Abstract
Rsp5 is a homologous to E6AP C terminus (HECT) ubiquitin ligase (E3) that controls many different cellular processes in budding yeast. Although Rsp5 targets a number of different substrates for ubiquitination, the mechanisms that regulate Rsp5 activity remain poorly understood. Here we demonstrate that Rsp5 carries a noncovalent ubiquitin-binding site in its catalytic HECT domain. The N-terminal lobe of the HECT domain mediates binding to ubiquitin, and point mutations that disrupt interactions with ubiquitin alter the ability of the Rsp5 HECT domain to assemble polyubiquitin chains in vitro. Point mutations that disrupt ubiquitin binding also result in temperature-sensitive growth defects in yeast, indicating that the Rsp5 ubiquitin-binding site is important for Rsp5 function in vivo. The Nedd4 HECT domain N-lobe also contains ubiquitin-binding activity, suggesting that interactions between the N-lobe and ubiquitin are conserved within the Nedd4 family of ubiquitin ligases. We propose that a subset of HECT E3s are regulated by a conserved ubiquitin-binding site that functions to restrict the length of polyubiquitin chains synthesized by the HECT domain.
Modification of proteins with ubiquitin plays an important role in controlling and modulating a number of cellular processes. Ubiquitination is the primary signal used to target cellular proteins for degradation by 26 S proteasomes (1) and it is an important nonproteolytic signal in many other biological pathways (2–4). The ability of ubiquitin to function in a variety of cellular processes can be explained by the existence of structurally distinct ubiquitin modifications and the recognition of ubiquitination signals by a diverse set of ubiquitin-binding domains (UBDs)3 found within a host of cellular proteins (5, 6).
Ubiquitin is conjugated to proteins by a series of enzymes that act in a well defined, sequential manner (7). The final transfer of ubiquitin to a cellular protein is usually carried out by an E3 ubiquitin ligase. E3s contain the primary determinants for substrate recognition and generally belong to one of two families. Really interesting new gene (RING) E3s are thought to act primarily as molecular scaffolds to bring together a ubiquitin-conjugating enzyme (E2) charged with ubiquitin and a substrate targeted for ubiquitination (8, 9). In contrast, HECT E3s form a covalent thioester intermediate with ubiquitin before transferring ubiquitin to the substrate. A conserved cysteine residue located within the active site of the HECT domain is required for formation of this intermediate (10, 11).
Different types of ubiquitin modifications adopt distinct structural conformations, and these structural differences are functionally important because they target proteins for different fates in the cell. Protein monoubiquitination is an important regulatory signal in a variety of basic cellular processes, including endocytosis, histone remodeling, and viral budding (12). Polyubiquitin chains act as signals specialized for other cellular functions. For example, chains linked through Lys-48 of ubiquitin play a well characterized role in targeting proteins for degradation by the proteasome, whereas chains linked through Lys-63 of ubiquitin act as nonproteolytic signals in DNA repair, kinase activation, and endocytosis (13). Despite the known importance of diverse ubiquitination signals, little is known about the mechanisms used by E3 enzymes to ensure that substrates targeted for ubiquitination are modified with the appropriate type of signal.
Rps5 is a HECT E3 that controls a broad range of cellular processes in Saccharomyces cerevisiae and is part of a large family of proteins that control analogous processes in mammalian cells (14). For example, Rsp5 and its well characterized mammalian homologue, Nedd4, are both required for efficient internalization of cell surface receptors from the plasma membrane (3, 15). Both E3s also play a role in regulating the stability of RNA polymerase II by targeting the polymerase for ubiquitination and degradation (16–18). In yeast, Rsp5 has been implicated in many other cellular processes, including the nuclear export of mRNA, mitochondrial inheritance, and the activation of endoplasmic reticulum-bound transcription factors (19–22).
The ability of Rsp5 to act as a multifunctional E3 in yeast is due, at least in part, to its capacity to modify different substrates with distinct mono- and polyubiquitin signals. Several proteins that function in endocytosis are monoubiquitinated in an Rsp5-dependent manner (23–25). In contrast, Rsp5 targets a number of cellular proteins for polyubiquitination, including the large subunit of RNA polymerase II, the vacuolar membrane protein Sna3, and the mRNA nuclear export factor Hpr1 (18, 20, 26, 27). Although Rsp5 possesses an intrinsic preference for Lys-63-linked chain synthesis in vitro (28) and modifies a number of its substrates with Lys-63-linked chains in vivo (29–31), several lines of evidence suggest that the enzyme can assemble chains linked through Lys-48 as well. In general, however, the mechanisms that control the linkage and length of polyubiquitin chains synthesized by Rsp5 remain poorly defined. Here we identify a previously unknown noncovalent ubiquitin-binding site located in the Rsp5 HECT domain that regulates the length of Lys-63- and Lys-48-linked polyubiquitin chains assembled by the Rsp5 HECT domain.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Mutagenesis—Plasmids encoding fragments of Rps5 fused to glutathione S-transferase (GST) were constructed in pGEX vectors (GE Healthcare). The GST (LHP497), GST-3xWW (aa 228–430, LHP703), and GST-HECT C-terminal lobe (C-lobe) (aa 691–809, LHP2468) plasmids were constructed by PCR amplifying the relevant DNA sequence and ligating into the pGEX-6P-2 vector. The GST-HECT domain plasmid (aa 425–809, LHP1434) was generated in a similar fashion but with ligation into the pGEX-4T-3 vector. A plasmid encoding the GST-HECT N-terminal lobe (N-lobe) (aa 425–691, LHP2325) was created by introducing a STOP codon after amino acid 691 in LHP1434 using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). The GST-C2 domain plasmid was obtained from Hilary Godwin (University of California, Los Angeles, Los Angeles, CA). All point mutations in LHP2325 and LHP1434 were introduced by QuikChange mutagenesis and verified by automated sequencing.
Plasmids encoding hexahistidine (His6)-tagged ubiquitin (LHP1404), Rvs167 SH3 domain (aa 428–482, LHP1496), and the HECT domain N-lobes from Rsp5 (aa 426–691, LHP2381), Nedd4 (aa 501–767, LHP2443), and Tom1 (aa 2880–3148, LHP2442) were generated by ligation-independent cloning of the relevant PCR-amplified fragment into the pET-30 vector (EMD Chemicals, La Jolla, CA). All point mutations in LHP1404 were introduced by QuikChange mutagenesis. A yeast expression vector carrying untagged wild-type RSP5 under the control of its endogenous promoter was provided by Jon Huibregtse (University of Texas, Austin, TX) and modified to remove a NotI site in the multiple cloning region, generating LHP472. QuikChange mutagenesis was used to introduce the Y516A (LHP2735) and F618A (LHP2737) mutations into this plasmid. Construction of the hemagglutinin (HA)-tagged Rsp5 plasmid (LHP478) has been described previously (32). Multicopy plasmids encoding wild-type (LHP308) or lysine-less (0K) (LHP306) ubiquitin were adapted from plasmids pES7 and pTER62 obtained from Mike Ellison (University of Alberta, Edmonton, Canada) by removing the c-myc epitope tag.
