (MOI) of 0.1. After the incubation period of 60 min. at 4°C to allow virus adsorption, inocula were removed and temperature was shifted to 37°C in the presence or absence of 50 μM of the phospholipase A2 (PLA2) inhibitor 2-(p-amylcinnamoyl)amino-4-chlorobenzoic acid (ONO-RS-082), dissolved in ethanol. Viral titers were determined after 48 hrs by plaque assay on BHK-21 cells. Values are means – SD from three independent experiments. The asterisk indicates a statistically significant difference with P = 0.0000011 (Student's t-test).
The replication of herpes simplex virus 1 (HSV-1) in Vero cells is inhibited by siRNA- down-regulation of sybl1
Finally, we tested whether our findings are restricted to MHV-68, or if they may be also relevant for other herpesviruses. As mentioned above, studies on the productive replication of the human gammaherpesviruses EBV and KSHV are hampered by the lack of cell culture systems which support productive replication. The human alphaherpesvirus HSV-1 replicates, like MHV-68, efficiently in various tissue cultured cells, including Vero cells. Since the siRNA directed against sybl1 was active in Vero cells this provided us with the possibility to analyse the effect on the replication of HSV-1. As for MHV-68, down-regulation of sybl1 resulted in a significant reduction in virus yield also for HSV-1 (Fig. 6).
Fig 6.
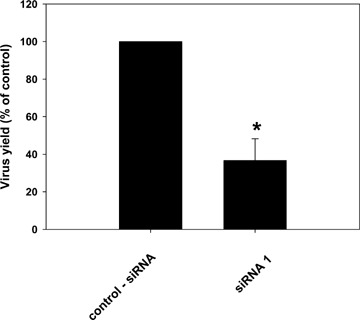
Effect of siRNA on herpes simplex virus 1 (HSV-1). Vero cells were transfected with 50 pmoles of siRNA1 directed against sybl1. 24 hrs after transfection, cells were infected with HSV-1 at an MOI of 1. After 60 min. at 37°C, the inoculum was removed and fresh medium was added. Cells and supernatants were harvested 48 hrs after infection, and virus titres were determined by plaque assay. Titres are expressed as percentages of the virus yield, where 100% represents the titre of virus grown in wells transfected with control siRNA. Values are means ± SD from three independent experiments. The asterisk indicates a statistically significant difference with P= 0.0007 (Student's t-test).
Discussion
Microarrays are widely used to determine the complex transcriptional programs of larger viruses and for global monitoring of host cell transcriptional changes in response to virus infection or expression of viral proteins [32–47]. Transcriptional changes in virally infected cells can be either the result of anti-viral, pro-viral or bystander host responses [48]. In contrast to antiviral response pathways of the host, other host pathways might be beneficial or even essential for viral replication. Inhibition of these pathways may lead to the discovery of novel anti-viral drugs [48]. For example, cyclooxygenase-2 was identified in microarray experiments as a host gene induced by human cytomegalovirus infection [49]. Specific inhibition of cyclooxygenase-2 leads to a significant decrease in virus titers [50]. Similar results were also reported for MHV-68 infection [51]. For KSHV, up-regulation of the proto-oncogene c-kit was identified by gene expression profiling and shown to be essential for the transformation of endothelial cells [36]. Concomitantly, inhibition of c-kit activity could reverse the KSHV-induced morphological transformation of endothelial cells [44].
