Abstract
Expression of a proof-reading deficient form of mitochondrial DNA (mtDNA) polymerase γ, POLG, causes early death accompanied by features of premature ageing in mouse. However, the mechanism of cellular senescence remains unresolved. In addition to high levels of point mutations of mtDNA, the POLG mutator mouse harbours linear mtDNAs. Using one- and two-dimensional agarose gel electrophoresis, we show that the linear mtDNAs derive from replication intermediates and are indicative of replication pausing and chromosomal breakage at the accompanying fragile sites. Replication fork arrest is not random but occurs at specific sites close to two cis-elements known as OH and OL. Pausing at these sites may be enhanced in the case of exonuclease-deficient POLG owing to delayed resumption of DNA replication, or replisome instability. In either case, the mtDNA replication cycle is perturbed and this might explain the progeroid features of the POLG mutator mouse.
INTRODUCTION
Mitochondria have been proposed to play an important role in longevity and ageing. Ablation of the mitochondrial (1), but not the cytosolic (2) form of superoxide dismutase has profound and widespread deleterious effects on organismal health; and over-expression of catalase in mice increases longevity (3). The case for mitochondrial DNA (mtDNA) involvement in ageing is uncertain, as many reports have detected only low levels of mutant mtDNA (4); an exception is the accumulation of partially deleted (or partially duplicated) mtDNAs in the Substantia nigra of aged individuals, particularly those with Parkinson's disease (5,6). The creation of a mouse with a mutant mtDNA polymerase lacking a functional proof-reading domain demonstrated that perturbing mtDNA metabolism is highly deleterious and shortens the life-span of the mouse (7,8). However, the high levels of point mutations found in the mutator mouse are never reached in the course of normal ageing (9). The first mice with the premature ageing phenotype also had high levels of sub-genomic linear pieces of mtDNA, (‘deleted’ mtDNAs), in addition to full-length circular molecules of 16 kb (8). The most prominent linear DNA species’ spanned ∼11 kb, from the major non-coding region (NCR) to the cluster of five tRNA genes that are interrupted by a short spacer region, which functions as a prominent start site of lagging strand DNA synthesis (OL) (8). These fragments are not archetypal mtDNA deletions, mapping as they do to the reciprocal region of most pathological deletions and being linear as oppose to circular molecules (10,11).
Initiation and termination of replication occur frequently in the NCR (12,13), and OL appears to be a replication pause site (14). Replication fork arrest in the NCR (nt 15 424–16 300) and at OL (nt 5160–5191) predicts a prominent replication intermediate with a bubble spanning ∼11 kb, with junctions susceptible to nicking by single-strand (S1) nuclease, which could result in the release of one branch of the bubble (illustrated schematically in Figure 1). If the abundant linear DNAs of the mutator mouse were broken replication intermediates then this would offer an explanation of the phenotypic consequences of POLG exonuclease deficiency. In the absence of repair, chromosomal breakage is effectively futile replication, and a high steady-state level of linear DNAs would imply that whatever repair systems exist in mitochondria for dealing with strand breaks, they are overwhelmed in the mutator mouse. As well as being wasteful, chromosomal breaks are hazardous, as the resultant DNA ends can invade other molecules precipitating illegitimate recombination, and genome rearrangement. Even if they remain intact, paused or stalled replication forks can reverse creating ends which are equally recombinogenic, and so cells go to considerable lengths to rescue or remove stalled replication forks (15,16).
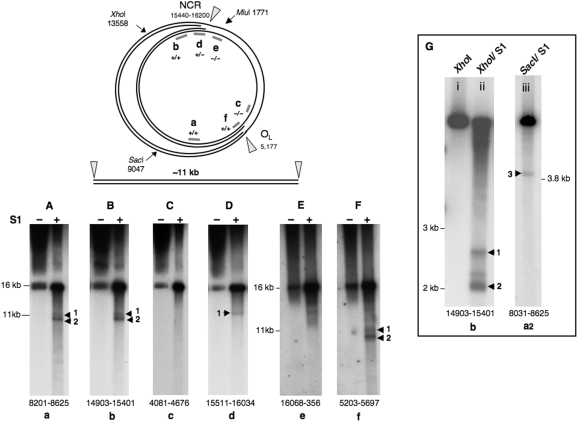
Figure 1.