Yeast Strains and Growth Media—All yeast strains were propagated in synthetic minimal medium (yeast nitrogen base; U. S. Biological, Swampscott, MA) or rich yeast extract peptone dextrose medium (2% bactopeptone, 1% yeast extract, 2% glucose supplemented with 10 mg/liter adenine, uracil, and tryptophan). Yeast transformations were performed using standard techniques (33). Strains carrying RSP5 (LHY5653), rsp5Y516A (LHY5655), or rsp5F618A (LHY5657) as the sole source of Rsp5 were constructed by transforming the wild-type or mutant plasmids into LHY4507 (rsp5Δ pGFP-Rsp5[URA3]) and selecting for loss of pGFP-RSP5[URA3] on media containing 5-fluoroorotic acid as described previously (32).
Ubiquitin and E2-binding Assays—Binding assays carried out with ubiquitin-agarose beads (Boston Biochem, Cambridge, MA) and lysates from yeast cells expressing HA-tagged Rsp5 were performed as previously described (34), except that 7.5 mg of lysate protein was incubated with the beads for 1 h at 4 °C. Recombinant proteins for all other binding experiments were expressed in Escherichia coli (BL21 CodonPlus cells, Stratagene) by inducing cultures with 1 mm isopropyl β-d-thiogalactopyranoside for 3–5 h at 18–37 °C. Preparation of bacterial cell lysates has been described previously (34). Immobilization of His6-tagged proteins on TALON metal affinity resin (Clontech) and GST-tagged proteins on glutathione resin (GE Healthcare) was performed according to the manufacturer's instructions.
Binding of bacterially expressed GST-tagged Rsp5 fragments to ubiquitin-agarose or agarose beads was performed by incubating the lysates and beads for 1 h at 4 °C. The beads were then washed four times in phosphate-buffered saline lysis buffer (115 mm NaCl, 16 mm Na2HPO4, 4 mm KH2PO4, 1% Triton X-100, 5% glycerol, pH 7.3), and bound proteins were eluted by boiling in 1× Laemmli sample buffer (125 mm Tris-HCl, pH 6.8, 4% SDS, 500 mm 2-mercaptoethanol, 20% glycerol, 0.02% bromphenol blue). Binding of bacterially expressed GST-tagged HECT fragments or purified N-lobes to immobilized His6-ubiquitin or His6-Rvs167SH3 was carried out in the same manner, except that the beads were washed twice in phosphate-buffered saline lysis buffer containing 10 mm imidazole and twice in phosphate-buffered saline lysis buffer containing 20 mm imidazole. Bound proteins were resolved by SDS-PAGE and analyzed by either immunoblotting with anti-GST antiserum (GE Healthcare) or staining with Coomassie Brilliant Blue G-250 (Bio-Rad Laboratories).
Binding to polyubiquitin chains was assayed by incubating 10 μg of purified Lys-63- or Lys-48-linked chains (Boston Biochem) with immobilized GST, GST-HECT, or GST-N-lobe proteins for 1 h at 4 °C. The beads were washed four times in phosphate-buffered saline lysis buffer, and bound chains were eluted by boiling in Tris-Tricine sample loading buffer (10 mm Tris-HCl, pH 6.8, 3% SDS, 500 mm 2-mercaptoethanol, 2 mm EDTA, 25% glycerol, 0.02% bromphenol blue). Bound chains were resolved by Tris-Tricine gel electrophoresis and analyzed by anti-His immunoblotting (Bethyl Laboratories, Montgomery, TX). UbcH5a binding assays were carried out in the same manner, except that the beads carrying GST-HECT domain mutants were incubated with 25 nm to 0.1 μm purified UbcH5a (Boston Biochem) and bound proteins were analyzed by anti-UbcH5a immunoblotting (Boston Biochem).
Analysis of Protein Expression Levels in Yeast Lysate—Protein expression levels of Rsp5 and free ubiquitin were analyzed as follows: 2 × 108 cells from each strain were harvested before and after shifting cultures to 37 °C for 1 h. Cells were lysed under denaturing conditions in 1 ml of lysis buffer (10 mm Tris-HCl, pH 7.0, 1 mm EDTA, 0.2 N NaOH, 0.5% 2-mercaptoethanol) and then precipitated with 10% trichlororoacetic acid. Protein precipitates were collected by centrifugation, resuspended in 1× Laemmli sample buffer, and analyzed by SDS-PAGE and immunoblotting with Rsp5 antiserum (25) or ubiquitin antiserum (Millipore, Temecula, CA).
In Vitro Ubiquitination Assays—Yeast E1, UbcH5a, ubiquitin, and all ubiquitin mutants were purchased from Boston Biochem. All GST-HECT proteins were expressed in E. coli (BL21-CodonPlus cells), purified on glutathione resin according to the manufacturer's instructions, and eluted from the resin in 10 mm Tris-HCl, pH 8.0, buffer containing 10 mm glutathione. Recovered proteins were assayed for purity by SDS-PAGE and Coomassie staining, and protein concentrations were determined using the Bradford assay. HECT domain autoubiquitination assays were carried out with 0.1 μm yeast E1, 0.2 μm UbcH5a, 0.3 μm GST-HECT, and 75 μm of the indicated ubiquitin. Reactions were initiated by adding buffer containing ATP (final concentrations: 25 mm Tris, pH 7.5, 50 mm NaCl, 4 mm MgCl2, 0.1 μm dithiothreitol, 4 mm ATP). After brief mixing, the zero time point was withdrawn and placed on ice, and reactions were immediately transferred to a 30 °C water bath. Reaction aliquots were removed at the indicated times, added to an equal volume of 2× Laemmli sample buffer, and analyzed by SDS-PAGE and anti-GST immunoblotting.
RESULTS
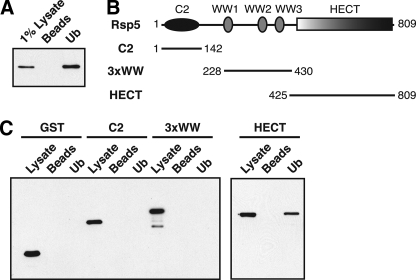
The Rsp5 HECT Domain Binds to Ubiquitin—To identify cellular proteins that bind to ubiquitin, a yeast genomic two-hybrid library was screened using ubiquitin as the bait. A fragment of Rsp5 encoding residues 195–809 was identified in this screen.4 To verify that the full-length Rsp5 protein binds to ubiquitin, we tested the ability of an HA-tagged version of Rsp5 expressed in yeast to bind to ubiquitin-agarose beads. HA-Rsp5 bound to ubiquitin-agarose beads but not to control agarose beads (Fig. 1A), confirming that full-length Rsp5 is a ubiquitin-binding protein.
