Abstract
Topoisomerase IIα (topo IIα) is a nuclear enzyme involved in several critical processes, including chromosome replication, segregation and recombination. Previously we have shown that chromosomal protein HMGB1 interacts with topo IIα, and stimulates its catalytic activity. Here we show the effect of HMGB1 on the activity of the human topo IIα gene promoter in different cell lines. We demonstrate that HMGB1, but not a mutant of HMGB1 incapable of DNA bending, up-regulates the activity of the topo IIα promoter in human cells that lack functional retinoblastoma protein pRb. Transient over-expression of pRb in pRb-negative Saos-2 cells inhibits the ability of HMGB1 to activate the topo IIα promoter. The involvement of HMGB1 and its close relative, HMGB2, in modulation of activity of the topo IIα gene is further supported by knock-down of HMGB1/2, as evidenced by significantly decreased levels of topo IIα mRNA and protein. Our experiments suggest a mechanism of up-regulation of cellular expression of topo IIα by HMGB1/2 in pRb-negative cells by modulation of binding of transcription factor NF-Y to the topo IIα promoter, and the results are discussed in the framework of previously observed pRb-inactivation, and increased levels of HMGB1/2 and topo IIα in tumors.
INTRODUCTION
DNA topoisomerase II (topo II) is an essential and ubiquitous enzyme for proliferation of eukaryotic cells (1). It can alter the topological state of DNA and untangle DNA knots and catenanes (interlocked rings) via ATP-dependent passing of an intact double helix through a transient double-stranded break generated in a separate DNA segment, followed by religation and enzyme turnover (2). In mammalian cells, topo II exists in two isoforms, α (170 kDa) and β (180 kDa), both having similar primary structure and almost identical catalytic properties, but differing in their production during the cell cycle (1,3). Topo II is the target of a number of drugs currently used in the treatment of human malignancies, such as etoposide, teniposide, doxorubicine and mitoxantrone (3). These drugs (also termed topo II poisons) can stabilize the covalent enzyme-associated complexes and shift the DNA cleavage/religation equilibrium of the enzyme reaction toward the cleavage state, converting biological intermediates of topo II activity into lethal ones ultimately leading to triggering of programmed cell death pathways (1,3,4).
HMGB-type proteins are relatively abundant and evolutionarily highly conserved non-histone chromatin-associated proteins in mammals. There are three HMGB variants in human and mice, HMGB1, HMGB2 and HMGB3. While HMGB1-3 proteins are expressed in early mice embryos, HMGB2 and HMGB3 are down-regulated during embryonic development (5). The abundant HMGB1 protein (∼1 molecule per 10–15 nucleosomes) is highly conserved among mammals, and it continues to be ubiquitously expressed in adults. HMGB1 and HMGB2 function in a number of fundamental cellular processes such as transcription, replication, DNA repair and recombination (5–8). HMGB1 is associated with chromosomes in mitosis and due to its extreme mobility in the cell the protein is continuously exchanged between nucleus and cytoplasm [(8) and refs therein]. HMGB1, but not HMGB2, also exhibits an important extracellular function in mediation of inflammation mechanisms, tumor growth and metastasis (6,8). HMGB1, like HMGB2-3, has a tripartite domain organization, consisting of two DNA-binding domains, the HMG-boxes A and B, and acidic C-terminal tails of variable length. While the two HMG-boxes interact with DNA (exhibiting a high affinity for distorted DNA conformations (9–12), the C-tail usually decreases the affinity of the protein for DNA (5,7). Binding of HMGB1 to DNA causes local distortions by bending/looping and changes in DNA topology (7,13,14). HMGB1 also interacts weakly with a number of proteins, including transcriptional factors, site-specific recombination and DNA repair proteins (8). The importance of HMGB1 for life is supported by the phenotype of the HMGB1 knockout mice, which die 24 h after birth due to hypoglycemia and exhibit a defect in the transcriptional function of the glucocorticoid receptor (15). Lack of HMGB1 in primary mouse embryonic fibroblasts correlates with higher rates of DNA damage after UV irradiation, and the cytogenetic analyses revealed high levels of aneuploidy and spontaneous chromosome aberrations, decreased activity of telomerase and shortening of telomere lengths, suggesting that HMGB1 plays an important role in promoting genomic stability (5,16).
Previously we have reported that HMGB1 could interact with topo IIα and stimulate its enzymatic activity (11). In the present study we studied an impact of over-expression of HMGB1 and its close relative, HMGB2, on the activity of human topo IIα promoter. Using luciferase gene reporter assay we have demonstrated that HMGB1, but not a mutant of HMGB1 incapable of DNA bending, up-regulated the activity of human topo IIα promoter in human cells that lack functional retinoblastoma protein pRb. Transient over-expression of pRb in Rb-minus cells inhibited the transactivation potential of HMGB1 over the topo IIα promoter. In agreement with the above data, up-regulation of the topo IIα promoter by HMGB1/2 was very low in cells with functional pRb. The involvement of HMGB1 and HMGB2 in modulation of cellular activity of the topo IIα gene was also supported by silencing of HMGB1/2 expression by plasmid-encoded specific shRNA resulting in diminished expression of topo IIα. Our experiments allowed us to propose a mechanism of HMGB1-mediated transactivation of the topo IIα promoter by modulation of transcriptional factor NF-Y binding to the promoter. The obtained results are discussed in the framework of previously observed increased levels of HMGB1 and topo IIα in tumors (17).
MATERIALS AND METHODS
Plasmids
Each of the supercoiled DNA plasmids was isolated by alkaline lysis method, followed by purification by two rounds of cesium chloride gradients or by the Qiagen plasmid kits. All purified plasmids exhibited ratios A260/A280 higher than 1.85.
Antibodies
Anti-HMGB1 or anti-HMGB2 (affinity purified rabbit polyclonal, BD Pharmingen), α-HMGB1 (monoclonal IgG2a, clone #KS1, Stressgen), α-topo IIα (mouse monoclonal or rabbit polyclonal, Topogen), α-pRb (rabbit polyclonal C-15, Santa-Cruz), α-pan-actin (mouse monoclonal, Dako), α-Flag (mouse monoclonal M2 clone, Sigma), α-NF-YB (rabbit polyclonal, a gift from Roberto Mantovani, Italy) or from GeneSpin (PAb001, Milano, Italy).
DNA circularization assay
T4 DNA ligase-mediated circularization was carried out as described earlier (18) with the following modifications. Briefly, ligation was carried out at DNA concentration of ∼1 nM and the concentrations of HMGB1 protein and peptides were 0.5–6 μM. The DNA probe was a gel-purified 123-bp AvaI DNA fragment (prepared by AvaI digestion of the 123-bp DNA lather, BRL) that was 32P-labeled at 5′-ends by T4 DNA kinase and [γ-32P]ATP (Amersham Biotech). Ligation reactions were initiated by addition of 0.1 U of T4 DNA ligase (Promega) and were allowed to proceed at 30°C for 30 min. The ligation products were then deproteinised, followed by their resolution by electrophoresis on 5% polyacrylamide gels in 0.5× TBE as detailed in (19). The amount of monomer DNA circles was determined from quantitative analyses of dried gels on a Molecular Dynamics Storm PhosphorImager using the ImageQuant software.
Electrophoreic mobility shift experiments (EMSA)
The oligonucleotides for EMSA were derived from human topo IIα promoter (5′ to 3′): ICE1(WT)-F:CTAGGAGCGAGTCAGGGATTGGCTGGTCTG; ICE1(WT)-R: CAGACCAGCCAATCCCTGACTCGCTCCTAG; ICE1(MUT)-F: CTAGGAGCGAGTCAGGGCTGGACTGGTCTG; ICE1(MUT)-R: CAGACCAGTCCAGCCCTGACTCGCTCCTAG; ICE2(WT)-F: CTAGAAGGCAAGCTACGATTGGTTCTT; ICE2(WT)-R: AAGAACCAATCGTAGCTTGCCTTCTA G-3′; ICE2(MUT)-F: CTAGAAGGCAAGCTACGCTGGATTCTT-3′; ICE2(MUT)-R: AAGAATCCAGCGTAGCTTGCCTTCTAG-3′; ICE3(WT)-F: CTAGCTCCCTAACCTGATTGGTTTATTCAAA-3′; ICE3(WT)-R: TTTGAATAAACCAATCAGGTTAGGGAGCTAG-3′; ICE3(MUT)-F: CTAGCTCCCTAACCTGCTGGATTTATTCAAA-3′; ICE3(MUT)-R: TTTGAATAAATCCAGCAGGTTAGGGAGCTAG-3′.
The oligonucleotides were 32P-labeled at their 5′-termini by T4 DNA kinase and [γ-32P]ATP (Amersham Biotech) and annealed with their complementary strands to form blunt-ended DNA duplexes. Reaction mixtures for EMSA contained control nuclear extract (5–10 μg of proteins) or extract pre-incubated with HMGB1 (wild-type or F38A/F103A mutant, typically 1–4 μM) and/or His-pRb(wt) [0.1–0.4 μg, purified from BL21(DE3)pLysS Escherichia coli cells harboring the plasmid pRSET(his-Rb); the plasmid was kindly provided by Ronen Marmorstein from the Wistar Institute, Philadelphia, USA], 1 μg of poly (dI.dC) as a non-specific competitor, and ∼3 ng of 32P-labeled DNA duplexes in EMSA buffer (20 mM HEPES, pH 7.6, 4% Ficoll, 50 mM KCl, 0.1% Nonidet P-40, 0.2 mM EDTA and 0.5 mM DTT) in a total volume of 20 μl. DNA and proteins from nuclear extract were pre-incubated on ice for 20 min. Identity of transcription factor NF-Y within the retarded DNA–protein complexes was verified by pre-incubation of the protein–DNA complexes with 0.5 μg of polyclonal α-NF-YB antibodies [kindly provided by Roberto Mantovani, Dipartimento di Scienze Biomolecolari e Biotecnologie, Università di Milano, Milano, Italy; (9)] at 30°C for 20 min. The protein–DNA complexes were resolved on 5% polyacrylamide gels in 0.5× TBE buffer at 200 V for ∼2–3 h (4°C). The gels were dried and the labeled DNA was imagined by PhosporImager Storm (Molecular Probes).
Cloning of HMGB1 plasmids
HMGB1 (residues 1–215), HMGB1 domain A (HMG-box A, residues 1–88), HMGB1 domain B (HMG-box B, residues 85–180) and HMGB1 mutants were derived from rat or human HMGB1 cDNAs (the amino-acid sequence of the rat HMGB1 protein is identical to that of the human HMGB1 protein). The DNA sequences coding for the HMGB1 protein, mutants and domains were inserted into BamHI and SalI sites of the vector pQE-80L (Qiagen), which allows tightly regulated N-terminal 6× His-tagged protein expression in E. coli. Alanine mutagenesis of intercalating residues F38, F103 or I122 of isolated HMGB1 domains A or B, and mutagenesis of F38/F103 (double mutant or F/F) of the full-length HMGB1 was carried out by the protocol using ‘chimeras’ (Štros, unpublished results). The introduced mutations were verified by dideoxi-sequencing of both strands. For protein expression in E. coli, the HMGB1 and truncated HMGB1 cDNAs were re-cloned into the pQE-80L vector (His-tagged proteins). For transfections, the HMGB1 and HMGB2 cDNAs were cloned into the Flag-pcDNA3 or pcDNA3 vectors (20).
Cell culture
The human osteosarcoma Saos-2 cells (p53−/−, Rb−/−), the breast cancer MCF-7 cells (p53+/+, Rb+/+) and the non-small lung carcinoma H1299 cells (p53−/−, Rb+/+) were grown in 1640 RPMI (Sigma). The 5637 cells (p53−/−, Rb−/−), originally established from the primary bladder carcinoma of a 68-year-old man, was purchased from DSMZ (No: ACC 35). Stable Saos-2 derivatives in which the expression of pRb was controlled by the tetracykline regulated promoter were kindly obtained from Liang Zhu [Department Developmental and Molecular Biology, Albert Einstein College of Medicine, NY; (21)]. The pRb-inducible Saos-2 cells were grown in the presence of 1 μg/ml of tetracycline to prevent pRb expression. All media were supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and the cells were grown in 5% C02 at 37°C.