In this study, we infected three different cell types (fibroblasts, endothelial precursor cells and macrophages) with MHV-68 and surveyed the host cell response for changes, either common to all, or unique to a particular cell type using oligonucleotide microar-rays covering 12,488 probe sets. A similar approach, analysing the modulation of host cell gene expression early during infection, has been described for KSHV [33]. In that study, modulation of host cell gene expression was analysed by oligonucleotide arrays immediately (2 and 4 hrs) after infection of endothelial, fibroblast and B cells. Among the genes found to be differentially expressed, 33 were shared by all three cell types [33]. Studies with KSHV during later time points of lytic infection are difficult due to the lack of cell culture systems that support KSHV replication. In contrast, MHV-68 efficiently replicates in various tissue culture cells. Thus, we were able to analyse the host cell response of different cell types during lytic replication of a gammaherpesvirus. We found that MHV-68 infection induced 231 transcripts and repressed 965 probe sets. The remarkable repression of mRNA transcription in MEF cells is consistent with the profound shutoff of host gene expression during the KSHV lytic cycle, induced by superinfection of latently infected endothelial cells with an adenovirus encoding the lytic switch protein replication and transcriptional activator (RTA) [24, 52], and also consistent with the reduced expression of cellular transcripts after infection of murine epithelial cells with MHV-68 [35].
To detect genotype specific patterns, Balb/c-derived MEF cells were compared with BL/6-derived MEF cells. In both cases, we observed a weak induction of transcription and a general transcrip-tional shutdown, which was more pronounced in Balb/c MEF cells. There was a substantial overlap of probe sets showing down-regulation between Balb/c and BL/6 MEFs, suggesting that the pattern of suppressed genes is not random. It is noteworthy that none of the chemokines, cytokines and genes associated with innate immune function induced in the BL/6 background was induced in Balb/c MEF cells. This suggests that, besides the well-recognized differences in the response of immune system cells between BL/6 and Balb/c [53], there is also an important role of the genetic background in the fibroblast-associated immune response. However, it should be noted that MEF cells are not pure fibroblast cultures and can be contaminated by small percentages of other cell types. Therefore, the findings with regard to differences in the innate response need to be corroborated in future studies.
In our study, the transcription of only two genes, Lman1 (also known as ERGIC-53) and sybl1, was found to be modulated in all three cell types, indicating that they might be critical for the virus life cycle independently of the cell type. Both proteins are implicated in cellular exocytosis, transporting glycoproteins through the secretory pathway. Lack of functional ERGIC-53 leads to a selective defect in secretion of glycoproteins and to haemophilia [54–57]. Sybl1 encodes a member of the synaptobrevin family of proteins involved in exocytosis and membrane transport [58, 59].
To test whether these two proteins are required for transport of viral proteins, and thus virus production, we used siRNA technology to knockdown the expression of either gene. Inhibition of either ERGIC-53 or sybl1 resulted in a significant reduction in the yield of MHV-68. In addition, inhibition of sybl1 resulted in a significant reduction in the yield of the human alphaherpesvirus HSV-1. Importantly, in VERO cells which have a defect in the interferon system [29, 30], the same effect was seen, arguing against interferon effects which can be induced by siRNAs under certain conditions [60].
Recently, siRNAs targeted against viral or host cell genes have been used to inhibit a variety of viruses both in vitro and in vivo [61–65]. Using siRNAs directed against the host cell genes sybl1 or ERGIC-53, we were now able to decrease virus production up to 10-fold, in one experiment even up to 400-fold. The extent of inhibition (approx. 10-fold) is in a range comparable to reports on pharmacological inhibitors or siRNAs directed against host cell genes. For example, a 10-fold reduction of poliovirus titres was observed in human cells transfected with siRNA against the ribo-somal protein S6 (RpS6) [66]. Treatment of cells with siRNAs against a host cell kinase caused a nearly 10-fold decrease in production of West Nile virus [48]. Pharmacological treatment of cells with the cyclooxygenase-2 inhibitor NS-398 resulted in an inhibition of MHV-68 production of about 50%[51], comparable to the approx. 80% of inhibition in our experiments using the PLA2 inhibitor ONO-RS-082.
It was not surprising that the siRNA-based approach resulted in only modest inhibition of virus production since the achieved knockdown of gene expression was far from complete and, in particular, since the targeted proteins have a very long half-life, which is in case of ERGIC-53 of several days [67]. Nevertheless, we think that the relevance of our results is not to suggest the use of siRNAs for inhibition of sybl1 or ERGIC-53 but is the validation that these and possibly other proteins of the secretory pathway are important for virus replication. This deserves further studies on the mechanisms which should lead to potential targets for antiviral therapy.