FIGURE 1.
The Rsp5 HECT domain binds directly to ubiquitin. A, a lysate prepared from yeast cells expressing HA-tagged Rps5 (LHY856) was incubated with ubiquitin-agarose beads (Ub) or agarose beads alone (Beads). Bound proteins were eluted, and Rsp5 was detected on an anti-HA immunoblot. B, schematic representation of Rsp5 indicating the position of its functional domains. Fragments tested for ubiquitin binding in C are shown. C, bacterial lysates from cells expressing the indicated GST-tagged Rsp5 domains were incubated with ubiquitin-agarose beads or agarose beads alone. Total lysates and bound proteins were analyzed by anti-GST immunoblotting.
Rsp5 is part of a large family of proteins found throughout eukaryotes that share a common modular domain architecture (14). All family members contain an N-terminal C2 domain, one to four central WW domains, and a large ∼350 amino acid C-terminal HECT domain (Fig. 1B). Rsp5 does not carry any of the numerous UBDs that have been described to date (6, 35–37). To test which region of the Rsp5 protein is responsible for its ubiquitin-binding activity, we assayed different fragments of Rsp5 for binding to ubiquitin-agarose beads. A GST-HECT domain fusion protein expressed in E. coli bound specifically to the ubiquitin-agarose beads in this assay, whereas fragments of Rsp5 containing the C2 or three WW domains did not (Fig. 1C). These observations indicate that the Rsp5 HECT domain binds directly to monoubiquitin.
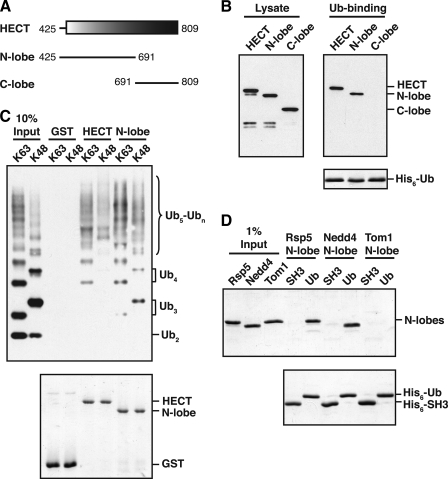
The Rsp5 and Nedd4 HECT Domain N-lobes Bind to Ubiquitin—The HECT domain is highly structured, with an elongated, α-helical N-terminal lobe (N-lobe) and a smaller, globular C-terminal lobe (C-lobe) (38–40). The C-lobe contains the conserved cysteine residue that forms a thioester with ubiquitin, whereas the N-lobe interacts with an E2 enzyme to enable the transfer of ubiquitin from the E2 to the HECT active site cysteine. To determine which lobe of the Rsp5 HECT domain is responsible for ubiquitin binding, we expressed the isolated N- and C-lobes (Fig. 2A) in bacteria and tested them for interaction with immobilized His6-tagged ubiquitin. Ubiquitin bound equally well to a GST-N-lobe fusion protein and a GST-HECT domain fusion, but did not bind to the isolated GST-C-lobe fusion (Fig. 2B). The HECT domain N-lobe also bound to polyubiquitin chains linked through either Lys-63 or Lys-48 of ubiquitin, with a preference for binding to longer chains (Ub4-Ubn) over shorter di- and triubiquitin chains (Fig. 2C). From these experiments, we conclude that all Rsp5 ubiquitin-binding activity resides in the N-lobe of its HECT domain.
FIGURE 2.
The Rsp5 and Nedd4 HECT domain N-lobes bind to ubiquitin. A, schematic representation of the Rsp5 HECT domain. Fragments tested for ubiquitin binding in B are shown. B, bacterial lysates from cells expressing the indicated GST-tagged HECT domain fragments were incubated with beads carrying immobilized His6-tagged ubiquitin (His6-Ub). Lysates and proteins eluted from the beads were analyzed by anti-GST immunoblotting (top panels) or Coomassie staining (bottom panel). C, GST-HECT domain and GST-HECT N-lobe fusions were immobilized on beads, and the beads were incubated with purified His6-tagged Lys-63- or Lys-48-linked polyubiquitin chains. Purified chains (10% Input) and proteins eluted from the beads were analyzed by anti-His immunoblotting (top panel) or Coomassie staining (bottom panel). D, the indicated N-lobes were purified from an E. coli lysate and incubated with equivalent amounts of immobilized His6-Ub or a control His6-tagged SH3 domain from Rvs167 (His6-SH3). Purified N-lobes (1% Input) and proteins eluted from Ub or SH3 beads were detected by Coomassie staining.
To test if the ability of the N-lobe to bind ubiquitin is a general property of HECT domains, we assayed the N-lobes from the mammalian Nedd4 and yeast Tom1 HECT domains for ubiquitin binding. Nedd4 is both structurally and functionally related to Rsp5, and its HECT domain N-lobe is 55% identical to the Rsp5 N-lobe in sequence. In contrast, Tom1 is unrelated to Rsp5, and its HECT domain N-lobe is only 40% identical to the Rsp5 N-lobe. In a ubiquitin-binding assay carried out with both N-lobes, the Nedd4 N-lobe bound specifically to immobilized His6-tagged ubiquitin, whereas the Tom1 N-lobe did not (Fig. 2D). These observations indicate that the N-lobe ubiquitin-binding site is conserved in the Nedd4 HECT domain, but is not a feature common to all HECT domains.