Cell transfections
Cells were detached by trypsine treatment at 80–90% confluence, and transfected with plasmids either by the use of Amaxa Nucleofector II (Amaxa, Germany) or Fugene HD (Roche). To select for stably transfected cells, Geneticin (BRL) or Zeocin (Invitrogen) were added following 48 h of transfection.
shRNA-mediated gene silencing of HMGB1 and HMGB2
Plasmid pcDNA-Zeo(−)-U6 under the control of a human U6 promoter was used to express two different short hairpin RNAs (shRNAs) that specifically cleave HMGB1 (construct #A) and HMGB1/2 (construct #B) mRNAs in human cells (the plasmids were kindly provided by Stephen Lippard, Dept. Chemistry, MIT, Cambridge). The following hHMGB1 (#A: GGAGAACATCCTGGCCTGT) or hHMGB2 (#B: AGTGAACACCCTGGCCTAT) sequences were used to create DNA cassettes in pcDNA-Zeo(−)-U6 vector expressing specific shRNAs for silencing of HMGB1 or HMGB1/2 expression in human cells, respectively. Scrambled human HMGB2 sequence (not related to any human sequence) was used as a control (GAGAGGACAAGAGATGTATT). For the purpose of preparation of stably transfected cells, the growth media contained 300 μg/ml Zeocin (Invitrogen) and the cells were selected for 2–3 weeks.
siRNA-mediated gene silencing of pRb
Specific siRNAs for knock-down of pRb in H1299 cells were designed as reported in ref. (44). Rb-positive H1299 cells were transfected with 0.25 μg of either a control duplex siRNA [all sequences were 5′-to-3′; GGGCGUCGAUCCUAACCGGdTdT (top) and CCGGUUAGGAUCGACGCCCdTdT (low)] or a mixture of two Rb-specific siRNA duplexes [#1: GAUACCAGAUCAUGUCAGAdTdT (top) and UCUGACAUGAUCUGGUAUCdTdT (low); #2: CCCAGCAGUUCGAUAUCUAdTdT (top) and UAGAUAUCGAACUGCUGGGdTdT (low); Eurogentec S.A. (Liège)] using the X-tremeGENE siRNA transfection reagent (Roche, Mannheim) as recommended by the manufacturer. For the luciferase gene reporter assays, the corresponding siRNA duplexes were transiently co-transfected with plasmid pTIIα-617 and either plasmid pcDNA3 or plasmid pcDNA-HMGB1 as detailed below.
Luciferase reporter gene assays
Transient transfections for luciferease gene reporter assays were carried out in 24-well plates using Fugene HD according to manufacturer instructions (Roche). When using the Amaxa Nucleofector II (Amaxa, Germany), transfections were carried out in six-well plates (2 × 105 cells/well) using the plasmid amounts as indicated below. Transfection mixtures contained Flag-pcDNA3 expression vectors encoding full-length HMGB1 (residues 1–214), HMGB1-ΔC (residues 1–179) or double-mutant HMGB1 (F38A/F103A), and/or human wild-type pRb [pBC-Rb(379–928)] and mutant pRb [pBC-Rb(379–928), amino-acid 706 C→F]; [kindly provided by Pradip Raychaudhuri, Department of Biochemistry, College of Medicine, University of Illinois, Chicago; (22)], typically 0.5 μg (Rb) and 2 μg (HMGB1 or HMGB2) of the plasmid DNA. The following reporter constructs (each 0.4 μg) were used: plasmid pGL3-Basic containing firefly Photinus pyralis cDNA (Promega) linked with sequences derived from human topo IIα gene promoter: plasmid pTIIα-617 containing either wild-type human topo IIα promoter or mutated promoter within the ICE2 sequence [the promoter sequences were from bp −617 to +90 and the plasmids were kindly provided by Karin M. Stowell, Institute of Molecular BioSciences, Palmerston North, New Zealand; (23)]; or one of the plasmids pTIIα-32, pTIIα-90, pTIIα-142, pTIIα-252 and pTIIα-557 containing varying lengths of the human topo IIα promoter as indicated [kindly provided by D. Parker Suttle, Department of Pharmacology, College of Medicine, University of Tennessee, Tennessee, USA; (24)]. Mutation of the ICE2 within the plasmid pTIIα-142 was carried out by the QuickChange II Site-Directed Mutagenesis Kit (Stratagene) using the following primers: top, 5′-GATATAAAAGGCAAGCTACGGTGGATTCTTCTGGACGGAGACG-3′; low, 5′-CGTCTCCGTCCAGAAGAATCCACCGTAGCTTGCCTTTTATATC-3′ (the mutated ICE2 is highlighted). The introduced mutations within the ICE2 (ATTGG mutated to GTGGA) of the topo IIα promoter (plasmid pTIIα-142_ICE2 mut) were confirmed by dideoxy sequencing. In control luciferase assays, the topo IIα promoter-pGL3 constructs were replaced by the empty vector pGL3-Basic. The equal amount of plasmid DNAs in each transfection mixtures was achieved by adding corresponding amounts of empty (promoter-less) plasmid vectors to final 3.6 μg of the transfection mixture when using Amaxa Nucleofector II for transfection. The luciferase activity was measured 40–44 h after transfection using a dual-luciferase reporter assay system (Promega). Results are presented as changes of transactivation of the topo IIα promoter constructs relative to the original activity of these promoter constructs in cells not transfected with plasmids expressing (HMGB or pRb) proteins. In some luciferase reporter gene assays, the pRL family Renilla luciferase control reporter vector (0.4 μg) with the cDNA encoding Renilla luciferase under the control of the herpes simplex virus thymidine kinase promoter (pRL-TK, Promega) was used. However, measuring the luciferase activity of the firefly Photinus pyralis proved to be unreliable due to partial activation of the herpes simplex virus thymidine kinase promoter of the pRL-TK by HMGB1/2 [in agreement with reported effects of HMGB1 on transactivation of various gene promoters (8)]. Therefore, the luciferase activity in each cellular lysates was normalized by determining the protein concentrations using the Coomassie G-250 protein-dye assay (Bio-Rad). The involvement of HMGB1/HMGB2 proteins on activity of the human topo IIβ gene promoter was studied using plasmid -700TOP2B-pGL3 (plasmid pGL3-Basic containing firefly Photinus pyralis cDNA linked with sequence −700 to +193 nt of the human topo IIβ gene promoter; kindly provided by Susan P.C. Colle (Queen's University, Cancer Research Institute, Division of Cancer Biology & Genetics, Kingston, Canada).
Quantitative real-time PCR of TOP2A gene
Total RNA was isolated from 2 × 105 Saos-2 cells by RNeasy Mini Kit (Qiagen). First-strand cDNA synthesis was performed using the SuperScriptTM II with oligo(dT)12–18 primer (Invitrogen). Approximatedly five-hundred nanograms of RNA was used for each 20 μl of RT-reaction. Each cDNA sample was analyzed in triplicates using TaqMan® Gene Expression Assay (Applied Biosystems) according to manufacturer's instructions. Amplification was detected using 7300 Real Time PCR System (Applied Biosystems). The TaqMan probe used hybridized between exons 10 and 11 of the TOP2A gene. Data were analyzed using Sequence Detection System (SDS) software, version 1.3.1. Results were obtained as the cycle number when the fluorescence reaches a set threshold (CT). The relative expression of TOP2A was quantified as a percentage of HPRT expression. The difference in CT values between the TOP2A and HPRT reactions (ΔCT) was converted into relative TOP2A expression using a formula 2−ΔCT × 100%.
Chromatin immunoprecipitation (ChIP) assays
Stably transfected Saos-2 cells (control or HMGB1/2 silencing; see shRNA-mediated gene silencing of HMGB1 and HMGB2 in the Materials and methods section) were grown to ∼80% confluence in 150 cm2 tissue culture flasks. Cells were then washed in minimal culture medium (without FBS and antibiotics) and fixed with 1% formaldehyde in the same medium on a shaking platform at room temperature for 10 min. Preparation of chromatin and immunoprecipitation (∼1.5 × 106 cells/reaction) was carried out using the ChiP-IT Enzymatic Express Kit (Active Motif, Rixensart, Belgium). ChIP-grade NF-YB rabbit polyclonal antibody (PAb001 from GeneSpin, Milano, Italy) were at 1 μg/reaction. Purified immunoprecipitated DNA was subjected to semi-quantitative PCR with primers (Fw: 5′-GGTGCCTTTTGAAGCCTCTCTAG-3′, Rev: 5′-GCTCCACTTGAACCTTCCTTTAGC-3′) specific for a region −215 to −21 of the human topo IIα promoter, generating a 195 bp product encompassing three NF-Y-binding sites (ICEs1-3). GoTaq Hot Start DNA polymerase (Promega; 2.5 U/100 μl reaction) was used in 1× Green Flexi buffer/2 mM MgCl2 (Promega). The amplification program consisted of denaturation at 94°C for 2 min, followed by 30–34 cycles of 94°C for 60 s, 58°C for 30 s and 72°C for 30 s. As a negative control, 1 μg of control IgG polyclonal antibody (or 1 μg of α-RNA pol II antibody using primers specific for human GADPH gene as a positive control) was used (ChIP-IT Control Kit-Human, Active Motif).
Preparation of nuclear extract
Nuclear extract was prepared using the published protocol (25) with some modifications. Cells were twice washed with 1× phosphate buffered saline (PBS), trypsinized, followed by washing with 1× PBS containing 10% FCS. After final wash with PBS, the pellet was re-suspended in 1 ml of ice-cold buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT) containing protease inhibitors (1 μg/ul aprotinin, 10 μg/ul leupeptin, 1 μg/ul pepstatin A, 100 μg/ul trypsin inhibitor, 0.1 mM TLCK and 20 mM benzamidine) and allowed to swell on ice for 15 min. Then 10% Nonidet P-40 was added to final 0.5% and the incubation was continued for more 5 min. Following the incubation, the cellular suspension was spun down (1200 g, 5 min), the nuclei (pellet) were then re-suspended in 1 ml of buffer A (lacking Nonidet P-40), and spun-down as above. The pellet (nuclei) was re-suspended in ice-cold buffer B (20 mM HEPES pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA) containing protease inhibitors, vortexed for 20 s and rocked at 4°C for 30 min. The suspension was clarified by centrifugation (15 000 × g, 10 min) at 4°C, and the clear supernatant containing nuclear extract was aliquoted, quick frozen in liquid nitrogen and stored at −70°C. For the purpose of the immunoprecipitation, the nuclear extracts were not frozen and used immediately.
Western blotting and immunological detection of proteins
Nuclear extract was prepared from transfected or untransfected cells as above. The protein concentrations were determined by the Bradford Coomassie G-250 assay (Bio-Rad) using bovine serum albumin (BSA) as a standard. Equal amounts of proteins from different nuclear extracts were then separated by SDS–PAGE on SDS/7.5% or 10%-polyacrylamide gels, and the resolved proteins were transferred onto the PVDF membrane (Bio-Rad) using a semi-dry blotting apparatus (Biometra). Detection of human topo IIα was carried out by overnight incubation of the membrane with monoclonal anti-topo IIα antibody (1:500 dilution, Topogen), followed by extensive washing the membrane in 1× PBS-T (PBS with 0.1% Tween-20) and incubation of the membrane with horseradish peroxidase conjugated anti-mouse antibody (IgG-HRP) (1:2000 dilution, GE Healthcare) for 1 h. Detection of HMGB1 or HMGB2 was carried out by overnight incubation of the membrane with polyclonal α-HMGB1 or HMGB2 antibodies (1:500 dilution, BD Pharmingen or Aviva), followed by washing the membrane in 1× PBS-T and incubation of the membrane with goat anti-rabbit IgG-HRP antibody (1:2000 dilution). Equal loading of transferred proteins was verified by detection of actin using mouse monoclonal Pan-actin antibody (Dako, 1:2000 dilution), followed by incubation of the membrane with horseradish peroxidase conjugated anti-mouse antibody (IgG-HRP) (1:2000 dilution, GE Healthcare) as detailed above. Visualization of proteins was performed using West Dura Extended Duration Signal Kit (Pierce). The intensities of signals were quantified using Multi Gauge software of the imaging system LAS-3000 (Fuji).