Our study was designed to investigate lytic replication of MHV-68 as a model for a gammaherpesvirus, and thus the question remains how this relates to KSHV. It is believed that products of lytic infection may act in a paracrine fashion to promote KS tumourigenesis [68], and that chronic lytic infection is required for KS tumourigenesis since interruption of lytic replication by ganci-clovir appears to prevent KS development [69]. Thus, it is likely that interfering with lytic replication may attenuate infection.
Our data suggest that the specific inhibition of proteins of the secretory pathway has an antiviral potential. Herpesviruses require the host cell secretory pathway for transport and processing of membrane glycoproteins during the course of virus assembly, and it has been suggested that specific host cell factors facilitate viral egress [70–73]. For example, HSV-1 maturation and egress are inhibited after treatment of infected cells with Brefeldin A or Monensin [74, 75]. Both Brefeldin A and Monensin block protein transport from the ER to the Golgi apparatus. Since the endoplasmic reticulum-golgi intermediate compartment (ERGIC) participates in the maturation of, or is target for several viruses, it has been suggested that understanding the targeting of viruses and viral proteins to the ERGIC could lead to the development of general approaches for viral interference [54]. Both Brefeldin A and Monensin have a profound effect and disassemble the Golgi complex and thus affect both infected and uninfected cells. In contrast, targeting cellular genes being expressed at low level in uninfected cells and up-regulated only during infection, like sybl1 and ERGIC-53, might allow a more subtle approach where virus production can be inhibited without interfering with the viability of both infected and uninfected cells. It might suffice to reduce the level of induction back to the pre-infec-tion state [48]. Even if inhibition of virus replication by targeting specific host cell pathways would not be complete, it may be thera-peutically beneficial especially in combination with other anti-viral compounds, which target other steps in the viral life cycle [50].
Acknowledgments
We are grateful to M. Semisch and Angela Servatius for expert technical assistance and to Dr. B. Adler for critical reading of the manuscript. This work was funded by grants from the Deutsche Forschungsgemeinschaft (DFG), Ad 121/2–1 to 2–4, and from the BMBF (NGFN-2, FKZ 01GS0405) to H.A., by the DFG (SFB 455) to U.H.K., and the National Genome Science Network (NGFN I) grant No. 01GS0113 to A.K.H.. The Microarray and Bioinformatics Core Unit at the Institute of Medical Microbiology, Immunology and Hygiene is supported by the Bundesministerium für Bildung und Forschung (NGFN Network Infection and Inflammation FKZ 01G0113, TP 37 to R. Lang, R. Hoffmann, and H. Wagner).
Footnotes
R Development Core Team (2006). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3–900051-07–0, URL http://www.R-project.org.