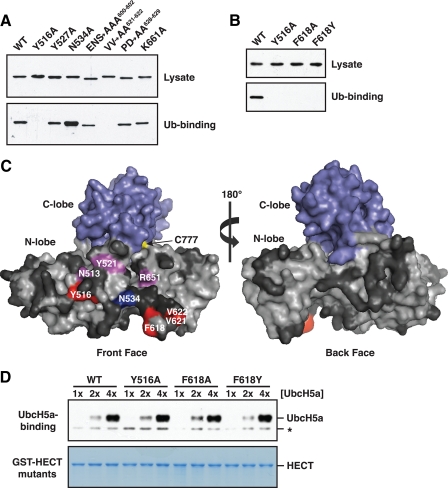
Interaction Surfaces on the Rsp5 HECT Domain N-lobe and Ubiquitin—To map the ubiquitin-binding surface on the Rsp5 HECT domain N-lobe, we used an alanine-scanning mutagenesis approach. Although the three-dimensional structure of the Rsp5 HECT domain has not been determined, modeling of the Rsp5 HECT domain onto the known structure of the closely related WWP1 HECT domain (40) allowed us to target surface-exposed residues on the Rsp5 N-lobe for mutagenesis. Individual residues and stretches of up to three contiguous residues were mutated to alanine, and the resulting GST-tagged N-lobe mutants were tested for binding to immobilized His6-tagged ubiquitin. Most of the ∼50 mutations we made had no effect on binding to ubiquitin. Three N-lobe mutations (Y516A, F618A, and V621A/V622A) completely abolished ubiquitin binding. An additional three mutations (N513A, Y521A, and R651A) caused a reduction in binding. Finally, one mutation (N534A) reproducibly enhanced binding ∼2–3-fold relative to wild-type (Fig. 3A, data not shown).
FIGURE 3.
Ubiquitin binds to a region on the front surface of the Rsp5 HECT domain N-lobe. A, a representative experiment from the alanine-scanning mutagenesis of residues in the Rsp5 HECT domain N-lobe. Bacterial lysates from cells expressing the indicated GST-tagged N-lobe mutants were incubated with beads carrying immobilized His6-tagged ubiquitin. Lysates and proteins bound to ubiquitin were analyzed by anti-GST immunoblotting. Mutation of the acidic residues in the E600A/N601A/S602A and P628A/D629A mutants resulted in slightly altered electrophoretic mobility. B, bacterial lysates from cells expressing the indicated GST-tagged HECT domain mutants were incubated with immobilized His6-tagged ubiquitin. Lysates and proteins bound to ubiquitin were analyzed by anti-GST immunoblotting. C, surface representation of the Rsp5 HECT domain, created by modeling onto the WWP1 HECT domain crystal structure (Protein Data Bank accession code 1ND7). Results of the alanine mutagenesis are summarized as follows: red, mutation abolished binding; magenta, mutation reduced binding; blue, mutation enhanced binding; dark gray, mutation had no effect. D, the indicated GST-HECT domain mutants were immobilized on beads, and the beads were incubated with increasing concentrations of purified UbcH5a: 25 (1×), 50 (2×), and 100 nm (4×). Proteins eluted from the beads were analyzed by anti-UbcH5a immunoblotting (top panel) or Coomassie staining (bottom panel). A nonspecific band unrelated to UbcH5a is represented by an asterisk.
To test if amino acids required for ubiquitin binding in the Rsp5 N-lobe are also important in the context of the entire HECT domain, we assayed GST-HECT domain mutants carrying the Y516A or F618A mutations for binding to immobilized His6-tagged ubiquitin. As in the N-lobe, these mutations abolished binding of the entire HECT domain to ubiquitin (Fig. 3B). We also constructed an F618Y HECT domain mutant for analysis because this residue is a Phe in the ubiquitin-binding Rsp5 and Nedd4 N-lobes, but is a Tyr in the non-binding Tom1 N-lobe. The F618Y mutation eliminated binding of the Rsp5 HECT domain to ubiquitin (Fig. 3B), indicating that this residue is a crucial component of the HECT domain ubiquitin-binding site.
Mapping of the results from the alanine mutagenesis onto the modeled three-dimensional structure of the Rsp5 HECT domain showed that ubiquitin binds to a region on the front surface of the N-lobe that lies ∼15–20 Å from the HECT domain active site cysteine. Importantly, none of the mutations made on the back face of the N-lobe had any effect on binding to ubiquitin, confirming the specificity of the alanine-scanning mutagenesis (Fig. 3C). Further analysis of the predicted ubiquitin-binding site revealed that ubiquitin binds to a region that is adjacent to the putative E2-binding site (38). To address the possibility that mutations that inactive the ubiquitin-binding site also affect interactions with an E2, we tested GST-HECT domain mutants carrying the Y516A, F618A, and F618Y mutations for their ability to interact with UbcH5a, a member of the highly homologous Ubc1/Ubc4/Ubc5 subfamily of E2s known to cooperate with Rsp5 in vitro (41). None of the mutations tested had any effect on the ability of the HECT domain to bind to UbcH5a over a series of different E2 concentrations (Fig. 3D), indicating that the Y516A, F618A, and F618Y mutations specifically impair binding to ubiquitin.
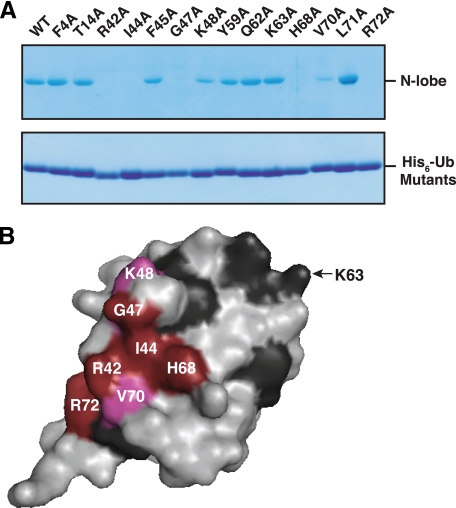
Most of the previously characterized UBDs bind to a hydrophobic surface patch on ubiquitin surrounding a key isoleucine residue, Ile-44. To test if the Rsp5 HECT domain N-lobe also uses this surface of ubiquitin for binding, we mutated a select number of ubiquitin surface residues to alanine and tested these mutants for their ability to bind to the N-lobe. Mutation of Ile-44 and its neighboring residues, Gly-47, His-68, Arg-42, and Arg-72, abolished binding to the N-lobe, and mutation of two additional residues, Lys-48 and Val-70, caused a reduction in binding. In contrast, mutations in and around K63 of ubiquitin (Y59A, Q62A, and K63A) and in residues encompassing another functionally important surface of ubiquitin (F4A, T14A) (42) had no effect on binding (Fig. 4, A and B). Thus, like most other UBDs characterized to date, the Rsp5 HECT domain N-lobe binds to the Ile-44 hydrophobic patch of ubiquitin.
FIGURE 4.
The Ile-44 hydrophobic patch of ubiquitin is required for binding. A, the indicated His6-tagged ubiquitin mutants were immobilized on beads and incubated with a bacterial lysate from cells expressing a wild-type GST-tagged N-lobe. Bound proteins and ubiquitin mutants eluted from the beads were detected by Coomassie staining. B, surface representation of ubiquitin based on its three-dimensional structure (Protein Data Bank accession code 1UBQ). Results from the alanine mutagenesis are summarized as follows: red, mutation abolished binding; magenta, mutation reduced binding; dark gray, mutation had no effect. The position of Lys-63 is shown for reference. No mutations were made on the back face of ubiquitin (not shown).