Pull-down assays
Plasmids encoding wild-type pRb (amino-acid residues 379–928) or Rb (C706Y) mutant (aa residues 379–928) fused at the N-terminus with glutathione-S-transferase (GST) were kindly provided by Sybille Mittnacht (Institute of Cancer Research, Chester Beatty Labs, London). GST, GST-pRb and GST-HMGB1 were isolated from BL21(DE3) cells as detailed in (20). GST or GST-tagged pRb (typically 1–2 μg) were bound to glutathione Sepharose 4B beads (40 μl of 1:1 v/v slurry), and incubated with purified HMGB1 from calf thymus or recombinant rat HMGB1 expressed in E. coli (0.5 μg) in buffer PD [20 mM Tris–HCl pH 7.6, 0.2 M NaCl, 10 mM DTT, 0.2% Triton X-100, 0.2% Tween-20, 20% glycerol (v/v), protease inhibitors: 1 μg/ul aprotinin, 10 μg/ul leupeptin, 1 μg/ul pepstatin A, 0.1 mM TLCK and 20 mM benzamidine] by rocking for 2 h at 4°C. In some pull-down experiments, GST-tagged HMGB1 or pRb were bound to glutathione Sepharose beads and rotated with nuclear extract from Saos-2 cells in the presence or absence of ethidium bromide (100 μg/ml) at 4°C for 2–3 h. After extensive washing of the beads in PD buffer, the proteins associated with the beads were separated on SDS/15% polyacrylamide gels. The proteins were then transferred onto the PVDF membrane by western blotting (as above) and incubated with specific primary antibodies overnight at room temperature. The polyclonal antibodies were: α-HMGB1 (1:1000 dilution, BD Pharmingen), α-pRb (1:500 dilution; C-15, Santa Cruz), and α-NF-YB (1:1000 dilution; a gift from Roberto Mantovani, Italy). Following incubation with the primary antibody, the washed membrane was further incubated with horseradish peroxidase conjugated anti-rabbit antibody (IgG-HRP) (1:2000 dilution, GE Healthcare) for 1 h. Visualization of detected proteins was performed as indicated above.
Immunoprecipitation of pRb and HMGB1
Nuclear extract from H1299 cells (0.5 mg per each experiment) was pre-cleared by rotation with 40 µl of protein–A+G (1:1) agarose beads (1:1 v/v slurry) in 1 ml of 1× ECB buffer (0.3 M NaCl, 50 mM HEPES, pH 7.9, 0.2 mM EDTA, 0. 5 mM DTT, 2% Nonidet P-40 plus freshly added protease and phophatase inhibitors) at 4°C for 1 h. The beads were removed by centrifugation (10 000 × g for 30 s), the supernatant (pre-cleared nuclear extract) was mixed with 10 μg of monoclonal anti-HMGB1 (IgG2a, clone #KS1, Stressgen) or 10 μg of pre-immune mouse IgG (Sigma), and rotated in 0.5 ml of ECB buffer at 4°C for 1 h. Then, 40 µl of pre-cleared 1:1 slurry of protein-A+G agarose beads (pre-incubated with acetylated bovine serum albumin at 0.1 mg/ml in ECB buffer for 1 h, followed by extensive washing in ECB buffer) was added and rotation was continued by more 2 h. The beads were then spin down (10 000 × g for 30 s) and extensively washed (five times) in 1 ml of ECB buffer containing 0.5% Nonidet P-40. The washed beads were then mixed with 20 μl of 10× loading buffer, boiled for 3–4 min and loaded onto SDS/7.5% polyacrylamide gels. The resolved proteins were transferred onto the PVDF membrane (Bio-Rad), and the membrane was incubated with polyclonal α-pRb antibody (1:500 dilution; C-15, Santa-Cruz), followed by washing the membrane in 1× PBS-T and incubation of the membrane with horseradish peroxidase conjugated anti-rabbit antibody (IgG-HRP) (1:2000 dilution, GE Healthcare) for 1 h. Visualization of detected proteins was performed by ECL detection kit (GE Healthcare).
RESULTS
HMGB1 up-regulates topo IIα gene promoter
To find out whether HMGB1 could modulate activity of the human topo IIα gene promoter, luciferase reporter gene assay was employed by using constructs containing sequences from bp −617 (or −557) to +90 of the human topo IIα gene promoter (referred as to the full-length promoter, pTIIα-617 or pTIIα-557). In some cases, HMGB1 was over-expressed from plasmid encoding the protein fused with a seven amino acids FLAG-tag at the N-terminus to distinguish it on western blots from the endogenous HMGB1 (Figure 1A, inset). However, the luciferase reporter assays using the Flag-tagged and untagged HMGB1 were indistinguishable (not shown). As shown in Figure 1, over-expression of HMGB1 resulted in ∼10–20-fold increase of luciferase activity, depending on individual transfection experiments, rather than on slight differences in the size of the topo IIα promoter constructs used (−617 or −557 bp). These findings demonstrated that HMGB1 could up-regulate the activity of the human topo IIα promoter, and that the activation of the topo IIα promoter increased with the amount of HMGB1 plasmid (Figure 2D; increased levels of ectopically expressed HMGB1 in nuclear lysates were confirmed by immunodetection using specific anti-HMGB1 antibody, not shown).
Figure 1.
HMGB1 up-regulates activity of human topo IIα promoter. (A) Identification of elements within human topo IIα gene promoter involved in HMGB1-mediated up-regulation of the promoter. Saos-2 cells were co-transfected with the plasmid constructs containing the –557 bp human topo IIα promoter (plasmid pTIIα-557) or one of the truncated topo IIα promoter constructs linked to the luciferase reporter gene (as detailed in Materials and methods section), and plasmid encoding HMGB1 (or empty vector). Inset, over-expression of Flag-tagged HMGB1. Lane 1, cells transfected with empty vector; lane 2, cells transfected with plasmid encoding Flag-HMGB1. Immunodetection was carried out using specific α–HMGB1 antibody. (B) Effect of mutation of ICE2 on transactivation of topo IIα promoter by HMGB1. Saos-2 cells were co-transfected with the plasmids harboring the full-length topo IIα promoter (plasmids pTIIα-617 or pTIIα-617_ICE2mt) or truncated topo IIα promoter (plasmid pTIIα-142 or pTIIα-142_ICE2mt) containing the wild-type (WT) or mutated ICE2 (ICE2mt), and either empty vector or plasmid encoding HMGB1. Luciferase activity (referred as to transactivation) in panels (A) and (B) is presented relative to the luciferase activity of the topo IIα promoter in cellular lysates without HMGB1 over-expression (taken as 1). Each transfection experiment was repeated at least five times with triplicate samples. Values are presented as the mean ± 1 SD (error bars) of luciferase activities from triplicate samples in a representative experiment.
Figure 2.
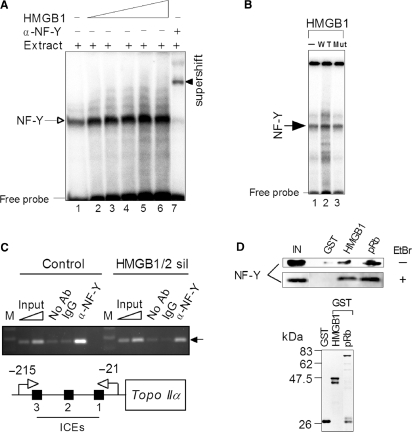
HMGB1 interacts with transcription factor NF-Y and modulates its binding to human topo IIα promoter. (A) HMGB1 enhances binding of NF-Y to ICE2. Radioactively labeled 30-bp DNA duplex containing ICE2 was mixed with nuclear extract from Saos-2 cells (lane 1) or nuclear extract containing increasing concentrations of HMGB1 (1, 2, 3, 4 and 6 μM, lanes 2-5, respectively), followed by separation of unbound ICE2 DNA duplex (free probe) and DNA–protein complexes on 5% non-denaturing polyacrylamide gels (EMSA). The presence of NF-Y within the retarded complex (arrow) was verified by super-shifting of the retarded complex (arrow head) with specific polyclonal antibodies against NF-YB. Specificity of NF-Y binding from nuclear extracts to DNA duplexes containing the wild-type ICE2 was also demonstrated by competition experiments with unlabeled DNA duplexes containing the wild-type or mutated sequence of the ICE2 (not shown). (B) HMGB1 mutant, incapable of DNA bending, cannot enhance binding of NF-Y to ICEs. EMSA experiment was carried out as in (A). Lane 1, no added HMGB1; lane 2, wild-type HMGB1 (2 μM); lane 3, HMGB1(F38A/F103A), 2 μM. Arrow indicate the mobility of (NF-Y)–DNA complexes. (C) ChIP assays using anti-NF-YB, control IgG antibodies or without antibodies were performed using chromatin from Saos-2 cells (control or upon silencing of HMGB1/2 expression, see Figure 6) as detailed in Materials and methods section. NF-Y binding to the topo IIα promoter was detected by gel staining (ethidium bromide) after PCR amplification using primers corresponding to the promoter as described in Materials and methods section. For semi-quantitative PCR, amplification reactions were carried out within the linear range of amplification (typically 30–34 cycles). The ChiP data were calculated as a ratio of PCR signal from ChIPed DNA to the input PCR signal, the final data were obtained from comparison of both cell lines (control or HMGB1/2 sil) and expressed as –fold change. Control, stably transfected Saos-2 cells (vector); HMGB1/2 sil, stably transfected Saos-2 cells with inhibited HMGB1/2 expression (shRNA-mediated gene silencing of HMGB1/2, Figure 6). The specific PCR product of 195-bp encompassing ICEs 1-3 is indicated by an arrow. M, DNA size marker; ICEs, inverted CCAAT elements. (D) Detection of NF-Y binding to HMGB1 or pRb in vitro. (Top) Equal amounts of GST, GST-HMGB1 or GST-pRb were immobilized on glutathione Sepharose beads and incubated with nuclear extract from Saos-2 cells. After extensive washings, the resin-bound proteins were eluted and separated by SDS-polyacrylamide gel electrophoresis, followed by W. blotting and immunological detection by anti-NF-YB antibody as detailed in the Material and methods section. EtBr, ethidium bromide. IN, 25% of the nuclear extract from Saos-2 cells used for the ‘pull-down’ assay (input). (Bottom) GST or GST-tagged proteins immobilized on agarose beads for the ‘pull-down’assay (Coomassie blue R-250 staining).
The known regulatory elements of the −557 topo IIα gene promoter (pTIIα-557) construct involve five inverted CCAAT boxes (referred as to ICEs, Inverted CCAAT Elements), an Activating TF-binding site (ATF), and one GC1 box (the second GC box, GC2, is present only within the −pTIIα-617) and no functional TATA box (26), Figure 1A. To identify sequences of the human topo IIα promoter involved in HMGB1-mediated up-regulation of the promoter, Saos-2 cells were co-transfected with the HMGB1 expression plasmid and the luciferase reporter constructs containing varying lengths of the topo IIα gene promoter. Deletions of the sequences from bp −557 to −142 (encompassing ICE5, ICE4, ICE3 and ATF) had very little, if any, effect on the ability of HMGB1 to up-regulate the activity of the promoter [albeit stepwise deletion of the 5′-promoter sequences from bp −557 to −142 of the full promoter resulted in up to ∼20-fold decrease in luciferase activity in the absence of over-expressed HMGB1, in agreement with previously published data (24)]. However, truncation of the topo IIα promoter from bp −142 to −90 including the ICE2 (pTIIα-90 containing only ICE1) markedly decreased the ability of HMGB1 to up-regulate the topo IIα gene promoter (Figure 1A, pTIIα-142 or pTIIα-90). A slight activation of the topo IIα promoter lacking all known regulatory sequences (pTIIα-32) by HMGB1 was to be explained by the reported effect of the protein on the basal transcription (5,8,27). The above data suggested the importance of the ICE2 for the HMGB1-mediated transactivation of the topo IIα promoter. The latter conclusion seemed to be confirmed by mutation of the ICE2 (ATTGG mutated to GTGGA) within the −142 promoter construct resulting in inability of HMGB1 to transactivate the −142 bp topo IIα promoter (pTIIα-142_ICE2mt, Figure 1B, right). However, while the ICE2 was crucial for the HMGB1-mediated transactivation of the −142 promoter (Figure 1A), it was much less important in the context of the full-length topo IIα promoter as revealed by the luciferase gene reporter assay using the −617 promoter construct with mutated ICE2 (pTIIα-617_ICE2mt, Figure 1B, left). These results questioned the central role of the ICE2 in the HMGB1-mediated up-regulation of the human topo IIα promoter, and pointed out the possible importance of multiple ICEs for the HMGB1-mediated up-regulation of the topo IIα promoter (see Discussion section).