References
- 1.Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. Isolation of five strains of her-pesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. [PubMed] [Google Scholar]
- 2.Virgin HWIV, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. Complete sequence and genomic analysis of murine gammaher-pesvirus 68. J Virol. 1997;71:5894–904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virgin HWIV, Speck SH. Unraveling immunity to -y-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr Opin Immunol. 1999;11:371–9. doi: 10.1016/s0952-7915(99)80063-6. [DOI] [PubMed] [Google Scholar]
- 4.Speck SH, Virgin HWIV. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Current Opinion in Microbiology. 1999;2:403–9. doi: 10.1016/s1369-5274(99)80071-x. [DOI] [PubMed] [Google Scholar]
- 5.Simas JP, Efstathiou S. Murine gamma-herpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends in Microbiology. 1998;6:276–82. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- 6.Nash AA, Dutia BM, Stewart JP, Davison AJ. Natural history of murine -y-herpesvirus infection. Phil Trans R Soc Lond B. 2001;356:569–79. doi: 10.1098/rstb.2000.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flano E, Woodland DL, Blackman MA. A mouse model for infectious mononucleosis. Immunol Res. 2002;25:201–17. doi: 10.1385/IR:25:3:201. [DOI] [PubMed] [Google Scholar]
- 8.Doherty PC, Trlpp RA, Hamilton-Easton A-M, Cardln RD, Woodland DL, Blackman MA. Tuning into immunological dissonance: an experimental model for infectious mononucleosis. Curr Opin Immunol. 1997;9:477–83. doi: 10.1016/s0952-7915(97)80098-2. [DOI] [PubMed] [Google Scholar]
- 9.Blackman MA, Flano E. Persistent -y-her-pesvirus infections: What can we learn from an experimental mouse model? J Exp Med. 2002;195:F29–32. doi: 10.1084/jem.20020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chain P, Kurtz S, Ohlebusch E, Slezak T. An applications-focused review of comparative genomics tools: capabilities, limitations and future challenges. Brief Bioinform. 2003;4:105–23. doi: 10.1093/bib/4.2.105. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Gersteln M. Of mice and men: phylogenetic footprinting aids the discovery of regulatory elements. J Biol. 2003;2:11. doi: 10.1186/1475-4924-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolzman B, Pellett PE. In: The Family Herpesviridae: A brief introduction. Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, et al., editors. Lippincott Williams & Wilkins; 2001. pp. 2381–97. In:., editors. Fields -Virology. Philadelphia:; [Google Scholar]
- 13.Adler H, Messerle M, Wagner M, Koszinowskl UH. Cloning and mutagene-sis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol. 2000;74:6964–74. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox GW, Mathleson BJ, Gandlno L, Blasi E, Radzloch D, Varesio L. Heterogeneity of hematopoietic cells immortalized by v-myc/v-raf recombinant retrovirus infection of bone marrow or fetal liver. J Natl Cancer Inst. 1989;81:1492–6. doi: 10.1093/jnci/81.19.1492. [DOI] [PubMed] [Google Scholar]
- 15.Hatzopoulos AK, Folkman J, Vaslle E, Eiselen GK, Rosenberg RD. Isolation and characterization of endothelial progenitor cells from mouse embryos. Development. 1998;125:1457–68. doi: 10.1242/dev.125.8.1457. [DOI] [PubMed] [Google Scholar]
- 16.Vajkoczy P, Blum S, Lamparter M, Mallhammer R, Erber R, Engelhardt B, Vestweber D, Hatzopoulos AK. Multistep nature of microvascular recruitment of ex vivo-expanded embryonic endothelial progenitor cells during tumor angiogene-sis. J Exp Med. 2003;197:1755–65. doi: 10.1084/jem.20021659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adler H, Beland JL, Del-Pan NC, Kobzlk L, Brewer JP, Martin TR, Rlmm IJ. Suppression of Herpes simplex virus type 1 (HSV-1)-induced pneumonia in mice by inhibition of inducible nitric oxide synthase (iNOS, NOS2) J Exp Med. 1997;185:1533–40. doi: 10.1084/jem.185.9.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborn JE, Walker DL. Enhancement of infectivity of murine cytomegalovirus in vitro by centrifugal inoculation. J Virol. 1968;2:853–8. doi: 10.1128/jvi.2.9.853-858.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–8. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 22.Hosack DA, Dennis G., Jr Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brum LM, Lopez MC, Varela JC, Baker HV, Moyer RW. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology. 2003;315:322–34. doi: 10.1016/s0042-6822(03)00532-4. [DOI] [PubMed] [Google Scholar]