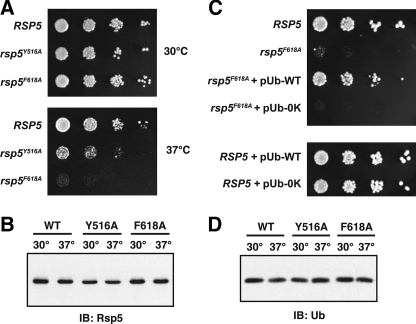
The Rsp5 Ubiquitin-binding Site Is Important for Rsp5 Activity in Vivo—Rsp5 controls a number of cellular processes in yeast, and many, if not all, of these processes are dependent on the catalytic activity of the ligase. To test if mutations that disrupt the Rsp5 ubiquitin-binding site affect Rsp5 activity in yeast, we constructed yeast strains expressing the Rsp5Y516A or Rsp5F618A mutants as the only source of Rsp5 in the cell. Both of these strains grew normally at 30 °C, but exhibited modest to severe temperature-sensitive growth defects at 37 °C (Fig. 5A). The observed growth defects were not due to altered expression or stability of the Rsp5 proteins because both mutants were expressed and stable, even at the elevated temperature (Fig. 5B). We conclude that the Rsp5 ubiquitin-binding site is important for Rsp5 function in yeast cells.
FIGURE 5.
Phenotypic analysis of the rsp5Y516A and rsp5F618A mutants. A, RSP5 (LHY5653), rsp5Y516A (LHY5655), and rsp5F618A (LHY5657) cells were serially diluted, plated onto rich media and grown at 30 or 37 °C for 2 days. B, yeast strains described in A were grown to mid-log phase at 30 °C and then shifted to 37 °C for 1 h. Cells were harvested before and after the temperature shift. Cell lysates were prepared and analyzed for Rsp5 expression by immunoblotting with Rsp5 antiserum. C, RSP5 and rsp5F618A yeast strains used in A were transformed with multicopy plasmids encoding either wild-type (WT) ubiquitin (pUb-WT) or 0K ubiquitin (pUb-0K). Serial dilutions of each strain were plated onto rich media and grown at 37 °C for 2 days. D, yeast strains tested in A were treated as described in B, except that cell lysates were analyzed for free ubiquitin levels by antiubiquitin immunoblotting.
To test if the growth defects of the rsp5Y516A and rsp5F618A mutants could be suppressed by overexpression of ubiquitin, we expressed ubiquitin from a multicopy plasmid in each of the mutant backgrounds and assayed growth at 37 °C. Strikingly, overexpression of ubiquitin almost completely rescued the temperature-sensitive growth defect of the rsp5F618A mutant (Fig. 5C); a similar result was obtained with the rsp5Y516A mutant (data not shown). To test whether these observations are simply due to a reduced ability of the rsp5F618A and rsp5Y516A mutants to synthesize free ubiquitin at the elevated temperature, as has been observed for the rsp5-1 mutant (43), we analyzed free ubiquitin levels in the rsp5F618A and rsp5Y516A mutants. Both of these mutants expressed ubiquitin at levels comparable to wild-type cells, even after shifting the cells to 37 °C (Fig. 5D). Thus, the rsp5F618A and rsp5Y516A growth defects cannot simply be due to limiting free ubiquitin. Instead, these observations suggest that the rsp5F618A and rsp5Y516A phenotypes are due to an effect on Rsp5 catalytic activity that alters the ubiquitination of one or more substrates required for growth at 37 °C.
To determine whether the ability of ubiquitin to form chains is required for the rescue of the rsp5F618A temperature-sensitive growth defect, we overexpressed lysine-less ubiquitin, in which all seven lysines have been mutated to arginine, in rsp5F618A cells. Overexpression of 0K ubiquitin did not rescue the growth defect of rsp5F618A cells (Fig. 5C), indicating that the ability of ubiquitin to form chains is required for this effect. This observation strongly suggests that the rsp5F618A growth phenotype is due to a specific effect on Rsp5-catalyzed polyubiquitination of one or more cellular substrates required for growth at 37 °C.
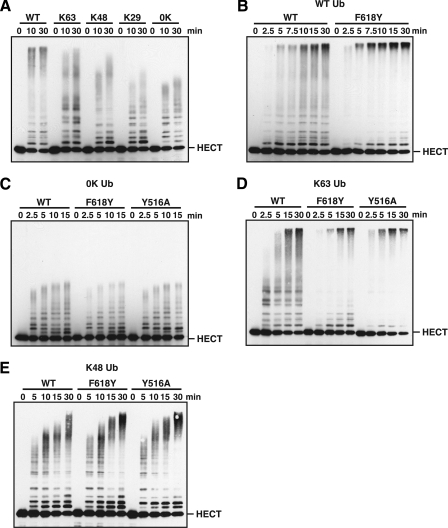
The Rsp5 Ubiquitin-binding Site Regulates the Length of Polyubiquitin Chains Assembled by the HECT Domain—To directly test the role of the Rsp5 ubiquitin-binding site in Rsp5-catalyzed ubiquitination, we used an in vitro autoubiquitination assay with the purified Rsp5 HECT domain. We first evaluated the types of polyubiquitin chains assembled by the Rsp5 HECT domain by performing ubiquitination assays with a series of ubiquitin mutants carrying a single lysine residue (K63-, K48-, or K29-ubiquitin). The Rsp5 HECT domain preferentially assembled chains linked through Lys-63 of ubiquitin, consistent with previous observations for full-length Rsp5 (28). The synthesis of Lys-48-linked chains was noticeably less efficient, and there was little to no chain synthesis activity through Lys-29 because the conjugation pattern observed with K29-ubiquitin was similar to the conjugation pattern observed with 0K ubiquitin (Fig. 6A).
FIGURE 6.
The Rsp5 ubiquitin-binding site regulates the length of polyubiquitin chains assembled by the HECT domain. A, in vitro autoubiquitination assays were carried out with a wild-type GST-HECT fusion protein and wild-type ubiquitin (WT), lysine-less ubiquitin (0K), or one of the indicated single lysine ubiquitins (K63, K48, or K29). Reactions were quenched at the indicated times and ubiquitin conjugates were detected by anti-GST immunoblotting. The position of the unmodified HECT domain is indicated. B–E, reactions were carried out as described in A but in the presence of either a wild-type or mutant GST-HECT fusion protein (WT, F618Y, or Y516A) and the indicated ubiquitin: WT ubiquitin for B, 0K ubiquitin for C, K63-ubiquitin for D, and K48-ubiquitin for E.