It is generally assumed that HMGB1 could facilitate binding of a number of transcription factors (such as steroid hormone receptors, homeitic HOX proteins, recombination activating gene protein RAG1, octamer transcription factors Oct-1/Oct-2, human TATA-binding protein TBP, TFIID and proteins of p53 family) to their cognate sites by binding to the TFs and delivering them to the corresponding DNA sequences (5,7,8). NF-Y (nuclear factor-Y) is a transcription factor comprising NF-YA, NF-YB, and NF-YC subunits. NF-Y specifically recognizes a CCAAT box motif which is found in the promoter and enhancer regions of many genes, including the human topo IIα gene (26,29). The NF-YA subunit associates only with the NF-YB:NF-YC heterodimer creating a functional NF-Y (trimer) CCAAT box DNA-binding complex. NF-Y binds to the human ICE1-4, but not to ICE5 (30). The effect of NF-Y on cellular levels of the human topo IIα (and subsequently on the regulation of the topo IIα gene promoter) is elusive due to conflicting reports (23,31,32). Thus, a simple explanation for the enhancement of the topo IIα promoter activity by HMGB1 could be that HMGB1 could promote binding of crucial activators, such as NF-Y, to their DNA-binding sites. Electrophoretic Mobility Shift Assay (EMSA) was therefore used to determine whether HMGB1 could enhance binding of NF-Y to short DNA duplexes containing ICE1, ICE2 or ICE3 sequences derived from the human topo IIα promoter. Addition of HMGB1 to nuclear lysate from Saos-2 cells could enhance (∼2–3-fold) binding of NF-Y to the ICE2 (Figure 2A), and also to the ICE1 and ICE3 (data not shown), demonstrating the ability of HMGB1 to promote NF-Y binding to various ICEs.
The ability of HMGB1 to promote in vitro binding of NF-Y to DNA duplexes containing inverted CCAAT elements (Figure 2A) could indicate that the protein could modulate NF-Y binding to the human topo IIα gene promoter in vivo. To find out whether HMGB1/2 could affect binding of NF-Y to the topo IIα promoter, chromatin immunoprecipitation (ChIP) was used. For this purpose, control Saos-2 cells and cells with inhibited HMGB1/2 expression were fixed with formaldehyde, chromatin sheared by enzymatic digestion and immunoprecipitated with anti-NF-YB or control antibodies. Purified immunoprecipitated DNA was subjected to semi-quantitative PCR with primers specific for a region −215 to −21 of the human topo IIα promoter, amplifying a 195 bp product encompassing three NF-Y-binding sites (ICEs1-3). As shown in Figure 2C, the ChIP analysis revealed that inhibition of HMGB1/2 expression resulted in up to ∼3-fold reduction in NF-Y binding to the topo IIα gene promoter relative to the control cells. These data provided evidence that HMGB1/2 could modulate NF-Y binding to the human topo IIα promoter in vivo.
To find out whether HMGB1 could interact with NF-Y, in vitro ‘pull-down’ (PD) assay was used. GST or GST-HMGB1 were immobilized on glutathione Sepharose beads and incubated with nuclear extract from Saos-2 cells. After extensive washings, the resin-bound proteins were separated by electrophoresis, followed by western blotting and immunological detection by anti-NF-YB antibody. As shown in Figure 2D (top), a significant fraction of NF-YB was found to be associated with HMGB1 relative to control GST-bound beads (equal amounts of GST or GST-HMGB1 immobilized on agarose beads were verified by gel electrophoresis, Figure 2D, bottom). Interaction between HMGB1 and NF-Y was not facilitated via DNA as similar results were obtained from PD assay in the presence of DNA intercalator ethidium bromide. The above results demonstrated the ability of HMGB1 to interact with NF-Y, and provided a possible explanation for the observed HMGB1-mediated enhancement of NF-Y binding to ICEs (Figure 2A), a basis for understanding of the effect of the protein on the activity of the topo IIα gene promoter (Figure 1).
Intercalating residues of HMGB1 are required for DNA bending and up-regulation of topo IIα gene promoter
Previous experiments with HMGB proteins from Sacharomyces cerevisae (NHP6A) and Drosophila melanogaster (HMG-D), which are highly related to human HMGB1 protein, revealed that mutation of DNA intercalating residues impaired their DNA-bending properties (33,34). Using ligase-mediated circularization assay we have studied whether the intercalating residues (Phe38 of the domain A, and Phe103/Ile122 of the domain B, Figure 3A) are also required for DNA bending by human HMGB1. This assay measures the efficiency with which T4 DNA ligase forms minicircles from fragments of DNA that are shorter than ∼150 bp. In the absence of internal curvature, the stiffness of a short DNA fragment (<150 bp) prevents intra-molecular alignment of its ends so that minicircles are detected only in the presence a protein that bends DNA.
Figure 3.
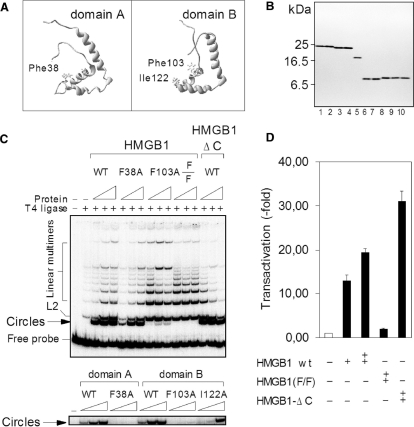
HMGB1 incapable of DNA bending cannot up-regulate activity of human the topo IIα gene promoter. (A) Position of intercalating residues within the HMGB1 domains A (Phe38) and B (Phe103 and Ile122). (B) Purity of HMGB1, domains A and B, and mutants as revealed by electrophoresis on an SDS/18% polyacrylamide gel and Coomassie blue R-250 staining. Lane 1, wild-type HMGB1; lane 2, HMGB1(F38A); lane 3, HMGB1(F103A); lane 4, HMGB1(F38A/F103A) [designated as F/F in panels (C) and (D)]; lane 5, HMGB1 lacking the C-tail (HMGB1-ΔC or A+B di-domain); lane 6, the wild-type domain A; lane 7, domain A (F38A); lane 8, the wild-type domain B; lane 9, domain B (F103A); lane 10, domain B (F103A/I122A). (C) Effect of mutated intercalating amino acids of HMGB1 or HMGB1 domains A and B on T4 DNA ligase-mediated DNA cyclization. Ligase-mediated circularization assay was carried out with a 32P-labeled 123-bp DNA duplex, and HMGB1 or HMGB1-ΔC at concentrations of 0.05, 1 and 1.5 μM (left to right, upper panel). For experiments with individual HMGB1 domains, the concentrations of the domain A or mutants were 1.5, 3 and 6 μM, and the concentrations of the domain B or mutants were 0.5, 1 and 1.5 μM (left to right, lower panel). The DNA–protein complexes were ligated with T4 DNA ligase and the deproteinised DNA samples were then separated on 5% non-denaturing polyacrylamide gels. (D) Transactivation of the topo IIα gene promoter by the double-mutant of HMGB1 or HMGB1-ΔC. Plasmids encoding HMGB1 or mutants were co-transformed with the plasmid pTIIα-617 into the Saos-2 cells, and the luciferase activity (transactivation) was measured as in Figure 1.
HMGB1 and individual domains, A and B (Figure 3A), have been mutated by alanine mutagenesis, and the proteins were expressed in E. coli and purified to near homogeneity (Figure 3B). To understand the importance of individual intercalating amino acids of HMGB1 for the ability of the protein to bend DNA, single HMG-boxes, domains A and B, and their mutants were first investigated. In agreement with previous studies (9,10,18,35,36), the domain B seemed to be more efficient than the domain A (Figure 3C, lower panel). Mutation of Phe38 (domain A) or Phe103 (domain B) to alanine completely abolished the ability of the both single domains to form DNA minicircles. On the other hand, mutation of the second intercalating residue of domain B, Ile122, had only a little effect on the ability of the domain to bend DNA. Interesting results were obtained upon mutagenesis of the intercalating residues in the context of the full-length HMGB1. Whereas mutation of Phe38 (domain A) exhibited only a little effect on the ability of the HMGB1 protein to bend DNA, mutation of Phe103 (domain B) nearly abrogated the ability of the protein to bend DNA. A complete inhibition of the ability of HMGB1 to bend DNA was to be achieved only when both intercalating phenylalanine residues, Phe38 and Phe103, had been mutated to alanine (Figure 3C, a double mutant F38/F103, upper panel). These results indicated that the domain B could largely substitute for the DNA-bending potential of the domain A within the full-length HMGB1, suggesting the major role of the domain B in mediating DNA bending by HMGB1 [see also (37)]. In agreement with previous studies (18,36), the DNA-bending potential of the two HMG-box domains of HMGB1 was down-regulated by its acidic C-tail as revealed by enhanced formation of minicircles by HMGB1 lacking the acidic C-tail (referred as to HMGB1-ΔC), Figure 3C (upper panel). We have found that single or multiple alanine mutagenesis of Phe38 and Phe103 had only a relative small effect on the binding properties of the full-length HMGB1 protein to linear DNA [see also (12,34,38,39,56)].
In order to find out whether the ability of HMGB1 to bend DNA was essential for the observed up-regulation of human topo IIα promoter, the impact of over-expression of either HMGB1 lacking the acidic C-tail (HMGB1-ΔC) or a double mutant HMGB1(F38A/F103A) has been investigated. As shown in Figure 3D, over-expression of HMGB1-ΔC in Saos-2 cells resulted in higher transactivation of the topo IIα promoter than that by the full-length HMGB1 (∼30-fold and ∼20-fold, respectively), suggesting that the acidic C-tail of HMGB1 inhibited the ability of the protein to activate the topo IIα promoter. The higher transactivation potential of HMGB1-ΔC was consistent with reported enhanced DNA-bending and -binding properties of the A+B di-domain (14,18,35,37). The inability of the HMGB1 mutant to transactivate the topo IIα promoter (Figure 3D) may be related to impaired DNA-bending properties of the mutant (Figure 3C, top; similar levels of ectopically expressed HMGB1 and mutants were verified by gel electrophoresis and immunodetection using specific antibodies, not shown). The latter idea was in agreement with the EMSA experiments demonstrating that HMGB1 incapable of DNA bending could no longer enhance NF-Y binding to ICEs (Figure 2B).
Up-regulation of topo IIα gene promoter by HMGB1 is inhibited by retinoblastoma protein pRb
In order to find out whether HMGB1 modulates activity of the human topo IIα gene promoter in other cell lines than the Saos-2 (p53−/−/Rb−/−) cells, comparative transfection experiments were performed with non-small lung cells carcinoma H1299 (p53−/−/Rb+/+), human breast cancer MCF-7 (p53+/+/Rb+/+) or human bladder carcinoma 5637 (Rb−/−) cells. While over-expression of HMGB1 could significantly up-regulate the topo IIα promoter both in Saos-2 (Figures 1A and 4A) and 5637 cells (data not shown), over-expression of HMGB1 had a relatively little (<2–3-fold) effect on the activity of the −617 bp human gene topo IIα promoter in MCF-7 or H1299 cells (Figure 4A).
Figure 4.
