Abstract
Replication protein A (RPA) is a heterotrimeric (70, 32 and 14 kDa subunits), single-stranded DNA-binding protein required for cellular DNA metabolism. All subunits of RPA are essential for life, but the specific functions of the 32 and 14 kDa subunits remains unknown. The 32 kDa subunit (RPA2) has multiple domains, but only the central DNA-binding domain (called DBD D) is essential for life in Saccharomyces cerevisiae. To define the essential function(s) of RPA2 in S. cerevisiae, a series of site-directed mutant forms of DBD D were generated. These mutant constructs were then characterized in vitro and in vivo. The mutations had minimal effects on the overall structure and activity of the RPA complex. However, several mutants were shown to disrupt crosslinking of RPA2 to DNA and to dramatically lower the DNA-binding affinity of a RPA2-containing subcomplex. When introduced into S. cerevisiae, all DBD D mutants were viable and supported normal growth rates and DNA replication. These findings indicate that RPA2–DNA interactions are not essential for viability and growth in S. cerevisiae. We conclude that DNA-binding activity of RPA2 is dispensable in yeast and that the essential function of DBD D is intra- and/or inter-protein interactions.
INTRODUCTION
Replication protein A (RPA) is a eukaryotic, single-stranded DNA (ssDNA)-binding protein; RPA binds ssDNA with high affinity and low specificity (1–3). RPA is highly conserved throughout evolution and has been found in all eukaryotes examined (1,4). RPA's two major functions are to bind ssDNA and to interact with proteins. These interactions are essential for cellular DNA replication, repair and recombination. RPA has also been implicated in the regulation of transcription and in the coordination of DNA metabolism with other cellular processes (1–3).
RPA is a heterotrimer composed of three structurally related subunits of 70, 32 and 14 kDa (RPA1, RPA2 and RPA3, respectively). All three subunits are essential for formation of a stable, functional RPA complex, and all three genes have been shown to be essential in yeast (the gene for RPA1 is referred to as RFA1, RPA2 is RFA2 and RPA3 is RFA3 in yeast) (5). The 70 kDa subunit is composed of four OB fold domains required for both DNA and protein interactions (6). The 32 kDa subunit contains three domains: an N-terminal flexible domain (7,8), a central, essential, OB-fold domain designated DNA-binding domain (DBD) D and a C-terminal protein-interaction domain. The 14 kDa subunit contains only one domain; this OB-fold domain (DBD E) appears to be primarily a structural component of the RPA complex (9) though contacts between it and ssDNA have recently been detected (10–12).
All of the OB folds in RPA can interact with ssDNA. Structural and crosslinking analyses suggest that the domains bind with a 5′ → 3′ polarity (13–15). Of the four domains in RPA1 (F, A, B and C), DBD A and B form a high-affinity DNA-binding core (16,17), while DBD F and C both bind with weaker affinity (3,18). DBD D in RPA2 has been shown to interact with DNA in the context of the heterotrimer (19–22), but can also weakly bind DNA as a subcomplex with RPA3 (23,24). DBD E in RPA3 has recently been shown to bind to telomere sequences and to contact ssDNA when the RPA complex binds to ssDNA (10,11).
The exact mechanism by which RPA modulates its interactions with various DNA substrates is unknown. RPA undergoes multiple conformational changes in the presence of ssDNA (25), which is in part dependent on the length of the DNA substrate (26). Analysis of RPA mutations in the conserved aromatic residues in each DBD imply that the different DBDs are involved in interactions with different lengths of ssDNA (19). This is supported by cross-linking studies, where RPA1 associates more with DNA duplexes containing short (10 nt) ssDNA tails, but RPA2 is primarily crosslinked to DNA duplexes containing long (30 nt) ssDNA tails (21,27,28). This may be functionally important, as RPA2 has been shown to initially cross-link to the growing DNA chain during DNA replication in isolated nuclei (29,30).
The data discussed earlier have lead to several models of DNA binding by RPA and implicated RPA2–DNA interactions in regulation of RPA function. It has been suggested that DBDs A and then B, the high-affinity core, first bind to a small region of DNA. This is followed by binding of DBDs C and D to form a stable, 30-nt binding mode (3). There is also evidence for a tighter packing of RPA on ssDNA at high RPA to DNA ratios (19,21,27,28). In this 10-nt binding mode, it is thought that DBDs A and B are primarily interacting with the DNA. It has also been suggested that the interactions of DBD C and D with DNA are modulated by interactions with other proteins and that this regulates RPA function and promotes the departure of RPA from ssDNA (3,30). This model, called the handoff model, predicts that RPA2–DNA interactions are important for the cellular function of RPA.
There is little direct evidence on the role of RPA2–DNA interactions in vivo. There have been two previous studies analyzing the function of RPA2 mutants (rfa2) in Saccharomyces cerevisiae (31,32). Mutations in RFA2 have significant phenotypes including reduced growth, temperature sensitivity, DNA damage sensitivity and increased rates of spontaneous mutation. The most severe phenotypes have been observed with mutations in DBD D and DBD D domain is the only region RPA2 shown to be essential for viability in yeast (33). However, several of the rfa2 mutants with strong phenotypes also caused decreased protein stability and/or complex formation. Therefore, it is unresolved whether the phenotypes resulted from altered DBD D function or decreased structural stability. It is currently not known what activities of RPA2 are essential for RPA function in the cell.
The goal of these studies was to determine the role of DBD D of RPA2. Current studies have suggested three possible, but not mutually exclusive, functions for this domain: DNA-binding, regulation (protein interactions) and structural (complex formation). Using existing structures for RPA domains and the sequence homology between RPA domains, we have generated a series of mutant forms of yeast RPA2 with changes in predicted DNA-binding site of DBD D. These mutants were then analyzed for biochemical function and their ability to function in the cell. All mutant forms were able to form complexes with the other subunits of RPA in vitro and were viable in vivo. Several of the mutants had altered conformations based on cross-linking analysis and were sensitive to DNA damaging agents. However, these phenotypes did not correlate with reduced RPA2–DNA interactions. These studies demonstrate that RPA2–DNA interactions are not an essential function of this subunit. This analysis suggests that RPA2 is necessary for structural integrity and may also be needed for intra- and/or inter-protein interactions.
MATERIALS AND METHODS
Materials
Oligonucleotides were synthesized by either Sigma (St Louis, MO) or Integrated DNA Technologies (Coralville, IA). Hydroxyurea (HU) was purchased from Sigma-Aldrich (St Louis, MO). The parent vector YCp52 was a gift from Bob Deschenes (Medical College of Wisconsin). S4-dUTP (4-thio-2′-deoxyuridine-5′-triphosphate) was kind gift of V. Bogachev (Institute of Chemical Biology and Fundamental Medicine, Russia). DNA polymerase β isolated according to (34) was a kind gift of Dr S Khodyreva (Institute of Chemical Biology and Fundamental Medicine, Russia).
The HI buffers used for protein purification and fluorescence polarization (FP) contained 30 mM HEPES (diluted from a 1 M stock, pH 7.8), 5 mM MgCl2, 0.5% (w/v) inositol, 1 mM dithiothreitol and indicated concentration of KCl (e.g. HI-100 contains 100 mM KCl).
Nomenclature
This manuscript will refer to the mutant forms of yeast RPA2 according to the abbreviations listed in Table 1. Mutant RPA subunits will be referred to as RPA2 (mutation), the trimeric RPA complex containing this mutation will be listed as RPA•2(mutation), the dimeric subcomplex containing this mutation will be RPA2(mutation)/3 and the gene will be referred to as rfa2-mutation. For example, the forms of the K135A, F137A mutation (subunit, complexes and yeast gene) will be referred to as RPA2(KF), RPA•2(KF), RPA2(KF)/3 and rfa2-KF, respectively.
Table 1.