We next analyzed the effect of mutations that disrupt the Rsp5 ubiquitin-binding site on HECT domain autoubiquitination. In an assay carried out with the F618Y HECT domain and wild-type ubiquitin, the pattern of conjugates observed with the F618Y mutant was markedly different from the pattern observed with the wild-type HECT domain. Specifically, there was a strong accumulation of high molecular weight polyubiquitin conjugates at the top of the gel and a concomitant decrease in low molecular weight conjugates in the lower to middle region of the gel (Fig. 6B). A similar effect was observed with the Y516A and F618A HECT domain mutants (data not shown). These observations indicate that HECT domains carrying mutations in the ubiquitin-binding site are enzymatically active and suggest that these mutations alter the length of polyubiquitin chains assembled by the Rsp5 HECT domain.
To test if mutations that inactivate the Rsp5 ubiquitin-binding site affect the ability of the HECT domain to catalyze monoubiquitination, we assayed the F618Y and Y516A HECT domain mutants for autoubiquitination in the presence of 0K ubiquitin. The pattern of 0K ubiquitin conjugates observed with both of these mutants was virtually indistinguishable from the pattern observed with the wild-type HECT domain (Fig. 6C). We conclude that the F618Y and Y516A mutations have no effect on the ability of the HECT domain to accept monoubiquitin from an E2 enzyme or to transfer monoubiquitin to a lysine residue targeted for ubiquitination. Together, the results presented in Fig. 6, B and C, indicate that the Rsp5 ubiquitin-binding site plays a specific role in regulating the assembly of a polyubiquitin chain.
To determine whether the Rsp5 ubiquitin-binding site plays a role in regulating the assembly of a specific type of polyubiquitin chain, we next assayed the F618Y and Y516A HECT domain mutants in autoubiquitination assays in the presence of either K63- or K48-ubiquitin. Both the F618Y and Y516A mutations significantly altered the distribution of Lys-63-linked conjugates in a manner similar to that observed in reactions carried out with wild-type ubiquitin (Fig. 6D). In contrast, there was a more modest effect on the distribution of Lys-48-linked conjugates, with the most significant differences in conjugation patterns appearing at the 15- and 30-min time points (Fig. 6E). We conclude that the F618Y and Y516A mutations alter the distribution of both Lys-63-linked and, to a lesser extent, Lys-48-linked polyubiquitin chains. The accumulation of high molecular weight polyubiquitinated species observed with the F618Y and Y516A HECT domains suggests a role for the Rsp5 ubiquitin-binding site in limiting the length of Lys-63- and Lys-48-linked polyubiquitin chains.
DISCUSSION
UBDs are often found in proteins that recognize ubiquitinated targets, where they interpret the information carried by ubiquitin signals to regulate downstream events. UBDs are also found in enzymes that catalyze the attachment or removal of ubiquitin to other proteins, where they presumably aid in the catalytic steps required for ubiquitination or deubiquitination (5, 6). Here we identify a previously unknown UBD in the HECT domain of Rsp5 and demonstrate that interactions with ubiquitin play a critical role in the regulation of Rsp5 activity in vitro and in vivo.
Ubiquitin binds to a region on the front surface of the Rsp5 HECT domain N-lobe that lies ∼15–20 Å from the conserved active site cysteine residue in the modeled structure. The site of interaction is adjacent to the putative E2-binding site (38), however, the results of our studies suggest that the ubiquitin and E2 binding sites do not overlap because mutations that severely inhibited ubiquitin binding had no effect on E2 binding. The binding site on ubiquitin is centered around the Ile-44 hydrophobic patch, the site of interaction for almost every UBD characterized to date. Mutations in and around Lys-63 of ubiquitin had no effect on binding, whereas a subset of mutations in and around Lys-48 disrupted binding. These observations are consistent with a model in which the N-lobe binds to ubiquitin in an orientation that favors polyubiquitin chain linkage through Lys-63. This model is also supported by the finding that mutations that inactivate the ubiquitin-binding site have a more pronounced effect on the distribution of Lys-63-linked chains than they do on the distribution of Lys-48-linked chains.
Although the chain synthesis activities of several E2 enzymes, including E2-25K, Ubc1, and the Mms2/Ubc13 complex, are known to be regulated by UBDs (44–48), it has been hypothesized that HECT E3s might also use a noncovalent ubiquitin-binding site to aid in polyubiquitin chain synthesis. The existence of a ubiquitin-binding site within the KIAA10 HECT domain has been inferred from mechanistic studies (49, 50), but direct interactions with ubiquitin have not been confirmed experimentally. Furthermore, the Rsp5 and KIAA10 HECT domains probably carry distinct UBDs, because a 60-amino acid sequence upstream of the KIAA10 HECT domain is required for presumed interaction with ubiquitin (50, 51). Here we demonstrate that the Rsp5 and Nedd4 HECT domain N-lobes carry a ubiquitin-binding site, but the N-lobe of the more distantly related Tom1 HECT domain does not. Thus, the N-lobe UBD is likely to be a specific feature of a subset of HECT E3s within the Nedd4/Rsp5 family of ubiquitin ligases.
Mechanistic Role of the Rsp5 Ubiquitin-binding Site in Polyubiquitin Chain Assembly—The results presented here demonstrate that the Rsp5 ubiquitin-binding site plays a specific role in regulating the assembly of a polyubiquitin chain. Mutations that disrupt the Rsp5 ubiquitin-binding site alter the ability of the HECT domain to assemble Lys-63-linked and Lys-48-linked polyubiquitin chains. These same mutations have no effect on the conjugation of lysine-less ubiquitin, indicating that the Rsp5 ubiquitin-binding site does not influence the ability of the HECT domain to transfer monoubiquitin to lysine residues targeted for ubiquitination. Consistent with the idea that the Rsp5 ubiquitin-binding site is specifically important for polyubiquitination, the temperature-sensitive growth defects of rsp5F618A and rsp5Y516A cells could be rescued by overexpression of wild-type but not lysine-less ubiquitin. Thus, the rsp5F618A and rsp5Y516A in vivo phenotypes are likely due to an effect on Rsp5 catalytic activity that alters the polyubiquitination of one or more cellular substrates important for growth at the restrictive temperature. We speculate that overexpression of ubiquitin rescues the rsp5F618A and rsp5Y516A growth phenotypes by restoring the normal distribution of polyubiquitin conjugates attached to one or several key physiological substrates of Rsp5.