HMGB1 up-regulates human topo IIα gene promoter in pRb-minus cells. (A) The plasmid pTIIα-617 bearing the human topo IIα gene promoter was co-transfected with either vector (empty bars) or plasmid encoding HMGB1 (black bars) into pRb-negative Saos-2 cells or pRb-positive H1299 or MCF-7 cells. (B) Up-regulation of activity of human topo IIα promoter by HMGB1 is inhibited by wild-type but not mutant pRb. Saos-2 cells were transiently transfected with the plasmid pTopoIIα-617 and plasmids encoding the wild-type retinoblastoma protein pRb, mutant pRb (amino-acid 706 C→F) or human HMGB2. Luciferase activity (transactivation) in (A) and (B) was measured and presented as in Figure 1. (C) Knock-down of pRb in H1299 cells. Nuclear lysates from untransfected cells (lane 1), cells transfected with control siRNA (lane 2) or Rb specific siRNAs (lane 3) were resolved by SDS-polyacrylamide gel electrophoresis, and the proteins were then transferred onto the membrane and immunodetected using anti-pRb or anti-actin antibodies. (D) HMGB1 significantly stimulates activity of topo IIα gene promoter in H1299 cells upon knock-down of pRb. Control siRNA or Rb specific siRNAs were transiently co-transfected with plasmid pTIIα-617 and either empty pcDNA3 vector or plasmid encoding HMGB1 into pRb-positive H1299 cells as detailed in the Material and methods section. Luciferase activity (transactivation) was measured and presented as in Figure 1. (E) Interaction of HMGB1 with pRb in H1299 cells. Cellular lysates from pRb-positive Saos-2 cells were immunoprecipitated (IP) with monoclonal α-HMGB1 antibody or control (pre-immune) IgG, followed by separation of the precipitated proteins by SDS-polyacrylamide gel electrophoresis, western blotting and immunodetection with polyclonal anti-pRb antibody as detailed in the Materials and methods section. (F) Schematic structures of pRb and pRb constructs (upper panel) used for the ‘pull-down’ assay (upper panel). GST or GST-pRb (wild-type or mutant) were immobilized on glutathione Sepharose beads and incubated with purified HMGB1 protein. After extensive washing of the beads, the proteins associated with the beads were resolved electrophoresis, followed by western blotting and immunological detection by α-HMGB1 antibody as detailed in Material and methods section. Equal amounts of GST-pRb immobilized on beads were confirmed by immunodetection using anti-pRb (C-15) antibody (lanes 2 and 3). Input, 10% of the HMGB1 used for the pull-down assay.
The above results led us to hypothesize that the distinct effects of HMGB1 on the activity of the topo IIα promoter in different cell lines could be explained by the presence of retinoblastoma susceptibility protein pRb. pRb is a negative regulator of cellular proliferation and a prototypic tumor suppressor protein (40). pRb is also required for efficient activation of cell cycle checkpoints in response to a variety of DNA lesions, including those induced by topo IIα poisons (41).
To find out whether the absence of functional pRb could explain why HMGB1 up-regulated the topo IIα promoter much more efficiently in pRb-negative as compared to pRb-positive cells, the impact of HMGB1 over-expression on the activity of the promoter was studied in the luciferase gene reporter assays using two different approaches: pRb was (i) over-expressed in pRb-negative Saos-2 cells or (ii) silenced by specific siRNAs in pRb-positive H1299 cells. (i) pRb over-expression. pRb was ectopically expressed in Saos-2 cells and the luciferase activity was measured. As shown in Figure 4B, the activity of the topo IIα promoter was significantly inhibited (up to ∼3–4-fold) when pRb alone was over-expressed in Saos-2 cells. When HMGB1 (plasmid-encoded) was over-expressed in the Saos-2 cells transiently expressing pRb, very little, if any, transactivation of the topo IIα promoter was observed (Figure 4B). Similar results were obtained when HMGB1 was transiently over-expressed in stable Saos-2 cell derivatives in which the activity of the Rb gene was controlled by the tetracycline regulated promoter (data not shown). These results indicated that ectopically-expressed pRb could counteract the stimulatory effect of HMGB1 over the topo IIα promoter. (ii) Knock-down of pRb. Silencing of pRb expression in H1299 cells by specific siRNAs (Figure 4C) could significantly enhance (up to ∼3-fold) the activity of the topo IIα promoter in a luciferase gene reporter assay (Figure 4D), in agreement with previously published microarray study demonstrating an enhanced activity of the topo IIα gene upon silencing of pRb in H1299 cells (44). Co-transfection of the cells with silenced expression of pRb with plasmid encoding HMGB1 could increase the activity of the promoter in a luciferase gene reporter assay up to ∼12-fold (Figure 4D). This was in contrast to the gene reporter assay using either no siRNA or control siRNA. While the control siRNA had no effect on the activity of the topo IIα promoter, co-transfection of these cells with plasmid encoding HMGB1 could increase the activity of the promoter only ∼2-fold (Figure 4D). The above results provided strong evidence that the up-regulation of the topo IIα promoter by HMGB1 was inhibited by pRb.
pRb protein is functionally inactivated in most human neoplasms either by direct mutation/deletion (such as occurs in osteosarcoma, retinoblastoma and small cell lung carcinoma) or indirectly through altered expression/activity of upstream regulators (42). pRb is a 928 amino-acid protein with functionally distinct protein-binding domains. The A/B pocket (residues 379–792), comprised of structural domains A and B (Figure 4F, upper panel), is a hot-spot for mutation in human cancers, and is the binding site of several viral oncoproteins that inactivate pRb (42). We have therefore tested whether mutation of pRb within the A/B pocket (amino-acid 706 C → F) could abolish the inhibitory effect of pRb on the HMGB1-mediated up-regulation of the topo IIα promoter. As shown in Figure 4B, the transcriptional activity of the topo IIα promoter in Saos-2 cells (in the absence of over-expressed HMGB1) was very little (if any) affected by transient expression of the mutant pRb. Similarly, the mutant pRb could not inhibit the HMGB1-mediated up-regulation of the activity of the topo IIα promoter in Saos-2 cells (Figure 4B).
A large number of proteins have been demonstrated to interact with pRb, the majority of which are involved in transcriptional repression such as histone deacetylases, components of the mammalian SWI/SNF chromatin-remodeling complex, histone methyltransferases, heterochromatin proteins, DNA methyltransferases and Polycomb group proteins (42). Therefore, we have used PD assay to study whether pRb could interact with factors affecting the activity of the human topo IIα promoter, HMGB1 or NF-Y. Previous PD experiments revealed that HMGB1 and recombinant pRb (unphosphorylated form) physically interact in free solution (28). In order to clarify whether HMGB1 could interact with pRb in the cell, nuclear lysate from pRb-positive H1299 cells was immunoprecipitated by specific anti-HMGB1 antibody. As shown in Figure 4E, a weak band of co-immunoprecipitated pRb from H1299 lysate with HMGB1 was apparent, suggesting an interaction of HMGB1 and pRb in vivo (co-immunoprecipitation of pRb with HMGB1 was also evident by immunoprecipitation experiments using nuclear lysate from human erythroleukemia K562 cells, unpublished). pRb was not co-immunoprecipitated with pre-immune rabbit IgG (Figure 4E). The electrophoretic mobility of the co-immunoprecipitated pRb corresponded to the non-phosphorylated form of the pRb protein [Figure 4E, see also (43)], although we have not yet determined directly the phosphorylation state of pRb involved in the interaction with HMGB1.
To find out whether the different effect of the wild-type and mutant pRb on the inhibition of the HMGB1-mediated transactivation of the topo IIα promoter (Figure 4B) could be explained by the different interaction of HMGB1 with the wild-type pRb and mutant pRb, the PD assay was performed. Although HMGB1 could bind to both the wild-type and mutant pRb (amino-acid 706 C → Y within the A/B pocket domain) (Figure 4F), a higher fraction of HMGB1 (∼2–3-fold) was associated with the wild-type pRb than with the mutant pRb (equal amounts of GST-pRb immobilized on the glutathione-Sepharose beads were confirmed by immunodetection using anti-pRb antibody; lanes 1 and 2 in Figure 4E). Thus, the distinct effects of HMGB1 on the activity of the topo IIα promoter in pRb-negative or pRb-positive cells (Figure 4A) could not be explained solely by different affinities of the wild-type and mutant pRb for HMGB1 (Figure 4F).
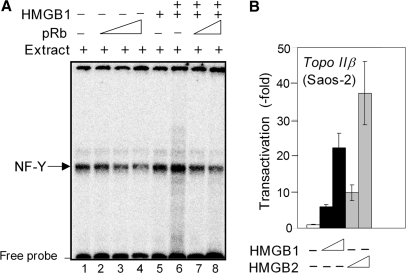
In order to understand the mechanism of pRb-mediated inhibition of the activity of the human topo IIα promoter, we have asked whether pRb could interact with NF-Y in vitro. As shown in Figure 2D (top), incubation of nuclear lysate from Saos-2 cells with GST-pRb immobilized on glutathione-agarose beads resulted in association of NF-Y with pRb. Similarly to the NF-Y binding to HMGB1, the interaction between NF-Y and pRb was not facilitated via DNA as similar results were obtained by PD assays in the presence or absence of ethidium bromide. The detection of pRb-(NF-Y) interactions prompted us to investigate using EMSA whether pRb could affect NF-Y binding to the specific ICE sites in vitro. Results shown in Figure 5A (lanes 2–5) demonstrated that binding of NF-Y to a short DNA duplex containing the ICE2 was visibly compromised by preincubation of nuclear lysate from Saos-2 cells with pRb (similar results were obtained from EMSA experiments using the ICE1, not shown). In addition, the HMGB1-mediated enhancement of NF-Y binding to the ICE2 was also reduced by preincubation of HMGB1 with pRb (Figure 5A; compare lanes 6, 7 and 8). We have verified that the observed reduction of NF-Y binding by pRb (Figure 5A) was not unspecific, as titration with un unrelated protein (GST) in comparable amounts to pRb had no effect (not shown). The above results suggested that pRb could counteract NF-Y binding to DNA duplexes containing inverted CCAAT elements.
Figure 5.
(A) pRb can reduce NF-Y binding to ICEs. Radioactively labeled 30-bp DNA duplex containing the ICE2 was mixed with nuclear extract from Saos-2 cells (lane 1) or nuclear extract containing increasing amounts of recombinant His-pRb [lane 2 (0.1 μg), lanes 3 and 7 (0.2 μg), lanes 4 and 8 (0.4 μg)]. In some reactions, HMGB1 was added [lane 5 (1 μM), lanes 6-8 (4 μM)]. Unbound DNA duplexes (free probe) and the DNA–protein complexes were resolved on 5% non-denaturing polyacrylamide gels (EMSA). (B) Up-regulation of human topo IIβ gene promoter by HMGB1 and HMGB2. The plasmid -700TOP2B-pGL3 bearing the human topo IIβ gene promoter was co-transfected with either pGL3 vector (empty bars), plasmid encoding HMGB1 (black bars) or plasmid encoding HMGB2 (gray bars) into the Saos-2 cells. Luciferase activity is presented as indicated in Figure 1.
HMGB-type family consists of three member—HMGB1, HMGB2 and HMGB3 with an 80% amino-acid identity among the three proteins. While HMGB1 is ubiquitously expressed in all mammalian cells (∼106 molecules per cell), expression of the other two family members is more restricted: HMGB2 is widely expressed during embryonic development, and HMGB3 is only expressed to a significant amount during embryogenesis (5,8). As both HMGB1 and HMGB2 relatively abundant in human cells (including Saos-2 cells), we have studied whether HMGB2 could also up-regulate the topo IIα gene promoter. As shown in Figure 4B, over-expression of HMGB2 in Saos-2 cells resulted in up to ∼2-fold higher transactivation of the topo IIα gene promoter relative to the effect of HMGB1. The latter was not due to different over-expression of HMGB1 and HMGB2 as similar levels of the two ectopically expressed proteins were immunodetected in transfected Saos-2 cells (not shown). Thus, it is likely that both HMGB1 and HMGB2 could transactivate the topo IIα gene in the cell.