Nomenclature of mutants
| Mutations in yeast RPA2 | Abbreviation | Primers | Restriction site |
|---|---|---|---|
| S159A,N161A | SN | AAG CCC ATA GAT GCA TTC GCA GAA GTG TTG ACG CAT CAC | NsiI |
| GTG ATG CGT CAA CAC TTC TGC GAA TGC ATC TAT GGG CTT | |||
| V134V,K135A,F137A | KF | GAA ATT GGC GGT TAC GTC GCA GTT GCA GGT GCT TTG AAA GAG | [SnaB1] |
| CTC TTT CAA AGC ACC TGC AAC TGC GAC GTA ACC GCC AAT TTC | |||
| T79T,D80A,T82A | DT | GGT GTG GTG AGA AAC ATT ACT GCG CAT GCT GCA AAT ATT | FspI |
| AAT ATT TGC AGC ATG CGC AGT AAT GTT TCT CAC CAC ACC | |||
| L96I,E97A,R99A,K100A | ERK | GA ACT GGT CAA TTG GCA GTG AGA AAA TGG AGC GAA GAT G | MfeI |
| CAT CTT CGC TCC ATT TTC TCA CTG CCA ATT GAC CAG TTC | |||
| GA ACT GGT CAA TTA GCA GTG GCA GCA TGG AGC GAA GAT G | [MfeI] | ||
| CAT CTT CGC TCC ATG CTG CCA CTG CTA ATT GAC CAG TTC | |||
| G145G,K146A,K147A | KK | GCT TTG AAA GAG TTT GGT GGC GCC GCA AAT ATA CAG | KasI |
| CTG TAT ATT TGC GGC GCC ACC AAA CTC TTT CAA AGC | |||
| D158A,E162A,V163V | DE | AAG CCC ATA GCT TCA TTC AAT GCA GTT TTG ACG CAT | [HincII] |
| GAT GCG TCA AAA CTG CAT TGA ATG AAG CTA TGG GCT T | |||
| E103A,D104A | ED | GA AAA TGG AGC GCG GCC GCA AAT GAC TTG GC | EagI |
| GCC AAG TCA TTT GCG GCC GCG CTC CAT TTT C | |||
| W101A,F143A,G144G | WF | GAA GTG AGA AAA GCT AGC GAA GAT GCA | NheI |
| TGC ATC TTC GCT AGC TTT TCT CAC TTC | |||
| GGT GCT TTG AAA GAG GCC GGC GGT AAG AAA AAT A | NgoMIV | ||
| TAT TTT TCT TAC CGC CGG CCT CTT TCA AAG CAC C | |||
| T79T,D80A,T82A, L96I,E97A,R99A,K100A, G145G,K146A, K147A | MM1 | same as for DT, ERK, KK | FspI, MfeI, KasI |
| T79T,D80A,T82A,D158A, E162A,V163V, E103A, D104A | MM2 | same as for DT, DE, ED | FspI, EagI, NheI |
Desired mutations shown in bold, conservative mutations used to aid cloning shown in plain text. Nucleotide changes in primers indicated in bold. Restriction site created or [eliminated] indicated in final column.
Construction of mutations in RPA2
Site-directed mutagenesis was carried out using the QuikChange Site-Directed Mutagenesis kit or the QuikChange Multi Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Primers contained changes that would either result in the conversion of polar/charged/aromatic residues to alanine residues or result in silent or conserved mutations that generated/eliminated restriction sites (Table 1). All constructs were sequenced at the DNA Core Facility (University of Iowa).
All RPA2 mutants in trimeric RPA, with the exception of RPA•32(W101A, F143A), were generated by site-directed mutagenesis of the plasmids p11d-tscRPA (23), pET28a-RFA2,RFA3 (35) or prs315-RFA2 (35). To generate constructs containing all three RPA genes, RFA1 was cloned into pET28a-RFA2,RFA3 containing the desired mutation or by site-directed mutagenesis of plld-scRPA (35). Mutant RPA•2(W101A, F143A) was expressed from the plasmid pSAS203, a gift from Steve Brill (Rutgers University, NJ). RPA2/3 dimers were expressed from the vector pET28a-RFA2/RFA3 (35). Mutants were made by direct mutagenesis of pET28a-RFA2/RFA3 using the QuikChange II Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) with the respective primers (Table 1) or by mutagenesis in additional vectors followed by subcloning into pET28a-RFA2,RFA3.
All yeast expression constructs containing RPA2 mutants were made by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) with the respective primers (Table 1) directly in prs315-RFA2 or in were subcloned into prs315-RFA2 (35).
Expression and protein purification of RPA complexes
Heterotrimeric RPA complexes were expressed and purified as described previously (36).
RPA dimeric subcomplexes were expressed using pET28a constructs that contained wild-type RFA3 and wild-type or mutant RFA2 containing a His-tag on the N-terminus. Each expression plasmid was transformed into BL21(DE3) cells, grown, induced and lysed as described for trimeric RPA (36). After cell lysis, the soluble fraction (60 ml at 30 mg/ml) was loaded onto 60 ml of Nickel resin (Qiagen, Valencia, CA) that had been washed in HI-100 mM KCl. The column was washed in HI-100 mM KCl buffer with a gradient of 0–10 mM imidazole for three column volumes. It was then washed in HI-100 mM KCl buffer with a gradient of 10–300 mM imidazole for 10 column volumes followed by two column volumes of HI-100 mM KCl, 500 mM imidazole. RPA2/3 eluted at a concentration of ∼135 mM imidazole, 100 mM KCl. RPA-containing fractions were loaded onto a Mono-Q column equilibrated with HI-50 mM KCl. The column was washed sequentially with four column volumes of HI-50 mM KCl and HI-100 mM KCl prior to elution with 10 column volumes of linear salt gradient in HI-buffer containing 200–400 mM KCl. RPA 2/3 eluted as a single peak at ∼200 mM KCl. RPA2/3 was then concentrated to 25–30 mg/ml in HI-100 buffer using Centricon tubes with a 3000 molecular weight cut off. All protein concentrations were determined by a Bradford assay using BSA as a standard (37).
Fluorescence polarization (FP) assays
DNA-binding assays were carried out as described previously (36). Eighty-microliter reactions containing HI buffer with the indicated salt, RPA and fluorescein-conjugated ssDNA were assembled at room temperature in the wells of black 96-well FluoroNunc plates (Nalge Nunc International, Rochester, NY). Trimeric RPA complexes were assayed over a concentration range of 0–1000 nM. The high salt concentrations were required to obtain equilibrium-binding conditions with the RPA trimer; therefore, RPA trimer-binding assays were carried out in 1.25 M KCl unless otherwise indicated. DNA (oligo-deoxythymidine 30 nt in length with a 5′ fluorescein moiety) was added last to a final concentration of 5 nM. Plates were covered with aluminum foil and gently rocked for 10 min to allow sample equilibration. The plates were then uncovered and assayed for polarization. The excitation wavelength of fluorescein is 480 nm with a 30 nm bandwidth, and the emission wavelength is 535 nm with a 40 nm bandwidth. Data were collected using a measurement time of 0.2 s per well. Anisotropy is defined as r or R = (Ivv − Ivh)/(Ivv + 2GIvh), where Ivv is the vertically excited and vertically emitted light and Ivh is the vertically excited and horizontally emitted light. G is an instrumental correction factor. Polarization is defined as P = 3r/2 + r. Data were analyzed by fitting to the Langmuir-binding equation as described by Kim et al. (38). All binding experiments were performed three to six times.
Experiments with RPA dimers were performed as described earlier with the following modifications. Dimeric RPA was titrated from 0 to 100 µM. Viscosity was also controlled in these experiments by adding BSA so that the BSA and RPA together equaled a total protein content of 3.65 μg. The final salt concentration was 100 mM NaCl, 21 mM KCl and 5 mM MgCl2. Plates were covered with aluminum foil and gently rocked for 60 min to allow sample mixing and protein binding.
Yeast strains
TWY177 (mec1-1; MAT a, leu2, ura3, his3, trp1) was a gift from Ted Weinert (University of Arizona). WY2 (MAT a, leu2, ura3, ade2, ade8, his3, met4) was a gift from Bob Deschenes (Medical College of Wisconsin). AMSY-1 (MAT a, leu2, ura3, ade2, ade8, his3, met4, rfa2:Kmr) was derived from WY2 and has a chromosomal knockout of the RPA2 gene (35). The rad52-1 (MAT α, ade2, lys2, tyr1, ura3, cyn2, his7) construct was a gift from Bob Malone [University of Iowa (39)].