HECT E3s can use at least two distinct mechanisms of polyubiquitin chain synthesis (50). The KIAA10 HECT domain builds up chains by catalyzing the sequential addition of ubiquitin monomers onto the end of a free or substrate-anchored polyubiquitin chain. The key feature of this model is the existence of a putative noncovalent ubiquitin-binding site in KIAA10, which nucleates the formation of chains by positioning a lysine residue within the bound “acceptor” ubiquitin in an orientation that facilitates attack on the HECT-ubiquitin thioester. In contrast, the E6AP HECT domain builds up chains on its active site cysteine prior to transferring the chain to a substrate. This mode of chain assembly requires an E3-E2 heterodimer and involves an attack by the HECT thioester-linked ubiquitin on the E2-ubiquitin thioester. Although the mechanism of polyubiquitin chain synthesis employed by the Rsp5 HECT domain is currently unknown, both the Rsp5 HECT domain and full-length Rsp5 assemble free ubiquitin chains inefficiently in vitro.4 This is diagnostic of an E6AP-like mode of chain synthesis, suggesting that Rsp5 assembles chains on its active site cysteine. However, the presence of a noncovalent ubiquitin-binding site within the Rsp5 HECT domain suggests that a KIAA10-like mode of chain synthesis might also be possible. Further work is needed to determine whether Rsp5 uses one or both modes of chain assembly and to determine the predominant mechanism of chain synthesis used on Rsp5 substrates.
The results presented here are consistent with a model in which the Rsp5 ubiquitin-binding site restricts the length of polyubiquitin chains assembled by the Rsp5 HECT domain. The basis for this model is the observation that mutations that inactivate the Rsp5 ubiquitin-binding site result in the increased synthesis of high molecular weight Lys-63- and Lys-48-linked polyubiquitinated species. We cannot formally exclude the possibility that these high molecular weight conjugates represent an accumulation of many short polyubiquitin chains attached to multiple sites of ubiquitination. However, the observation that HECT domain ubiquitin-binding mutants transfer lysine-less ubiquitin to the same number of ubiquitination sites as the wild-type HECT domain argues against this possibility. A role for the Rsp5 ubiquitin-binding site in limiting chain length is also supported by the finding that the Rsp5 HECT domain N-lobe binds preferentially to longer polyubiquitin chains (Ub4-Ubn) over shorter di- and triubiquitin chains. Consequently, we propose a model in which the Rsp5 N-lobe binds to the distal ubiquitin on the end of a growing chain to limit chain elongation. A similar model has been proposed to explain the role of the Ubc1 ubiquitin-associated domain in restricting polyubiquitin chain length (44, 46) and to explain the role of the Met-4 ubiquitin-interacting motif in preventing the extension of a polyubiquitin chain on Met-4 (52).
Here we describe a previously unknown noncovalent ubiquitin-binding site located in the Rsp5 HECT domain that plays a role in the regulation of polyubiquitin chain length. Rsp5 is part of a large family of proteins that control diverse cellular processes in both yeast and mammalian cells (14). The existence of a ubiquitin-binding site within the Nedd4 HECT domain suggests that the chain synthesis activities of other family members are likely to be regulated by an analogous ubiquitin-binding site. Thus, these studies reveal a new mode of regulation for HECT E3s within the Nedd4 family of ubiquitin ligases and shed light on the diverse role of UBDs in the dynamic assembly of polyubiquitin chains.
Acknowledgments
We are grateful to Susan Shih and Myra Sutanto for identifying Rsp5 as a ubiquitin-interacting protein in the yeast two-hybrid screen and Rebecca Dunn for plasmid construction. We thank Mike Ellison, Hilary Godwin, and Jon Huibregtse for providing plasmids, Andreas Matouschek and Erik Sontheimer for numerous helpful discussions and critical comments on the manuscript, and members of the Hicke lab for constructive feedback and advice.
This work was supported, in whole or in part, by National Institutes of Health Grants DK53257 and DK61299.
Footnotes
The abbreviations used are: UBDs, ubiquitin-binding domains; E3, ubiquitin ligase; RING, really interesting new gene; E2, ubiquitin-conjugating enzyme; HECT, homologous to E6AP C terminus; GST, glutathione S-transferase; aa, amino acid; C-lobe, C-terminal lobe; N-lobe, N-terminal lobe; HA, hemagglutinin; 0K, lysine-less; E1, ubiquitin-activating enzyme; Ub4-Ubn, polyubiquitin chains longer than four subunits in length; Tricine, N-(tris(hydroxymethyl)methyl)glycine; SH3, Src homology domain 3.
M. Sutanto, S. Shih, and L. Hicke, unpublished data.
References
- 1.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425-479 [DOI] [PubMed] [Google Scholar]
- 2.Chen, Z. J. (2005) Nat. Cell Biol. 7 758-765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hicke, L., and Dunn, R. (2003) Annu. Rev. Cell Dev. Biol. 19 141-172 [DOI] [PubMed] [Google Scholar]
- 4.Huang, T. T., and D'Andrea, A. D. (2006) Nat. Rev. Mol. Cell Biol. 7 323-334 [DOI] [PubMed] [Google Scholar]
- 5.Hicke, L., Schubert, H. L., and Hill, C. P. (2005) Nat. Rev. Mol. Cell Biol. 6 610-621 [DOI] [PubMed] [Google Scholar]
- 6.Hurley, J. H., Lee, S., and Prag, G. (2006) Biochem. J. 399 361-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickart, C. M., and Eddins, M. J. (2004) Biochim. Biophys. Acta 1695 55-72 [DOI] [PubMed] [Google Scholar]
- 8.Zheng, N., Wang, P., Jeffrey, P. D., and Pavletich, N. P. (2000) Cell 102 533-539 [DOI] [PubMed] [Google Scholar]
- 9.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., Conaway, R. C., Conaway, J. W., Harper, J. W., and Pavletich, N. P. (2002) Nature 416 703-709 [DOI] [PubMed] [Google Scholar]