Linear 11 kb fragments of mtDNA are released from control mouse liver mtDNA by single-stranded nuclease treatment. The figure shows a schematic diagram of a replication intermediate of mammalian mtDNA, where fork arrest has occurred in the NCR and near OL; nicking by S1, or other, nuclease can in theory release a linear fragment of mtDNA of ∼11 kb as illustrated. S1 nuclease treatment of Balb C mouse liver mtDNA yielded just such fragments based on a series of probes from around the mouse mitochondrial genome (Panels A–F). (Panels A–D) represent a single gel: 0.62% agarose, 60 V, 30 h; whereas, the mtDNA of (Panel E) and (Panel F) was separated on a 0.5% agarose gel, at 65 V for 30 h. Treating mouse liver mtDNA with XhoI or SacI, in addition to S1 nuclease, produced shorter fragments (Panel G, i–iii), after separation at 100 V for 4 h on a 0.8% agarose gel. Probes were assigned lowercase letters (a–f), and the numbers at the foot of the gel panels indicate the span of the probes, based on the revised mouse mtDNA reference sequence (17).
Here we show that linear mtDNAs, of the type documented previously in the mutator mouse (8), can indeed be generated from mtDNA of control mice by means of single-strand nuclease treatment, and so they are inferred to derive from paused replication intermediates. In solid tissue of mutator mouse, paused replication intermediates were highly abundant indicating that expression of a proof-reading defective mtDNA polymerase leads to elevated or prolonged replication pausing. These findings suggest that perturbed mtDNA replication is an important feature of the mutator mouse that is likely to contribute to its pathophysiology.
MATERIALS AND METHODS
Solid tissue analysis utilized control female BALB/CJ mice aged 4–6 weeks (Figure 1) or mutator mice POLG D257A, of a mixed 129/ICR/B6 genetic background (7) and wild-type littermates (Figures 2–5). Mouse liver mtDNA isolation, restriction digestion and 2D-AGE were as described previously (12). 1D-AGE conditions are indicated in the figure legends. S1 nuclease treatment was 1 U for 10 min at 37°C (Figures 1 and 2), or 1 U for 1 min at 37°C (Figure 5). From (17), mouse mtDNA probes were 5′–3′: a nt 8201–8625; ACGCCTAATCAACAACCGTCTC and CATGGACTTGGATTAACTATGTGATATGC; a2 8031–8625 TTACACCTACTACCCAACTAT CCATAAATC and CATGGACTTGGATTAACTATGTGATATGC; b nt 14 903–15 401 CAGACAACTACATACCAGCTAATCCAC and ACCAGCTTTGGGTGCTGGTG; c nt 4081–4676 AACAAAATACTTCGTCACACAAGCAACAGC and GAAGGCCTCCTAGGGATAGTAATATCA; d nt 15 511–16 034 ATCAATGGTTC AGGTCATAAAATAATCATCAAC and GCCTTAGGTGATTGGGTTTTGC; e nt 16 068–356 TCAATACCAAATTTTAACTCTCCAAACCCCCCA and ACGCCGAAGATAATTAGTTTGGGTTAATCG; f nt 5203–5697 GAGATTTCTCTACACCTTCGAATTTGC and TTCCTGCTCCTGCTTCTACTATTGATG.
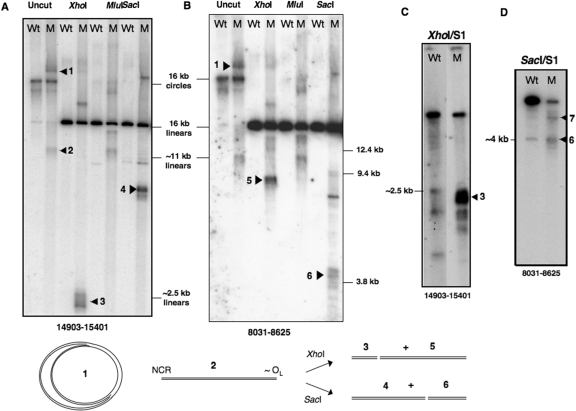
Figure 2.