Yeast methods
The plasmid YCp52-RFA2, containing a wild-type copy of the RFA2, ADE8 and URA3 genes, was transformed into AMSY-1. The RPA2 mutant genes used for in vitro assays were cloned into the yeast vector prs-315 that contained a LEU2 biosynthesis gene and were then transformed into the yeast strain AMSY1 by using the Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, CA). The plasmid YCp52-RFA2 that contained the wild-type copy of RFA2 was then exchanged for the pRS315 plasmid that contained the mutant form of RFA2 by growing the cells in leucine-deficient media. When plated on YEPD, cells that had lost the YCp52-RFA2 plasmid were white and cells that retained the plasmid were red. White cells were selected and streaked on sc-Ura/glc and a master plate of sc-Leu/glc. Cells that had lost the wild-type RFA2 plasmid could not grow on sc-Ura/glc but could grow on the master plate of sc-Leu/glc and were used for further analysis.
Plasmid stability was determined based on the rate of loss of a non-essential plasmid (31). AMSY-1 cells containing the various prs315-mutant rfa2 plasmids were transformed with YCp50. Cultures were then grown to log phase in sc-Ura media, transferred to the rich, nonselective YEPD media and grown overnight. Cells were then plated on YEPD and –Ura media. Plasmid loss rates were calculated by comparing the number of colonies on –Ura plates to the number of colonies on YEPD plates. Plasmid loss rate is compared to the loss rate of strain containing wild-type RFA2.
Sensitivity to DNA damaging agents
AMSY1cells were grown to log phase and then serial dilutions (10 µl of 103, 104, 105 and 106 cells/ml) were plated on YEPD and grown for 3 days at 30°C. HU sensitivity was determined using YEPD plates containing 0.1 M HU. UV sensitivity was determined by exposing plates to UV doses of 0–200 J/M2 and incubating in the dark to prevent repair by photolyase. After growth, plates were assessed for the number of remaining viable colony-forming units. All mutants were examined in two or more independent experiments.
Synthesis of photoreactive DNA structures
Photoreactive DNA structures were made by using the activity of DNA polymerase β to add a photoreactive 4-thio-dUMP residue to the 3′-end of a primer in a primer-template (27). Ten microliters of reaction mixtures containing 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, 20 μM DNA-duplex, 5.6 μM DNA polymerase β and 1 μM 4-thio-dUTP (4-thio-2′-deoxyuridine-5′-triphosphate) were incubated for 20 min at 37°C to allow elongation of the primers. Subsequently, the mixtures were incubated at 70°C for 5 min, cooled slowly and centrifuged to remove precipitated DNA polymerase β. To check the resulting annealing efficiency, aliquots 1 μM of the sample volume were brought to a final concentration of 5% Ficoll-400 and 0.05% bromophenol blue and electrophoresed on a 12% polyacrylamide gel containing 89 mM Tris–HCl pH 8.8, 89 mM boric acid and 2 mM EDTA at 100 V/cm. Radioactive bands were visualized by autoradiography.
Photochemical crosslinking
Reaction mixtures (10 μl) contained 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, 2.5 μg BSA, 0.01% Tween, 0.1 μM 5′-[32P]-labeled photoreactive DNA and the indicated amounts of RPA or one of its mutant forms (from 0.3 × 10−7 to 2.5 × 10−7 M). The reaction mixtures were preincubated in micro-Eppendorf tubes for 10 min at 37°C and UV-irradiated on ice through the open top. The probes were irradiated for 45 min. A Lomo VIO-1 UV-crosslinker (St Petersburg, Russia) equipped with lamps producing UV-light of 334–365 nm was used as light source. Reactions were stopped by adding Laemmli loading buffer and heating for 5 min at 95°C. The photochemically crosslinked protein–DNA samples were separated by 12.5% SDS–PAGE. Gels were dried and subjected to autoradiography using a ‘Molecular imager FX Pro+’ (BioRad) screen.
RESULTS
Sequence alignments and identification of putative DNA-interacting residues in RPA2
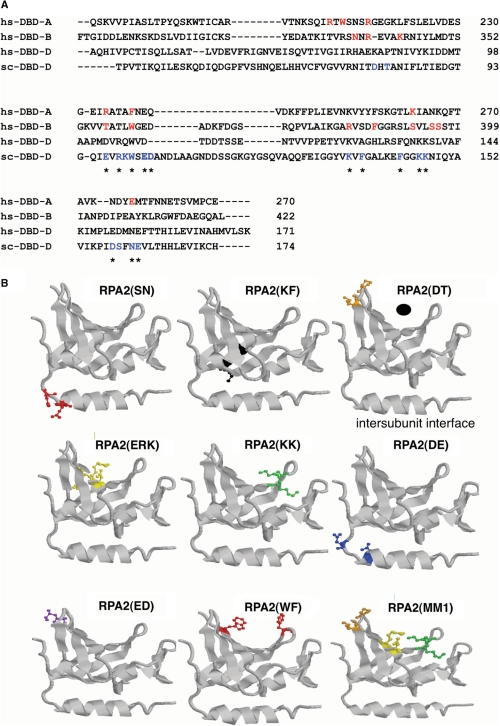
The structure of yeast RPA2 has not been determined and there are no structures of any RPA2 subunit interacting with ssDNA. Overall, the amino acid sequences of human and yeast RPA are 44% similar with the DBDs having the highest level of conservation (1,5). Therefore, sequence homology was used to identify the putative DNA-binding interface of yeast RPA2. The crystal structure of the high-affinity DNA-binding core (DBD A and DBD B) of human RPA1 bound to DNA has been determined (13). The structure of DBD A from the yeast 70 kDa subunit is similar to the homologous domain of human RPA and the residues that interact with DNA are similar (40). Mutation of individual polar residues that interact with DNA dramatically and additively decreased RPA's affinity for ssDNA (16,41). By analogy, we hypothesized that the DNA-interaction site of RPA2 would contain a combination of polar and aromatic residues that interact directly with DNA. Furthermore, we predicted that mutation of these residues would disrupt binding of RPA2 to DNA.
The amino acid sequences of human RPA1 core DBDs (DBD A and DBD B) were aligned with the DBD D sequences from yeast and human RPA2 (Figure 1A). The residues known to interact with DNA in RPA1 are in red (Figure 1A) and polar or aromatic residues in similar positions in RPA2 in blue (Figure 1A). All the identified residues were highly conserved between the six sequenced Saccharomyces genomes and showed partial conservations between more distantly related fungal species (data not shown). These residues were targeted as putative DNA-binding residues in our mutagenesis and each was mutated to alanines to disrupt the putative DNA-binding site of RPA2. Eight mutant genes were generated with each containing two to three of the putative DNA-interacting residues mutated to alanine (Table 1). In addition, two combination mutants, termed multi-mutants, were generated that contained seven (D80A, T82A, E97A, R99A, K100A, K146A, K147A) and six alanine (D80A, T82A, E103A, D104A, D158A, E162A) substitutions, these will be referred to as RPA2(MM1) and RPA2(MM2), respectively (Table 1).
Figure 1.
RPA DBD D mutants. (A) Sequence alignment of protein sequence of human DBD A (residues 181–289), B (residues 302–442) and D (residues 45–171) and yeast DBD D (residues 40–174). The amino acid sequences aligned by Clustal W. Residues from human RPA known from the crystal structure of DBD A and B to interact with DNA are shown in red. Mutations made in S. cerevisiae DBD D are indicated in blue. Mutated residues that are conserved between humans and yeast RPA2 are starred. Dashes indicate a gap in the alignment. (B) Models of DBD mutant forms. The crystal structure of human DBD D was used to model equivalent primary residues (aligned by Clustal W) that had been mutated in yeast DBD D using RasMol. In all the models, the DNA-binding cleft is on the top and the intersubunit interface (which includes the horizontal alpha helix) is on the bottom. To help the reader orient the models, the upper right model has the putative position of DNA indicated by a black dot and the intersubunit interface labeled. The mutated yeast residue side chains were modeled onto the homologous residue in the human DBD D structure and shown in color (stick form). All residues were mutated to alanine. Mutants shown: RPA2(SN), homologous human residues D151,N153; RPA2(KF), human residues K127,A129; RPA2(DT), human residues K85,P87; RPA2(ERK), human residues D103,R105,Q106; RPA2(KK), human residues K138,K139; RPA2(DE), human residues E150,E159; RPA2(ED), human residues D109,T110; RPA2(WF), human residues W107,F135; RPA2(MM1) is composed of DT, ERK and KK, human residues K85,P87,D103,R105,Q106,K138,K139; RPA2(MM2) is not shown.