- 10.Huibregtse, J. M., Scheffner, M., Beaudenon, S., and Howley, P. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92 2563-2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheffner, M., Nuber, U., and Huibregtse, J. M. (1995) Nature 373 81-83 [DOI] [PubMed] [Google Scholar]
- 12.Hicke, L. (2001) Nat. Rev. Mol. Cell Biol. 2 195-201 [DOI] [PubMed] [Google Scholar]
- 13.Pickart, C. M., and Fushman, D. (2004) Curr. Opin. Chem. Biol. 8 610-616 [DOI] [PubMed] [Google Scholar]
- 14.Ingham, R. J., Gish, G., and Pawson, T. (2004) Oncogene 23 1972-1984 [DOI] [PubMed] [Google Scholar]
- 15.Staub, O., and Rotin, D. (2006) Physiol. Rev. 86 669-707 [DOI] [PubMed] [Google Scholar]
- 16.Anindya, R., Aygun, O., and Svejstrup, J. Q. (2007) Mol. Cell 28 386-397 [DOI] [PubMed] [Google Scholar]
- 17.Beaudenon, S. L., Huacani, M. R., Wang, G., McDonnell, D. P., and Huibregtse, J. M. (1999) Mol. Cell. Biol. 19 6972-6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huibregtse, J. M., Yang, J. C., and Beaudenon, S. L. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3656-3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisk, H. A., and Yaffe, M. P. (1999) J. Cell Biol. 145 1199-1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwizdek, C., Hobeika, M., Kus, B., Ossareh-Nazari, B., Dargemont, C., and Rodriguez, M. S. (2005) J. Biol. Chem. 280 13401-13405 [DOI] [PubMed] [Google Scholar]
- 21.Hoppe, T., Matuschewski, K., Rape, M., Schlenker, S., Ulrich, H. D., and Jentsch, S. (2000) Cell 102 577-586 [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez, M. S., Gwizdek, C., Haguenauer-Tsapis, R., and Dargemont, C. (2003) Traffic 4 566-575 [DOI] [PubMed] [Google Scholar]
- 23.Rotin, D., Staub, O., and Haguenauer-Tsapis, R. (2000) J. Membr. Biol. 176 1-17 [DOI] [PubMed] [Google Scholar]
- 24.Shih, S. C., Prag, G., Francis, S. A., Sutanto, M. A., Hurley, J. H., and Hicke, L. (2003) EMBO J. 22 1273-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamenova, S. D., Dunn, R., Adler, A. S., and Hicke, L. (2004) J. Biol. Chem. 279 16017-16025 [DOI] [PubMed] [Google Scholar]
- 26.McNatt, M. W., McKittrick, I., West, M., and Odorizzi, G. (2007) Mol. Biol. Cell 18 697-706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oestreich, A. J., Aboian, M., Lee, J., Azmi, I., Payne, J., Issaka, R., Davies, B. A., and Katzmann, D. J. (2007) Mol. Biol. Cell 18 707-720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kee, Y., Lyon, N., and Huibregtse, J. M. (2005) EMBO J. 24 2414-2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galan, J. M., and Haguenauer-Tsapis, R. (1997) EMBO J. 16 5847-5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soetens, O., De Craene, J. O., and Andre, B. (2001) J. Biol. Chem. 276 43949-43957 [DOI] [PubMed] [Google Scholar]
- 31.Stawiecka-Mirota, M., Pokrzywa, W., Morvan, J., Zoladek, T., Haguenauer-Tsapis, R., Urban-Grimal, D., and Morsomme, P. (2007) Traffic 8 1280-1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn, R., and Hicke, L. (2001) Mol. Biol. Cell 12 421-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman, F. (1991) Methods Enzymol. 194 3-21 [DOI] [PubMed] [Google Scholar]
- 34.Shih, S. C., Katzmann, D. J., Schnell, J. D., Sutanto, M., Emr, S. D., and Hicke, L. (2002) Nat. Cell Biol. 4 389-393 [DOI] [PubMed] [Google Scholar]
- 35.Messick, T. E., Russell, N. S., Iwata, A. J., Sarachan, K. L., Shiekhattar, R., Shanks, J. R., Reyes-Turcu, F. E., Wilkinson, K. D., and Marmorstein, R. (2008) J. Biol. Chem. 283 11038-11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamenova, S. D., French, M. E., He, Y., Francis, S. A., Kramer, Z. B., and Hicke, L. (2007) Mol. Cell 25 273-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner, S., Carpentier, I., Rogov, V., Kreike, M., Ikeda, F., Lohr, F., Wu, C. J., Ashwell, J. D., Dotsch, V., Dikic, I., and Beyaert, R. (2008) Oncogene 27 3739-3745 [DOI] [PubMed] [Google Scholar]
- 38.Huang, L., Kinnucan, E., Wang, G., Beaudenon, S., Howley, P. M., Huibregtse, J. M., and Pavletich, N. P. (1999) Science 286 1321-1326 [DOI] [PubMed] [Google Scholar]
- 39.Ogunjimi, A. A., Briant, D. J., Pece-Barbara, N., Le Roy, C., Di Guglielmo, G. M., Kavsak, P., Rasmussen, R. K., Seet, B. T., Sicheri, F., and Wrana, J. L. (2005) Mol. Cell 19 297-308 [DOI] [PubMed] [Google Scholar]
- 40.Verdecia, M. A., Joazeiro, C. A., Wells, N. J., Ferrer, J. L., Bowman, M. E., Hunter, T., and Noel, J. P. (2003) Mol. Cell 11 249-259 [DOI] [PubMed] [Google Scholar]
- 41.Nuber, U., and Scheffner, M. (1999) J. Biol. Chem. 274 7576-7582 [DOI] [PubMed] [Google Scholar]
- 42.Sloper-Mould, K. E., Jemc, J. C., Pickart, C. M., and Hicke, L. (2001) J. Biol. Chem. 276 30483-30489 [DOI] [PubMed] [Google Scholar]
- 43.Krsmanovic, T., and Kolling, R. (2004) FEBS Lett. 577 215-219 [DOI] [PubMed] [Google Scholar]
- 44.Haldeman, M. T., Xia, G., Kasperek, E. M., and Pickart, C. M. (1997) Biochemistry 36 10526-10537 [DOI] [PubMed] [Google Scholar]
- 45.Hodgins, R., Gwozd, C., Arnason, T., Cummings, M., and Ellison, M. J. (1996) J. Biol. Chem. 271 28766-28771 [DOI] [PubMed] [Google Scholar]
- 46.McKenna, S., Spyracopoulos, L., Moraes, T., Pastushok, L., Ptak, C., Xiao, W., and Ellison, M. J. (2001) J. Biol. Chem. 276 40120-40126 [DOI] [PubMed] [Google Scholar]
- 47.Merkley, N., and Shaw, G. S. (2004) J. Biol. Chem. 279 47139-47147 [DOI] [PubMed] [Google Scholar]
- 48.VanDemark, A. P., Hofmann, R. M., Tsui, C., Pickart, C. M., and Wolberger, C. (2001) Cell 105 711-720 [DOI] [PubMed] [Google Scholar]
- 49.Wang, M., Cheng, D., Peng, J., and Pickart, C. M. (2006) EMBO J. 25 1710-1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, M., and Pickart, C. M. (2005) EMBO J. 24 4324-4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You, J., and Pickart, C. M. (2001) J. Biol. Chem. 276 19871-19878 [DOI] [PubMed] [Google Scholar]
- 52.Flick, K., Raasi, S., Zhang, H., Yen, J. L., and Kaiser, P. (2006) Nat. Cell Biol. 8 509-515 [DOI] [PubMed] [Google Scholar]