- 24.Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell. 2004;13:713–23. doi: 10.1016/s1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig H, Mages J, Staib C, Lehmann MH, Lang R, Sutter G. Role of viral factor E3L in modified vaccinia virus ankara infection of human HeLa Cells: regulation of the virus life cycle and identification of differentially expressed host genes. J Virol. 2005;79:2584–96. doi: 10.1128/JVI.79.4.2584-2596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epstein DA, Czarniecki CW, Jacobsen H, Friedman RM, Panet A. A mouse cell line, which is unprotected by interferon against lytic virus infection, lacks ribonuclease F activity. Eur J Biochem. 1981;118:9–15. doi: 10.1111/j.1432-1033.1981.tb05479.x. [DOI] [PubMed] [Google Scholar]
- 27.Mittnacht S, Jacobsen H. Selection and characterization of interferon-sensitive cells derived from an interferon-resistant NIH 3T3 line. J Gen Virol. 1987;68:2945–51. doi: 10.1099/0022-1317-68-11-2945. [DOI] [PubMed] [Google Scholar]
- 28.Mittnacht S, Jacobsen H. Studies on interferon-sensitive cells derived from the interferon-resistant NIH 3T3 clone 1 line. Prog Clin Biol Res. 1985;202:237–45. [PubMed] [Google Scholar]
- 29.Desmyter J, Melnick JL, Rawls WE. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J Virol. 1968;2:955–61. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosca JD, Pitha PM. Transcriptional and posttranscriptional regulation of exoge-nous human beta interferon gene in simian cells defective in interferon synthesis. Mol Cell Biol. 1986;6:2279–83. doi: 10.1128/mcb.6.6.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Figueiredo P, Drecktrah D, Polizotto RS, Cole NB, Lippincott-Schwartz J, Brown WJ. Phospholipase A2 antagonists inhibit constitutive retrograde membrane traffic to the endoplasmic reticulum. Traffic. 2000;1:504–11. doi: 10.1034/j.1600-0854.2000.010608.x. [DOI] [PubMed] [Google Scholar]
- 32.Dittmer DP, Gonzalez CM, Vahrson W, DeWire SM, Hines-Boykin R, Damania B. Whole-genome transcription profiling of rhesus monkey rhadinovirus. J Virol. 2005;79:8637–50. doi: 10.1128/JVI.79.13.8637-8650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naranatt PP, Krishnan HH, Svojanovsky SR, Bloomer C, Mathur S, Chandran B. Host gene induction and transcriptional reprogramming in Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8)-infected endothelial, fibroblast, and B cells: insights into modulation events early during infection. Cancer Res. 2004;64:72–84. doi: 10.1158/0008-5472.can-03-2767. [DOI] [PubMed] [Google Scholar]
- 34.Yang WC, Devi-Rao GV, Ghazal P, Wagner EK, Triezenberg SJ. General and specific alterations in programming of global viral gene expression during infection by VP16 activation-deficient mutants of herpes simplex virus type 1. J Virol. 2002;76:12758–74. doi: 10.1128/JVI.76.24.12758-12774.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebrahimi B, Dutia BM, Roberts KL, Garcia-Ramirez JJ, Dickinson P, Stewart JP, Ghazal P, Roy DJ, Nash AA. Transcriptome profile of murine gamma-herpesvirus-68 lytic infection. J Gen Virol. 2003;84:99–109. doi: 10.1099/vir.0.18639-0. [DOI] [PubMed] [Google Scholar]