Each of these residues was also mapped onto the structure of human RPA2 (Figure 1B). The structure of human RPA2 has a very shallow putative DNA-binding cleft relative to the OB-folds in the DNA-binding core of RPA1 (24,42) (see upper right model in Figure 1B). Nevertheless, most of the putative DNA-binding residues identified by homology with RPA1 were modeled to be in or near the putative binding cleft of the human RPA2 structure. Two mutants, RPA2(SN) and RPA2(DE), were predicted to be near to intersubunit interface of RPA2 (Figure 1B) (43). However, because the structure of yeast RPA2 is not known and the role of individual regions of RPA2 poorly understood, these mutants were also analyzed to determine their effect on RPA2 DNA-binding and cellular function. Taken together, this set of mutations represents a directed alanine-scan of the DBD of RPA2.
Biochemical characterization of RPA2 mutants
Each of the mutant forms of RPA2 was expressed with the two other RPA subunits in E. coli and the resulting RPA complexes were purified (Supplementary Figure 1). All the mutant forms purified as heterotrimeric complexes with yields similar to that of wild-type RPA. This shows that all of the mutants were stable and interacted with the other two RPA subunits to from heterotrimeric complexes.
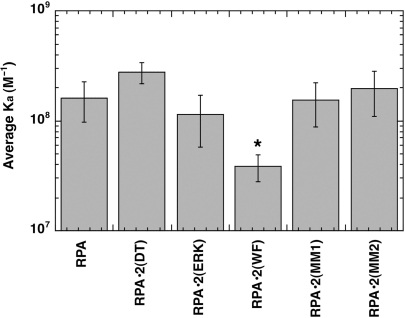
The DNA binding of the mutant RPA complexes was analyzed by gel mobility shift (GMSA) and FP assays. All the forms showed high-affinity binding in GMSA to oligonucleotides between 20 and 40 nt. The association constants determined for each of the heterotrimeric mutant complexes were not statistically different from that of wild-type RPA (data not shown). To try to define any subtle differences in DNA-binding activity, a subset of the mutants was also analyzed by fluorescence polarization (FP). FP is a direct technique that measures the anisotropy of a fluorescently labeled DNA probe when it is titrated with protein. An advantage of FP is that it can be used to directly determine apparent binding constants under a variety of solution conditions. Thus, it was possible to directly determine the binding constant under equilibrium conditions. RPA•2(ERK), RPA•2(DT), RPA•2(MM1) and RPA•2(MM2) had binding constants equivalent to wild-type RPA (Figure 2). In contrast, RPA•2(WF) had slightly reduced binding to dT30 (Ka of RPA = 1.6 × 108 and Ka RPA•2(WF) = 3.85 × 107; P = 0.018). RPA•2(MM1) and RPA•2(MM2) had wild-type binding in all assays examined. Each of the multi-mutants contained three sets of mutations (Table 1). These results strongly indicate that none of the mutations contained in the two multi-mutants (DT, ERK, KK, DE and ED) had a significant effect on binding either individually or in combination. We conclude that none of the mutant forms of RPA2 except for RPA•2(WF) effect binding of the RPA complex to ssDNA but that the RPA complex containing RPA•2(WF) has only a slightly reduced affinity for DNA. These data confirm previous work that showed that mutations in DBD D have only subtle effects on overall binding to DNA but that the aromatic mutant (W101A,F143F) reduced the affinity of DBD D (19).
Figure 2.
Affinity of yeast heterotrimeric RPA complexes containing mutations in RPA2 for dT30. Binding constants of wild-type or mutant forms of RPA were determined by FP assays at 1.25 M KCl. The average association constant (Ka) for each mutant is presented. The experiments were performed at least three times with the standard deviation shown. *The only mutant that had an association constant that was statistically different than wild-type (using a one-way ANOVA with a Tukey post hoc analysis) was RPA•2(WF).
Viability, growth and plasmid stability of rfa2 mutants
We next analyzed their properties in yeast cells. Each mutant form of RPA2 was placed under the control of the endogenous RFA2 promoter on a low copy yeast plasmid. These genes are each designated rfa2-‘mutation’ using standard yeast terminology. Each was then analyzed in a RPA2 knockout strain, AMSY1 (generated by disrupting the RFA2 genomic locus with a kanamycin resistance gene).
All of the DNA-binding domain mutants were capable of supporting life; all were able to support growth when they were the only form of rfa2 in the cell (Table 2). Growth was also observed at 37°C or 25°C (Table 2). All the mutants had wild-type growth rates at 30°C (Table 2). We conclude that all of these mutants were able to support chromosomal DNA replication and other essential cellular processes.
Table 2.
Summary of mutant phenotypes
| Yeast mutant | Growth |
Doubling time 30°C (min) | Growth HU | Growth UV | Plasmid loss (%) | Dimer binding Ka (M−1) | ||
|---|---|---|---|---|---|---|---|---|
| 25° | 30° | 36° | ||||||
| RFA2 | + | + | + | 171 ± 14 | + | + | 31 ± 3 | 1.5 × 104 |
| rfa2-SN | + | + | + | 148 | + | + | ||
| rfa2-KF | + | + | + | 160 ± 11 | + | + | ≤0.1 × 104 | |
| rfa2-DT | + | + | + | 163 ± 15 | + | + | 36 ± 5 | |
| rfa2-ERK | + | + | + | 168 ± 22 | +/– | +/– | 40 ± 5 | |
| rfa2-KK | + | + | + | 166 ± 5 | + | + | ||
| rfa2-DE | + | + | + | 167 ± 9 | + | + | ||
| rfa2-ED | + | + | + | 162 ± 12 | + | + | ||
| rfa2-WF | + | + | + | 165 ± 10 | +/– | +/– | 32 ± 3 | ≤0.15 × 104 |
| rfa2-MM1 | + | + | + | 181 ± 23 | +/– | +/– | 44 ± 9 | ≤0.6 × 104 |
| rfa2-MM2 | + | + | + | 179 ± 20 | + | + | ||
| sec 23 | + | – | – | |||||
| rfa2-Δ wh | + | +/– | – | +/– | ||||
| mec1-1 | – | |||||||
| rad52-1 | – | |||||||
Phenotypes of RPA mutants indicated. Growth of AMSY1 cells containing the indicated rfa2 gene on a plasmid is indicated at different temperatures or with the indicated DNA damaging agent (+ growth, +/– reduced or slow growth, – no growth). Data for selected control strains is also shown: sec23 is temperature sensitive, mec1-1 is HU sensitive and rad52-1 is weakly UV sensitive; rfa2-Δwh [deletion of the C-terminal winged-helix domain of RPA2; (31)] has been shown to have a weak UV sensitive phenotype. The ability of selected strains to support the replication of a non-selected plasmid (plasmid loss) is also shown. Estimated DNA binding constants for selected RPA2/3 complexes is also shown. Blank—not determined.
rfa2 mutant responses to ultraviolet irradiation
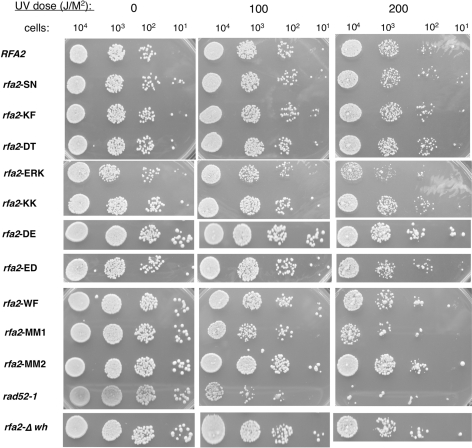
Previously, RPA2 mutations have been shown to cause defects in DNA repair after exposure to UV light (31,32). Cells containing the mutant forms of rfa2 were exposed to different levels of UV radiation and the number of viable cells determined. Two strains were used as controls: a deletion of the C-terminal winged-helix domain of RPA2 [rfa2-Δwh; (31)] and rad52-1. The RAD52 epistasis group is involved in homologous recombination, and it is not the major pathway by which cells repair UV-induced lesions. Thus, mutants with defects in the RAD52 epistasis group are only partially sensitive to UV; they show sensitivity only at high doses of radiation as observed in the experiment shown in Figure 3. Cells containing rfa2-Δwh are temperature sensitive and sensitive to methyl methane sulfonate but are not sensitive to UV damage at 25°C (31). Only three of the new rfa2 mutant strains were found to have minimal UV-sensitivity: rfa2-WF, rfa2-ERK and rfa2-MM1, which contains rfa-ERK (Figure 3). Strain rfa2-ERK and rfa2-MM1 had an intermediate UV phenotype, with sensitivity between wild-type RFA2 and the rad52-1 control (Figure 3). A similar phenotype was observed in a second RFA2 knockout strain, AWY1 (data not shown). Mutant strain rfa2-WF also had a weak but reproducible decrease in colony number indicated a slight UV sensitivity in both AMSY1 and AWY1 strains (Figure 3 and data not shown). All other DBD-D mutants showed no increase in UV sensitivity (Figure 3).