The two isoforms of topoisomerase II, α and β, exhibit very similar catalytic activities, but differ in their production during the cell cycle (1–4). While the activity of the human topo IIα gene promoter was low in resting cells and enhanced during proliferation (such as in tumors), the activity of the human topo IIβ gene promoter remained constant throughout the cell cycle (49). Despite a distinct regulation of the two topo II gene promoters (49), NF-Y could bind to ICEs of both types of promoters [(49) and refs therein]. Therefore we were wondering whether HMGB1 and/or HMGB2 could also modulate transcriptional activity of the topo IIβ promoter. As shown in Figure 5B, the activity of the topo IIβ promoter in Saos-2 cells was dose-dependently up-regulated by HMGB1. Similarly to the topo IIα promoter (Figure 4B), the transactivation of the topo IIβ promoter was more prominent upon over-expression of HMGB2 (Figure 5B). Thus, the activity of both the topo IIα and topo IIβ promoters could be up-regulated by HMGB1/2.
HMGB1 and HMGB2 modulate cellular expression of topoisomerase IIα
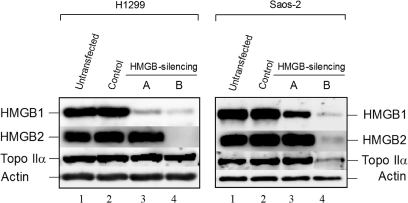
Results of this article indicated that HMGB1 and 2 could up-regulate the topo IIα promoter in the luciferase gene reporter assay (Figures 1 and 4), suggesting a possibility that HMGB1 and HMGB2 could affect cellular expression of topo IIα. To find out whether the cellular levels of topo IIα were affected by HMGB1 and/or HMGB2 in Saos-2 or H1299 cells, stable cell lines were generated producing plasmid-encoded short hairpin RNAs (shRNAs) specific for either HMGB1 or HMGB1/HMGB2 mRNAs. In order to verify silencing of HMGB1 and/or HMGB2 expression, proteins from nuclear lysates were resolved by electrophoresis, transferred onto the membrane and detected with specific antibodies recognizing HMGB1 or HMGB2. As shown in Figure 6, the expression of HMGB1 was inhibited in H1299 cells by >90% by shRNAs specific for HMGB1 mRNA (lane 3; H1299) or for HMGB1 and HMGB2 mRNAs (lane 4; H1299). The latter approach resulted in simultaneous knock-down of HMGB1 and HMGB2 expression. Similar results were obtained in Saos-2 cells although silencing of HMGB1 expression was only ∼60–70% (Figure 6, lane 3; Saos-2). Control silencing experiments revealed that the levels of HMGB1 and HMGB2 were unaffected in both H1299 and Saos-2 cells transfected with plasmid producing unrelated shRNA (Figure 6, lanes 2 as compared to lanes 1 for untransfected cells).
Figure 6.
Knock-down of HMGB1 and HMGB2 results in inhibition of topo IIα expression. Nuclear lysates from untransfected (lanes 1) and mock-transfected (lanes 2) cells, as well as from cells transfected with plasmids producing shRNAs specific for HMGB1 (lanes 3, construct #A) or HMGB1/HMGB2 (lanes 4, construct #B) were resolved on SDS-polyacrylamide gels and transferred onto the membranes by western blotting. Proteins were detected by specific antibodies as detailed in the Materials and methods section.
The cellular levels of topo IIα in cells with silenced HMGB1 or HMGB1/2 expression were then determined by western blotting and immunodetection. No significant changes in topo IIα protein levels were observed in pRb-positive H1299 cells upon silencing of HMGB1 or HMGB1/2 expression (Figure 6, lanes 3 and 4; H1299), albeit QRT-PCR analysis revealed minor decrease in topo IIα mRNA levels in H1299 cells with silenced HMGB1/2 expression (not shown). On the other hand, the topo IIα protein levels in pRb-negative Saos-2 cells with silenced HMGB1/HMGB2 expression were decreased up to ∼3-fold relative to control cells (Figure 6, lane 4; Saos-2). This finding was also supported by QRT-PCR analysis revealing ∼3-fold decrease in topo IIα mRNA levels in Saos-2 cells with silenced HMGB1/2 expression (data not shown). The fact that only very little (<20%) decrease in topo IIα protein levels was observed when solely HMGB1 expression was silenced in Saos-2 cells (Figure 6, lane 3; Saos-2) may be related to possible functional redundancy of these two closely related HMGB-type proteins (see Discussion section).
In conclusion, we have found that chromosomal proteins HMGB1 and HMGB2 could modulate cellular expression of topo IIα in cells lacking functional retinoblastoma gene susceptibility protein pRb by promoting binding of NF-Y to the topo IIα gene promoter. Our findings are discussed in the view of reported over-expression of HMGB1/2 proteins in tumors [(17) and refs therein], as well as the fact that the Rb gene is deleted/mutated in >50% human cancers (42).
DISCUSSION
Recently we have reported that chromosomal protein HMGB1 could stimulate catalytic activities of human topoisomerase IIα by enhancing DNA binding and cleavage by the enzyme (11). Here we report that HMGB1 and its close relative, HMGB2, could also up-regulate the activity of the human topo IIα gene promoter and cellular expression of the enzyme. Our results suggest that HMGB1 and HMGB2 proteins may serve as positive regulators of the cellular activity of the topo IIα gene (this article) as well as that of the enzyme (11).
HMGB1 and HMGB2 proteins have been implicated in regulation of chromatin structure and DNA metabolic processes such as transcription, replication, recombination and repair (5,8). The lack of HMGB1 in knockout mice results in death within a few hours after birth due to inefficient activation of glucocorticoid receptor responsive genes, but HMGB2 knockout mice are viable (15,47). HMGB2 protein could not substitute in mice for the loss of HMGB1 (15,47), suggesting that HMGB1 and HMGB2 proteins may have distinct roles, albeit most of the reported DNA-binding studies indicated that the in vitro DNA-binding properties of HMGB1 and HMGB2 were indistinguishable (5,7,8). Murine and human cells were viable without functional HMGB1, indicating that HMGB2 could partially compensate for the loss of HMGB1 in the cell (48). The possible functional redundancy of HMGB1 and HMGB2 (at least in some biological processes) may also explain our finding that the double knock-down of HMGB1 and HMGB2 in pRb-negative cells was necessary in order to observe a significant decrease in cellular levels of topoisomerase IIα. However, as we have not succeeded in silencing of HMGB2 expression only and/or without concomitant silencing of HMGB1 expression (using various shRNA constructs directed against different parts of hHMGB2 mRNA, unpublished results) we could not determine the exact contribution of the individual HMGB-type proteins to the cellular expression of topo IIα.
HMGB1 is an abundant and most mobile nuclear protein (∼1 × 106 molecules per nucleus), but despite its abundance the protein may be limiting within cells as evident from transient over-expression of HMGB1 resulting in transactivation of numerous genes (5,48). Previous transfection experiments revealed that HMGB1, but not HMGB1 lacking the acidic C-tail (HMGB1-ΔC), could stimulate the Gal4VP16-mediated reporter gene expression (52,53), or the p53-dependent transactivation of the p53-responsive promoter in H1299 cells (54). Transactivation of the human topoIIα gene promoter by HMGB1 did not require the acidic C-tail of HMGB1 suggesting that the C-tail did not function as an activator of transcription of the topoIIα promoter (this article). Higher DNA binding and/or bending/looping by HMGB1-ΔC relative to the full-length HMGB1 [see also (14,18,35)] may explain why the truncated HMGB1 protein activated the topo IIα gene promoter more efficiently than the full-length HMGB1. Thus, shielding of positively charged A+B di-domain of HMGB1 by the acidic C-tail (13,14,55) may account for lower ability of the full-length protein to up-regulate the topo IIα gene promoter.
We have demonstrated that intercalating residues of human HMGB1 (Phe 38 and Phe 103)—that were essential for the ability of HMGB1 to bend DNA [see also (12,33,34,56)]—were required for the ability of HMGB1 to enhance binding of NF-Y to ICEs and for the up-regulation of the topo IIα gene promoter (this article). Interestingly, the intercalating residues of HMGB1 proved to be dispensable for protein–protein interactions such as binding of HMGB1 to topoisomerase IIα (11 and unpublished results) or progesterone receptor (56), as well as for a specific binding of progesterone receptor to its cognate sites (56). The mechanism by which HMGB1/2 proteins stimulate activity of the topo IIα promoter most likely involves enhancement of transcription factors binding, possibly by pre-bending of the DNA promoter sequences. Numerous transcription factors have been reported to interact with HMGB1 and/or HMGB2, including TBP, Oct and Sp1 (8,27). Binding of NF-Y to short DNA duplexes containing ICEs1-4-binding sites (this article), as well as Oct-1/2 (74) and Sp1 (20) to their DNA cognate sites was enhanced by HMGB1. NF-Y, Sp1 and ICBP90 have previously been shown to be required for efficient transcription from human topo IIα and topo IIβ promoters [(23,49,50,51,58) and refs therein). The ability of HMGB1/2 to enhance NF-Y binding to the topo IIα promoter in vivo was to be explained by HMGB1-(NF-Y) interactions, and possibly also by HMGB1-mediated DNA pre-bending of the DNA-binding sites for NF-Y. The fact that no ternary HMGB1-(NF-Y)-DNA complex was detected did not necessarily mean the lack of existence of such a complex. It is possible that HMGB1 could ‘deliver’ NF-Y to its DNA-binding site by forming a ternary complex which is transient and unstable. This idea is in agreement with previous reports demonstrating the ability of HMGB1 to enhance binding a plethora of sequence-specific proteins to their cognate DNA sites without formation of ternary complexes, despite the detection of the corresponding protein–HMGB1 interactions in ‘pull-down’ assays (5,8,11). The existence of interactions of pRb with NF-Y (and also with HMGB1) may constitute a basis for understanding of pRb-mediated inhibition of the activity of the topo IIα promoter. The observed decrease in binding of NF-Y to ICEs by pRb, as well as inhibition of the ability of HMGB1 to promote binding of NF-Y binding to ICEs (this report), suggested that pRb could down-regulate the activity of the topo IIα promoter by preventing NF-Y binding to inverted CCAAT elements. The exact nature of the interactions involving NF-Y, HMGB1, pRb (phosphorylated or non-phosphorylated?) as well as other binding partners on the the topo IIα promoter is unknown. In addition, interaction of HMGB1 to NF-Y, as well as stimulation of NF-Y binding to several ICEs in vitro, may also explain our finding that sequential deletion of ICEs5-3 from the topo IIα promoter had very little effect on the ability of HMGB1 to up-regulate the promoter. Thus, our results could indicate possible functional redundancy of ICEs for the activation of the topo IIα promoter by HMGB1.
Results of this article demonstrated that HMGB1 and HMGB2 proteins could significantly up-regulate the human topo IIα gene promoter only in cells lacking functional retinoblastoma protein pRb. The exact reason for the inability of HMGB1/2 to significantly up-regulate the activity of the human topo IIα gene promoter in pRb-positive cells is unknown. However, the detectable (albeit slight) transactivation of the topo IIα promoter in pRb-positive cells could be related to weakening of the inhibitory effect of pRb on the promoter. This could be due to HMGB1 binding to pRb which could partially counteract the inhibitory effect of pRb on NF-Y binding to ICEs of the topo IIα promoter (see also the above paragraph). On the other hand, the fact that HMGB1 could significantly (>10-fold) enhance activity of the topo IIα promoter in Saos-2 cells transiently expressing mutant pRb (within the A/B pocket) was most likely related to the inability of the mutant to inhibit the activity of the topo IIα promoter, rather than to the pRb(mut)-HMGB1 interactions [see also (43)]. The possibility that the failure of the pRb mutant to inhibit the activity of the topo IIα promoter was related to the inability of the mutant to interfere with NF-Y binding to ICEs remains to be investigated.