- 36.Moses AV, Jarvis MA, Raggo C, Bell YC, Ruhl R, Luukkonen BG, Griffith DJ, Wait CL, Druker BJ, Heinrich MC, Nelson JA, Fruh K. Kaposi's sarcoma-associated her-pesvirus-induced upregulation of the c-kit proto-oncogene, as identified by gene expression profiling, is essential for the transformation of endothelial cells. J Virol. 2002;76:8383–99. doi: 10.1128/JVI.76.16.8383-8399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karaca G, Anobile J, Downs D, Burnside J, Schmidt CJ. Herpesvirus of turkeys: microarray analysis of host gene responses to infection. Virol. 2004;318:102–11. doi: 10.1016/j.virol.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 38.Hertel L, Mocarski ES. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of Pseudomitosis independent of US28 function. J Virol. 2004;78:11988–2011. doi: 10.1128/JVI.78.21.11988-12011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray N, Enquist LW. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J Virol. 2004;78:3489–501. doi: 10.1128/JVI.78.7.3489-3501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slobedman B, Stern JL, Cunningham AL, Abendroth A, Abate DA, Mocarski ES. Impact of human cytomegalovirus latent infection on myeloid progenitor cell gene expression. J Virol. 2004;78:4054–62. doi: 10.1128/JVI.78.8.4054-4062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmen KA, Singh J, Luukkonen BG, Lopper M, Bittner A, Miller NE, Jackson MR, Compton T, Fruh K. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glyco-protein B. Proc Natl Acad Sci USA. 2001;98:7140–45. doi: 10.1073/pnas.121177598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–5. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, Elliman S, Flanagan AM, Alitalo K, Boshoff C. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36:687–93. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- 44.Moses AV, Jarvis MA, Raggo C, Bell YC, Ruhl R, Luukkonen BG, Griffith DJ, Wait CL, Druker BJ, Heinrich MC, Nelson JA, Fruh K. A functional genomics approach to Kaposi's sarcoma. Ann N Y Acad Sci. 2002;975:180–91. doi: 10.1111/j.1749-6632.2002.tb05951.x. [DOI] [PubMed] [Google Scholar]
- 45.Mossman KL, Macgregor PF, Rozmus JJ, Goryachev AB, Edwards AM, Smiley JR. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol. 2001;75:750–8. doi: 10.1128/JVI.75.2.750-758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter KL, Cahir-McFarland E, Kieff E. Epstein-barr virus-induced changes in B-lymphocyte gene expression. J Virol. 2002;76:10427–36. doi: 10.1128/JVI.76.20.10427-10436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones JO, Arvin AM. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J Virol. 2003;77:1268–80. doi: 10.1128/JVI.77.2.1268-1280.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeFilippis V, Raggo C, Moses A, Fruh K. Functional genomics in virology and antiviral drug discovery. Trends Biotechnol. 2003;21:452–57. doi: 10.1016/S0167-7799(03)00207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75:12319–30. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu H, Cong JP, Yu D, Bresnahan WA, Shenk TE. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci USA. 2002;99:3932–7. doi: 10.1073/pnas.052713799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Symensma TL, Martinez-Guzman D, Jia Q, Bortz E, Wu TT, Rudra-Ganguly N, Cole S, Herschman H, Sun R. COX-2 induction during murine gammaher-pesvirus 68 infection leads to enhancement of viral gene expression. J Virol. 2003;77:12753–63. doi: 10.1128/JVI.77.23.12753-12763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glaunsinger B, Ganem D. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral patho-genesis. J Exp Med. 2004;200:391–8. doi: 10.1084/jem.20031881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinberg JB, Lutzke ML, Alfinito R, Rochford R. Mouse strain differences in the chemokine response to acute lung infection with a murine gammaher-pesvirus. Viral Immunol. 2004;17:69–77. doi: 10.1089/088282404322875467. [DOI] [PubMed] [Google Scholar]
- 54.Hauri HP, Kappeler F, Andersson H, Appenzeller C. ERGIC-53 and traffic in the secretory pathway. J Cell Sci. 2000;113:587–96. doi: 10.1242/jcs.113.4.587. [DOI] [PubMed] [Google Scholar]
- 55.Zhang B, Cunningham MA, Nichols WC, Bernat JA, Seligsohn U, Pipe SW, McVey JH, Schulte-Overberg U, de Bosch NB, Ruiz-Saez A, White GC, Tuddenham EG, Kaufman RJ, Ginsburg D. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat Genet. 2003;34:220–5. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