Linear 11 kb fragments of mtDNA are present at high abundance in mutator mouse liver mtDNA, in the absence of single-strand nuclease treatment. Liver mtDNA samples isolated from wild-type (Wt) and mutator (M) mice were digested with restriction enzymes, XhoI, MluI or SacI, or left uncut, and fractionated by 1D-AGE in TBE buffer. Separation conditions were 55 V for 20 h in 0.4% agarose (Panels A and B) or 100 V for 4 h in 0.8% agarose (Panels C and D). After Southern blotting, membranes were probed with PCR products corresponding to nt 14 903–15 401 (Panels A and C); and nt 8031–8625 (Panels B and D) of mouse mtDNA. The samples in panels (C) and (D) were treated additionally with single-strand specific (S1) nuclease after restriction digestion (see ‘Materials and Methods’ section). The two lanes in panel (C) are different exposures of the same gel; a longer exposure of the digest of Wt mtDNA was needed to show that S1 nuclease generated some fragments of similar size to the abundant short fragments seen in mutator mouse mtDNA digests. The species that distinguished mutator mouse mtDNA from wild-type mtDNA are interpreted as follows: 1—replicating theta structures, or eyebrows, as illustrated at the base of the figure. Wild-type mtDNA samples also include theta structures but their low abundance means they are difficult to detect unless 2D-AGE is applied (12); 2—linear mtDNA fragments of approximately 11 kb; 3—XhoI digested mtDNA fragments with one end corresponding to the restriction site at nt 13 558 and the other mapping to the major non-coding region (NCR); 4—SacI fragments ∼7 kb, spanning nt 9047 to the NCR; 5—fragments ∼8 kb, with one end close to OL the other nt 13 558; 6—fragments ∼4 kb, with termini at nt 9047 and near OL. The fragments labelled 7 in panel D were gel-extracted, converted to circles, cloned and sequenced, without recourse to S1 nuclease treatment, and found to contain point mutations, which indicated that the novel fragments were the result of SacI site gains (see Supplementary Figure 3 for details).
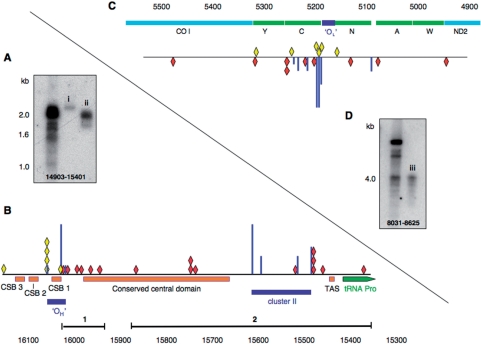
Figure 3.
The ends of the prominent linear molecules of mutator mouse mtDNA map to the NCR and in the vicinity of OL. S1 products of mutator mouse mtDNA were recovered from specific regions of agarose gels using Recochips™; repeated separation of some of the material by 1D-AGE (0.8% agarose, 4 h, 100 V) was used to confirm successful recovery (panel A, i and ii; panel D, iii). Fragments were circularized, and the junction amplified, cloned and sequenced. Panel (B) A partial linear map of the mouse mitochondrial genome encompassing the NCR. The ends (red diamonds) identified by direct sequencing were nt 16 033, 16 016, 15 997, 15 995, 15 978, 15 956 (Recochip 1); and 15 875, 15 751, 15 751, 15 750, 15 747, 15 529, 15 489, 15 487, 15 486, 15 467 and 15 389 (Recochip 2). Scarcer ends mapping to the NCR were also cloned and sequenced from control littermates (yellow diamonds). The NCR of mouse mtDNA includes three so-called conserved sequenced boxes (CSBs 1–3), a conserved central domain, and a predicted clover-leaf structure, TAS, which is believed to effect termination of 7S DNA (D-loop) synthesis. Three of the ends (15 751, 15 751 and 15 750) in the conserved central domain are flanked by a GC-rich sequence CCGGGCCC and GGGGG, which is extremely rare in the L-strand of mammalian mtDNAs, suggesting that this may represent a cis-element. Free 5′ ends of DNA identified previously by LM-PCR are represented as vertical blue lines; in panel B the free 5′ ends comprise two clusters, cluster I (OH) and cluster II (12,13,38). The height of the most prominent free end (nt 16 034) was set arbitrarily and the others expressed as a fraction of its height. Panel (C) The sequenced SacI/S1 products with an end mapping close to OL (nt 5160–5191) were nt 4946, 5082, 5133, 5201, 5217, 5255, 5257, 5318, 5477 (red diamonds) in mutator mouse samples and nt 5169, 5193, 5196, 5199, 5244 and 5362 (yellow diamonds) in controls. In humans, free 5′ ends are concentrated in the tRNACys gene (C), which is adjacent to OL (38,39) and free 5′ ends map to similar positions in mouse mtDNA (see Supplementary Figure 2); in panel (C) they are again represented as blue vertical lines. Panel (D) SacI/S1-treated mutator mouse mtDNA analysed by 1D-AGE, iii is the material that was used to define the ends of DNA represented as red diamonds in panel (C).