Figure 3.
Sensitivity of rfa2 mutant strains to UV. AMSY1 cells containing the indicated rfa2 gene on a plasmid were grown at 30°C to log phase (OD600 ∼0.5) and the indicated number plated on YEPD plates. The plates were irradiated with the respective doses of UV light and incubated in the dark at 25°C for 3 days. Only a subset of the doses is shown. The rad52-1 strain (Bob Malone, University of Iowa) and rfa2-Δwh [deletion of the C-terminal winged-helix domain; (31)] were used as controls.
One explanation for the UV sensitive phenotype could be that these mutations affect protein stability. Indeed, characterization of most published rfa2 mutants suggests that the lethal phenotypes at 37°C were due to disrupted complex formation or decreased RPA2 stability (31,32). To rule out altered protein levels as the cause of the observed phenotypes of the mutants, western analysis was performed with yeast extracts from cells containing rfa2-WF, rfa2-ERK and rfa2-MM1. Other strains wild-type RFA2, rfa2-DT and rfa2-MM2 were included as controls. The membranes were incubated with antibodies against actin and RPA2 (Supplementary Figure 2). Quantification of the bands observed followed by statistical analysis indicated that all mutants had wild-type protein levels. This suggests that the modest phenotypes present in these mutants were not caused by either underexpression or overexpression of the mutated proteins.
Since the most sensitive DBD D mutants were less sensitive to UV than rad52-1, a protein not directly involved in UV damage recovery, we conclude that DBD D only makes a minor contribution to UV damage recovery. This modest phenotype appears to be caused by a change in protein activity with these mutants.
rfa2 mutant effects on DNA replication
HU is a potent inhibitor of ribonucleotide reductase, which inhibits DNA replication and causes replication fork stalling by reducing the deoxynucleotide pools in the cell. We examined the sensitivity of rfa2 strains to HU treatment, as compared to a Mec1 kinase mutant mec1-1. Mec1 is the yeast homologue of the damage response kinase ATR and is the primary sensor for collapsed replication forks.
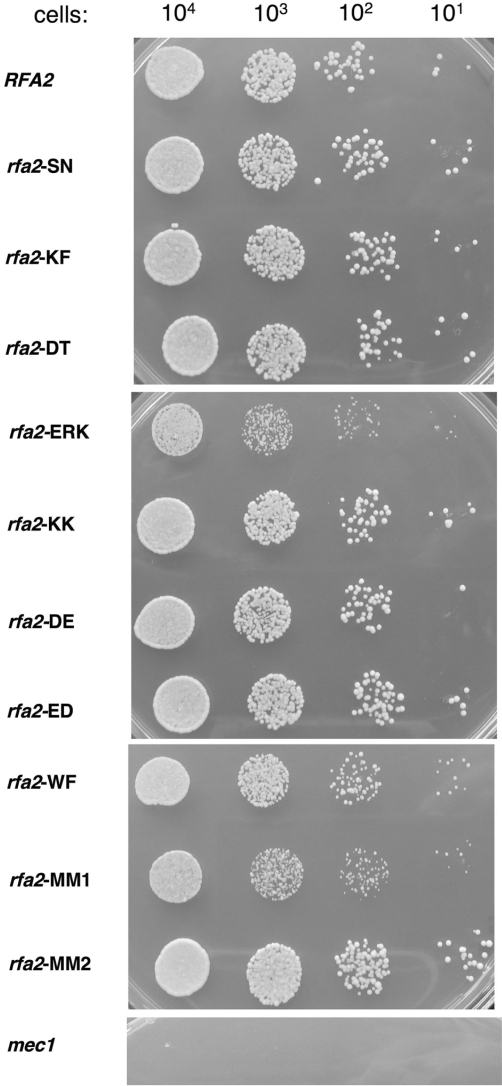
Of the 10 mutant rfa2 genes examined in AMSY1, only mutant strains rfa2-WF, rfa2-ERK and its derivative rfa2-MM1 had modest slow-growth phenotypes when grown on plates containing 0.1 M HU (Figure 4). Similar numbers of colonies were observed with these mutant strains but in all cases the colonies observed were much smaller that wild-type or other rfa2 mutant strains. This phenotype was modest compared with the mec1-1 strain, which was inviable at this concentration of HU (Figure 4). A subset of the mutants was examined in a different strain, AWY1. They were less sensitive to HU in this background, with only the mutant strain rfa2-(ERK) having detectable HU sensitivity (data not shown). These findings suggest that the sites mutated in DBD-D are not essential but may play a modest role in the cellular response to collapsed DNA replication forks in yeast.
Figure 4.
Growth phenotype of rfa2 strains on HU. AMSY1 cells containing the indicated rfa2 gene on a plasmid were grown at 30°C to log phase (OD600 ∼0.5) and the indicated number plated on YEPD plates containing 0.1 M HU. The plates were incubated at 30°C for 3 days. The mec1-1 strain was used as a control (55).
All yeast chromosomes have multiple origins of replication and contain essential genes: therefore, it is difficult to detect partial defects in DNA replication by chromosome loss or viability assays. To examine DNA replication of a non-essential episome, we determined plasmid loss rates for selected mutant strains including wild-type, rfa2-DT, rfa2-WF, rfa2-ERK and its derivative rfa2-MM1. In these experiments, cells were grown with a plasmid under non-selecting conditions and the rate at which the plasmid was lost determined. Elevated plasmid loss rates indicate reduced efficiency of DNA replication. All the strains lost the non-selected plasmid at rates comparable to that of a strain containing wild-type RPA2 (data not shown, summarized in Table 2). These findings indicate that none of the mutations have an effect on plasmid DNA replication under these growth conditions.
Cumulatively, these analyses (summarized in Table 2) suggest that the mutations made in the putative DNA-binding site of DBD D have no effect on growth and DNA replication, but in some cases, show minor defects in DNA damage response. We conclude that these mutations have minimal effects on RPA function in the cell.
Effects of mutations on the intrinsic affinity of RPA2
The findings that these mutations have minimal effects on the biochemical activity of the RPA complex and no or modest phenotypes in vivo raise the question of to what extent the mutations have directly affected the DNA-binding activity of the RPA2 subunit. RPA2 has a low affinity for DNA in comparison with the high affinity of the RPA complex, making it difficult to determine the effects of RPA2 mutations on DNA binding of the trimer complex (23,44). In order to quantitate the DNA-binding activity of RPA2 directly, it was necessary to analyze binding of a subcomplex containing only the 32 and 14 kDa subunits. Therefore, we expressed and purified from E. coli RPA2/3 sub-complexes of two forms that had no phenotype in vivo [wild type and RPA2(KF)] and two forms that had a phenotype in vivo [RPA2(MM1) and RPA2(WF) (Supplementary Figure 1)]. All four of these subcomplexes forms were expressed at high levels in E. coli and purified with similar yields to wild-type RPA2/3 (data not shown). The affinity of each complex was then analyzed using FP assays.
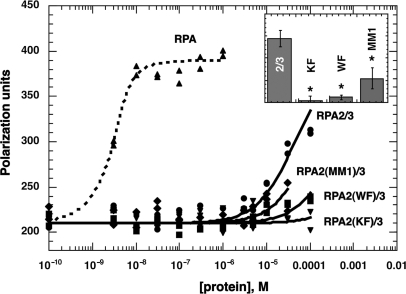
Under near-physiological salt conditions (100 mM NaCl, 21 mM KCl and 5 mM MgCl2), the binding of heterotrimeric RPA to dT30 was near stoichiometric; it saturated quickly at ∼10 nM protein and reached a maximal polarization value of ∼400 U (Figure 5). Under the same conditions, a wild-type RPA2/3 subcomplex required more than four orders of magnitude higher protein concentrations to bind the same amount of DNA; binding reached near saturation at a protein concentration of 100 µM (Figure 5). The physical limitations of the assay prevented analysis of higher concentrations of protein. This result indicated that the affinity of RPA2/3 for DNA is at least four orders of magnitude lower than that of the trimeric RPA complex.