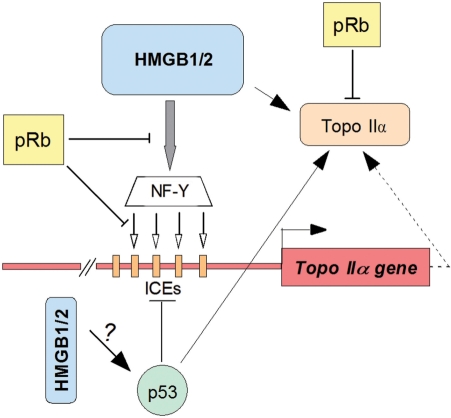
Previous reports indicated that p53 could also inhibit activity of the topo IIα gene promoter despite the lack of clear consensus p53-binding sites within the human topo IIα gene promoter (26). This inhibition was explained as consequence of an interference of p53 with NF-Y binding to regulatory sequences of the promoter (25,46). HMGB1 and HMGB2 proteins have previously been reported to promote binding of p53 (or its close relative, p73) to their specific DNA-binding sites by bending and specific p53–HMGB1/2 interactions (20,59). Whether the latter interactions could counteract or even enhance the reported p53-mediated inhibition of the topo IIα gene promoter activity (20,24,45,60) is unclear. Interestingly, the catalytic activity of topo IIα was stimulated by p53 (46), albeit by a mechanism different from that reported by HMGB1 (11). Contrary to the action of pRb or p53, the HMGB-type proteins could both transactivate the topo IIα promoter and stimulate the catalytic activity of the enzyme (11). A possible mechanism summarizing interplay of pRb, p53 and HMGB1/2 on the activity of the topo IIα gene and topoisomerase IIα, is outlined in Figure 7.
Figure 7.
Modulation of activity of human topo IIα gene and topoisomerase IIα by pRb and HMGB1/2 (a hypothesis). HMGB1 and HMGB2 proteins up-regulate the expression of the human topo IIα gene in pRb-negative cells by enhancement of binding of transcription factor NF-Y to its specific DNA-binding sites (ICEs) within the topo IIα gene promoter. Binding of NF-Y to the ICEs is facilitated by pre-bending the DNA sequences by HMGB1/2. pRb can inhibit the activity of the topo IIα gene, as well as the ability of HMGB1 to up-regulate the gene, possibly by reducing binding of NF-Y to ICEs (this article). HMGB1/2 promote cellular expression of topoisomerase IIα (this article), and the proteins have also the potential of enhancing catalytic activity of the enzyme, as previously demonstrated in vitro (11 and unpublished results). p53 inhibits activity of the topo IIα gene by compromising NF-Y binding (24,45). Unlike the inhibitory effect of pRb on the catalytic activity of topo IIα (22), the enzyme is stimulated by p53 (46).
The two isoforms of topoisomerase II, α and β, are encoded by distinct genes on chromosomes 17q21-22 and 3p24, respectively (61,62). While the activity of the human topo IIα gene promoter is low in resting cells and enhanced during proliferation (such as in tumors), the activity of the human topo IIβ gene promoter is constant throughout the cell cycle (49). Both isoforms differ in their production during the cell cycle (1–4), but they exhibit very similar catalytic activities, including the sensitivity to anticancer drugs specifically targeting topo II. The latter may be interesting in the view of our finding that the activities of both the topo IIα and topo IIβ gene promoters were up-regulated by HMGB1/2. Whether the HMGB1/2-mediated activation of the topo IIβ gene promoter is also affected by pRb, awaits further investigation.
Although HMGB1 is expressed throughout the cell cycle with no significant variations (5), the protein is clearly over-expressed in most human tumors, including breast carcinoma, melanoma, gastrointestinal stromal tumors, colon carcinomas and acute myeloblastic leukaemia [(5,8,17,63) and references herein]. HMGB1 over-expression has also been observed upon administration of hormones (estrogen and/or progesterone), resulting in increased potency of anticancer drugs cisplatin and its analogue carboplatin (64). Recently HMGB2 was also found to be over-expressed in cancer (http://expression.gnf.org/cgi-bin/index.cgi#Q; Gene Expression Atlas, 38065_at).
Most of the human tumors have excess DNA copies of the TOP2A (encoding topoisomerase IIα) gene [(65,66) and refs therein], and the TOP2A gene amplification may explain the significantly enhanced cellular levels of topo IIα in tumors (67,68). Topo IIα over-expression has been reported to be associated with poor cancer-specific survival and presence of metastases (66). Up-regulation of topo IIα could significantly affect responsiveness of tumors to drugs specifically targeting the enzyme (topo II poisons) (69,70). The fact that the HMGB1/2-mediated up-regulation of cellular levels of topo IIα was most prominent in Rb-minus cells (this article) raises the question of a possible relevance of simultaneous HMGB1/2 overexpression and Rb deletions in tumors in respect to clinical prognosis of patients treated with topo II poisons. While over-expression of HMGB1/2 proteins could enhance cellular levels (and/or activity) of topo IIα, reduced transcriptional activity of HMGB1/2 genes could diminish the cellular levels (and/or activity) of the enzyme. Several polymorphisms and/or mutations were recently identified in the HMGB1 gene with a potential regulatory impact on HMGB1 transcription (71). In this respect, the B-cell chronic lymphocytic leukemia (B-CLL), a malignant disease with highly variable clinical course, could be of interest due to deletions of the 13q14 locus (72). About 30–50% of patients with 13q14 deletions bear Rb deletions (some of them even biallelic, unpublished results). Interestingly, the HMGB1 gene is localized in the 13q12 locus (5,73). Furthermore, research is necessary to fully understand the involvement of HMGB-type proteins and their possible interplay with tumor suppressor proteins, pRb and p53, in modulation of cellular activity of topo IIα.
FUNDING
This research was supported by grants to M.Š. by the Grant Agency of the Czech Republic (204/08/1530), the Grant Agency of the Academy of Sciences of the Czech Republic (IAA400040702), and by the Academy of Sciences of the Czech Republic (grants no AV0Z50040507 and AV0Z50040702). Š.P. was supported by grant IGA MZ ČR NR/9293-3. Funding for open access charge: GA ČR 204/08/1530 and GA AS CR (IAA400040702).
Conflict of interest statement. None declared
ACKNOWLEDGEMENTS
We greatly appreciate the gifts of the following materials: stable Saos-2 derivatives expressing pRb under the tetracycline regulated promoter from Liang Zhu (Department Developmental and Molecular Biology, Albert Einstein College of Medicine, New York, USA); α-NF-YB antibodies from Roberto Mantovani (Dipartimento di Scienze Biomolecolari e Biotecnologie, Università di Milano, Milano, Italy); pcDNA-Zeo(-)-U6 contructs expressing specific shRNAs for silencing of HMGB1 or HMGB1/2 expression in human cells from Stephen J. Lippard (Department of Chemistry, MIT, Cambridge, USA); human wild-type and mutant pRb in pCMV vector from Pradip Raychaudhuri (Department of Biochemistry, College of Medicine, University of Illinois, Chicago, USA); plasmids encoding GST-fused wild-type pRb and mutant pRb from Sybille Mittnacht (Institute of Cancer Research, Chester Beatty Labs, London, UK); plasmid pRSET(his-Rb) encoding human wild-type His-pRb was kindly provided by Ronen Marmorstein (The Wistar Institute, Philadelphia, USA); plasmids pTIIα-617 and pTIIα-617(ICE2mt) from Kathryn M. Stowell (Institute of Molecular BioSciences, Palmerston North, New Zealand); plasmid pTIIα-557 and its truncated forms from Parker D. Suttle (Department of Pharmacology, College of Medicine, University of Tennessee, Tennessee, USA); human topo IIβ gene promoter plasmid construct from Susan P.C. Colle (Queen's University, Cancer Research Institute, Division of Cancer Biology & Genetics, Kingston, Canada). A skilful assistance of Alena Bačíková and Lucia Gulášová in some experiments is greatly appreciated. We also thank Lenka Juračková for excellent technical assistance, Jitka Malčíková for QRT-PCR results evaluation, and François Strauss (Université Pierre et Marie Curie, Paris) for critical reading of the manuscript and many helpful suggestions.
REFERENCES
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Osheroff N, Zechiedrich EL, Gale KC. Catalytic function of DNA topoisomerase II. Bioessays. 1991;13:269–273. doi: 10.1002/bies.950130603. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. Anticancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 4.Denny WA. Emerging DNA topoisomerase inhibitors as anticancer drugs. Expert Opin. Emerg. Drugs. 2004;9:105–133. doi: 10.1517/eoed.9.1.105.32948. [DOI] [PubMed] [Google Scholar]
- 5.Štros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol. Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi ME. Significant (re)location: how to use chromatin and/or abundant proteins as messages of life and death. Trends Cell Biol. 2004;14:287–293. doi: 10.1016/j.tcb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 8.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 9.Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 10.Pil PM, Chow CS, Lippard SJ. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc. Natl Acad. Sci. USA. 1993;90:9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Štros M, Bačíková A, Polanská E, Štokrová J, Strauss F. HMGB1 interacts with human topoisomerase IIalpha and stimulates its catalytic activity. Nucleic Acids Res. 2007;35:5001–5013. doi: 10.1093/nar/gkm525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaouen S, de Koning L, Gaillard C, Muselíková-Polanská E, Štros M, Strauss F. Determinants of specific binding of HMGB1 protein to hemicatenated DNA loops. J. Mol. Biol. 2005;353:822–837. doi: 10.1016/j.jmb.2005.08.073. [DOI] [PubMed] [Google Scholar]
- 13.Sheflin LG, Fucile NW, Spaulding SW. The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry. 1993;32:3238–3248. doi: 10.1021/bi00064a005. [DOI] [PubMed] [Google Scholar]
- 14.Štros M, Štokrová J, Thomas JO. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 1994;22:1044–1051. doi: 10.1093/nar/22.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 16.Giavara S, Kosmidou E, Hande MP, Bianchi ME, Morgan A, d'Adda di Fagagna F, Jackson SP. Yeast Nhp6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Curr. Biol. 2005;15:68–72. doi: 10.1016/j.cub.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 17.Völp K, Brezniceanu M-L, Bösser S, Brabletz T, Kirchner T, Göttel D, Joos S, Zörnig M. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut. 2006;55:234–242. doi: 10.1136/gut.2004.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Testa A, Donati G, Yan P, Romani F, Huang TH, Viganò MA, Mantovani R. Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J. Biol. Chem. 2005;280:13606–13615. doi: 10.1074/jbc.M414039200. [DOI] [PubMed] [Google Scholar]
- 19.Štros M. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J. Biol. Chem. 1998;273:10355–10361. [PubMed] [Google Scholar]
- 20.Štros M, Ozaki T, Bačíková A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J. Biol. Chem. 2002;277:7157–7164. doi: 10.1074/jbc.M110233200. [DOI] [PubMed] [Google Scholar]
- 21.Jiang H, Karnezis AN, Tao M, Guida PM, Zhu L. pRB and p107 have distinct effects when expressed in pRB-deficient tumor cells at physiologically relevant levels. Oncogene. 2000;19:3878–3887. doi: 10.1038/sj.onc.1203722. [DOI] [PubMed] [Google Scholar]