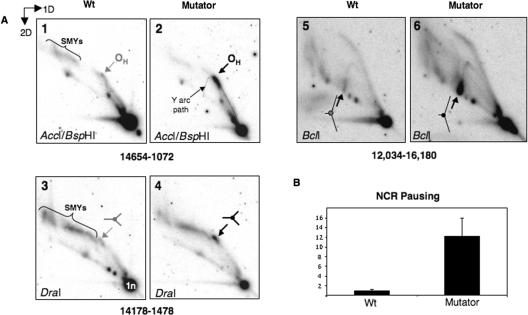
Figure 4.
Enhanced replication pausing in the NCR of POLG mutator mouse mtDNA. (A) 2D-AGE analysis of mtDNA of control (WT) and mutator mouse liver was performed after digestion with AccI and BspHI (panels 1 and 2), DraI (panels 3 and 4) or BclI (panels 5 and 6). Arrows indicate the position on a standard Y arc where replication forks frequently arrest [for details of replication fork arrest, see (40)]. Restriction site blockage accounts for the high molecular mass fragments of mtDNA (slow-moving Y-like arcs or SMYs), which were previously attributed to RNA incorporation during mtDNA replication (12). (B) The abundance of paused replication forks mapping to the NCR was 12 times higher than controls based on Typhoon™ phosphorimager (GE Healthcare) quantification of paused RIs. n = 3 experiments using mtDNA derived from two distinct groups of control and mutator mouse livers.
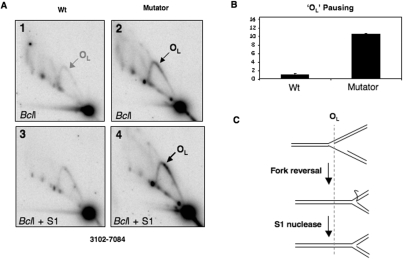
Figure 5.
Increased replication pausing in the vicinity of OL in mutator mouse mtDNA. (A) Liver mtDNA samples from mutator mouse and controls (WT) were digested with BclI and probed with a PCR product spanning nt 5568–6044 after N2D-AGE, to reveal a fragment encompassing the OL region. (B) Pausing at OL was increased 11-fold based on phosphorimager analysis of gels such as the ones shown in panels 1 and 2. Although increased pausing on the Y arc was more apparent after a brief single-strand (S1) nuclease treatment (1 U for 1 min at 37°C) (panels 3 and 4), quantification indicated that the S1 treatment decreased the relative abundance of paused RIs between POLG mutator mouse and control samples (data not shown), and so the data in B relate to samples that were not treated with S1 nuclease. At least some of the increase in signal on the Y arc produced by S1 nuclease (panel A-4) can be attributed to the removal of single-stranded DNA from regressed forks, as illustrated in (C).
XhoI/S1 treated mouse mtDNA was recovered from 1D agarose gels by electrophoresis at 70 V for 2 h into Recochips™ (TaKaRa, Japan) and ligated overnight with 200 U of T4 ligase (New England Biolabs), at 22°C; the junction of the ligated DNA fragments was amplified over the course of 30 cycles of 94°C for 30 s, 68°C for 30 s and 72°C for 30 s using Biotaq (Bioline), or KOD HiFi DNA polymerase (Novagen). SacI/S1 digested mtDNA was gel-extracted, cloned and sequenced using the same approach. Primers were 5′–3′: TGACTTGTCCCACTAATAATCGGAG nt 5568–5592 and CCCAAAGAATCAGAACAGATGCTG nt 6044–6021; GGATATACGACTGCTATAGCTACTGAGG, nt 13 843–13 870 combined with CCTTAAATAAGACATCTCGATGGTATCG, nt 15 840–15 867 or CCATGTCTTGATAGTATAAACATTACTCTGGTC, nt 15 286–15 318. In the case of material eluted into Recochip 2 (see Figure 3A) a second round of PCR was employed using nested primers: GAGGAATATCCAGAGACTTGGGGATCTAACTG nt 13 815–13 846 and GGTCTTGTAAACCTGAAATGAAGATCTTCTCTTCTC nt 15 314–15 350. In all cases there was a final extension step of 72°C for 10 min with Biotaq polymerase to ensure addition of a terminal adenine nucleotide. After cloning into pcr2.1 (Invitrogen) the products were sequenced by Geneservices.