Figure 5.
Affinity of yeast dimeric RPA2/3 complexes containing mutations in RPA2 for dT30 under physiological salt conditions. Increasing amounts of yeast RPA and yeast dimeric RPA2/3 complexes containing mutations in RPA2 were incubated with 5 nM fluorescein-conjugated dT30 in 5 mM MgCl2, 21 mM KCl and 100 mM NaCl buffer and fluorescence polarization determined. The raw data from wild-type, trimeric RPA (triangles); RPA2/3 (circles); RPA2(WF)/3 (squares); RPA2(KF)/3 (inverted triangles), and RPA2(MM1)/3 (diamonds) are shown. Data were fit to a langmuir binding isotherm (curves shown). Insert. The association constant for wild-type RPA2/3 (2/3) and upper limit estimated association constants for RPA2(KF)/3 (KF), RPA2(WF)/3 (WF) and RPA2(MM1)/3 (MM1) are shown. Reproducibility of the assay is indicated by error bars. *The upper limit estimates of all three mutants were statistically different than wild-type (using a one-way ANOVA with a Tukey post hoc analysis).
To confirm this finding, DNA binding was examined at other ionic strengths. RPA binding is ionic strength dependent (45,46). At higher ionic strength, essentially no binding of the RPA2/3 complex was observed (data not shown). At low ionic strength (30 mM KCl), saturation of binding was obtained at slightly lower protein concentrations and the polarization maximum obtained at saturation was similar to the level of polarization obtained with wild-type trimeric RPA (data not shown). These studies demonstrated that similar anisotropy values were obtained with both trimer and dimer RPA complexes under stoichiometric-binding conditions. Therefore, we fit the RPA2/3 subcomplex binding data (Figure 5) assuming that polarization would have saturated at the same maximum polarization observed with the trimeric RPA. This allowed us to estimate the association constant of wild-type RPA2/3 for dT30 to be ∼1.5 × 104 M−1 (Figure 5). This association constant is five orders of magnitude lower than that of the trimeric yeast RPA complex (23).
All three mutant subcomplexes appeared to bind to DNA with a lower affinity than the wild-type subcomplex. Addition of up to 100 µM of RPA2(KF)/3, RPA2(WF)/3 or RPA2(MM1)/3 resulted in polarization signals either at or slightly above background (Figure 5). It was not possible to determine the absolute binding affinity for these mutant complexes. However, assuming (1) that the small changes in polarization signal observed with the mutants were caused by binding and (2) that the isotherms would have saturated if sufficient protein was added, we estimated the maximum binding constants for each of the mutant subcomplexes (curves shown for mutant forms in Figure 5). This analysis provided estimates of the upper limit for the binding constants of these mutant forms: 10, 10 and 40% that of wild-type dimer for RPA2(KF)/3, RPA2(WF)/3 and RPA2(MM1)/3, respectively (Table 2; Figure 5, insert). These differences are statistically significant from wild-type (P < 0.001, P < 0.001 and P = 0.002, respectively, using a one-way ANOVA followed by a post hoc Tukey test). We conclude that all three of these mutants have dramatically lower binding than the wild-type dimeric complex. These results demonstrate that these mutations in RPA2 significantly decreased RPA2–DNA interactions.
There was no correlation between in vivo phenotype observed with these mutants and the DNA-binding affinity of DBD D. Of the three mutant forms examined, RPA2(MM1) had the strongest phenotype; however, this mutant also was estimated to have the highest affinity of the three mutant subcomplexes. RPA2(WF) had a phenotype in vivo and effected binding of the trimeric RPA complex (Figure 2) but appeared to have an intrinsic affinity for DNA that was similar to RPA2(KF), which has no phenotype in vivo. These data argue that the ssDNA-binding activity of RPA2 is not essential for RPA function in the cell.
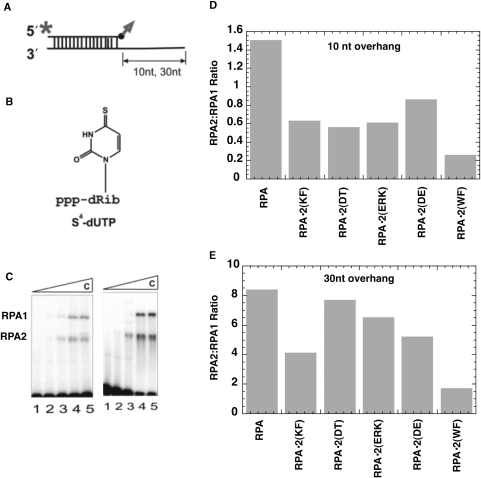
RPA2 interactions with primer-template junctions
When RPA binds to a partially duplex primed single-stranded DNA, RPA2 has been shown to crosslink to the 3′ end of a primer strand (21,27,47). This interaction is believed to reflect binding of DBD D with the primer strand and is dependent on the binding and conformation of the RPA complex (14,21,27,28). To try to understand the effects of the RPA2 mutation on the biochemical function of the RPA complex, we also examined the interactions of the trimeric RPA mutant complexes with primer template junctions. A single-stranded template strand was annealed with a 5′ radiolabel primer strand that then had a 4-thio-deoxyuridine incorporated on the 3′ terminus (Figure 6A and B). Using different length template strands allowed us to create primer template junctions with either a 10 or 30-nt ssDNA tail. The DNA was then incubated with RPA at different protein:DNA ratios, UV irradiated to activate the photolabel, separated on an SDS–PAGE gel, and autoradiography was performed to visualize crosslinked complexes. The extent of RPA2 cross-link is always compared with crosslinking of RPA1 as an internal control (Figure 6C).
Figure 6.
Photoaffinity labeling of yeast trimeric RPA complexes containing mutations in RPA2. (A) Schematic of DNA duplexes used in crosslinking assays, with radiolabel (asterisk) and photoreactive group (arrow) shown. The upper strand is the primer strand and the bottom strand is the template. (B) 4-Thio-dUMP photoreactive moiety that is conjugated to the 3′ end of the primer. (C) Example of raw data from crosslinking experiments with wild-type RPA and partial DNA duplexes: left, 10-nt ssDNA tail; right, 30-nt ssDNA tail. For both gels, lane 1: no protein; lane 2: 1:2 molar ratio of RPA:DNA; lane 3: 1:1 molar ratio; lane 4: 2:1 molar ratio; and lane 5: 2.5:1 molar ratio. (D and E) Quantification of a representative crosslinking experiment. The bars indicate the ratio of intensities of RPA2 to RPA1 crosslinking to DNA duplexes with a 10-nt overhang (D) or 30-nt (E) overhang at a molar RPA:DNA of 1:1.
In previous studies, the amount of crosslinking of RPA2 observed has depended on both the affinity of the complex and length of the single-stranded region available for binding (21,27,28). This is thought to reflect intrinsic DNA-binding activity of RPA2 and its position in the RPA–DNA complex. When crosslinking of the mutant complexes were examined with a 10-nt overhang, all showed reduced crosslinking of RPA2 (Figure 6D). It has been observed previously that human wild-type RPA2 is crosslinked more efficiently with a 30-nt overhang (21,27,28). This increase was observed with all of the mutants examined (compare ratios in Figures 6C and 6D). In addition, we observed more variation in crosslinking with the 30-nt overhang. RPA•2(DT) crosslinked at levels near wild-type RPA and the two mutant RPA•2(WF) and RPA•2(KF) had the lowest levels of crosslinking to the DNA (∼20% and ∼40% of wild-type, respectively; Figure 6E and data not shown). These data do not distinguish whether these differences reflect a difference in the conformation of the RPA–DNA complex or a change in the interactions of the mutant RPA2 subunit and the DNA (see also ‘Discussion’ section). RPA•2(WF) had the lowest level of cross-linking of any of the mutant forms with both DNA substrates. This is consistent with RPA•2(WF) being the only trimeric complex that had reduced affinity for ssDNA.