- 22.Bhat UG, Raychaudhuri P, Beck WT. Functional interaction between human topoisomerase IIα and retinoblastoma protein. Proc. Natl Acad. Sci. USA. 1999;96:7859–7864. doi: 10.1073/pnas.96.14.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magan N, Szremska AP, Isaacs RJ, Stowell KM. Modulation of DNA topoisomerase IIα promoter activity by members of the Sp (specificity protein) and NF-Y (nuclear factor Y) families of transcription factors. Biochem. J. 2003;374:723–729. doi: 10.1042/BJ20030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Zambetti GP, Suttle DP. Inhibition of DNA topoisomerase IIα gene expression by the p53 tumor suppressor. Mol. Cell Biol. 1997;17:389–397. doi: 10.1128/mcb.17.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamanaka R, Kohno K, Seguchi T, Okamura K, Morimoto A, Ono M, Ogata J, Kuwano M. Induction of low density lipoprotein receptor and a transcription factor SP-1 by tumor necrosis factor in human microvascular endothelial cells. J. Biol. Chem. 1992;267:13160–13165. [PubMed] [Google Scholar]
- 26.Bronner C, Hopfner R, Mousli M. Transcriptional regulation of the human topoisomerase IIα gene. Anticancer Res. 2002;22:605–612. [PubMed] [Google Scholar]
- 27.Das D, Scovell WM. The binding interaction of HMG-1 with the TATA-binding protein/TATA complex. J. Biol. Chem. 2001;276:32597–32605. doi: 10.1074/jbc.M011792200. [DOI] [PubMed] [Google Scholar]
- 28.Dintilhac A, Bernués J. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J. Biol. Chem. 2002;277:7021–7028. doi: 10.1074/jbc.M108417200. [DOI] [PubMed] [Google Scholar]
- 29.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 30.Herzog CE, Zwelling LA. Evaluation of a potential regulatory role for inverted CCAAT boxes in the human topoisomerase IIα promoter. Biochem. Biophys. Res. Commun. 1997;232:608–612. doi: 10.1006/bbrc.1997.6267. [DOI] [PubMed] [Google Scholar]
- 31.Isaacs RJ, Harris AL, Hickson ID. Regulation of the human topoisomerase IIα gene promoter in confluence-arrested cells. J. Biol. Chem. 1996;271:16741–16747. doi: 10.1074/jbc.271.28.16741. [DOI] [PubMed] [Google Scholar]
- 32.Morgan SE, Beck WT. Role of an inverted CCAAT element in human topoisomerase IIα gene expression in ICRF-187-sensitive and -resistant CEM leukemic cells. Mol. Pharmacol. 2001;59:203–211. doi: 10.1124/mol.59.2.203. [DOI] [PubMed] [Google Scholar]
- 33.Wong B, Masse JE, Yen YM, Giannikopoulos P, Feigon J, Johnson RC. Binding to cisplatin-modified DNA by the Saccharomyces cerevisiae HMGB protein Nhp6A. Biochemistry. 2002;41:5404–5414. doi: 10.1021/bi012077l. [DOI] [PubMed] [Google Scholar]
- 34.Klass J, Murphy FV, 4th, Fouts S, Serenil M, Changela A, Siple J, Churchill ME. The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity. Nucleic Acids Res. 2003;31:2852–2864. doi: 10.1093/nar/gkg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teo S-H, Grasser KD, Thomas JO. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur. J. Biochem. 1995;230:943–950. doi: 10.1111/j.1432-1033.1995.tb20640.x. [DOI] [PubMed] [Google Scholar]
- 36.Grasser KD, Teo S-H, Lee KB, Broadhurst RW, Rees C, Hardman CH, Thomas JO. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1) Eur. J. Biochem. 1998;253:787–795. doi: 10.1046/j.1432-1327.1998.2530787.x. [DOI] [PubMed] [Google Scholar]
- 37.Štros M. Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry. 2001;40:4769–4779. doi: 10.1021/bi002741i. [DOI] [PubMed] [Google Scholar]
- 38.He Q, Ohndorf UM, Lippard SJ. Intercalating residues determine the mode of HMGB1 domains A and B binding to cisplatin-modified DNA. Biochemistry. 2000;39:14426–14435. doi: 10.1021/bi001700j. [DOI] [PubMed] [Google Scholar]
- 39.Jung Y, Lippard SJ. Nature of full-length HMGB1 binding to cisplatin-modified DNA. Biochemistry. 2003;42:2664–2671. doi: 10.1021/bi026972w. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 41.Xiao H, Goodrich DW. The retinoblastoma tumor supressor protein is required for efficient processing and repair of trapped topoisomerase II-DNA-cleavable complexes. Oncogene. 2005;24:8105–8113. doi: 10.1038/sj.onc.1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, Dibling B, Spike B, Dirlam A, Macleod K. New roles for the RB tumor supressor protein. Curr. Opin. Genet. Dev. 2004;14:55–64. doi: 10.1016/j.gde.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Jiao Y, Wang HC, Fan SJ. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol. Sin. 2007;28:1957–1967. doi: 10.1111/j.1745-7254.2007.00669.x. [DOI] [PubMed] [Google Scholar]
- 44.Semizarov D, Kroeger P, Fesik S. siRNA-mediated gene silencing: a global genome view. Nucleic Acids Res. 2004;32:3836–3845. doi: 10.1093/nar/gkh714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandri MI, Isaacs RJ, Ongkeko WM, Harris AL, Hickson ID, Broggini M, Vikhanskaya F. p53 regulates the minimal promoter of the human topoisomerase IIα gene. Nucleic Acids Res. 1996;24:4464–4470. doi: 10.1093/nar/24.22.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon Y, Shin BS, Chung IK. The p53 tumor supressor stimulates the catalytic activity of human topoisomerase IIα by enhancing the rate of ATP hydrolysis. J. Biol. Chem. 2000;275:18503–18510. doi: 10.1074/jbc.M002081200. [DOI] [PubMed] [Google Scholar]
- 47.Ronfani L, Ferraguti M, Croci L, Ovitt CE, Schöler HR, Consalez GG, Bianchi ME. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development. 2001;128:1265–1273. doi: 10.1242/dev.128.8.1265. [DOI] [PubMed] [Google Scholar]
- 48.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Lok C-N, Lang AJ, Mirski SE, Cole SP. Characterization of the human topoisomerase IIβ (TOP2B) promoter activity: essential roles of the nuclear factor-Y (NF-Y)- and specificity protein-1 (Sp1)-binding sites. Biochem. J. 2002;368:741–751. doi: 10.1042/BJ20020791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon JH, Kim JK, Rha GB, Oh M, Park S-H, Seong RH, Hong SH, Park SD. Sp1 mediates cell proliferation-dependent regulation of rat DNA topoisomerase IIα gene promoter. Biochem. J. 1999;344:367–374. [PMC free article] [PubMed] [Google Scholar]
- 51.Hopfner R, Mousli M, Oudet P, Bronner C. Overexpression of ICBP90, a novel CCAAT-binding protein, overcomes cell contact inhibition by forcing topoisomerase II alpha expression. Anticancer Res. 2002;22:3165–3170. [PubMed] [Google Scholar]
- 52.Aizawa S, Nishino H, Saito K, Kimura K, Shirakawa H, Yoshida M. Stimulation of transcription in cultured cells by high mobility group protein 1: essential role of the acidic carboxyl-terminal region. Biochemistry. 1994;33:14690–14695. doi: 10.1021/bi00253a006. [DOI] [PubMed] [Google Scholar]
- 53.Ueda T, Chou H, Kawase T, Shirakawa H, Yoshida M. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transciption stimulation. Biochemistry. 2004;43:9901–9908. doi: 10.1021/bi035975l. [DOI] [PubMed] [Google Scholar]
- 54.Banerjee S, Kundu TK. The acidic C-terminal domain and A-box of HMGB-1 regulates p53-mediated transcription. Nucleic Acids Res. 2003;31:3236–3247. doi: 10.1093/nar/gkg412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watson M, Stott K, Thomas JO. Mapping intramolecular interactions between domains in HMGB1 using a tail-truncation approach. J. Mol. Biol. 2007;374:1286–1297. doi: 10.1016/j.jmb.2007.09.075. [DOI] [PubMed] [Google Scholar]
- 56.Roemer SC, Adelman J, Churchill ME, Edwards DP. Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding. Nucleic Acids Res. 2008;36:3655–3666. doi: 10.1093/nar/gkn249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beck WT, Mo YY, Bhat UG. Cytotoxic signalling by inhibitors of DNA topoisomerase II. Biochem. Soc. Trans. 2001;29:702–703. doi: 10.1042/0300-5127:0290702. [DOI] [PubMed] [Google Scholar]
- 58.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- 59.Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu D, Huang CL, Kameyama K, Hayashi E, Yamauchi A, Sumitomo S, Yokomise H. Topoisomerase II alpha gene expression is regulated by the p53 tumor supressor gene in nonsmall cell lung carcinoma patients. Cancer. 2002;94:2239–2247. doi: 10.1002/cncr.10450. [DOI] [PubMed] [Google Scholar]
- 61.Tsai-Pflugfelder M, Liu LF, Liu AA, Tewey KM, Whang-Peng J, Knutsen T, Huebner K, Croce CM, Wang JC. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21-22. Proc. Natl Acad. Sci. USA. 1988;85:7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jenkins JR, Ayton P, Jones T, Davies SL, Simmons DL, Harris AL, Sheer D, Hickson ID. Isolation of cDNA clones encoding the β isozyme of human DNA topoisomerase II and localisation of the gene to chromosome 3p24. Nucleic Acids Res. 1992;20:5587–5592. doi: 10.1093/nar/20.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brezniceanu ML, Völp K, Bösser S, Solbach C, Lichter P, Joos S, Zörnig M. HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. FASEB J. 2003;17:1295–1297. doi: 10.1096/fj.02-0621fje. [DOI] [PubMed] [Google Scholar]
- 64.He Q, Liang CH, Lippard SJ. Steroid hormones induce HMG1 overexpression and sensitize breast cancer cells to cisplatin and carboplatin. Proc. Natl Acad. Sci. USA. 2000;97:5768–5772. doi: 10.1073/pnas.100108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Washiro M, Ohtsuka M, Kimura F, Shimizu H, Yoshidome H, Sugimoto T, Seki N, Miyazaki M. Upregulation of topoisomerase IIalpha expression in advanced gallbladder carcinoma: a potential chemotherapeutic target. J. Cancer Res. Clin. Oncol. 2008;134:793–801. doi: 10.1007/s00432-007-0348-0. [DOI] [PubMed] [Google Scholar]
- 66.Järvinen TA, Liu ET. Topoisomerase IIalpha gene (TOP2A) amplification and deletion in cancer – more common than anticipated. Cytopathology. 2003;14:309–313. doi: 10.1046/j.0956-5507.2003.00105.x. [DOI] [PubMed] [Google Scholar]
- 67.Rody A, Karn T, Ruckhäberle E, Müller V, Gehrmann M, Solbach C, Ahr A, Gätje R, Holtrich U, Kaufmann M. (March 14, 2008) Gene expression of topoisomerase II alpha (TOP2A) by microarray analysis is highly prognostic in estrogen receptor (ER) positive breast cancer. Breast Cancer Res. Treat. doi: 10.1007/s10549-008-9964-x. 10.1007/s10549-008-9964-x. [DOI] [PubMed] [Google Scholar]
- 68.Guérin E, Entz-Werlé N, Eyer D, Pencreac’h E, Schneider A, Falkenrodt A, Uettwiller F, Babin A, Voegeli AC, Lessard M, Gaub MP, Lutz P, Oudet P. Modification of topoisomerase genes copy number in newly diagnosed childhood acute lymphoblastic leukemia. Leukemia. 2003;17:532–540. doi: 10.1038/sj.leu.2402774. [DOI] [PubMed] [Google Scholar]
- 69.Pritchard KI, Messersmith H, Elavathil L, Trudeau M, O'Malley F, Dhesy-Thind B. HER-2 and topoisomerase II as predictors of response to chemotherapy. J. Clin. Oncol. 2008;26:736–744. doi: 10.1200/JCO.2007.15.4716. [DOI] [PubMed] [Google Scholar]
- 70.Arriola E, Rodriguez-Pinilla SM, Lambros MBK, Jones RL, James M, Savage K, Smith IE, Dowsett M, Reis-Filho JS. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res. Treat. 2007;106:181–189. doi: 10.1007/s10549-006-9492-5. [DOI] [PubMed] [Google Scholar]
- 71.Kornblit B, Munthe-Fog L, Petersen SL, Madsen HO, Vindeløv L, Garred P. The genetic variation of the human HMGB1 gene. Tissue Antigens. 2007;70:151–156. doi: 10.1111/j.1399-0039.2007.00854.x. [DOI] [PubMed] [Google Scholar]
- 72.Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 73.Ferrari S, Finelli P, Rocchi M, Bianchi ME. The active gene that encodes human high mobility group 1 protein (HMG1) contains introns and maps to chromosome 13. Genomics. 1996;35:367–371. doi: 10.1006/geno.1996.0369. [DOI] [PubMed] [Google Scholar]