Neutral 2D agarose gel electrophoresis (2D-AGE) was essentially as described previously (12). Typical conditions were: first dimension electrophoresis, 28 V for 15 h at room temperature in a 0.4% agarose gel; second dimension electrophoresis 1% agarose, 260 mA for 6 h at 4°C. After electrophoresis, gels were alkaline blotted onto nylon membranes and hybridized to radiolabelled probes corresponding to nt 15 511–16 034 and nt 5568–6044 of the mouse mitochondrial genome, by overnight incubation at 65°C in 7% SDS, 0.25 M sodium phosphate pH 7.4. Post-hybridization washes were 1× SSC three times, followed by 1× SSC, 0.1% SDS twice, each for 20 min at 65°C. Filters were exposed to X-ray film and developed after 3–7 days, or exposed to phosphor imaging plates for quantification of spots of RIs and unit length fragments (1 n), using a Typhoon™ phosphorimager (GE Healthcare).
RESULTS
Single-strand nuclease treatment of control mouse liver mtDNA generates linear DNA molecules of ∼11 kb
To test the idea that replication forks paused in the vicinity of the NCR and OL are fragile, purified liver mtDNA of normal control mice was treated with single-strand specific S1 nuclease and the products of the reaction fractionated in 1D on agarose gels. S1 nuclease-treated mtDNA revealed a doublet not detectable in untreated samples (Figure 1A–F). The products of S1 nuclease treatment were very similar to the sub-genomic linear mtDNAs reported for the POLG mutator mouse (8), as they covered approximately the region from the NCR to OL, based on the results of a series of hybridizations to probes scattered around the mouse mitochondrial genome (Figure 1A–F). However, the shorter of the two bands lacked at least some of the NCR, as it was not detected with a probe spanning nucleotide numbers 15 511–16 034 (Figure 1D). To improve the resolution of the S1 nuclease products, mouse liver mtDNA was first digested with XhoI, which cuts mouse mtDNA once only at nt 13 558. A probe corresponding to nt 14 903–15 401 of mouse mtDNA revealed a series of bands of ∼2–2.5 kb when control mtDNA samples were treated with S1 nuclease (Figure 1G-ii). On the basis of their mobility and the probe applied, such fragments extend from the XhoI site at nt 13 558 to positions in the NCR. Another series of restriction fragments modified by S1 nuclease treatment mapped near OL (Figure 1G-iii). These findings corroborate the earlier data from 2D agarose gels suggesting that mitochondrial replication pauses in the vicinity of the NCR and OL (12–14). Furthermore, the analysis of control mouse liver mtDNA demonstrates that linear mtDNAs can be derived from replication intermediates, by single-strand nuclease digestion.
Mapping and sequencing of the 11 kb linear fragments of mouse mtDNA from control and mutator mice
Mitochondrial DNA was prepared from liver mitochondria of mutator mice and age-matched controls (littermates) purified on sucrose-gradients and analysed by 1D-AGE. Restriction digested and undigested mutator mouse mtDNA yielded bands barely detectable in controls (Figure 2A and B). The majority of these bands were shorter than the expected restriction fragment, and the short XhoI digestion products (3 in Figure 2A) were similar to those produced in control mice by the combined XhoI/S1 nuclease treatment (Figure 1G-ii). The fragments ∼2.5 kb in size were detected by a probe covering nt 14 903–15 401 (Figure 2A), but not by probes corresponding to nt 8031–8625 (Figure 2B) or 12 605–12 963 (not shown), and so they must logically extend from the XhoI restriction site at nt 13 558 to the NCR. S1 nuclease treatment of XhoI digested mutator mouse mtDNA increased the abundance of the 13 558-NCR fragments (Figure 2C), suggesting that at least some of them were derivatives of replication intermediates. Gel-extraction of S1/XhoI digested fragments ∼2–2.5 kb was followed by circularization, PCR amplification and sequencing, to define the junctions. This provided confirmation that one end (depicted as red diamonds in Figure 3B) frequently mapped to the NCR. The S1 nuclease treatment removed only the single-stranded overhang created by XhoI and so there was no suggestion that the nuclease treatment removed any uninterrupted duplex DNA from the NCR end of the fragments. Several of the fragment ends were close to, yet rarely coincident with, 5′-ends of H-strand DNA mapped previously to the NCR (blue vertical lines in Figure 3B), which are putative initiation, pause or termination sites (12,13,18). However, similar-sized fragments of much lower abundance in control samples (yellow diamonds, Figure 3B) mapped at or near the most prominent 5′ ends previously designated as OH, albeit that nt 16 065 was the most common end of linear dsDNA (this report), whereas 16 034 was the most abundant free 5′ DNA end (12). The other terminus of the linear mtDNA molecules was often in the vicinity of OL (Figure 3C and D), based on sequence analysis of SacI digested products of ∼4 kb (Figure 2D, species 6). Here again the DNA ends were dispersed across a region of several hundred nucleotides (red diamonds in Figure 3D). The ends of the cloned fragments near OL were less dispersed in control mouse samples and half of them coincided with prominent free 5′ ends of DNA mapped by ligation-mediated PCR (Figure 3D and Supplementary Figure 2). The minor differences in map positions notwithstanding, the end mapping data indicate that the linear DNA species of mutator mouse mitochondria are similar to single-stranded nuclease generated fragments of controls, and so support the view that they are related to replication pausing.