Two general conclusions can be drawn from the crosslinking experiments. First, the reduction in crosslinking observed with the 10-nt overhang suggests that these mutants are affecting the DNA-binding activity of DBD D or are changing the conformation of the RPA–DNA complex in these RPA complexes. Second, the changes in crosslinking observed with both substrates do not correlate with activity in the cell: RPA•2(WF) had the lowest level of crosslinking with both substrates and a modest repair phenotype, RPA•2(KF) had reduced crosslinking with both DNA substrates but no phenotype, and RPA•2(ERK) and RPA•2(DT) had higher crosslinking with the 30-nt overhang but only (ERK) had a phenotype. This supports the general conclusion that the DNA-binding activity of RPA2 is not critical for RPA complex function in the cell.
DISCUSSION
RPA contains six DBDs. All these domains can interact with DNA (3,11,16–18). Previous studies provide evidence for multiple interactions between the different DBDs of RPA and DNA. A variety of deletion and mutational analyses have indicated that DBD A and DBD B are both necessary and sufficient for high-affinity DNA binding by RPA (16,17). Electron microscopic analysis showed that human RPA binds as a compact form with short ssDNA substrates and as an elongated form with long ssDNA substrates (48). Mutation of aromatic residues in four DBDs of yeast RPA indicate that DBD A and B are involved in binding to DNA of all lengths, whereas DBD C and D seem to contribute modestly to binding to long substrates (19). Two binding modes have been identified for yeast RPA; at low salt concentrations, a mode with an occluded binding site size of 18–21 nt was observed while at high salt concentrations, a larger site size of 26–28 nt was observed (46). Cross-linking analysis has also demonstrated that multiple domains of RPA are in close proximity to the primer-template junction when RPA binds to partially duplex-ssDNA (27,28,47). These types of interactions are also observed in situ, where RPA2 is initially crosslinked to nascent RNA–DNA primers but RPA1 becomes crosslinked as the primers are elongated into Okazaki fragments (29,30). Together these and other data have been used to suggest that different domains of RPA bind sequentially to form different complexes depending on the length of the DNA bound (3). Furthermore, it has been suggested that the positioning and weak DNA affinity of RPA2 is used to modulate RPA–DNA binding through interactions with other proteins in a ‘hand-off’ model of DNA binding (3).
To test these models and to gain a better understanding of the role of RPA2–DNA binding, we generated and characterized a series of polar or aromatic residue to alanine mutations in yeast DBD D with the intent of disrupting RPA2–DNA interactions. These mutations primarily affected residues in the putative DNA-binding cleft of DBD D but also included some residues predicted to be in other regions of the domain. These mutants were then analyzed in vitro and in vivo. None of these mutant forms affected the formation of the RPA complex in vitro or the stability of the protein in vivo. All these mutant rfa2 alleles were also viable and supported normal DNA replication in yeast. Three of the mutant rfa2 strains (rfa2-ERK, rfa2-MM1 and rfa2-WF) were found to have modest DNA-repair phenotypes in vivo. All three strains contained mutations of residues in the putative DNA-binding cleft and one mutant [RPA2(WF)] caused reduced binding of the trimeric RPA complex. However, other mutant forms examined also had mutations in the putative DNA-binding cleft (rfa2-DT, rfa2-KK, rfa2-ED and rfa2-MM2) and had no discernable phenotype in yeast. Thus, there was no simple correlation between the location of the mutations and their effect on cell function. Direct analysis of DNA-binding activity of the mutated subunits and crosslinking efficiency also did not simply correlate to the phenotypes of the mutants. For example, RPA2(KF), which had low ssDNA binding, reduced crosslinking to primer template junctions but no phenotype. In contrast, mutant RPA2(MM1) had a higher affinity for DNA but also had the strongest DNA damage phenotype. These data argue that RPA2–DNA interactions are not essential for RPA function in cell viability and growth.
The ‘hand-off’ model was originally proposed based on analysis of protein interactions in SV40 DNA replication (3). This model suggests that the winged helix of RPA2 recruits proteins and allows them to gain access to the DNA by displacing DBD C and D from the DNA. Although our findings do not rule out a role for the winged-helix domain of RPA2 in protein recruiting, they argue that interactions with DNA are not essential for RPA2 function and indicate that the ‘hand-off’ model is not occurring in yeast. This is not surprising as yeast cells with a deletion of the winged helix survive and have no or minimal phenotypes (31–33). Deletion of the winged helix in yeast also only has a marginal effect on the DNA damage response (31,32).
If the DNA-binding activity of RPA2 is not essential, what is the essential activity of this domain? At minimum, RPA2 is necessary for the formation of the RPA complex (9). In addition, a number of protein-interactions have been mapped to RPA2 (3) indicating that DBD D could also be participating in essential protein contacts. Other DBDs in RPA, DBD A and DBD F in RPA1 also interact with both proteins and DNA in their respective binding clefts (16,17,49–51). In the case of RPA2, protein interactions may be the predominant, critical function.
The model that RPA2 has an essential function in addition to complex formation is supported by analysis of RPA in plants. The deepwater rice, Oryza satvia, has multiple genes encoding RPA1 and RPA2 subunits but only one RPA3 gene. Three distinct forms of RPA are found in O. satvia, each contains one of the RPA1 and RPA2 gene products and all contain the common RPA3 subunit (52). These three RPA complexes have different expression and intracellular localization patterns suggesting that they have different roles in the cell. Since these functionally different RPA complexes have different RPA2 subunits but share a single RPA3 subunit, it is most likely that the RPA2 subunits have functions in addition to complex formation. Multiple RPA genes and complexes also seem to exist in Arabidopsis, where mutations in one gene for Arabidopsis RPA1 are lethal but mutations in another RPA1 gene only cause DNA damage sensitivity (53).
Humans also contain a second RPA2-related gene. This RPA2-like subunit (called RPA4) is expressed in certain human tissues and in some cell lines (54). RPA4 has 47% identity with RPA2 and can form a complex with RPA1 and RPA3 (54). It is also interesting to note that RPA4 is reported to be predominantly expressed in quiescent cells (54). This suggests that like plants, humans may have different RPA2/RPA4 in actively replicating versus mature tissues. This also supports DBD D having functions in non-complex related, tissue-specific functions. Further studies are needed to define which protein interactions are needed for RPA2 function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Research Grant [GM44721 to M.S.W.]; Russian Fund for Basic Research and the Program of Presidium of RAS ‘Molecular and Cellular Biology’ [N08-04-00704 to O.L.]; American Heart Association Predoctoral Fellowship [0310046 to A.M.D.]. Funding for open access charge: National Institutes of Health Research Grant [GM44721].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Wold Lab and Dr N Rechkunova (Lavrik Lab) for scientific discussions and critical reading of this manuscript.