- 56.Nichols WC, Terry VH, Wheatley MA, Yang A, Zivelin A, Ciavarella N, Stefanile C, Matsushita T, Saito H, de Bosch NB, Ruiz-Saez A, Torres A, Thompson AR, Feinstein DI, White GC, Negrier C, Vinciguerra C, Aktan M, Kaufman RJ, Ginsburg D, Seligsohn U. ERGIC-53 gene structure and mutation analysis in 19 combined factors V and VIII deficiency families. Blood. 1999;93:2261–6. [PubMed] [Google Scholar]
- 57.Nichols WC, Seligsohn U, Zivelin A, Terry VH, Hertel CE, Wheatley MA, Moussalli MJ, Hauri HP, Ciavarella N, Kaufman RJ, Ginsburg D. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell. 1998;93:61–70. doi: 10.1016/s0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- 58.Matarazzo MR, Cuccurese M, Strazzullo M, Vacca M, Curci A, Giuseppina MM, Cocchia M, Mercadante G, Torino A, D'Urso M, Ciccodicola A, D'Esposito M. Human and mouse SYBL1 gene structure and expression. Gene. 1999;240:233–8. doi: 10.1016/s0378-1119(99)00375-3. [DOI] [PubMed] [Google Scholar]
- 59.Gerst JE. SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell Mol Life Sci. 1999;55:707–34. doi: 10.1007/s000180050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, Hartmann G. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasma-cytoid dendritic cells through TLR7. Nat Med. 2005;11:263–70. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 61.Cuadras MA, Bordier BB, Zambrano JL, Ludert JE, Greenberg HB. Dissecting rotavirus particle-raft interaction with small interfering RNAs: insights into rotavirus transit through the secretory pathway. J Virol. 2006;80:3935–46. doi: 10.1128/JVI.80.8.3935-3946.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jia Q, Sun R. Inhibition of gammaher-pesvirus replication by RNA interference. J Virol. 2003;77:3301–6. doi: 10.1128/JVI.77.5.3301-3306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia Q, Wu TT, Liao HI, Chernishof V, Sun R. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J Virol. 2004;78:6610–20. doi: 10.1128/JVI.78.12.6610-6620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–5. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 65.Zhang W, Yang H, Kong X, Mohapatra S, Juan-Vergara H, Hellermann G, Behera S, Singam R, Lockey RF, Mohapatra SS. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparti-cles targeting the viral NS1 gene. Nat Med. 2005;11:56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
- 66.Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–52. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schweizer A, Fransen JA, Bachi T, Ginsel L, Hauri HP. Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J Cell Biol. 1988;107:1643–53. doi: 10.1083/jcb.107.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–60. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 69.Martin DF, Kuppermann BD, Wolitz RA, Palestine AG, Li H, Robinson CA. Oral gan-ciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med. 1999;340:1063–70. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 70.Harley CA, Dasgupta A, Wilson DW. Characterization of herpes simplex virus-containing organelles by sub-cellular fractionation: role for organ elle acidification in assembly of infectious particles. J Virol. 2001;75:1236–51. doi: 10.1128/JVI.75.3.1236-1251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hambleton S, Gershon MD, Gershon AA. The role of the trans-Golgi network in varicella zoster virus biology. Cell Mol Life Sci. 2004;61:3047–56. doi: 10.1007/s00018-004-4269-7. [DOI] [PubMed] [Google Scholar]
- 72.Heineman TC, Krudwig N, Hall SL. Cytoplasmic domain signal sequences that mediate transport of varicella-zoster virus gB from the endoplasmic reticulum to the Golgi. J Virol. 2000;74:9421–30. doi: 10.1128/jvi.74.20.9421-9430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mettenleiter TC. Herpesvirus assembly and egress. J Virol. 2002;76:1537–47. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheung P, Banfield BW, Tufaro F. Brefeldin A arrests the maturation and egress of herpes simplex virus particles during infection. J Virol. 1991;65:1893–904. doi: 10.1128/jvi.65.4.1893-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson DC, Spear PG. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–12. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