Enhanced replication pausing at defined sites in mutator mouse mtDNA
Neutral 2D-AGE has been widely used to characterize replication intermediates (19,20). If, as suggested above, replication pausing in the NCR and near OL is much more frequent or prolonged in the mutator mouse then this should manifest as accumulated replication intermediates. Therefore, control and mutator mouse mtDNA was digested with DraI, or AccI and BspHI, or BclI, and the products separated by 2D-AGE. Southern hybridization to probes detecting the fragment of mtDNA containing the NCR region revealed a prominent spot on the standard replication fork (Y) arc in mutator mouse samples (Figure 4A). A prominent spot on an arc of replication intermediates is indicative of replication pausing (21); the position of the pause on the three arcs (Figure 4A) allied to previous 2D-AGE analyses (13,22) mapped the pause to the NCR, not far from OH. Thus, this pause site mapped to the same position as one of the ends of the linear mtDNA fragments (Figures 2 and 3). Quantification by phosphorimager analysis indicated that the pause was 12-fold stronger in mutator mouse samples than wild-type littermates (Figure 4B). The other end of the linear fragments mapped approximately to OL (Figure 3C) and so the region spanning nt 3102–7084 with OL near its centre was analysed, revealing an 11-fold increase in signal towards the apex of the ascending Y arc, in mutator mouse liver samples compared to controls (Figure 5A and B). This is the portion of the arc where replication intermediates paused in the vicinity of OL would be expected to resolve. Thus, both termini of the linear fragments correspond to narrow regions of prominent replication pausing. Nevertheless, the cloned fragments need not be perfectly representative of the paused replication intermediates; for instance, fork regression and nuclease digestion in vivo could generate junctions that were no longer partially single stranded (23), thereby decreasing their fragility. Hence, regressed forks would be more resistant to S1 nuclease and so under-represented among the cloned fragments. After 2D-AGE, nuclease-digested, regressed forks would resolve lower on the ascending Y arc, i.e. closer to the unit length fragment (1 n), than forks where regression had not occurred, and this could explain why there was a smear on the ascending Y arc, rather than a more tightly defined spot (Figure 5A-2). Logically there must also have been molecules that had regressed and yet retained some single-stranded DNA, as S1 nuclease treatment enhanced the pausing on the Y arc (Figure 5A and illustrated in 5C). Although almost nothing is known about the reactivation of replication at pause sites in mitochondria, the apparatus to re-start replication is ubiquitous in systems where faithful replication is required, and so it is unlikely to be absent from mitochondria. If pausing is prolonged, rather than elevated, in the mutator mouse, then this would implicate POLG in re-starting replication. Alternatively or additionally, exonuclease-deficient POLG of mouse may be less processive than the wild-type enzyme; in vitro this is true of proof-reading deficient forms of yeast (24), but not mammalian (25), POLG. The exonuclease-deficient POLG of the mutator mouse may, nevertheless, make the mitochondrial replisome prone to collapse on encountering an obstacle to replication, such as that presented by a pause site. In turn, this might increase the amount of single-strandedness at the fork thereby creating fragile sites; chromosomal breakage at such sites could thereby account for the high levels of linear DNA fragments in mutator mouse tissues.
Other fragments of mtDNA present in mutator mouse, at higher abundance than wild-type littermates, were incompatible with pausing near OH and OL. Some of the most prominent of these fragments were products of SacI digestion (species 7, Figure 2D). Cloning and sequencing indicated that such fragments were the result of point mutations creating new restriction sites (Supplementary Figure 3), rather than replication pausing, which provides further evidence of POLG-generated mutational hotspots in mammalian mtDNA (26,27).