REFERENCES
- 1.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. CRC Crit. Rev. Biochem. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 3.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishibashi T, Kimura S, Sakaguchi K. A higher plant has three different types of RPA heterotrimeric complex. J. Biochem. 2006;139:99–104. doi: 10.1093/jb/mvj014. [DOI] [PubMed] [Google Scholar]
- 5.Brill SJ, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 6.Walther AP, Gomes XV, Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. I. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry. 1999;38:3963–3973. doi: 10.1021/bi982370u. [DOI] [PubMed] [Google Scholar]
- 7.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 10.Pestryakov PE, Krasikova YS, Petruseva IO, Khodyreva SN, Lavrik OI. The role of p14 subunit of replication protein A in binding to single-stranded DNA. Dokl. Biochem. Biophys. 2007;412:4–7. doi: 10.1134/s1607672907010024. [DOI] [PubMed] [Google Scholar]
- 11.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 12.Romero Salas T, Petruseva I, Lavrik O, Saintome C. Evidence for direct contact between the RPA3 subunit of the human replication protein A and single-stranded DNA. Nucleic Acids Res. 2008;37:38–46. doi: 10.1093/nar/gkn895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 14.Kolpashchikov DM, Khodyreva SN, Khlimankov DY, Wold MS, Favre A, Lavrik OI. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 2001;29:373–379. doi: 10.1093/nar/29.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NGJ, Hoeijmakers JHJ. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyka IM, Dhar K, Binz SK, Wold MS. Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry. 2003;42:12909–12918. doi: 10.1021/bi034930h. [DOI] [PubMed] [Google Scholar]
- 17.Arunkumar AI, Stauffer ME, Bochkareva E, Bochkarev A, Chazin WJ. Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J. Biol. Chem. 2003;278:41077–41082. doi: 10.1074/jbc.M305871200. [DOI] [PubMed] [Google Scholar]
- 18.Daughdrill GW, Ackerman J, Isern NG, Botuyan MV, Arrowsmith C, Wold MS, Lowry DF. The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res. 2001;29:3270–3276. doi: 10.1093/nar/29.15.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastin-Shanower SA, Brill SJ. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA in ssDNA binding. J. Biol. Chem. 2001;276:36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolpashchikov DM, Weisshart K, Nasheuer HP, Khodyreva SN, Fanning E, Favre A, Lavrik OI. Interaction of the p70 subunit of RPA with a DNA template directs p32 to the 3′-end of nascent DNA. FEBS Lett. 1999;450:131–134. doi: 10.1016/s0014-5793(99)00484-6. [DOI] [PubMed] [Google Scholar]
- 21.Pestryakov PE, Weisshart K, Schlott B, Khodyreva SN, Kremmer E, Grosse F, Lavrik OI, Nasheuer HP. Human replication protein A: the C-terminal RPA70 and the central RPA32 domains are involved in the interactions with the 3′-end of a primer-template DNA. J. Biol. Chem. 2003;278:17515–17524. doi: 10.1074/jbc.M301265200. [DOI] [PubMed] [Google Scholar]
- 22.Weisshart K, Pestryakov P, Smith RW, Hartmann H, Kremmer E, Lavrik O, Nasheuer HP. Coordinated regulation of replication protein A activities by its subunits p14 and p32. J. Biol. Chem. 2004;279:35368–35376. doi: 10.1074/jbc.M403825200. [DOI] [PubMed] [Google Scholar]
- 23.Sibenaller ZA, Sorensen BR, Wold MS. The 32- and 14-kDa subunits of replication protein A are responsible for species-specific interactions with ssDNA. Biochemistry. 1998;37:12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- 24.Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes XV, Henricksen LA, Wold MS. Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry. 1996;35:5586–5595. doi: 10.1021/bi9526995. [DOI] [PubMed] [Google Scholar]
- 26.Blackwell LJ, Borowiec JA. Human replication protien A binds single-stranded DNA in two distinct complexes. Mol. Cell Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavrik OI, Kolpashchikov DM, Nasheuer HP, Weisshart K, Favre A. Alternative conformations of human replication protein A are detected by crosslinks with primers carrying a photoreactive group at the 3′-end. FEBS Lett. 1998;441:186–190. doi: 10.1016/s0014-5793(98)01544-0. [DOI] [PubMed] [Google Scholar]
- 28.Lavrik OI, Kolpashchikov DM, Weisshart K, Nasheuer HP, Khodyreva SN, Favre A. RPA subunit arrangement near the 3′-end of the primer is modulated by the length of the template strand and cooperative protein interactions. Nucleic Acids Res. 1999;27:4235–4240. doi: 10.1093/nar/27.21.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mass G, Nethanel T, Kaufmann G. The middle subunit of replication protein A contacts growing RNA-DNA primers in replicating simian virus 40 chromosomes. Mol. Cell Biol. 1998;18:6399–6407. doi: 10.1128/mcb.18.11.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mass G, Nethanel T, Lavrik OI, Wold MS, Kaufmann G. Replication protein A modulates its interface with the primed DNA template during RNA-DNA primer elongation in replicating SV40 chromosomes. Nucleic Acids Res. 2001;29:3892–3899. doi: 10.1093/nar/29.18.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santocanale C, Neecke H, Longhese MP, Lucchini G, Plevani P. Mutations in the gene encoding the 34 kDa subunit of yeast replication protein A cause defective S phase progression. J. Mol. Biol. 1995;254:595–607. doi: 10.1006/jmbi.1995.0641. [DOI] [PubMed] [Google Scholar]
- 32.Maniar HS, Wilson R, Brill SJ. Roles of replication protein-A subunits 2 and 3 in DNA replication fork movement in S. cerevisiae. Genetics. 1997;145:891–902. doi: 10.1093/genetics/145.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philipova D, Mullen JR, Maniar HS, Gu C, Brill SJ. A hierachachy of SSB protomers in Replication Protein-A. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 34.Drachkova IA, Petruseva IO, Safronov IV, Zakharenko AL, Shishkin GV, Lavrik OI, Khodyreva SN. [Reagents for modification of protein-nucleic acids complexes. II. Site-specific photomodification of DNA-polymerase β complexes with primers elongated by the dCTP exo-N-substituted arylazido derivatives] Bioorg. Khim. 2001;27:197–204. doi: 10.1023/a:1011329220632. [DOI] [PubMed] [Google Scholar]
- 35.Dickson AM. University of Iowa; 2007. The role of RPA32-DNA interactions in Saccharomyces Cerevisiae. Ph.D. Dissertation. [Google Scholar]
- 36.Binz SK, Dickson AM, Haring SJ, Wold MS. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 38.Kim C, Paulus BF, Wold MS. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 39.Malone RE, Montelone BA, Edwards C, Carney K, Hoekstra MF. A reexamination of the role of the RAD52 gene in spontaneous mitotic recombination. Curr. Genet. 1988;14:211–223. doi: 10.1007/BF00376741. [DOI] [PubMed] [Google Scholar]
- 40.Park CJ, Lee JH, Choi BS. NMR assignment of the DNA binding domain A of RPA from S. cerevisiae. J. Biomol. NMR. 2005;33:75. doi: 10.1007/s10858-005-1616-4. [DOI] [PubMed] [Google Scholar]
- 41.Haring SJ, Mason AC, Binz SK, Wold MS. Cellular functions of human RPA1: multiple roles of domains in replication, repair, and checkpoints. J. Biol. Chem. 2008;283:19095–19111. doi: 10.1074/jbc.M800881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng X, Habel JE, Kabaleeswaran V, Snell EH, Wold MS, Borgstahl GE. Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation. J. Mol. Biol. 2007;374:865–876. doi: 10.1016/j.jmb.2007.09.074. [DOI] [PubMed] [Google Scholar]
- 43.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestryakov PE, Khlimankov DY, Bochkareva E, Bochkarev A, Lavrik OI. Human replication protein A (RPA) binds a primer-template junction in the absence of its major ssDNA-binding domains. Nucleic Acids Res. 2004;32:1894–1903. doi: 10.1093/nar/gkh346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim C, Wold MS. Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 46.Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lavrik OI, Nasheuer HP, Weisshart K, Wold MS, Prasad R, Beard WA, Wilson SH, Favre A. Subunits of human replication protein A are crosslinked by photoreactive primers synthesized by DNA polymerases. Nucleic Acids Res. 1998;26:602–607. doi: 10.1093/nar/26.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blackwell LJ, Borowiec JA, Mastrangelo IA. Single-stranded DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binz SK, Lao Y, Lowry DF, Wold MS. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions: evidence for an intersubunit interaction. J. Biol. Chem. 2003;278:35584–35591. doi: 10.1074/jbc.M305388200. [DOI] [PubMed] [Google Scholar]
- 50.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl Acad. Sci. USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X, Vaithiyalingam S, Glick GG, Mordes DA, Chazin WJ, Cortez D. The basic cleft of RPA70N binds multiple checkpoint proteins including RAD9 to regulate ATR signaling. Mol. Cell Biol. 2008;28:7345–7353. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishibashi T, Kimura S, Furukawa T, Hatanaka M, Hashimoto J, Sakaguchia K. Two types of replication protein A 70 kDa subunit in rice, Oryza sativa: molecular cloning, characterization, and cellular and tissue distribution. Gene. 2001;272:335–343.c. doi: 10.1016/s0378-1119(01)00555-8. [DOI] [PubMed] [Google Scholar]
- 53.Ishibashi T, Koga A, Yamamoto T, Uchiyama Y, Mori Y, Hashimoto J, Kimura S, Sakaguchi K. Two types of replication protein A in seed plants. FEBS J. 2005;272:3270–3281. doi: 10.1111/j.1742-4658.2005.04719.x. [DOI] [PubMed] [Google Scholar]
- 54.Keshav KF, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex. Mol. Cell Biol. 1995;15:3119–3128. doi: 10.1128/mcb.15.6.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.