DISCUSSION
Replication pausing may explain the progeria-like features of the mutator mouse
That the high incidence of point mutations in the mtDNA of the mutator mouse (8) is a manifestation of the crippled exonuclease function of POLG is not a doubt. However, it remains unclear whether the impact of the point mutations on oxidative phosphorylation is responsible for the progeria-like features. Respiratory enzyme activities were reduced by up to 50% of control values in the mutator mouse, yet the respiratory enzyme capacity of heart was barely below normal (8). In any case, decreased respiratory chain capacity is a well-recognized feature of mitochondrial disease in man, and yet there is scant evidence of progeria in such disorders (10,28). The accumulated paused replication intermediates and linear mtDNA fragments characterized here are a key distinguishing feature of the mutator mouse: could they account for the premature ageing phenotype of the mutator mouse? The high steady-state level of particular replication intermediates (Figures 4 and 5) indicates that replication is subject to long delays at specific pause sites, or else pausing occurs at much higher frequency than normal during the replication of mtDNA in mice expressing a proof-reading deficient POLG. However, the linear 11 kb DNA species generated by breakage of the DNA at the pause sites is present at a similar level throughout the life of the mutator mouse, from embryonic development to its early death: it does not increase with age. Nevertheless, enhanced pausing and the failure of innumerable rounds of mtDNA replication could well prove deleterious over time, especially when one considers the close parallels between the mutator mouse phenotype and the impact of mutations in the RecQ helicase WRN, which are responsible for progeria-like syndromes (see below). Hence, we hypothesize that it is the long-term impact of chromosomal breakage and aborted replication that leads to premature ageing of the mutator mouse.
The problem of chromosome breakage could be exacerbated by the free ends of the linear DNAs precipitating recombination, via strand-invasion; thereby giving rise to deletions (29). Even if the fragile sites remained intact in vivo, free DNA ends could be generated by fork reversal. Hence, recombination between the two branches of replication intermediates paused near OH and OL could account for the preponderance of deletions in the so-called major arc of human mtDNA (30). By the same token, prolonged replication pausing could also underlie the elevated level of mtDNA deletions in the mutator mouse that was reported recently (31). However, we question whether such a low level of deletions (only detectable using a PCR-based assay) is, of itself, relevant to the progeria-like features of the mutator mouse, particularly as high levels of single or multiple deletions fail to elicit progeria in humans (10,28).
Parallels between mutant forms of POLG and WRN suggest DNA replication defects often underpin progeria-like syndromes
Cultured cells from human premature aging syndromes show replicative senescence. For instance, mutations in the WRN gene, a RecQ helicase, and in XPD, a helicase involved in nucleotide excision repair, each cause a progeria-like syndrome with some features that are similar to the mutator mouse. WRN protein is implicated in replication fork recovery (32), and S phase is prolonged in cultured fibroblasts lacking functional WRN. As mentioned above, replication may well be prolonged in the mitochondria of the mutator mouse, based on the high steady-state level of mitochondrial replication intermediates (Figures 4 and 5). Fragile sites are prevalent in nuclear DNA where there is a dearth of the RecQ helicase WRN, leading to chromosomal breakage (33). The sensitivity of mtRIs to single-strand nuclease in controls and the high level of sub-genomic linear mtDNA fragments (∼11 kb) in mutator mouse (Figures 1 and 2, respectively), coupled with the high abundance of specific replication intermediates, suggest that replication pausing in mitochondria produces fragile sites, and that breakage at such sites occurs at high frequency in the mutator mouse. Thus, here again mutant WRN and POLG produce similar molecular phenotypes. Furthermore, WRN is the only RecQ helicase with 3′–5′ exonuclease activity (34), and it has been shown to be involved in DNA repair resulting from oxidative damage (35). By analogy with WRN, POLG may be directly involved in DNA repair and depend on its 3′–5′ exonuclease activity for this function. Therefore 3′–5′ exonuclease-deficient POLG may represent double jeopardy: increased errors during DNA synthesis and reduced DNA repair capacity. As such, prolonged pausing may reflect an effort to execute DNA repair, in which case replication pausing would form part of a mitochondrial fidelity-surveillance system, and one function of pause sites would be to act as DNA repair checkpoints. The formation of replication intermediates with fragile sites could therefore be envisaged as a mechanism for rejecting defunct copies of mtDNA; if pausing is extended due to a high mutant load that cannot be repaired efficiently then strand breakage occurs, which effectively aborts replication. The 11 kb linear mtDNAs would therefore be a manifestation of aborted replication.
While, in this scenario, the underlying problem is after all the high level of point mutations generated by the exonuclease-deficient POLG, the effect on cell fitness is indirect: essential factors involved in DNA metabolism eventually become exhausted in mitochondria of affected cells due to increased replication activity that is itself the result of inadequate DNA repair capacity, owing to the mutant form of POLG creating many more mutations than normal and being unable to assist in their repair. In any event, the 11 kb linear mtDNAs are evidence of futile replication and so it is reasonable to suggest that this might underpin the progeria-like features of the mutator mouse, more especially as there is scant evidence that impairment of oxidative phosphorylation causes progeria. Indeed it would be remarkable if the life-long requirement for increased mtDNA replication in the mutator mouse came at no cost to the animal.
The ‘aberrant replication’ hypothesis can also explain the modest impact of mutant POLG on ROS production (36), which has puzzled many researchers, as elevated ROS is a frequently allied to mitochondrial diseases [(37) and references therein]. According to our hypothesis, progeria-like phenotypes of the mutator mouse are not primarily a consequence of the downstream effects of expressing mutant mtDNA, but an effect of trying to prevent mutations accumulating to such a level that ROS homeostasis and mtDNA expression are catastrophically compromised. Moreover, because both nuclear and mtDNA metabolism are co-dependent on numerous factors, including DNA ligase III, uracil DNA glycosylase, Flap endonuclease 1, Brca1, RNase H1 and the nucleotide precursor pool, the increased sequestration of such materials by mitochondria with deficient POLG might conceivably impact on nuclear DNA replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Medical Research Council [to I.J.H. and J.P.]; the Wellcome Trust [to J.P.]; the European Union Framework 6 Integrated Programme – EU Mitocombat [to I.H. and A.R.]; National Institutes of Health grants AG021905 [to T.A.P.], AG17994 and AG21042 [to C.L.]. Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr M. Yang for preparing mouse mtDNA.
REFERENCES
- 1.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 2.Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, Siwek DF, Wilcox HM, Flood DG, Beal MF, Brown RH, Jr., et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 3.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs HT. The mitochondrial theory of aging: dead or alive? Aging Cell. 2003;2:11–17. doi: 10.1046/j.1474-9728.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 5.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 6.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 7.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 8.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 9.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat. Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 10.Holt IJ, Harding AE, Cooper JM, Schapira AH, Toscano A, Clark JB, Morgan-Hughes JA. Mitochondrial myopathies: clinical and biochemical features of 30 patients with major deletions of muscle mitochondrial DNA. Ann. Neurol. 1989;26:699–708. doi: 10.1002/ana.410260603. [DOI] [PubMed] [Google Scholar]
- 11.Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 12.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell. 2005;18:651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 15.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 16.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat. Rev. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 17.Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fish J, Raule N, Attardi G. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science. 2004;306:2098–2101. doi: 10.1126/science.1102077. [DOI] [PubMed] [Google Scholar]
- 19.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 20.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 21.Brewer BJ, Lockshon D, Fangman WL. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 22.Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 23.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 24.Foury F, Vanderstraeten S. Yeast mitochondrial DNA mutators with deficient proofreading exonucleolytic activity. EMBO J. 1992;11:2717–2726. doi: 10.1002/j.1460-2075.1992.tb05337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longley MJ, Ropp PA, Lim SE, Copeland WC. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 26.Khrapko K, Coller HA, Andre PC, Li XC, Hanekamp JS, Thilly WG. Mitochondrial mutational spectra in human cells and tissues. Proc. Natl Acad. Sci. USA. 1997;94:13798–13803. doi: 10.1073/pnas.94.25.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng W, Khrapko K, Coller HA, Thilly WG, Copeland WC. Origins of human mitochondrial point mutations as DNA polymerase gamma-mediated errors. Mutat. Res. 2006;599:11–20. doi: 10.1016/j.mrfmmm.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Suomalainen A, Kaukonen J. Diseases caused by nuclear genes affecting mtDNA stability. Am. J. Med. Genet. 2001;106:53–61. doi: 10.1002/ajmg.1379. [DOI] [PubMed] [Google Scholar]
- 29.Labib K, Hodgson B. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 2007;8:346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuels DC, Schon EA, Chinnery PF. Two direct repeats cause most human mtDNA deletions. Trends Genet. 2004;20:393–398. doi: 10.1016/j.tig.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat. Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 32.Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): Holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′→5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J. Biol. Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 36.Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl Acad. Sci. USA. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vergani L, Malena A, Sabatelli P, Loro E, Cavallini L, Magalhaes P, Valente L, Bragantini F, Carrara F, Leger B, et al. Cultured muscle cells display defects of mitochondrial myopathy ameliorated by anti-oxidants. Brain. 2007;130:2715–2724. doi: 10.1093/brain/awm151. [DOI] [PubMed] [Google Scholar]
- 38.Kang D, Miyako K, Kai Y, Irie T, Takeshige K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 1997;272:15275–15279. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 39.Hyvarinen AK, Pohjoismaki JL, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.